Abstract
Histone/protein deacetylases play multiple roles in regulating gene expression and protein activation and stability. Their deregulation during cancer initiation and progression cause resistance to therapy. Here, we review the role of histone deacetylases (HDACs) and the NAD+ dependent sirtuins (SIRTs) in the DNA damage response (DDR). These lysine deacetylases contribute to DNA repair by base excision repair (BER), nucleotide excision repair (NER), mismatch repair (MMR), non-homologous end joining (NHEJ), homologous recombination (HR) and interstrand crosslink (ICL) repair. Furthermore, we discuss possible mechanisms whereby these histone/protein deacetylases facilitate the switch between DNA double-strand break (DSB) repair pathways, how SIRTs play a central role in the crosstalk between DNA repair and cell death pathways due to their dependence on NAD+, and the influence of small molecule HDAC inhibitors (HDACi) on cancer cell resistance to genotoxin based therapies. Throughout the review, we endeavor to identify the specific HDAC targeted by HDACi leading to therapy sensitization.
INTRODUCTION
The contribution of posttranslational protein modification by acetylation to the regulation of DNA repair is of expanding interest. Here, we review the multiple levels of control that lysine deacetylases exert on the induction and reversal of genomic insult. Lysine deacetylases either depend on Zn2+, such as class I histone deacetylases (HDACs), comprising HDAC1, 2, 3 and 8, class IIA, comprising HDAC4, 5, 7 and 9, class IIB, comprising HDAC6 and 10 and the class IV enzyme HDAC11, or they depend on NAD+, a characteristic of the class III sirtuin like deacetylases SIRT1-7 (1). They remove acetyl-marks from lysines of histones and non-histone proteins and thereby contribute to widely diverse biological processes such as epigenetic regulation of gene expression, protein stability, enzyme and transcription factor activity and protein–protein interactions (1). Due to the numeral targets of these lysine deacetylases, they have been shown to play roles in apoptosis (2), differentiation (3) and DNA repair (4), to name a few.
Lysine deacetylases are deregulated in numerous cancers (Table 1). Some HDACs are upregulated in colorectal, stomach, oesophagus, breast, ovary, lung, pancreas, thyroid, prostate, melanoma, neuroblastoma and oral cancers (5–28), while some SIRTs are upregulated in myeloma, breast, liver and thyroid cancers (29–32). Downregulation of HDAC4 in glioblastoma (33), SIRT1 and SIRT6 in colorectal cancer (34,35) and SIRT6 in pancreas cancer (35) has also been reported. Consequently, the deregulation of these deacetylases could contribute to cancer formation/progression and the response of cancer cells to genotoxin based therapeutics. The introduction of small molecule HDAC inhibitors (HDACi) in cancer therapy has already shown promise. Inhibition of HDACs has been shown to abolish drug-resistance in cancer cells (36). In support of this finding, HDAC inhibition sensitizes neuroblastoma cells to the genotoxins etoposide, melphalan, carboplatin and vincristine (37), melanoma cells to temozolomide, psoralen and UVA, fotemustine and ionizing radiation (16,38,39), prostate cancer cells to bleomycin, doxorubicin, etoposide, hydroxyurea, cisplatin and ionizing radiation (40,41), breast cancer cells to etoposide and olaparib (42,43) and head and neck cancer cells to cisplatin (44). In addition, HDAC inhibition sensitizes squamous cell carcinoma, non-small cell lung cancer, osteosarcoma, rhabdomyosarcoma and cervical cancer cells to ionizing radiation (45–48).
Table 1. Expression of histone deacetylases in cancers.
| Histone deacetylase | Upregulated in cancer | Downregulated in cancer | |
|---|---|---|---|
| Class I | HDAC1 | Colorectal (5–7), Stomach (5,8,9), Oesophagus (5,10), Breast (5,11,12), Ovaries (5), Lung (5), Pancreas (5), Thyroid (5), Prostate (13–15), Melanoma (16) | |
| HDAC2 | Colorectal (5,6,17–19), Stomach (5,20), Oesophagus (5), Breast (5), Ovaries (5), Lung (5), Pancreas (5), Thyroid (5), Prostate (15), Melanoma (16) | ||
| HDAC3 | Colorectal (5), Stomach (5), Oesophagus (5), Breast (5,11), Ovaries (5), Lung (5,21), Pancreas (5), Thyroid (5), Prostate (15) | ||
| HDAC8 | Neuroblastoma (22) | ||
| Class IIA | HDAC4 | Colorectal (5), Stomach (5,23), Oesophagus (5), Breast (5,7), Ovaries (5,24), Lung (5), Pancreas (5), Thyroid (5) | Glioblastoma (33) |
| HDAC5 | Colorectal (7,25), Melanoma (26), Liver (27) | ||
| HDAC7 | Colorectal (7) | ||
| Class IIB | HDAC6 | Oral (28), Melanoma (26) | |
| Class III | SIRT1 | Skin (81), Colorectal (34) | |
| SIRT6 | Myeloma (29) | Pancreas (35), Colorectal (35) | |
| SIRT7 | Breast (30), Thyroid (31), Liver (32) | ||
The causes ascribed for the sensitization of cancer cells to genotoxic therapeutics by the inhibition of HDACs are many and various. Firstly, and the topic of this review, exposing cancer cells to HDAC inhibitors has been shown to decrease the catalytic activity, as well as the expression of proteins, involved in the detection, removal and reversal of DNA damage induced by genotoxic based chemotherapeutics. HDACi, therefore, prevents the effective reversal of DNA damage, the persistent damage interferes or prevents cellular processes like transcription or DNA replication and the cancer cell initiates a cell death mechanism. Secondly, overexpressed HDACs cause resistance by enhancing the detoxification of chemotherapeutic drugs in cancer cells. This has been shown as HDAC overexpression leads to more effective detoxification due to the HDAC2 and 4 dependent expression of the efflux pump p-glycoprotein. Consequently, colorectal and lung cancer cells resist doxorubicin and etoposide, respectively, which could be reversed upon HDACi application (17,49). HDACi furthermore influence detoxification by decreasing the levels of glutathione, thereby sensitising squamous cell carcinoma cells to cisplatin (50), a known target of detoxification by glutathione (51). Thirdly, as HDACs compact chromatin by removing acetyl groups from histones, inhibition of HDACs could theoretically open chromatin, thereby allowing access of chemotherapeutic genotoxins to the DNA. HDACi were shown to open up chromatin and increase the level of DNA–protein crosslinks upon etoposide and camptothecin exposure (52). The roles of HDACs in p-glycoprotein and glutathione expression as well as their role in compacting chromatin support a role for HDACi in influencing the amount of DNA damage induced by a given concentration of genotoxin. Additionally, HDACs increase the ‘barrier of entry’ into death pathways such as apoptosis (53), which means that upon HDACi less DNA damage is required for triggering apoptosis. One protein that prevents the entry of cells into apoptosis by inhibiting the catalytic activity of caspases is survivin (54). HDAC2 stimulates the expression of survivin (55) while HDACi decreases survivin expression (56,57), which sensitizes cancer cells to anti-cancer drugs.
Here, we review how lysine deacetylases contribute to the removal of DNA adducts as a mechanism dictating the cellular response of cancer cells to DNA-damaging therapies. The influence of HDACs and SIRTs on base excision repair (BER), nucleotide excision repair (NER), mismatch repair (MMR), DNA damage signalling, the choice between DNA double-strand break repair pathways as well as their role in non-homologous end joining (NHEJ), homologous recombination (HR) and interstrand crosslink (ICL) repair will be reviewed. This review, furthermore, discusses which HDACs in the DNA damage response (DDR) are targeted by HDACi and aims at increasing our understanding of the complexity of the cellular processes lysine deacetylases play roles in.
DNA repair
DNA repair plays a central role in protecting cells from the damage induced by extracellular sources and intracellularly generated DNA reactive molecules. By removing adducts from DNA, or resealing DNA strand breaks, DNA repair ensures the faithful propagation of the template for life through generations. Unrepaired DNA damage leads to genomic instability, ageing, carcinogenesis and/or cell death. HDACs and SIRTs play roles in preventing all of these endpoints, for example, SIRT6 deficiency increases genomic instability, premature ageing and cancer formation (35,58,59). As genotoxin-based cancer therapy specifically exploits the fact that DNA damage induces cell death, and HDACs and SIRTs are deregulated in cancer (Table 1), it is important to understand the interplay between these deacetylases and DNA repair. In the following sections, the literature demonstrating the effect of lysine deacetylases on the expression of, or catalytic activity of, DNA repair proteins will be discussed (Table 2).
Table 2. Effect of HDACs and SIRTs on proteins involved in the DNA damage response and DNA repair.
| Deacetylase | HDACi | Influence on expression | Influence on activity | |
|---|---|---|---|---|
| Class I | HDAC1 | H3K56Ac (111), p53 (143,148) | ||
| HDAC2 | p-Glycoprotein (17), RAD51 (16) | H3K56Ac (111) | ||
| HDAC3 | TIP60 (123) | |||
| Class IIA | HDAC4 | p-Glycoprotein (49) | ||
| Class IIB | HDAC6 | MSH2 (89) | ||
| HDAC10 | MSH2 (90) | |||
| Class III | SIRT1 | TDG (63), APE1 (75), WRN (78), XPA (83), TIP60 (124), p53 (149), KU70 (171), NBS1 (179) | ||
| SIRT6 | PARP1 (71), DNA-PKcs (173), CtIP (181) | |||
| Trichostatin A (pan-HDAC inhibitor) | BRCA1 (45), KU70 (46), KU80 (46), DNA-PKcs (46) | KU70 (41) | ||
| SAHA (pan-HDAC inhibitor) | Glutathione (50), KU80 (47), RAD50 (180), MRE11 (180), RAD51 (47) | KU70 (41) | ||
| Sodium butyrate (Class I and IIA HDAC inhibitor) | KU70 (39), KU80 (39), DNA-PKcs (39) | |||
| Valproic acid (Class I HDAC inhibitor) | FANCD2 (16), CHK1 (40), BRCA1 (40), RAD51 (16,40) | |||
| MS-275 (HDAC1/2/3 inhibitor) | FANCD2 (16,84), RAD51 (16) | KU70 (41) | ||
| ACY-957 (HDAC1/2 inhibitor) | H4K91Ac (112) | |||
Histone deacetylases and base excision repair
DNA base modifications can be cytotoxic, as some prevent replicative polymerases from synthesizing DNA during S-phase, or they can be mutagenic, because of base mispairing during DNA synthesis. Modified DNA bases are excised by BER (60), which protects cells against these cytotoxic and mutagenic lesions. The base modifications excised by BER arise from spontaneous deamination (e.g. uracil), base oxidization induced by endogenous ROS (e.g. 8-oxoguanine and thymine glycol) and base alkylation induced by endogenous or therapy-induced alkylating agents (e.g. N3-methyladenine and N7-methylguanine). In addition to damage removal, BER excises 5-formylcytosine and 5-carboxylcytosine (61), the oxidation products of the epigenetic mark 5-methylcytosine and therefore play a role in active demethylation during epigenetic regulation.
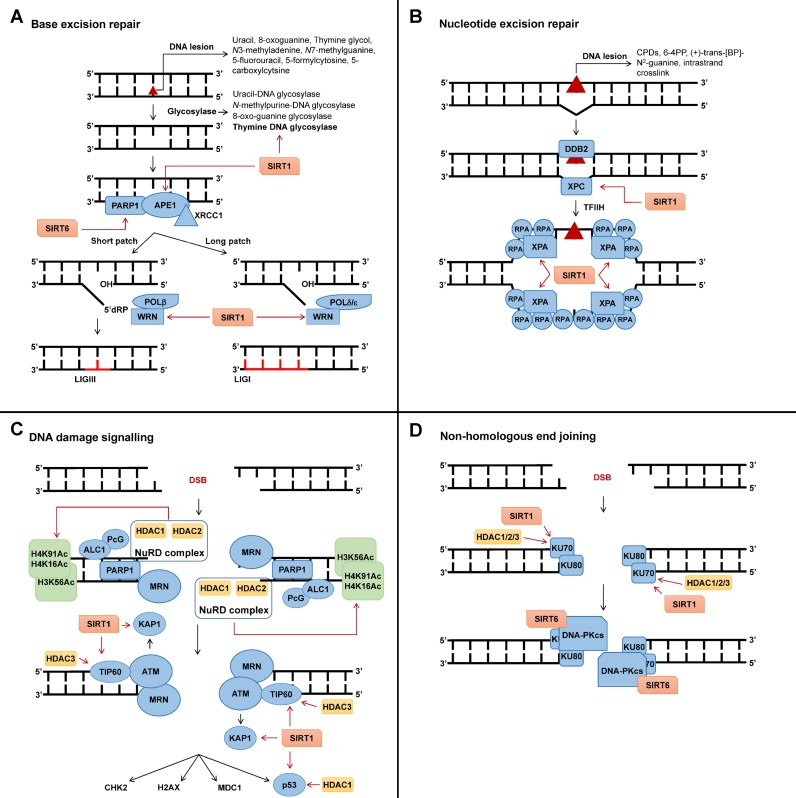
Repair by BER is initiated by excision of the modified base via hydrolysis of the N-glycosylic bond catalyzed by damage specific DNA glycosylases that leave an apurinic/apyrimidinic site (AP site) (60) (Figure 1A). DNA glycosylases differ in their substrate and enzymatic activity: the monofunctional uracil-DNA glycosylase or the N-methylpurine-DNA glycosylase only contain glycosylase activity, while bifunctional glycosylases such as 8-oxo-guanine glycosylase also contain lyase activity, allowing for the cleavage of the sugar–phosphate backbone 3′ to the AP site (62). The substrate specificity of the monofunctional thymine DNA glycosylase (TDG) is regulated via the deacetylase SIRT1 (63). Human SIRT1 is a nuclear SIRT (64–66). The deacetylation of TDG by SIRT1 stimulates active DNA demethylation as un-acetylated TDG excises 5-formylcytosine and 5-carboxylcytosine (61), while acetylated TDG has a higher substrate specificity toward 5-fluorouracil (63), induced by the chemotherapeutic 5-FU (Figure 1A). The low expression of SIRT1 in colorectal cancers (34) (Table 1), should, therefore, very likely contribute to this cancer's resistance to 5-FU.
Figure 1.
The role of histone deacetylases (HDACs) and SIRTs in the DNA damage response. Please refer to main text for detailed discussion. (A) Influence of NAD+ dependent sirtuin (SIRT1) and 6 on base excision repair (BER). SIRT1 deacetylates thymine DNA glycosylase (TDG), AP-endonuclease 1 (APE1) and RecQ protein Werner (WRN), thereby contributing to the excision of base lesions and repair by the short and long patch repair pathways. SIRT6 contribute to the activation of poly (ADP-ribose) polymerase 1 (PARP1). (B) The role of SIRT1 in nucleotide excision repair (NER). SIRT1 deacetylates xeroderma pigmentosum complementation group A (XPA), thereby contributing to the stabilization of the single-stranded DNA (ssDNA) NER intermediate, which is important for transcription-coupled and global genomic repair. SIRT1 furthermore stimulate the expression of XPC. (C) HDAC1, 2, 3 and SIRT1 mediate the effective activation of DNA damage signalling. HDAC1 and 2 deacetylate histone 3 (K56) and 4 (K16 and K91), while SIRT1 and HDAC3 deacetylate TIP60. These deacetylations facilitate the effective activation of ataxia telangiectasia mutated (ATM) at double-strand breaks (DSBs). (D) The role of HDAC1, 2, 3 and SIRT1 in the sealing of DSBs by non-homologous end joining (NHEJ). HDAC1, 2 and 3 and SIRT1 deacetylate KU70. These deacetylations stimulate the detection of DSBs by the KU70/KU80 heterodimer and the activation of DNA-dependent protein kinase catalytic subunit (DNA-PKcs).
Following excision of the base by glycosylases, AP sites and single-strand breaks (SSBs) are bound by poly (ADP-ribose) polymerase 1 (PARP1) (67,68), which modifies itself and the surrounding proteins in chromatin by covalently attaching poly-ADP-ribose (PAR) chains in a process termed PARylation. The negatively charged PAR chains, derived from NAD+ (69), and PARP1 allows for the recruitment of proteins required for BER (70). Extreme levels of base modifications, subject to repair by BER, will trigger cell death due to the PARP1-dependent catastrophic depletion of NAD+ (53). Following the induction of tolerable levels of base lesions, SIRT6 protects cells against these lesions by contributing to the activation of PARP1 (71,72). Similar to SIRT1, SIRT6 is also found in the nucleus of cells (65,73,74). Mouse embryonic fibroblasts and embryonic stem (ES) cells obtained from SIRT6-deficient mice are more sensitive to methyl methanesulfonate (MMS) and H2O2, that induce cytotoxic lesions like N3-methyladenine and thymine glycol, respectively (58). Interaction of PAR and PARP1 with X-ray repair cross-complementing protein (XRCC1) stimulates the BER factor AP-endonuclease 1 (APE1), which is required for the cleavage of the phosphate backbone of DNA 5′ to the monofunctional glycosylases generated AP site, giving rise to an SSB (60). Upon the induction of DNA damage, SIRT1 associates with, and, deacetylates lysines 6 and 7 of APE1 (75) (Figure 1A). The deacetylation of APE1 by SIRT1 stimulates its endonuclease activity by promoting its binding with XRCC1 and protects cells against base lesions induced by MMS and H2O2 (75). Following the cleavage by APE1, polymerase β (POLβ) inserts the new nucleotide (60). The RecQ protein Werner (WRN), which contains helicase and exonuclease activity, stimulates POLβ strand displacement during the nucleotide insertion by its helicase activity (76). WRN can be acetylated by the histone acetyltransferase p300 (77), which causes its inactivation. SIRT1 counteracts this by deacetylating WRN (78) and, therefore, stimulates the insertion of the new nucleotide by POLβ (79) (Figure 1A). POLβ additionally has phosphodiesterase activity, which hydrolyzes unmodified 5′ ends, thereby making them a substrate for DNA ligase III (LIGIII) that seals the ends (60). Under these conditions, only one nucleotide is inserted and is therefore referred to as short-patch BER. If the 5′ end is modified (oxidized, reduced), polymerase δ (POLδ) or polymerase ϵ continue inserting up to 10 nucleotides (60). The exonuclease activity of WRN has been shown to play a role in long patch BER (79). Here, the deacetylation of WRN by SIRT1 also contributes to effective BER as the deacetylation of WRN stimulates its exonuclease activity (78) (Figure 1A). Long patch BER leads to the displacement of the original strand by the newly synthesized strand, which is removed by flap endonuclease (FEN1) and the nick is sealed by DNA ligase I (LIGI) (60).
The role of nuclear SIRTs in regulating BER via TDG substrate specificity and APE1 and WRN enzymatic activity is telling as these deacetylases are NAD+ dependent (1). As stated, the activation of PARP1 during BER leads to the depletion of NAD+ due to the PARylation at the sites of damage. The level of damage, and consequently, the level of PARylation, will determine whether the cell has sufficient NAD+ for the completion of repair by BER as very high levels of DNA damage will deplete NAD+ and, therefore, inhibit the catalytic activity of the SIRTs.
Histone deacetylases and nucleotide excision repair
NER removes 22–30 nucleotides surrounding a helix-distorting DNA lesion (80). Examples of these bulky lesions are ultraviolet light-induced cyclobutane pyrimidine dimers and 6-4 pyrimidone photoproducts (6-4PP), adducts formed by epoxide intermediates during detoxification of polycyclic aromatic hydrocarbons (e.g. (+)-trans-benzo(a)pyrene 7,8,9 triol-N2-guanine) that are found in tobacco smoke and food, and intrastrand crosslinks formed by chemotherapeutics like cisplatin. Two NER pathways have been described, namely global genome repair (GG-NER), which removes bulky adducts independent of transcription, and transcription-coupled repair (TC-NER), which is limited to actively transcribed genes (62). In GG-NER, xeroderma pigmentosum complementation group C (XPC) binds to a small single-stranded piece of DNA opposite the actual lesion caused by lesion-related impaired base pairing (Figure 1B) (80). The deacetylase activity of SIRT1 stimulates the expression of XPC (81), thereby contributing to the initiation of GG-NER by XPC and the protection of cells against ultraviolet light, platinum drugs or other crosslinking agents (82).
Once XPC has bound, it recruits the multiprotein complex transcription factor IIH (TFIIH) and its subunits XPB and XPD mediate strand opening, unwinding as well as lesion verification (80). Next, XPA and RPA stabilize the structure and orchestrate the assembly of the XPG endonuclease responsible for the 3′ incision and the excision repair cross-complementation group 1 (ERCC1)-XPF endonuclease responsible for the 5′ incision (80). Here, the SIRT1 deacetylase has been shown to increase the interaction of XPA with NER proteins by deacetylating it (83) (Figure 1B). ERCC1-XPF incises 5′ and XPG 3′ to the lesion, respectively, releasing the DNA fragment containing the lesion (80). Nuclear SIRT1, therefore, plays a dual role in promoting NER. Firstly, it stimulates lesion recognition by promoting the expression of XPC on gene level (81) and, secondly, it stimulates lesion excision by promoting the assembly of NER endonucleases at the lesion site by the deacetylation of XPA (83).
Apart from the role of SIRT1 in NER, the overexpressed HDACs in melanoma cells (16,26) (Table 1) seem to also play a role in stimulating NER, as HDAC inhibition by sodium butyrate, an inhibitor of class I and class IIA HDACs (84), inhibits removal of bulky lesions by NER in these cells (38). Contrary to melanoma cells, HDAC inhibition with sodium butyrate in normal human fibroblasts enhances NER upon UV irradiation (85). These seemingly contradictory findings in cancer cells overexpressing HDACs and normal cells with normal HDAC expression illustrate the importance in determining the exact mechanism whereby HDAC inhibitors modulate DNA repair of different DNA lesions in different cell systems.
Histone deacetylases and mismatch repair
MMR corrects spontaneously occurring base mismatches (62) and has been shown to play a role in trinucleotide repeat (TNR) expansions (86). Lesions that do not follow Watson–Crick base pairing are detected and bound by either the MutSα heterodimer, comprised of MSH2 and MSH6 or the MutSβ heterodimer, comprised of MSH2 and MSH3 (62). MutSα detects mispairs while MutSβ detects insertion or deletion loops (87). MutSα not only binds to post-replicative base mismatches but also to DNA lesions such as O6-methylguanine, induced by methylating chemotherapeutics like temozolomide and dacarbazine, paired with cytosine or thymine (88). Acetylation of MSH2, by a hereto unknown mechanism, increases its stability and therefore contributes to mismatch detection and effective MMR. HDAC6 and 10 have been shown to deacetylate MSH2 (89,90). HDAC6 is predominantly found in the cytoplasm of cells (91,92), but upon the induction of DNA damage (89) or differentiation (93), a fraction of HDAC6 becomes nuclear. HDAC10 is found in both the cytoplasm and the nucleus of the cell (94,95). Deacetylation of MSH2 by HDAC6 and consequent ubiquitination causes MSH2's degradation by the proteasome (89). HDAC6 may, therefore, suppress effective MMR mediated by MutSα. For chemotherapeutic agents that induce O6-methylguanine in DNA, MutSα activity and O6-methylguanine cytotoxicity show an inverse relationship (96). The deacetylation of MSH2 by HDAC6 and its proteasomal degradation decreases MMR activity and causes the resistance of cells to this class of chemotherapeutics (89).
An example of insertions that are detected by MutSβ is trinucleotide repeat (TNR) expansions. TNRs (e.g. CAG) are found in many genes. Below a certain number, these trinucleotides are tolerated very well, but once they expand, the polyglutamine tract, coded by the CAG repeats, can cause the protein to aggregate. This invariably leads to TNR disorders such as Huntington's disease. MutSβ, HDAC3 and 5 promote TNR expansions, very likely through a common pathway (86,97).
Histone deacetylases and DNA damage signalling
Central players in the DDR are three phosphatidylinositol 3-kinase-related kinases, namely ataxia telangiectasia mutated (ATM), ataxia telangiectasia and Rad3-related (ATR) and DNA-dependent protein kinase (DNA-PK). The detection and initial remodelling of double-strand breaks (DSBs) and stalled replication forks fulfil the following fundamental functions: (i) downstream checkpoint activation that prevents cell cycle progression, (ii) recruitment of DNA repair proteins that facilitate DSB repair by NHEJ or lesion bypass in an HR and FA proteins dependent manner.
Strand breaks in the phosphate backbone of DNA, leading to DNA SSBs or DNA DSBs, are rapidly detected and bound by PARP1 (68,98) that leads to the PARylation of itself and proteins in the surrounding chromatin. SIRT6 promotes the activation of PARP1 at DSBs and the repair of DSBs by both NHEJ and HR (71,99). For DSBs, PARP1 and the PAR chains cause the recruitment of the nucleosome remodelling deacetylase (NuRD) complex (100,101) and the polycomb group (PcG) proteins (101–103), all of which are essential for the effective repair of DSBs (100,102–104) (Figure 1C). The NuRD complex, comprised of the deacetylases HDAC1 and 2, the histone-binding proteins RbAp46 and RbAp48, the metastasis-associated proteins (MTA1 or 2 or 3), the methyl-CpG-binding domain protein (MBD3 or 2) and the chromodomain-helicase-DNA-binding protein (CHD3 or 4), is recruited to the DSB via CHD4 in a PARP1 dependent manner (100,101). CHD4 and NuRD are required for the subsequent steps in DSB repair stimulated by the ring finger protein 8 (RNF8) and RNF168 (105–107). HDAC1 and 2, which are exclusively found in the nucleus of cells as they do not contain a nuclear export signal (108,109), deacetylate acetylated lysine 56 of histone 3 (H3K56Ac) (110,111) and acetylated lysine 16 of histone 4 (H4K16Ac) (110) (Figure 1C). In addition, inhibition of HDAC1 and 2 increases acetylation of histone 4 at lysine 91 (H4K91Ac) (112) (Figure 1C). Decreased H3K56Ac, H4K16Ac and H4K91Ac deacetylation due to the lack of or inhibition of, HDAC1 and 2 were paralleled by decreased survival of cells upon the induction of DSBs (110,112), and HDAC1 and 2 could, therefore, promote DSB repair by removing histone marks at DSBs. The PcG polycomb repressive complex 1 (PRC1) contributes to the ubiquitination of histone H2A following the induction of DSBs (103,113–115). Phosphorylation of KREB-associated protein 1 (KAP1) by ATM relaxes the chromatin structure and stimulates DSB repair (116) and, therefore, play a role in the repair of DSBs in heterochromatin. Here, SIRT1 has been shown to play a role as it can deacetylate KAP1 thereby positively regulating DSB repair (117). HDAC1 and 2, therefore, play a central role in preparing the chromatin for the activation of the DDR via ATM (118), while SIRT1 contributes to the repair of DSBs in heterochromatin (117).
The MRN complex, consisting of MRE11, RAD50 and NBS1, is recruited to DSBs (119) (Figure 1C). MRE11 has exo- and endonuclease activity, RAD50 has DNA binding capabilities and NBS1 is responsible for shuttling the MRN complex into the nucleus (120). The recruitment of the MRN complex to DSBs requires PARP1 (98). ATM in complex with the histone acetyltransferase (HAT) TIP60 is then recruited to the DSB whereupon ATM becomes activated in an acetylation-dependent autophosphorylation mechanism (121,122). Two nuclear deacetylases (108), namely HDAC3 and SIRT1, have been shown to deacetylate TIP60, thereby regulating its stability and activity (123,124) (Figure 1C). Whereas the deacetylation of TIP60 by SIRT1 negatively regulates its activity, at least in vitro (124), deacetylation by HDAC3 stabilizes TIP60 thereby increasing its half-life (123) and this might explain why HDAC3 is essential for the maintenance of chromatin structure and genomic stability (125). ATM interacts with HDAC1 both in vitro and in vivo and this interaction is stimulated by DSBs (126). ATM further increases the activity of HDAC1 by activating protein phosphatase 1 (PP1), which dephosphorylates HDAC1 (127). Once activated, ATM phosphorylates more than 700 downstream targets (128), including checkpoint kinase 2 (CHK2) (129), which plays a role in cell cycle progression, and histone 2AX (H2AX) and mediator of DNA damage checkpoint 1 (MDC1) (130,131), which play roles in DNA repair.
Apurinic/apyrimidinic sites, SSBs and bulky DNA adducts block replicative DNA polymerases that leads to the uncoupling of the polymerase from the replicative helicase and the formation of long stretches of single-stranded DNA (ssDNA) at the stalled DNA replication fork (132). These fragile structures are protected by replication protein A (RPA), and RPA bound to ssDNA then recruits ATR complexed with ATR-interacting protein (133–135). ATR is found in a complex with HDAC2 and CHD4, where HDAC2 may contribute to chromatin remodelling and consequently the downstream signalling of ATR (136). Important downstream targets of ATR are checkpoint kinase 1 (CHK1) that plays a role in cell cycle progression, H2AX and breast cancer 1 (BRCA1) (137) that contribute to DNA repair and also facilitate the switch from NHEJ to HR (138), among others. If the replication block cannot be bypassed due to excessive amounts of damage, it collapses into a DSB which is detected by ATM and DNA-PK (139). Inhibition of the overexpressed class I HDACs in prostate cancer (13–15) by valproic acid decreases the E2F1-dependent expression of CHK1 and BRCA1 (40) and could, therefore, attenuate the protective role of the DDR in this cancer.
The kinases ATM, ATR, CHK2 and CHK1 phosphorylate p53, thereby preventing its proteasomal degradation mediated by the E3 ubiquitin–protein ligase MDM2 (140,141). p53 is also acetylated in a DNA damage-dependent manner. The HATs PCAF and p300 acetylate p53 at K382 and K320, respectively (142,143), increasing its stability and DNA binding specificity. p53 is a transcription factor that transcribes DNA repair genes such as DDB2, XPC and POLH (144), cell cycle regulating genes such as CDKN1A (145) and apoptosis genes such as p53AIP1, PIG3, NOXA, PUMA and DR5 (146). HDAC1, 2 and 3 directly interact with the p53 protein (147), where HDAC1 modulates p53's transcriptional activity (148). SIRT1 deacetylates K382 of p53 thereby deactivating it, and preventing the initiation of p53 dependent apoptosis (149,150).
The role of histone deacetylases in the choice of DNA double-strand-break repair pathways
Non-homologous end joining versus homologous recombination
The MRN complex initiates the process whereby the cell regulates whether DSBs are repaired by NHEJ or HR. Simply put, if end-resection of the DSB by MRE11, CtBP-interacting protein (CtIP) and exonuclease 1 (EXO1) occurs then HR repairs the DSB, otherwise, NHEJ repairs it (151–154). ATM, bound to DSBs phosphorylates H2AX (forming γH2AX) (130). The protein MDC1 binds to γH2AX and in turn is also phosphorylated by ATM (131). MDC1 serves as a scaffold for the E3 ubiquitin ligase RNF8 that contributes to the ubiquitin signalling at H2A (155,156) very likely in a mechanism relying on the linker histone H1 (157). Ubiquitinated H2A then recruits RNF168 that amplifies the ubiquitin signal at sites of damage (158). The ubiquitin binding protein Rap80/Abraxas along with BRCA1 (159) and p53-binding protein 1 (53BP1) (160) are recruited to this site. The recruitment of 53BP1 to the DSB is mediated by the interaction of its methyl-lysine-binding Tudor domain with H4K20me2 (161) and its carboxy-terminal ubiquitination-dependent recruitment motif with ubiquitinated H2A (K15) (160). 53BP1 prevents the end-resection of DNA by MRE11, CtIP and EXO1 (162) in an RAP1 interacting factor homolog (RIF1)-dependent manner (163) and therefore 53BP1 stimulates NHEJ. The binding of 53BP1 to H4K20me2 can be prevented by the TIP60-dependent acetylation of H4K16 (164) and H2AK15 (165). Acetylation of H4K16 disrupts the interaction between H4K16 and Glu1551 in 53BP1 (164) while the acetylation of H2AK15 prevents this site from becoming ubiquitinated by RNF168 (165). TIP60, therefore, stimulates HR by preventing the localization of 53BP1 to DBSs (164). HDAC1 and 2 deacetylate H4K16 (110), leading to the stimulation of RNF8/RNF168-dependent ubiquitination at the DSB (107) and repair by NHEJ (110). In addition to the regulation of 53BP1 localization to the DSB by acetylation/deacetylation, the inhibitory effect of 53BP1-RIF1 on MRE11 can also be overcome in a cell cycle-dependent manner. This is accomplished by the cyclin dependent kinase (CDK)-dependent phosphorylation of CtIP (166), leading to increased activity and BRCA1 binding. BRCA1-CtIP is able to inhibit 53BP1, thereby allowing for the end-resection by MRE11, CtIP and EXO1 (167) and consequently for HR to occur (138). The pan-HDACi trichostatin A has been shown to downregulate BRCA1 in squamous carcinoma cells (45) thereby impeding the switch from NHEJ to HR.
Histone deacetylases and non-homologous end joining
The re-ligation of two DNA DSB ends, induced by, for example, ionizing radiation, occurs predominantly by NHEJ (168) (Figure 1D) that does not require homology. It should always be kept in mind that repair of DSBs occurs in chromatin, and chromatin-modifying enzymes are therefore centrally involved in this process. Consequently, it has been shown that the deacetylation of H3K56Ac and H4K16Ac by HDAC1 and 2 stimulates repair by NHEJ (110). Furthermore, knockout of Sirt7 in mice leads to an increase in H3K18Ac levels and impaired NHEJ activity (169). The deacetylation of KAP1, a protein required for the relaxation of the chromatin structure during DSB repair, by SIRT1 also stimulates NHEJ (117). NHEJ is initiated by the binding of a heterodimer, consisting of KU70 and KU80, to the DSB ends, which protects the DSB ends from degradation and attack from exonucleases (170). The class I histone deacetylases HDAC1, 2 and 3 and the class III SIRT1 have been shown to play a role in the deacetylation of KU70 (41,171) (Figure 1D). Inhibition of HDAC1, 2 and 3 in prostate cancer cells leads to increased acetylated KU70 levels, decreased KU70 DSB binding, decreased DSB repair and increased sensitivity to DSB inducing chemotherapeutics (41). The KU70/KU80 dimer serves as a scaffold for subsequent NHEJ repair factors. Interaction of KU70/80 with DNA-dependent protein kinase catalytic subunit (DNA-PKcs) activates the kinase activity of the complex, which allows for the autophosphorylation of DNA-PKcs itself and the phosphorylation of downstream NHEJ proteins (172). SIRT6 stabilizes and localizes DNA-PKcs at the site of the DSB (173) (Figure 1D). In addition to its roles in deacetylating KU70 and TIP60, SIRT1 deacetylates HDAC1, thereby stimulating HDAC1 activity and NHEJ repair (174). Inhibiting HDACs in non-small cell lung cancer with the pan-HDACi trichostatin A (84) downregulates KU70, KU80 and DNA-PKcs thereby sensitising to ionizing radiation (46), while HDAC inhibition using the pan-HDACi SAHA (84) in osteosarcoma and rhabdomyosarcoma cell lines downregulated KU80 that also sensitized to ionizing radiation (47). Using the more specific HDACi sodium butyrate, that only inhibits class I and class IIA HDACs (84), in melanoma cells the downregulation of KU70, KU80 and DNA-PKcs and the sensitization to ionizing radiation was still observed (39).
Histone deacetylases and homologous recombination
HR-mediated repair of DSBs (175) differs from NHEJ as it makes use of homologous sequences preferably on sister chromatids. It is therefore regarded as error free but limited to the late S and G2 phase of the cell cycle. It essentially contributes to the resolution of replication-coupled errors like stalled replication forks and repairs one-ended DSBs resulting from collapsed replication forks (176). HDAC3 has been shown to be required for the resolution of replication-coupled errors as knockout of Hdac3 in mice or inhibition of HDAC3 in lymphoma cells decreases the cells’ ability to deal with replication stress (177,178). As stated, HR is initiated by the end-resection of the DSB by MRE11, in complex with RAD50 and NBS1, and CtIP (152,153). SIRT1 can deacetylate NBS1 (179), which could stimulate HR. Inhibition of HDACs with the pan-HDACi SAHA downregulates RAD50 and MRE11 in cancer cells thereby contributing to the inhibition of repair upon damage induction (180). SIRT6 also promotes DNA end-resection as it deacetylates CtIP (181). The deacetylation of CtIP by SIRT6 stimulates HR, and, consequently, SIRT6 depletion sensitizes to chemical agents like camptothecin and PARP inhibitors that induce replication-dependent DSBs (181). End-resection of the DSB by MRE11 and CtIP leads to 3′ overhangs that can be extended by the 5′-3′ exonuclease EXO1 (154). The resulting 3′ ssDNA ends are bound by RPA and as SIRT6 promotes end-resection, SIRT6 depletion decreases the amount of RPA coated single-stranded DNA at sites of DSBs (181). Central proteins in HR are RAD51 and its paralogs XRCC3, XRCC2, RAD51D, RAD51C and RAD51B. RPA is displaced from the ssDNA by RAD51 with the help of breast cancer 2 (BRCA2) to form RAD51 filaments (182). Of interest, the expression of the HR proteins SIRT6, RAD51, RAD51C, RAD52 and NBS1 decrease with a cell's increasing replicative age, and is thought to contribute to the senescence-related decrease in repair efficiency by this pathway (183). Overexpression of SIRT6 prevents the loss of HR repair activity during this process (183).
The 3′ RAD51-nucleofilament then aligns with homologous regions of dsDNA followed by strand invasion of the dsDNA and displacement of the complementary strand, resulting in the displacement-loop (D-Loop). Inhibition of class I HDACs by MS-275 or knockdown of HDAC2 decreases RAD51 expression and impedes HR in melanoma cells (16). In osteosarcoma and rhabdomyosarcoma cells the pan-HDACi SAHA caused downregulation of RAD51 (47) while in prostate cancer cells inhibition of class I HDACs by valproic acid also decreased RAD51 expression, which the authors ascribed to a decrease in E2F1 promoter binding following HDAC inhibition (40). An unexpected finding in the cervical cancer cell line HeLa was that upon HDAC9 or 10 knockdown, HeLa cells showed decreased HR repair activity and became more sensitive to mitomycin C, a chemical agent inducing DNA interstrand crosslinks (184). Although a fraction of HDAC9 and 10 are localized in the nucleus (92,94,95,185), we should state that the mechanism whereby HDAC9 and 10 stimulate HR remains unclear. The 3′ end of the (broken) invaded strand can serve as a primer for DNA synthesis by POLδ in association with PCNA, which uses the intact homologous DNA strand as the template. PCNA interacts with HDAC1 in human cells in vitro and a considerable fraction of PCNA and HDAC1 co-localizes in the cell nucleus (186). The resulting HR structure can be resolved by either the synthesis-dependent strand annealing pathway or the double-strand break repair pathway (175,187,188).
Histone deacetylases and repair of interstrand crosslinks
The covalent joining of two strands of a DNA double helix is induced by platinum-based drugs, cyclophosphamide, mitomycin C and chloroethylating agents. ICLs are effective against cancer because they block RNA transcription and DNA replication and therefore kill cancer cells due to their high metabolic and replication rates. ICLs are detected in any cell cycle phase by the NER protein XPC if the crosslink distorts the DNA (189), or by CSB (190) if met by RNAPII. The deacetylase SIRT1 might, therefore, facilitate the detection of ICL due to it stimulating the expression of XPC (81), which could lead to the resistance of cells to interstrand crosslinkers like fotemustine (191). ERCC1-XPF unhooks the ICL by an incision close to the lesion and gap filling requires translesion synthesis (192). As the localization of ERCC1-XPF to the DNA lesion is XPA-dependent, and SIRT1 increases the interaction of XPA with ERCC1-XPF (83), deacetylation of XPA by SIRT1 could contribute to the unhooking of the ICL during repair. HDACs also promote the NER-dependent removal of ICLs as sodium butyrate decreases the repair rate of psoralen plus UVA-induced ICLs (38).
ICL repair at stalled replication forks makes use of proteins belonging to the Fanconi anaemia (FA) complementation group and they initiate replication-dependent repair (192). The replication fork-encountered ICL is recognized by FANCM, which mediates fork regression resulting, most likely, in a chicken foot DNA structure (192). Next, heterodimers of FANCD2 and FANCI are monoubiquitinated by the FANCL subunit of the FA core complex (FANCA, FANCB, FANCC, FANCE, FANCF, FANCG and FANCL) (192). Monoubiquitinated FANCD2 recruits nucleases that very likely promote dual incision and unhooking of the ICL by the structure-specific endonucleases SLX4-SLX1, ERCC1-XPF and MUS81-EME1, as well as the Fanconi-associated nuclease 1 (192). Exposing melanoma cells to the HDACi MS-275, that inhibits HDAC1, 2 and 3 (84), caused a transcriptional downregulation of FANCD2 and sensitized these cells to the chloroethylating agent fotemustine (16). The unhooking of the ICL allows for the HR-mediated restart of the replication fork that proceeds across the unhooked lesion with the help of the translesion polymerases REV1 and polymerase ζ (POLζ) (192). The regulation of HR by SIRT1 (179), SIRT6 (181), HDAC9 and 10 (184) may, therefore, play important roles in HR-mediated fork restart. Partial support for this is found in the observation that HDAC9 or 10 knockdown sensitizes cells to the interstrand crosslinker mitomycin C (184). Following fork restart, the unhooked lesion is removed enzymatically by NER or spontaneously by hydrolysis (192). Inhibiting class I and class IIA HDACs with phenylbutyrate interferes with the FA pathway and sensitizes head and neck cancer to cisplatin (44) in a to date unknown mechanism, but might be due to the downregulation of FANCD2, as seen upon inhibition of class I HDACs in melanoma cells (16).
Histone deacetylases and DNA repair in cancer
The class I HDACs contribute to DNA damage signalling, NHEJ and HR on multiple levels (Table 2). HDAC1 and 2 have a central role in preparing the chromatin surrounding the DSB for the activation of the DDR and sealing of the DSB by NHEJ or HR. During the DDR, HDAC3 deacetylates TIP60 (123) that leads to the activation and stimulation of ATM. HDAC2 and 3 play a hereto undefined role in how cells process replication blocking lesions (136,177,178). These deacetylases deacetylate p53 (143,147,148) and KU70 (41), thereby contributing to p53's and KU70's respective transcriptional and DNA binding activities. Apart from these direct deacetylation targets, HDAC2, in addition, promotes the expression of RAD51 (16) a central protein involved in HR. In colorectal (5–7,17–19), stomach (5,8,9,20), esophagus (5,10), breast (5,11,12), ovaries (5), lung (5,21), pancreatic (5), thyroid (5), prostate (13–15) and melanoma (16) cancers where these HDACs are overexpressed, combining genotoxin based therapies with small molecule inhibitors like sodium butyrate, valproic acid, trichostatin A, SAHA or MS-275 could be of therapeutic advantage. Butyrate based HDACi have been shown to sensitize melanoma cells (38,39) to interstrand crosslinking agents and ionizing radiation, breast cancer cells to etoposide (42) and head and neck cancer to cisplatin (44). Valproic acid has been shown to sensitize melanoma cells to alkylating chemotherapeutics and ionizing radiation (16,40) while sensitizing prostate cancer cells to cisplatin and ionizing radiation (40). Trichostatin A sensitizes lung cancer cells to ionizing radiation (46) and prostate cancer cells to bleomycin, 5-FU and doxorubicin (41). MS-275 sensitizes melanoma cells to alkylating agents, olaparib and ionizing radiation (16) and prostate cancer cells to bleomycin, 5-FU and doxorubicin (41), while SAHA sensitizes osteosarcoma and rhabdomyosarcoma cells to ionizing radiation (47), prostate cancer cells to bleomycin, 5-FU and doxorubicin (41) and breast cancer cells to olaparib (43).
In addition to the effect of the class I HDACs on DNA repair, the class IIB histone deacetylase HDAC6 affects MMR due to the HDAC6 mediated breakdown of MSH2 (Table 2). Overexpressed HDAC6 in melanoma cancers (26) could, therefore, contribute to the resistance of this cancer to methylating agent based therapies with dacarbazine and temozolomide as these chemotherapeutics require an intact MMR system to kill cells (193).
The deacetylation and resulting stimulation of the DNA repair factors WRN (78), TDG (63), APE1 (75), XPA (83), TIP60 (124), p53 (149,150), KU70 (41,171), HDAC1 (174) and NBS1 (179) by the class III lysine deacetylase SIRT1 show the importance of this NAD+ dependent histone deacetylase in the regulation of BER, NER, DNA damage signalling, NHEJ and HR (Table 2). Another NAD+ dependent histone deacetylase, namely SIRT6, deacetylates CtIP (181) and contributes to the recruitment of DNA-PKcs to DSBs (173) and the activation of PARP1 (71,99) and therefore plays a role in HR and NHEJ (71). BER, DNA damage signalling, NHEJ and HR are initiated, at least in part, by PARP1 as PARP1 binds to and is activated by SSBs (68), AP-sites (67) and DSBs (98). The process of PARylation, catalyzed by PARP1, consumes NAD+ and excessive levels of DNA damage will, therefore, prevent the stimulation of DNA repair by the NAD+ dependent histone deacetylases SIRT1 and 6. These findings points to the tempting speculation that PARP1, SIRT1 and SIRT6 regulate a switch between repairable DNA damage and cell death, as excessive DNA damage triggers a necroptotic form of cell death termed parthanatos (53).
The questions of how inhibition of histone deacetylases cause the transcriptional repression of the DNA repair genes BRCA1 (40,45), KU70 (39,46), KU80 (39,46,47), DNA-PKcs (39,46), RAD50 (180), MRE11 (180), RAD51 (16,40,47), FANCD2 (16), CHK1 (40), FANCD2 (16,84) or whether the underlying mechanism for this gene regulation is cancer specific remains unresolved.
SUMMARY
It is clear that HDACs and SIRTs are involved in almost every aspect of DNA repair. This ranges from the detection of, and signalling from, DNA lesions to the removal or reversal of the damage. HDACs and/or SIRTs regulate the expression, the activation and the degradation of key factors involved in DNA damage signalling, excision repair, strand break annealing and DNA replication block bypass pathways, while the interplay between SIRTs, PARP1 and NAD+ hints at a mechanism whereby a cell can differentiate between repairable and unrepairable damage. Targeting these lysine deacetylases during cancer therapy should, therefore, be ideal for sensitizing cancer cells to genotoxin based chemotherapeutics.
Acknowledgments
The authors thank Prof. Markus Christmann for critical reading of the manuscript. Work of the authors was supported by the German Research Foundation (DFG) RO3617.
FUNDING
German Research Foundation (DFG) [RO3617]. Funding for open access charge: Deutsche Forschungsgemeinschaft [RO3617].
Conflict of interest statement. None declared.
REFERENCES
- 1.Van Dyke M.W. Lysine deacetylase (KDAC) regulatory pathways: An alternative approach to selective modulation. ChemMedChem. 2014;9:511–522. doi: 10.1002/cmdc.201300444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fulda S. Histone deacetylase (HDAC) inhibitors and regulation of TRAIL-induced apoptosis. Exp. Cell Res. 2012;318:1208–1212. doi: 10.1016/j.yexcr.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Botrugno O.A., Santoro F., Minucci S. Histone deacetylase inhibitors as a new weapon in the arsenal of differentiation therapies of cancer. Cancer Lett. 2009;280:134–144. doi: 10.1016/j.canlet.2009.02.027. [DOI] [PubMed] [Google Scholar]
- 4.Gong F., Miller K.M. Mammalian DNA repair: HATs and HDACs make their mark through histone acetylation. Mutat Res. 2013;750:23–30. doi: 10.1016/j.mrfmmm.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Nakagawa M., Oda Y., Eguchi T., Aishima S., Yao T., Hosoi F., Basaki Y., Ono M., Kuwano M., Tanaka M., et al. Expression profile of class I histone deacetylases in human cancer tissues. Oncol. Rep. 2007;18:769–774. [PubMed] [Google Scholar]
- 6.Giannini R., Cavallini A. Expression analysis of a subset of coregulators and three nuclear receptors in human colorectal carcinoma. Anticancer Res. 2005;25:4287–4292. [PubMed] [Google Scholar]
- 7.Ozdag H., Teschendorff A.E., Ahmed A.A., Hyland S.J., Blenkiron C., Bobrow L., Veerakumarasivam A., Burtt G., Subkhankulova T., Arends M.J., et al. Differential expression of selected histone modifier genes in human solid cancers. BMC Genomics. 2006;7:90–115. doi: 10.1186/1471-2164-7-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi J.H., Kwon H.J., Yoon B.I., Kim J.H., Han S.U., Joo H.J., Kim D.Y. Expression profile of histone deacetylase 1 in gastric cancer tissues. Jpn. J. Cancer Res. 2001;92:1300–1304. doi: 10.1111/j.1349-7006.2001.tb02153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim J.H., Choi Y.K., Kwon H.J., Yang H.K., Choi J.H., Kim D.Y. Downregulation of gelsolin and retinoic acid receptor beta expression in gastric cancer tissues through histone deacetylase 1. J. Gastroenterol. Hepatol. 2004;19:218–224. doi: 10.1111/j.1440-1746.2004.03336.x. [DOI] [PubMed] [Google Scholar]
- 10.Toh Y., Yamamoto M., Endo K., Ikeda Y., Baba H., Kohnoe S., Yonemasu H., Hachitanda Y., Okamura T., Sugimachi K. Histone H4 acetylation and histone deacetylase 1 expression in esophageal squamous cell carcinoma. Oncol. Rep. 2003;10:333–338. [PubMed] [Google Scholar]
- 11.Krusche C.A., Wulfing P., Kersting C., Vloet A., Bocker W., Kiesel L., Beier H.M., Alfer J. Histone deacetylase-1 and -3 protein expression in human breast cancer: a tissue microarray analysis. Breast Cancer Res. Treat. 2005;90:15–23. doi: 10.1007/s10549-004-1668-2. [DOI] [PubMed] [Google Scholar]
- 12.Kawai H., Li H., Avraham S., Jiang S., Avraham H.K. Overexpression of histone deacetylase HDAC1 modulates breast cancer progression by negative regulation of estrogen receptor alpha. Int. J. Cancer. 2003;107:353–358. doi: 10.1002/ijc.11403. [DOI] [PubMed] [Google Scholar]
- 13.Patra S.K., Patra A., Dahiya R. Histone deacetylase and DNA methyltransferase in human prostate cancer. Biochem. Biophys. Res. Commun. 2001;287:705–713. doi: 10.1006/bbrc.2001.5639. [DOI] [PubMed] [Google Scholar]
- 14.Halkidou K., Gaughan L., Cook S., Leung H.Y., Neal D.E., Robson C.N. Upregulation and nuclear recruitment of HDAC1 in hormone refractory prostate cancer. Prostate. 2004;59:177–189. doi: 10.1002/pros.20022. [DOI] [PubMed] [Google Scholar]
- 15.Weichert W., Roske A., Gekeler V., Beckers T., Stephan C., Jung K., Fritzsche F.R., Niesporek S., Denkert C., Dietel M., et al. Histone deacetylases 1, 2 and 3 are highly expressed in prostate cancer and HDAC2 expression is associated with shorter PSA relapse time after radical prostatectomy. Br. J. Cancer. 2008;98:604–610. doi: 10.1038/sj.bjc.6604199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krumm A., Barckhausen C., Kucuk P., Tomaszowski K.H., Loquai C., Fahrer J., Kramer O.H., Kaina B., Roos W.P. Enhanced histone deacetylase activity in malignant melanoma provokes RAD51 and FANCD2-triggered drug resistance. Cancer Res. 2016;76:3067–3077. doi: 10.1158/0008-5472.CAN-15-2680. [DOI] [PubMed] [Google Scholar]
- 17.Ye P., Xing H., Lou F., Wang K., Pan Q., Zhou X., Gong L., Li D. Histone deacetylase 2 regulates doxorubicin (Dox) sensitivity of colorectal cancer cells by targeting ABCB1 transcription. Cancer Chemother. Pharmacol. 2016;77:613–621. doi: 10.1007/s00280-016-2979-9. [DOI] [PubMed] [Google Scholar]
- 18.Zhu P., Martin E., Mengwasser J., Schlag P., Janssen K.P., Gottlicher M. Induction of HDAC2 expression upon loss of APC in colorectal tumorigenesis. Cancer Cell. 2004;5:455–463. doi: 10.1016/s1535-6108(04)00114-x. [DOI] [PubMed] [Google Scholar]
- 19.Ashktorab H., Belgrave K., Hosseinkhah F., Brim H., Nouraie M., Takkikto M., Hewitt S., Lee E.L., Dashwood R.H., Smoot D. Global histone H4 acetylation and HDAC2 expression in colon adenoma and carcinoma. Dig. Dis. Sci. 2009;54:2109–2117. doi: 10.1007/s10620-008-0601-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song J., Noh J.H., Lee J.H., Eun J.W., Ahn Y.M., Kim S.Y., Lee S.H., Park W.S., Yoo N.J., Lee J.Y., et al. Increased expression of histone deacetylase 2 is found in human gastric cancer. APMIS. 2005;113:264–268. doi: 10.1111/j.1600-0463.2005.apm_04.x. [DOI] [PubMed] [Google Scholar]
- 21.Bartling B., Hofmann H.S., Boettger T., Hansen G., Burdach S., Silber R.E., Simm A. Comparative application of antibody and gene array for expression profiling in human squamous cell lung carcinoma. Lung Cancer. 2005;49:145–154. doi: 10.1016/j.lungcan.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 22.Oehme I., Deubzer H.E., Wegener D., Pickert D., Linke J.P., Hero B., Kopp-Schneider A., Westermann F., Ulrich S.M., von Deimling A., et al. Histone deacetylase 8 in neuroblastoma tumorigenesis. Clin. Cancer Res. 2009;15:91–99. doi: 10.1158/1078-0432.CCR-08-0684. [DOI] [PubMed] [Google Scholar]
- 23.Kang Z.H., Wang C.Y., Zhang W.L., Zhang J.T., Yuan C.H., Zhao P.W., Lin Y.Y., Hong S., Li C.Y., Wang L. Histone deacetylase HDAC4 promotes gastric cancer SGC-7901 cells progression via p21 repression. PLoS One. 2014;9:e98894. doi: 10.1371/journal.pone.0098894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen Y.F., Wei A.M., Kou Q., Zhu Q.Y., Zhang L. Histone deacetylase 4 increases progressive epithelial ovarian cancer cells via repression of p21 on fibrillar collagen matrices. Oncol. Rep. 2016;35:948–954. doi: 10.3892/or.2015.4423. [DOI] [PubMed] [Google Scholar]
- 25.He P., Liang J., Shao T., Guo Y., Hou Y., Li Y. HDAC5 promotes colorectal cancer cell proliferation by up-regulating DLL4 expression. Int. J. Clin. Exp. Med. 2015;8:6510–6516. [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J., Gu J., Feng Z., Yang Y., Zhu N., Lu W., Qi F. Both HDAC5 and HDAC6 are required for the proliferation and metastasis of melanoma cells. J. Transl. Med. 2016;14:7–20. doi: 10.1186/s12967-015-0753-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng G.W., Dong L.D., Shang W.J., Pang X.L., Li J.F., Liu L., Wang Y. HDAC5 promotes cell proliferation in human hepatocellular carcinoma by up-regulating Six1 expression. Eur. Rev. Med. Pharmacol. Sci. 2014;18:811–816. [PubMed] [Google Scholar]
- 28.Sakuma T., Uzawa K., Onda T., Shiiba M., Yokoe H., Shibahara T., Tanzawa H. Aberrant expression of histone deacetylase 6 in oral squamous cell carcinoma. Int. J. Oncol. 2006;29:117–124. [PubMed] [Google Scholar]
- 29.Cea M., Cagnetta A., Adamia S., Acharya C., Tai Y.T., Fulciniti M., Ohguchi H., Munshi A., Acharya P., Bhasin M.K., et al. Evidence for a role of the histone deacetylase SIRT6 in DNA damage response of multiple myeloma cells. Blood. 2016;127:1138–1150. doi: 10.1182/blood-2015-06-649970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ashraf N., Zino S., Macintyre A., Kingsmore D., Payne A.P., George W.D., Shiels P.G. Altered sirtuin expression is associated with node-positive breast cancer. Br. J. Cancer. 2006;95:1056–1061. doi: 10.1038/sj.bjc.6603384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Nigris F., Cerutti J., Morelli C., Califano D., Chiariotti L., Viglietto G., Santelli G., Fusco A. Isolation of a SIR-like gene, SIR-T8, that is overexpressed in thyroid carcinoma cell lines and tissues. Br. J. Cancer. 2002;87:1479–1479. doi: 10.1038/sj.bjc.6600636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim J.K., Noh J.H., Jung K.H., Eun J.W., Bae H.J., Kim M.G., Chang Y.G., Shen Q., Park W.S., Lee J.Y., et al. Sirtuin7 oncogenic potential in human hepatocellular carcinoma and its regulation by the tumor suppressors MiR-125a-5p and MiR-125b. Hepatology. 2013;57:1055–1067. doi: 10.1002/hep.26101. [DOI] [PubMed] [Google Scholar]
- 33.Cheng W., Li M., Cai J., Wang K., Zhang C., Bao Z., Liu Y., Wu A. HDAC4, a prognostic and chromosomal instability marker, refines the predictive value of MGMT promoter methylation. J. Neurooncol. 2015;122:303–312. doi: 10.1007/s11060-014-1709-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jang S.H., Min K.W., Paik S.S., Jang K.S. Loss of SIRT1 histone deacetylase expression associates with tumour progression in colorectal adenocarcinoma. J. Clin. Pathol. 2012;65:735–739. doi: 10.1136/jclinpath-2012-200685. [DOI] [PubMed] [Google Scholar]
- 35.Sebastian C., Zwaans B.M., Silberman D.M., Gymrek M., Goren A., Zhong L., Ram O., Truelove J., Guimaraes A.R., Toiber D., et al. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell. 2012;151:1185–1199. doi: 10.1016/j.cell.2012.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharma S.V., Lee D.Y., Li B., Quinlan M.P., Takahashi F., Maheswaran S., McDermott U., Azizian N., Zou L., Fischbach M.A., et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keshelava N., Davicioni E., Wan Z., Ji L., Sposto R., Triche T.J., Reynolds C.P. Histone deacetylase 1 gene expression and sensitization of multidrug-resistant neuroblastoma cell lines to cytotoxic agents by depsipeptide. J. Natl. Cancer Inst. 2007;99:1107–1119. doi: 10.1093/jnci/djm044. [DOI] [PubMed] [Google Scholar]
- 38.Toyooka T., Ibuki Y. Histone deacetylase inhibitor sodium butyrate enhances the cell killing effect of psoralen plus UVA by attenuating nucleotide excision repair. Cancer Res. 2009;69:3492–3500. doi: 10.1158/0008-5472.CAN-08-2546. [DOI] [PubMed] [Google Scholar]
- 39.Munshi A., Kurland J.F., Nishikawa T., Tanaka T., Hobbs M.L., Tucker S.L., Ismail S., Stevens C., Meyn R.E. Histone deacetylase inhibitors radiosensitize human melanoma cells by suppressing DNA repair activity. Clin. Cancer Res. 2005;11:4912–4922. doi: 10.1158/1078-0432.CCR-04-2088. [DOI] [PubMed] [Google Scholar]
- 40.Kachhap S.K., Rosmus N., Collis S.J., Kortenhorst M.S., Wissing M.D., Hedayati M., Shabbeer S., Mendonca J., Deangelis J., Marchionni L., et al. Downregulation of homologous recombination DNA repair genes by HDAC inhibition in prostate cancer is mediated through the E2F1 transcription factor. PLoS One. 2010;5:e11208. doi: 10.1371/journal.pone.0011208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen C.S., Wang Y.C., Yang H.C., Huang P.H., Kulp S.K., Yang C.C., Lu Y.S., Matsuyama S., Chen C.Y., Chen C.S. Histone deacetylase inhibitors sensitize prostate cancer cells to agents that produce DNA double-strand breaks by targeting Ku70 acetylation. Cancer Res. 2007;67:5318–5327. doi: 10.1158/0008-5472.CAN-06-3996. [DOI] [PubMed] [Google Scholar]
- 42.Li L., Sun Y., Liu J., Wu X., Chen L., Ma L., Wu P. Histone deacetylase inhibitor sodium butyrate suppresses DNA double strand break repair induced by etoposide more effectively in MCF-7 cells than in HEK293 cells. BMC Biochem. 2015;16:2–11. doi: 10.1186/s12858-014-0030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Min A., Im S.A., Kim D.K., Song S.H., Kim H.J., Lee K.H., Kim T.Y., Han S.W., Oh D.Y., Kim T.Y., et al. Histone deacetylase inhibitor, suberoylanilide hydroxamic acid (SAHA), enhances anti-tumor effects of the poly (ADP-ribose) polymerase (PARP) inhibitor olaparib in triple-negative breast cancer cells. Breast Cancer Res. 2015;17:33–46. doi: 10.1186/s13058-015-0534-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burkitt K., Ljungman M. Phenylbutyrate interferes with the Fanconi anemia and BRCA pathway and sensitizes head and neck cancer cells to cisplatin. Mol. Cancer. 2008;7:24–33. doi: 10.1186/1476-4598-7-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y., Carr T., Dimtchev A., Zaer N., Dritschilo A., Jung M. Attenuated DNA damage repair by trichostatin A through BRCA1 suppression. Radiat. Res. 2007;168:115–124. doi: 10.1667/RR0811.1. [DOI] [PubMed] [Google Scholar]
- 46.Zhang F., Zhang T., Teng Z.H., Zhang R., Wang J.B., Mei Q.B. Sensitization to gamma-irradiation-induced cell cycle arrest and apoptosis by the histone deacetylase inhibitor trichostatin A in non-small cell lung cancer (NSCLC) cells. Cancer Biol. Ther. 2009;8:823–831. doi: 10.4161/cbt.8.9.8143. [DOI] [PubMed] [Google Scholar]
- 47.Blattmann C., Oertel S., Ehemann V., Thiemann M., Huber P.E., Bischof M., Witt O., Deubzer H.E., Kulozik A.E., Debus J., et al. Enhancement of radiation response in osteosarcoma and rhabdomyosarcoma cell lines by histone deacetylase inhibition. Int. J. Radiat. Oncol. Biol. Phys. 2010;78:237–245. doi: 10.1016/j.ijrobp.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 48.Koprinarova M., Botev P., Russev G. Histone deacetylase inhibitor sodium butyrate enhances cellular radiosensitivity by inhibiting both DNA nonhomologous end joining and homologous recombination. DNA Repair (Amst) 2011;10:970–977. doi: 10.1016/j.dnarep.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 49.Kaewpiboon C., Srisuttee R., Malilas W., Moon J., Oh S., Jeong H.G., Johnston R.N., Assavalapsakul W., Chung Y.H. Upregulation of Stat1-HDAC4 confers resistance to etoposide through enhanced multidrug resistance 1 expression in human A549 lung cancer cells. Mol. Med. Rep. 2015;11:2315–2321. doi: 10.3892/mmr.2014.2949. [DOI] [PubMed] [Google Scholar]
- 50.Rikiishi H., Shinohara F., Sato T., Sato Y., Suzuki M., Echigo S. Chemosensitization of oral squamous cell carcinoma cells to cisplatin by histone deacetylase inhibitor, suberoylanilide hydroxamic acid. Int. J. Oncol. 2007;30:1181–1188. [PubMed] [Google Scholar]
- 51.Meijer C., Mulder N.H., Timmer-Bosscha H., Sluiter W.J., Meersma G.J., de Vries E.G. Relationship of cellular glutathione to the cytotoxicity and resistance of seven platinum compounds. Cancer Res. 1992;52:6885–6889. [PubMed] [Google Scholar]
- 52.Kim M.S., Blake M., Baek J.H., Kohlhagen G., Pommier Y., Carrier F. Inhibition of histone deacetylase increases cytotoxicity to anticancer drugs targeting DNA. Cancer Res. 2003;63:7291–7300. [PubMed] [Google Scholar]
- 53.Roos W.P., Thomas A.D., Kaina B. DNA damage and the balance between survival and death in cancer biology. Nat. Rev. Cancer. 2016;16:20–33. doi: 10.1038/nrc.2015.2. [DOI] [PubMed] [Google Scholar]
- 54.Tamm I., Wang Y., Sausville E., Scudiero D.A., Vigna N., Oltersdorf T., Reed J.C. IAP-family protein survivin inhibits caspase activity and apoptosis induced by Fas (CD95), Bax, caspases, and anticancer drugs. Cancer Res. 1998;58:5315–5320. [PubMed] [Google Scholar]
- 55.Seo S.K., Hwang C.S., Choe T.B., Hong S.I., Yi J.Y., Hwang S.G., Lee H.G., Oh S.T., Lee Y.H., Park I.C. Selective inhibition of histone deacetylase 2 induces p53-dependent survivin downregulation through MDM2 proteasomal degradation. Oncotarget. 2015;6:26528–26540. doi: 10.18632/oncotarget.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jin J.S., Tsao T.Y., Sun P.C., Yu C.P., Tzao C. SAHA inhibits the growth of colon tumors by decreasing histone deacetylase and the expression of cyclin D1 and survivin. Pathol. Oncol. Res. 2012;18:713–720. doi: 10.1007/s12253-012-9499-7. [DOI] [PubMed] [Google Scholar]
- 57.De Schepper S., Bruwiere H., Verhulst T., Steller U., Andries L., Wouters W., Janicot M., Arts J., Van Heusden J. Inhibition of histone deacetylases by chlamydocin induces apoptosis and proteasome-mediated degradation of survivin. J. Pharmacol. Exp. Ther. 2003;304:881–888. doi: 10.1124/jpet.102.042903. [DOI] [PubMed] [Google Scholar]
- 58.Mostoslavsky R., Chua K.F., Lombard D.B., Pang W.W., Fischer M.R., Gellon L., Liu P., Mostoslavsky G., Franco S., Murphy M.M., et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 59.Toiber D., Erdel F., Bouazoune K., Silberman D.M., Zhong L., Mulligan P., Sebastian C., Cosentino C., Martinez-Pastor B., Giacosa S., et al. SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol. Cell. 2013;51:454–468. doi: 10.1016/j.molcel.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Krokan H.E., Bjoras M. Base excision repair. Cold Spring Harb. Perspect. Biol. 2013;5:a012583. doi: 10.1101/cshperspect.a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maiti A., Drohat A.C. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J. Biol. Chem. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Christmann M., Tomicic M.T., Roos W.P., Kaina B. Mechanisms of human DNA repair: an update. Toxicology. 2003;193:3–34. doi: 10.1016/s0300-483x(03)00287-7. [DOI] [PubMed] [Google Scholar]
- 63.Madabushi A., Hwang B.J., Jin J., Lu A.L. Histone deacetylase SIRT1 modulates and deacetylates DNA base excision repair enzyme thymine DNA glycosylase. Biochem. J. 2013;456:89–98. doi: 10.1042/BJ20130670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Langley E., Pearson M., Faretta M., Bauer U.M., Frye R.A., Minucci S., Pelicci P.G., Kouzarides T. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 2002;21:2383–2396. doi: 10.1093/emboj/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Michishita E., Park J.Y., Burneskis J.M., Barrett J.C., Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol. Biol. Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tanno M., Sakamoto J., Miura T., Shimamoto K., Horio Y. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J. Biol. Chem. 2007;282:6823–6832. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- 67.Khodyreva S.N., Prasad R., Ilina E.S., Sukhanova M.V., Kutuzov M.M., Liu Y., Hou E.W., Wilson S.H., Lavrik O.I. Apurinic/apyrimidinic (AP) site recognition by the 5′-dRP/AP lyase in poly(ADP-ribose) polymerase-1 (PARP-1) Proc. Natl. Acad. Sci. U.S.A. 2010;107:22090–22095. doi: 10.1073/pnas.1009182107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Satoh M.S., Lindahl T. Role of poly(ADP-ribose) formation in DNA repair. Nature. 1992;356:356–358. doi: 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]
- 69.D'Amours D., Desnoyers S., D'Silva I., Poirier G.G. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999;342:249–268. [PMC free article] [PubMed] [Google Scholar]
- 70.El-Khamisy S.F., Masutani M., Suzuki H., Caldecott K.W. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31:5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mao Z., Hine C., Tian X., Van Meter M., Au M., Vaidya A., Seluanov A., Gorbunova V. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;332:1443–1446. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu Z., Zhang L., Zhang W., Meng D., Zhang H., Jiang Y., Xu X., Van Meter M., Seluanov A., Gorbunova V., et al. SIRT6 rescues the age related decline in base excision repair in a PARP1-dependent manner. Cell Cycle. 2015;14:269–276. doi: 10.4161/15384101.2014.980641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tennen R.I., Berber E., Chua K.F. Functional dissection of SIRT6: identification of domains that regulate histone deacetylase activity and chromatin localization. Mech. Ageing Dev. 2010;131:185–192. doi: 10.1016/j.mad.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liszt G., Ford E., Kurtev M., Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J. Biol. Chem. 2005;280:21313–21320. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- 75.Yamamori T., DeRicco J., Naqvi A., Hoffman T.A., Mattagajasingh I., Kasuno K., Jung S.B., Kim C.S., Irani K. SIRT1 deacetylates APE1 and regulates cellular base excision repair. Nucleic Acids Res. 2010;38:832–845. doi: 10.1093/nar/gkp1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Harrigan J.A., Opresko P.L., von Kobbe C., Kedar P.S., Prasad R., Wilson S.H., Bohr V.A. The Werner syndrome protein stimulates DNA polymerase beta strand displacement synthesis via its helicase activity. J. Biol. Chem. 2003;278:22686–22695. doi: 10.1074/jbc.M213103200. [DOI] [PubMed] [Google Scholar]
- 77.Blander G., Zalle N., Daniely Y., Taplick J., Gray M.D., Oren M. DNA damage-induced translocation of the Werner helicase is regulated by acetylation. J. Biol. Chem. 2002;277:50934–50940. doi: 10.1074/jbc.M210479200. [DOI] [PubMed] [Google Scholar]
- 78.Li K., Casta A., Wang R., Lozada E., Fan W., Kane S., Ge Q., Gu W., Orren D., Luo J. Regulation of WRN protein cellular localization and enzymatic activities by SIRT1-mediated deacetylation. J. Biol. Chem. 2008;283:7590–7598. doi: 10.1074/jbc.M709707200. [DOI] [PubMed] [Google Scholar]
- 79.Harrigan J.A., Wilson D.M., 3rd, Prasad R., Opresko P.L., Beck G., May A., Wilson S.H., Bohr V.A. The Werner syndrome protein operates in base excision repair and cooperates with DNA polymerase beta. Nucleic Acids Res. 2006;34:745–754. doi: 10.1093/nar/gkj475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Scharer O.D. Nucleotide excision repair in eukaryotes. Cold Spring Harb. Perspect. Biol. 2013;5:a012609. doi: 10.1101/cshperspect.a012609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ming M., Shea C.R., Guo X., Li X., Soltani K., Han W., He Y.Y. Regulation of global genome nucleotide excision repair by SIRT1 through xeroderma pigmentosum C. Proc. Natl. Acad. Sci. U.S.A. 2010;107:22623–22628. doi: 10.1073/pnas.1010377108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Clement F.C., Camenisch U., Fei J., Kaczmarek N., Mathieu N., Naegeli H. Dynamic two-stage mechanism of versatile DNA damage recognition by xeroderma pigmentosum group C protein. Mutat. Res. 2010;685:21–28. doi: 10.1016/j.mrfmmm.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 83.Fan W., Luo J. SIRT1 regulates UV-induced DNA repair through deacetylating XPA. Mol. Cell. 2010;39:247–258. doi: 10.1016/j.molcel.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 84.Bolden J.E., Peart M.J., Johnstone R.W. Anticancer activities of histone deacetylase inhibitors. Nat. Rev. Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 85.Smerdon M.J., Lan S.Y., Calza R.E., Reeves R. Sodium butyrate stimulates DNA repair in UV-irradiated normal and xeroderma pigmentosum human fibroblasts. J. Biol. Chem. 1982;257:13441–13447. [PubMed] [Google Scholar]
- 86.Gannon A.M., Frizzell A., Healy E., Lahue R.S. MutSbeta and histone deacetylase complexes promote expansions of trinucleotide repeats in human cells. Nucleic Acids Res. 2012;40:10324–10333. doi: 10.1093/nar/gks810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Genschel J., Littman S.J., Drummond J.T., Modrich P. Isolation of MutSbeta from human cells and comparison of the mismatch repair specificities of MutSbeta and MutSalpha. J. Biol. Chem. 1998;273:19895–19901. doi: 10.1074/jbc.273.31.19895. [DOI] [PubMed] [Google Scholar]
- 88.Duckett D.R., Drummond J.T., Murchie A.I., Reardon J.T., Sancar A., Lilley D.M., Modrich P. Human MutSalpha recognizes damaged DNA base pairs containing O6-methylguanine, O4-methylthymine, or the cisplatin-d(GpG) adduct. Proc. Natl. Acad. Sci. U.S.A. 1996;93:6443–6447. doi: 10.1073/pnas.93.13.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang M., Xiang S., Joo H.Y., Wang L., Williams K.A., Liu W., Hu C., Tong D., Haakenson J., Wang C., et al. HDAC6 deacetylates and ubiquitinates MSH2 to maintain proper levels of MutSalpha. Mol. Cell. 2014;55:31–46. doi: 10.1016/j.molcel.2014.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Radhakrishnan R., Li Y., Xiang S., Yuan F., Yuan Z., Telles E., Fang J., Coppola D., Shibata D., Lane W.S., et al. Histone deacetylase 10 regulates DNA mismatch repair and may involve the deacetylation of MutS homolog 2. J. Biol. Chem. 2015;290:22795–22804. doi: 10.1074/jbc.M114.612945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hubbert C., Guardiola A., Shao R., Kawaguchi Y., Ito A., Nixon A., Yoshida M., Wang X.F., Yao T.P. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 92.Bertos N.R., Wang A.H., Yang X.J. Class II histone deacetylases: structure, function, and regulation. Biochem. Cell Biol. 2001;79:243–252. [PubMed] [Google Scholar]
- 93.Verdel A., Curtet S., Brocard M.P., Rousseaux S., Lemercier C., Yoshida M., Khochbin S. Active maintenance of mHDA2/mHDAC6 histone-deacetylase in the cytoplasm. Curr. Biol. 2000;10:747–749. doi: 10.1016/s0960-9822(00)00542-x. [DOI] [PubMed] [Google Scholar]
- 94.Fischer D.D., Cai R., Bhatia U., Asselbergs F.A., Song C., Terry R., Trogani N., Widmer R., Atadja P., Cohen D. Isolation and characterization of a novel class II histone deacetylase, HDAC10. J. Biol. Chem. 2002;277:6656–6666. doi: 10.1074/jbc.M108055200. [DOI] [PubMed] [Google Scholar]
- 95.Kao H.Y., Lee C.H., Komarov A., Han C.C., Evans R.M. Isolation and characterization of mammalian HDAC10, a novel histone deacetylase. J. Biol. Chem. 2002;277:187–193. doi: 10.1074/jbc.M108931200. [DOI] [PubMed] [Google Scholar]
- 96.Dosch J., Christmann M., Kaina B. Mismatch G-T binding activity and MSH2 expression is quantitatively related to sensitivity of cells to methylating agents. Carcinogenesis. 1998;19:567–573. doi: 10.1093/carcin/19.4.567. [DOI] [PubMed] [Google Scholar]
- 97.Lahue R.S., Frizzell A. Histone deacetylase complexes as caretakers of genome stability. Epigenetics. 2012;7:806–810. doi: 10.4161/epi.20922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Haince J.F., McDonald D., Rodrigue A., Dery U., Masson J.Y., Hendzel M.J., Poirier G.G. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J. Biol. Chem. 2008;283:1197–1208. doi: 10.1074/jbc.M706734200. [DOI] [PubMed] [Google Scholar]
- 99.Van Meter M., Simon M., Tombline G., May A., Morello T.D., Hubbard B.P., Bredbenner K., Park R., Sinclair D.A., Bohr V.A., et al. JNK Phosphorylates SIRT6 to Stimulate DNA Double-Strand Break Repair in Response to Oxidative Stress by Recruiting PARP1 to DNA Breaks. Cell Rep. 2016;16:2641–2650. doi: 10.1016/j.celrep.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Polo S.E., Kaidi A., Baskcomb L., Galanty Y., Jackson S.P. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 2010;29:3130–3139. doi: 10.1038/emboj.2010.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chou D.M., Adamson B., Dephoure N.E., Tan X., Nottke A.C., Hurov K.E., Gygi S.P., Colaiacovo M.P., Elledge S.J. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc. Natl. Acad. Sci. U.S.A. 2010;107:18475–18480. doi: 10.1073/pnas.1012946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Facchino S., Abdouh M., Chatoo W., Bernier G. BMI1 confers radioresistance to normal and cancerous neural stem cells through recruitment of the DNA damage response machinery. J. Neurosci. 2010;30:10096–10111. doi: 10.1523/JNEUROSCI.1634-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ginjala V., Nacerddine K., Kulkarni A., Oza J., Hill S.J., Yao M., Citterio E., van Lohuizen M., Ganesan S. BMI1 is recruited to DNA breaks and contributes to DNA damage-induced H2A ubiquitination and repair. Mol. Cell Biol. 2011;31:1972–1982. doi: 10.1128/MCB.00981-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ismail I.H., Andrin C., McDonald D., Hendzel M.J. BMI1-mediated histone ubiquitylation promotes DNA double-strand break repair. J. Cell Biol. 2010;191:45–60. doi: 10.1083/jcb.201003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Luijsterburg M.S., Acs K., Ackermann L., Wiegant W.W., Bekker-Jensen S., Larsen D.H., Khanna K.K., van Attikum H., Mailand N., Dantuma N.P. A new non-catalytic role for ubiquitin ligase RNF8 in unfolding higher-order chromatin structure. EMBO J. 2012;31:2511–2527. doi: 10.1038/emboj.2012.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Larsen D.H., Poinsignon C., Gudjonsson T., Dinant C., Payne M.R., Hari F.J., Rendtlew Danielsen J.M., Menard P., Sand J.C., Stucki M., et al. The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. J. Cell Biol. 2010;190:731–740. doi: 10.1083/jcb.200912135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Smeenk G., Wiegant W.W., Vrolijk H., Solari A.P., Pastink A., van Attikum H. The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. J. Cell Biol. 2010;190:741–749. doi: 10.1083/jcb.201001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.de Ruijter A.J., van Gennip A.H., Caron H.N., Kemp S., van Kuilenburg A.B. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Johnstone R.W. Histone-deacetylase inhibitors: novel drugs for the treatment of cancer. Nat. Rev. Drug Discov. 2002;1:287–299. doi: 10.1038/nrd772. [DOI] [PubMed] [Google Scholar]
- 110.Miller K.M., Tjeertes J.V., Coates J., Legube G., Polo S.E., Britton S., Jackson S.P. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat. Struct. Mol. Biol. 2010;17:1144–1151. doi: 10.1038/nsmb.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhu Q., Battu A., Ray A., Wani G., Qian J., He J., Wang Q.E., Wani A.A. Damaged DNA-binding protein down-regulates epigenetic mark H3K56Ac through histone deacetylase 1 and 2. Mutat. Res. 2015;776:16–23. doi: 10.1016/j.mrfmmm.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Johnson D.P., Spitz G.S., Tharkar S., Quayle S.N., Shearstone J.R., Jones S., McDowell M.E., Wellman H., Tyler J.K., Cairns B.R., et al. HDAC1,2 inhibition impairs EZH2- and BBAP-mediated DNA repair to overcome chemoresistance in EZH2 gain-of-function mutant diffuse large B-cell lymphoma. Oncotarget. 2015;6:4863–4887. doi: 10.18632/oncotarget.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tavares L., Dimitrova E., Oxley D., Webster J., Poot R., Demmers J., Bezstarosti K., Taylor S., Ura H., Koide H., et al. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell. 2012;148:664–678. doi: 10.1016/j.cell.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang H., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R.S., Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 115.Cao R., Tsukada Y., Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol. Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 116.Ziv Y., Bielopolski D., Galanty Y., Lukas C., Taya Y., Schultz D.C., Lukas J., Bekker-Jensen S., Bartek J., Shiloh Y. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat. Cell Biol. 2006;8:870–876. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
- 117.Lin Y.H., Yuan J., Pei H., Liu T., Ann D.K., Lou Z. KAP1 Deacetylation by SIRT1 Promotes Non-Homologous End-Joining Repair. PLoS One. 2015;10:e0123935. doi: 10.1371/journal.pone.0123935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Thurn K.T., Thomas S., Raha P., Qureshi I., Munster P.N. Histone deacetylase regulation of ATM-mediated DNA damage signaling. Mol. Cancer Ther. 2013;12:2078–2087. doi: 10.1158/1535-7163.MCT-12-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mirzoeva O.K., Petrini J.H. DNA damage-dependent nuclear dynamics of the Mre11 complex. Mol. Cell. Biol. 2001;21:281–288. doi: 10.1128/MCB.21.1.281-288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.D'Amours D., Jackson S.P. The Mre11 complex: at the crossroads of dna repair and checkpoint signalling. Nat. Rev. Mol. Cell Biol. 2002;3:317–327. doi: 10.1038/nrm805. [DOI] [PubMed] [Google Scholar]
- 121.Bakkenist C.J., Kastan M.B. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 122.Sun Y., Jiang X., Chen S., Fernandes N., Price B.D. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc. Natl. Acad. Sci. U.S.A. 2005;102:13182–13187. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yi J., Huang X., Yang Y., Zhu W.G., Gu W., Luo J. Regulation of histone acetyltransferase TIP60 function by histone deacetylase 3. J. Biol. Chem. 2014;289:33878–33886. doi: 10.1074/jbc.M114.575266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wang J., Chen J. SIRT1 regulates autoacetylation and histone acetyltransferase activity of TIP60. J. Biol. Chem. 2010;285:11458–11464. doi: 10.1074/jbc.M109.087585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bhaskara S., Knutson S.K., Jiang G., Chandrasekharan M.B., Wilson A.J., Zheng S., Yenamandra A., Locke K., Yuan J.L., Bonine-Summers A.R., et al. Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer Cell. 2010;18:436–447. doi: 10.1016/j.ccr.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kim G.D., Choi Y.H., Dimtchev A., Jeong S.J., Dritschilo A., Jung M. Sensing of ionizing radiation-induced DNA damage by ATM through interaction with histone deacetylase. J. Biol. Chem. 1999;274:31127–31130. doi: 10.1074/jbc.274.44.31127. [DOI] [PubMed] [Google Scholar]
- 127.Guo C., Mi J., Brautigan D.L., Larner J.M. ATM regulates ionizing radiation-induced disruption of HDAC1:PP1:Rb complexes. Cell Signal. 2007;19:504–510. doi: 10.1016/j.cellsig.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 128.Shiloh Y., Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol. 2013;14:197–210. [PubMed] [Google Scholar]
- 129.Matsuoka S., Huang M., Elledge S.J. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science. 1998;282:1893–1897. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- 130.Burma S., Chen B.P., Murphy M., Kurimasa A., Chen D.J. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 2001;276:42462–42467. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- 131.Jungmichel S., Clapperton J.A., Lloyd J., Hari F.J., Spycher C., Pavic L., Li J., Haire L.F., Bonalli M., Larsen D.H., et al. The molecular basis of ATM-dependent dimerization of the Mdc1 DNA damage checkpoint mediator. Nucleic Acids Res. 2012;40:3913–3928. doi: 10.1093/nar/gkr1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zeman M.K., Cimprich K.A. Causes and consequences of replication stress. Nat. Cell Biol. 2014;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Byun T.S., Pacek M., Yee M.C., Walter J.C., Cimprich K.A. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zou L., Elledge S.J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 135.Ball H.L., Myers J.S., Cortez D. ATRIP binding to replication protein A-single-stranded DNA promotes ATR-ATRIP localization but is dispensable for Chk1 phosphorylation. Mol. Biol. Cell. 2005;16:2372–2381. doi: 10.1091/mbc.E04-11-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Schmidt D.R., Schreiber S.L. Molecular association between ATR and two components of the nucleosome remodeling and deacetylating complex, HDAC2 and CHD4. Biochemistry. 1999;38:14711–14717. doi: 10.1021/bi991614n. [DOI] [PubMed] [Google Scholar]
- 137.Tibbetts R.S., Cortez D., Brumbaugh K.M., Scully R., Livingston D., Elledge S.J., Abraham R.T. Functional interactions between BRCA1 and the checkpoint kinase ATR during genotoxic stress. Genes Dev. 2000;14:2989–3002. doi: 10.1101/gad.851000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chapman J.R., Sossick A.J., Boulton S.J., Jackson S.P. BRCA1-associated exclusion of 53BP1 from DNA damage sites underlies temporal control of DNA repair. J. Cell Sci. 2012;125:3529–3534. doi: 10.1242/jcs.105353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.McNeely S., Conti C., Sheikh T., Patel H., Zabludoff S., Pommier Y., Schwartz G., Tse A. Chk1 inhibition after replicative stress activates a double strand break response mediated by ATM and DNA-dependent protein kinase. Cell Cycle. 2010;9:995–1004. doi: 10.4161/cc.9.5.10935. [DOI] [PubMed] [Google Scholar]
- 140.Chehab N.H., Malikzay A., Appel M., Halazonetis T.D. Chk2/hCds1 functions as a DNA damage checkpoint in G(1) by stabilizing p53. Genes Dev. 2000;14:278–288. [PMC free article] [PubMed] [Google Scholar]
- 141.Honda R., Tanaka H., Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 142.Sakaguchi K., Herrera J.E., Saito S., Miki T., Bustin M., Vassilev A., Anderson C.W., Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gu W., Roeder R.G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 144.Christmann M., Kaina B. Transcriptional regulation of human DNA repair genes following genotoxic stress: Trigger mechanisms, inducible responses and genotoxic adaptation. Nucleic Acids Res. 2013;41:8403–8420. doi: 10.1093/nar/gkt635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jabbur J.R., Huang P., Zhang W. DNA damage-induced phosphorylation of p53 at serine 20 correlates with p21 and Mdm-2 induction in vivo. Oncogene. 2000;19:6203–6208. doi: 10.1038/sj.onc.1204017. [DOI] [PubMed] [Google Scholar]
- 146.Hopker K., Hagmann H., Khurshid S., Chen S., Schermer B., Benzing T., Reinhardt H.C. Putting the brakes on p53-driven apoptosis. Cell Cycle. 2012;11:4122–4128. doi: 10.4161/cc.21997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Juan L.J., Shia W.J., Chen M.H., Yang W.M., Seto E., Lin Y.S., Wu C.W. Histone deacetylases specifically down-regulate p53-dependent gene activation. J. Biol. Chem. 2000;275:20436–20443. doi: 10.1074/jbc.M000202200. [DOI] [PubMed] [Google Scholar]
- 148.Luo J., Su F., Chen D., Shiloh A., Gu W. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature. 2000;408:377–381. doi: 10.1038/35042612. [DOI] [PubMed] [Google Scholar]
- 149.Vaziri H., Dessain S.K., Ng Eaton E., Imai S.I., Frye R.A., Pandita T.K., Guarente L., Weinberg R.A. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 150.Luo J., Nikolaev A.Y., Imai S., Chen D., Su F., Shiloh A., Guarente L., Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 151.Symington L.S., Gautier J. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 152.Sartori A.A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., Jackson S.P. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Takeda S., Nakamura K., Taniguchi Y., Paull T.T. Ctp1/CtIP and the MRN complex collaborate in the initial steps of homologous recombination. Mol. Cell. 2007;28:351–352. doi: 10.1016/j.molcel.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 154.Liu T., Huang J. DNA End Resection: Facts and Mechanisms. Genomics Proteomics Bioinformatics. 2016;14:126–130. doi: 10.1016/j.gpb.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Marteijn J.A., Bekker-Jensen S., Mailand N., Lans H., Schwertman P., Gourdin A.M., Dantuma N.P., Lukas J., Vermeulen W. Nucleotide excision repair-induced H2A ubiquitination is dependent on MDC1 and RNF8 and reveals a universal DNA damage response. J. Cell Biol. 2009;186:835–847. doi: 10.1083/jcb.200902150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Huen M.S., Grant R., Manke I., Minn K., Yu X., Yaffe M.B., Chen J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Thorslund T., Ripplinger A., Hoffmann S., Wild T., Uckelmann M., Villumsen B., Narita T., Sixma T.K., Choudhary C., Bekker-Jensen S., et al. Histone H1 couples initiation and amplification of ubiquitin signalling after DNA damage. Nature. 2015;527:389–393. doi: 10.1038/nature15401. [DOI] [PubMed] [Google Scholar]
- 158.Doil C., Mailand N., Bekker-Jensen S., Menard P., Larsen D.H., Pepperkok R., Ellenberg J., Panier S., Durocher D., Bartek J., et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 159.Wang B., Elledge S.J. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc. Natl. Acad. Sci. U.S.A. 2007;104:20759–20763. doi: 10.1073/pnas.0710061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Fradet-Turcotte A., Canny M.D., Escribano-Diaz C., Orthwein A., Leung C.C., Huang H., Landry M.C., Kitevski-LeBlanc J., Noordermeer S.M., Sicheri F., et al. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature. 2013;499:50–54. doi: 10.1038/nature12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Botuyan M.V., Lee J., Ward I.M., Kim J.E., Thompson J.R., Chen J., Mer G. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chapman J.R., Taylor M.R., Boulton S.J. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell. 2012;47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 163.Chapman J.R., Barral P., Vannier J.B., Borel V., Steger M., Tomas-Loba A., Sartori A.A., Adams I.R., Batista F.D., Boulton S.J. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol. Cell. 2013;49:858–871. doi: 10.1016/j.molcel.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Tang J., Cho N.W., Cui G., Manion E.M., Shanbhag N.M., Botuyan M.V., Mer G., Greenberg R.A. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat. Struct. Mol. Biol. 2013;20:317–325. doi: 10.1038/nsmb.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Jacquet K., Fradet-Turcotte A., Avvakumov N., Lambert J.P., Roques C., Pandita R.K., Paquet E., Herst P., Gingras A.C., Pandita T.K., et al. The TIP60 Complex Regulates Bivalent Chromatin Recognition by 53BP1 through Direct H4K20me Binding and H2AK15 Acetylation. Mol. Cell. 2016;62:409–421. doi: 10.1016/j.molcel.2016.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yu X., Chen J. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol. Cell. Biol. 2004;24:9478–9486. doi: 10.1128/MCB.24.21.9478-9486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Mimitou E.P., Symington L.S. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Waters C.A., Strande N.T., Wyatt D.W., Pryor J.M., Ramsden D.A. Nonhomologous end joining: a good solution for bad ends. DNA Repair (Amst) 2014;17:39–51. doi: 10.1016/j.dnarep.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Vazquez B.N., Thackray J.K., Simonet N.G., Kane-Goldsmith N., Martinez-Redondo P., Nguyen T., Bunting S., Vaquero A., Tischfield J.A., Serrano L. SIRT7 promotes genome integrity and modulates non-homologous end joining DNA repair. EMBO J. 2016;35:1488–1503. doi: 10.15252/embj.201593499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Downs J.A., Jackson S.P. A means to a DNA end: The many roles of Ku. Nat. Rev. Mol. Cell Biol. 2004;5:367–378. doi: 10.1038/nrm1367. [DOI] [PubMed] [Google Scholar]
- 171.Cohen H.Y., Miller C., Bitterman K.J., Wall N.R., Hekking B., Kessler B., Howitz K.T., Gorospe M., de Cabo R., Sinclair D.A. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 172.Davis A.J., Chen B.P., Chen D.J. DNA-PK: a dynamic enzyme in a versatile DSB repair pathway. DNA Repair (Amst) 2014;17:21–29. doi: 10.1016/j.dnarep.2014.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.McCord R.A., Michishita E., Hong T., Berber E., Boxer L.D., Kusumoto R., Guan S., Shi X., Gozani O., Burlingame A.L., et al. SIRT6 stabilizes DNA-dependent protein kinase at chromatin for DNA double-strand break repair. Aging (Albany NY) 2009;1:109–121. doi: 10.18632/aging.100011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Dobbin M.M., Madabhushi R., Pan L., Chen Y., Kim D., Gao J., Ahanonu B., Pao P.C., Qiu Y., Zhao Y., et al. SIRT1 collaborates with ATM and HDAC1 to maintain genomic stability in neurons. Nat. Neurosci. 2013;16:1008–1015. doi: 10.1038/nn.3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Jasin M., Rothstein R. Repair of strand breaks by homologous recombination. Cold Spring Harb. Perspect. Biol. 2013;5:a012740. doi: 10.1101/cshperspect.a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Petermann E., Helleday T. Pathways of mammalian replication fork restart. Nat. Rev. Mol. Cell Biol. 2010;11:683–687. doi: 10.1038/nrm2974. [DOI] [PubMed] [Google Scholar]
- 177.Bhaskara S., Chyla B.J., Amann J.M., Knutson S.K., Cortez D., Sun Z.W., Hiebert S.W. Deletion of histone deacetylase 3 reveals critical roles in S phase progression and DNA damage control. Mol. Cell. 2008;30:61–72. doi: 10.1016/j.molcel.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wells C.E., Bhaskara S., Stengel K.R., Zhao Y., Sirbu B., Chagot B., Cortez D., Khabele D., Chazin W.J., Cooper A., et al. Inhibition of histone deacetylase 3 causes replication stress in cutaneous T cell lymphoma. PLoS One. 2013;8:e68915. doi: 10.1371/journal.pone.0068915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Uhl M., Csernok A., Aydin S., Kreienberg R., Wiesmuller L., Gatz S.A. Role of SIRT1 in homologous recombination. DNA Repair (Amst) 2010;9:383–393. doi: 10.1016/j.dnarep.2009.12.020. [DOI] [PubMed] [Google Scholar]
- 180.Lee J.H., Choy M.L., Ngo L., Foster S.S., Marks P.A. Histone deacetylase inhibitor induces DNA damage, which normal but not transformed cells can repair. Proc. Natl. Acad. Sci. U.S.A. 2010;107:14639–14644. doi: 10.1073/pnas.1008522107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kaidi A., Weinert B.T., Choudhary C., Jackson S.P. Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science. 2010;329:1348–1353. doi: 10.1126/science.1192049. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 182.Liu J., Doty T., Gibson B., Heyer W.D. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat. Struct. Mol. Biol. 2010;17:1260–1262. doi: 10.1038/nsmb.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Mao Z., Tian X., Van Meter M., Ke Z., Gorbunova V., Seluanov A. Sirtuin 6 (SIRT6) rescues the decline of homologous recombination repair during replicative senescence. Proc. Natl. Acad. Sci. U.S.A. 2012;109:11800–11805. doi: 10.1073/pnas.1200583109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kotian S., Liyanarachchi S., Zelent A., Parvin J.D. Histone deacetylases 9 and 10 are required for homologous recombination. J. Biol. Chem. 2011;286:7722–7726. doi: 10.1074/jbc.C110.194233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zhou X., Marks P.A., Rifkind R.A., Richon V.M. Cloning and characterization of a histone deacetylase, HDAC9. Proc. Natl. Acad. Sci. U.S.A. 2001;98:10572–10577. doi: 10.1073/pnas.191375098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Milutinovic S., Zhuang Q., Szyf M. Proliferating cell nuclear antigen associates with histone deacetylase activity, integrating DNA replication and chromatin modification. J. Biol. Chem. 2002;277:20974–20978. doi: 10.1074/jbc.M202504200. [DOI] [PubMed] [Google Scholar]
- 187.Paques F., Haber J.E. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Chandramouly G., Kwok A., Huang B., Willis N.A., Xie A., Scully R. BRCA1 and CtIP suppress long-tract gene conversion between sister chromatids. Nat. Commun. 2013;4:2404–2416. doi: 10.1038/ncomms3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Muniandy P.A., Thapa D., Thazhathveetil A.K., Liu S.T., Seidman M.M. Repair of laser-localized DNA interstrand cross-links in G1 phase mammalian cells. J. Biol. Chem. 2009;284:27908–27917. doi: 10.1074/jbc.M109.029025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Enoiu M., Jiricny J., Scharer O.D. Repair of cisplatin-induced DNA interstrand crosslinks by a replication-independent pathway involving transcription-coupled repair and translesion synthesis. Nucleic Acids Res. 2012;40:8953–8964. doi: 10.1093/nar/gks670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Barckhausen C., Roos W.P., Naumann S.C., Kaina B. Malignant melanoma cells acquire resistance to DNA interstrand cross-linking chemotherapeutics by p53-triggered upregulation of DDB2/XPC-mediated DNA repair. Oncogene. 2014;33:1964–1974. doi: 10.1038/onc.2013.141. [DOI] [PubMed] [Google Scholar]
- 192.Clauson C., Scharer O.D., Niedernhofer L. Advances in understanding the complex mechanisms of DNA interstrand cross-link repair. Cold Spring Harb. Perspect. Biol. 2013;5:a012732. doi: 10.1101/cshperspect.a012732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Roos W.P., Kaina B. DNA damage-induced cell death: From specific DNA lesions to the DNA damage response and apoptosis. Cancer Lett. 2013;332:237–248. doi: 10.1016/j.canlet.2012.01.007. [DOI] [PubMed] [Google Scholar]