Abstract
Endonuclease G (EndoG) is an evolutionarily conserved mitochondrial protein in eukaryotes that digests nucleus chromosomal DNA during apoptosis and paternal mitochondrial DNA during embryogenesis. Under oxidative stress, homodimeric EndoG becomes oxidized and converts to monomers with diminished nuclease activity. However, it remains unclear why EndoG has to function as a homodimer in DNA degradation. Here, we report the crystal structure of the Caenorhabditis elegans EndoG homologue, CPS-6, in complex with single-stranded DNA at a resolution of 2.3 Å. Two separate DNA strands are bound at the ββα-metal motifs in the homodimer with their nucleobases pointing away from the enzyme, explaining why CPS-6 degrades DNA without sequence specificity. Two obligatory monomeric CPS-6 mutants (P207E and K131D/F132N) were constructed, and they degrade DNA with diminished activity due to poorer DNA-binding affinity as compared to wild-type CPS-6. Moreover, the P207E mutant exhibits predominantly 3′-to-5′ exonuclease activity, indicating a possible endonuclease to exonuclease activity change. Thus, the dimer conformation of CPS-6 is essential for maintaining its optimal DNA-binding and endonuclease activity. Compared to other non-specific endonucleases, which are usually monomeric enzymes, EndoG is a unique dimeric endonuclease, whose activity hence can be modulated by oxidation to induce conformational changes.
INTRODUCTION
Endonuclease G is an evolutionarily conserved mitochondrial protein in eukaryotes, named Nuc1 in yeast, CPS-6 in Caenorhabditis elegans, and EndoG in mammals (1–3). The discovery of EndoG from mitochondria of rat liver and Neurospora crassa revealed that this protein is a Mg2+-dependent endonuclease that cleaves phosphodiester bonds in nucleic acids to produce small digested fragments with 3′-OH and 5′-phosphate ends (4,5). Subsequent studies identified a similar endonuclease in the mitochondria of Saccharomyces cerevisiae, mouse liver, chicken erythrocytes and bovine heart cells (see the review: (6)). EndoG isolated from chicken erythrocytes was shown to have a preference for cleaving G-tract sequences in DNA, so the enzyme was named Endonuclease G (7). Both DNA and RNA are substrates for EndoG, which typically has a marginal preference for single-stranded RNA or DNA with G-rich sequences (7–9).
EndoG has been extensively characterized over the past four decades, but it is still unclear what the molecular function of EndoG is in mitochondria, where it is located both in the matrix and intermembrane space (1,2). In unicellular eukaryotes, such as Saccharomyces cerevisiae, EndoG (referred to as Nuc1) is suggested to function in mitochondrial DNA recombination since inactivation of Nuc1 decreases the recombination frequency (3). Mammalian EndoG was first thought to digest RNA primers and generate the free 3′ hydroxyl ends for initiation of mitochondrial DNA replication by DNA polymerase in the mitochondrial matrix (10). Mitochondrial EndoG isolated from bovine heart was suggested to target on the opposite strand of DNA with single-strand breaks generated by active oxygen radicals, and is involved in maintenance of the integrity of mitochondrial DNA genome (11). Morevoer, EndoG has also been reported to be present in nuclei (10,12), and the nuclease activity of mammalian EndoG is suggested to be involved in nuclear DNA recombination by introducing double-strand DNA breaks in switch regions (13,14). Taken together, these results suggest that EndoG plays a role in DNA metabolism in mitochondria and nuclei and, therefore, is required for normal cellular proliferation (13,15).
EndoG was later identified to be an important pro-apoptotic protein in the caspase-independent cell death pathway (1,2). During apoptosis, a fraction of EndoG localized in the mitochondrial intermembrane space is trafficked to the nucleus to degrade chromosomal DNA. Human EndoG interacts with and is stimulated by Apoptosis Inducing Factor (AIF), which has a nuclear localization signal that can translocate with EndoG to the nucleus during apoptosis (16,17). Similarly, in C. elegans, the EndoG homolog CPS-6 is required for apoptotic DNA degradation and normal progression of cell death (2,18,19). WAH-1–the homolog of human AIF–interacts with CPS-6 to enhance its endonuclease activity in apoptotic DNA fragmentation (19–21). Recently, a new function has been characterized for CPS-6 associated with rapid removal of paternal mitochondrial DNA during early embryogenesis (22). Upon fertilization, CPS-6 relocates from the intermembrane space of paternal mitochondria to the matrix to degrade mitochondrial DNA. EndoG thus has two types of functions, playing a role in DNA metabolism in mitochondria and degrading DNA either during embryogenesis in paternal mitochondria or during apoptosis in nuclei.
The crystal structures of EndoG have been reported, including those of Drosophila EndoG (23) and C. elegans CPS-6 (8), revealing a similar dimeric structure consisting of two active sites with a conserved DRGH sequence located within the ββα-metal motifs. Interestingly, the two ββα-metal motifs of the CPS-6 dimer are distantly located and pointed away from each other, hinting that the two protomers of the EndoG dimer likely bind to two separate DNA substrates (8). Recently, we further discovered that the dimeric conformation of EndoG/CPS-6 is important for the maintenance and regulation of its nuclease activity since, under oxidative conditions, EndoG/CPS-6 becomes oxidized and is dissociated into monomers with diminished nuclease activity (24). We also demonstrated that WAH-1 acts as an antioxidant that binds to the CPS-6 dimer and stabilizes its dimeric conformation, which in turn stimulates the nuclease activity of CPS-6. Oxidative stress may impair EndoG functionality, leading to mitochondrial dysfunction and cell death defects. Hence, it is important to determine not only how the EndoG dimer binds and cleaves DNA substrates, but also why the dimeric conformation is important for maintaining its optimal endonuclease activity.
In this study, multiple attempts were made to co-crystallize the CPS-6 double mutant H148A/F122A that had a greatly reduced nuclease activity, with either double- or single-stranded DNA. However, CPS-6 H148A/F122A mutant was only co-crystallized with single-stranded DNA for X-ray crystallographic study. The crystal structure of the CPS-6–DNA complex reveals at a resolution of 2.3 Å how dimeric CPS-6 binds and cleaves two respective DNA substrates. Two obligatory monomeric CPS-6 mutants, P207E and K131D/F132N, were further purified and we found that the monomeric mutants are still capable of binding and cleaving DNA, but with diminished activity due to decreased DNA-binding affinity. Moreover, the monomeric CPS-6 P207E mutant exhibits a predominant 3′-to-5′ exonuclease (as opposed to endonuclease) activity. Our results provide a molecular basis to explain how EndoG degrades DNA substrates without a sequence preference and why EndoG exhibits optimal endonuclease activity as a homodimer. Stabilizing EndoG's dimeric conformation may provide a way to promote its endonuclease activity and fight against diseases induced by oxidative stress.
MATERIALS AND METHODS
Protein expression and purification
All of the wild-type and mutated CPS-6 proteins were cloned, expressed and purified according to the protocol described by Lin et al. (5). All the mutated CPS-6 proteins were generated by Quickchange site-directed mutagenesis kits (Stratagene). After purification of the His-tagged CPS-6 by a TALON metal affinity resin column (BD Biosciences), CPS-6 proteins were applied to Superdex-75 (S-75) in buffers of 50 mM Tris–HCl pH 7.4, 200 mM NaCl, and 2.5 mM DTT (Figures 1B and 5B). The purified protein samples were stored in -80°C until use. In the cases of CPS-6 H148A and CPS-6 H148A/P207E, the peak fraction collectables from S-75 were reapplied for the size exclusion chromatography (Figure 5B). The intensities of the ethidium bromide-stained DNA bands were quantitated by scanning with AlphaEaseFC™ software (Alpha Innotech Corporation).
Figure 1.
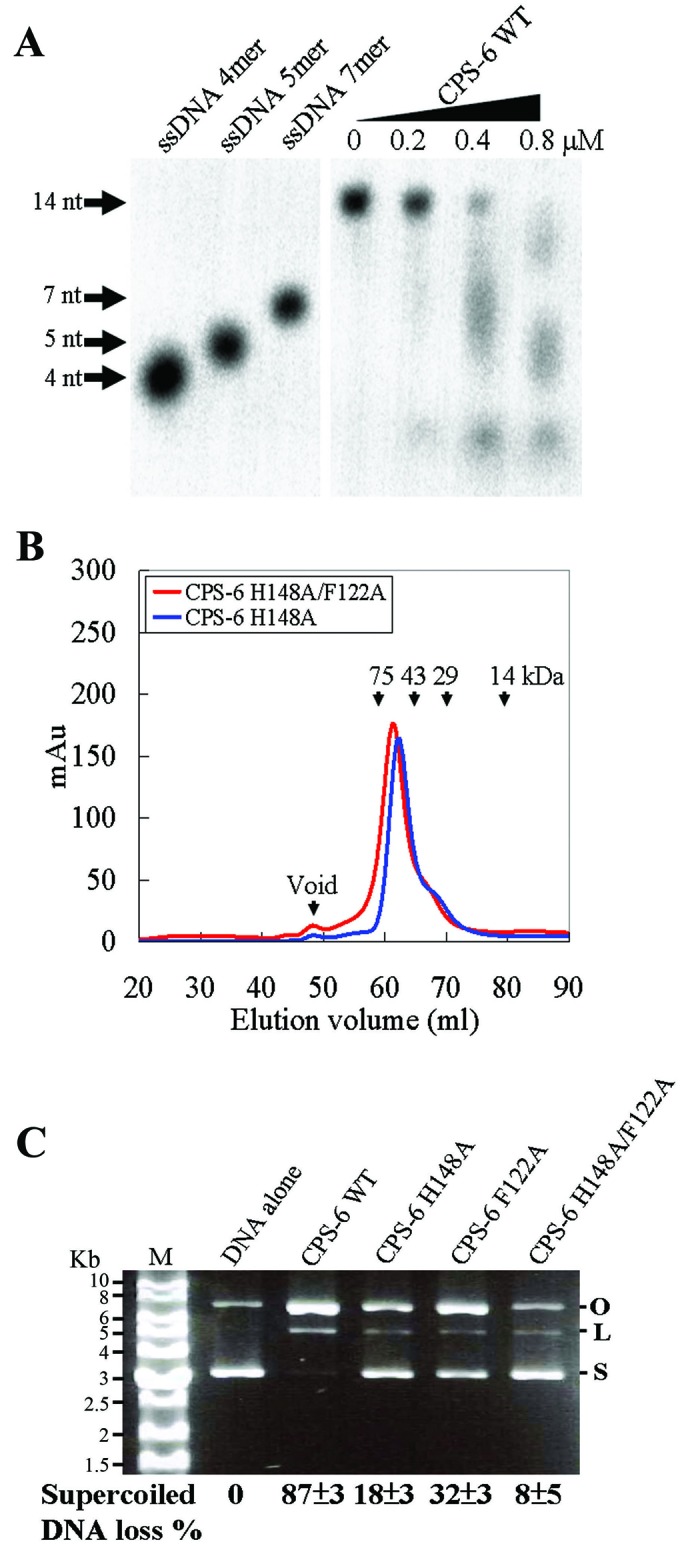
CPS-6 forms homodimers and degrades DNA into small fragments. (A) Wild-type CPS-6 was incubated with a 3′-end P32-labeled single-stranded 14-nucleotide DNA that was degraded into small fragments (<7 nucleotides) as the concentration of CPS-6 was increased up to 0.8 μM. (B) Both the CPS-6 H148A and H148A/F122A mutants were eluted as homodimers in Superdex-75 size exclusion chromatography. (C) Plasmid nicking assays show that 87%, 18%, 32% and 8% of the supercoiled DNA was digested by wild-type CPS-6, H148A, F122A and H148A/F122A mutants, respectively.
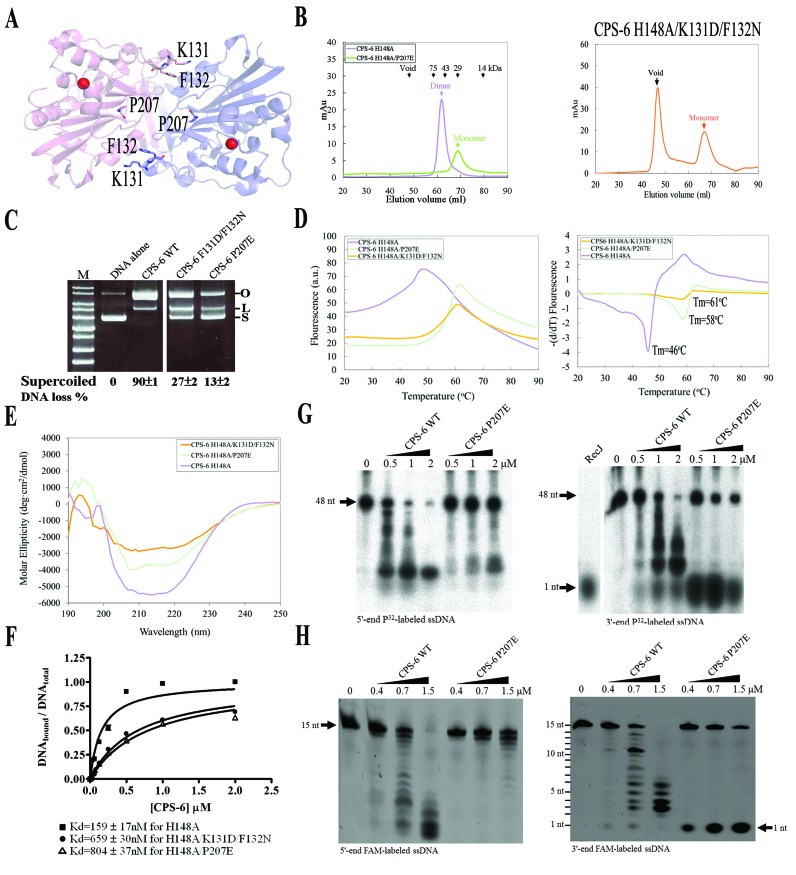
Figure 5.
Dimeric conformation of CPS-6 is critical for its optimal endonuclease activity. (A) P207, K131 and F132 are located in the dimeric interface in CPS-6 dimer based on the crystal structure of CPS-6(H148A) (PDB entry: 3S5B). (B) The CPS-6 H148A/P207E and H148A/K131D/F132N mutants were eluted as monomers in the size exclusion chromatography (Superdex 75), whereas the CPS-6 H148A mutant was eluted as a homodimer. (C) Plasmid-nicking assays show that 90%, 27% and 13% of supercoiled DNA was digested by wild-type CPS-6, K131D/F132N and P207E mutants, respectively. (D) The overall folding stability of CPS-6 mutants was assayed by differential scanning fluorimetry (left panel). The fluorescence intensities for three CPS-6 mutants were normalized for comparison. The melting points were estimated for the three CPS-6 mutant proteins (right panel). (E) Circular dichroism spectra (in molar ellipticity) for CPS-6 H148A, H148A/P207E and H148A/K131D/F132N mutants. (F) Filter binding assays were performed by incubating CPS-6 with a 5′-end P32-labeled single-stranded 48-nucleotide DNA. (G) Wild-type CPS-6 digests the 5′-end P32-labeled single-stranded DNA more efficiently than the P207E mutant (left panel). Wild-type CPS-6 digests the 3′-end P32-labeled single-stranded DNA into small fragments, whereas P207E mainly generated a 3′-end mononucleotide (right panel). (H) Distinct digesting patterns were observed for the wild-type CPS-6 and P207E mutant in digesting 5′-end or 3′-end FAM-labeled single-stranded DNA with a sequence of 5′-AATGTTGGGAGGAAA-3′. P207E digested the 3′-end FAM-labeled single-stranded DNA to yield an instant release of the 3′-end FAM-labeled mononucleotide (right panel).
Nuclease activity assays
For the DNase activity assays shown in Figure 1A, purified wild-type CPS-6 (0 to 0.8 μM) was incubated with 14-nucleotide single-stranded DNA at a concentration of 60 nM (5′-end 32P-labeled 5′-TCCCCCCTTTTTTT-3′) in the buffers containing 10 mM HEPES, pH7.0, 100 mM NaCl, 2 mM MgCl2, and 2 mM DTT for 3 h. For the DNase activity assays shown in Figure 1C, a quantity of 25 ng of pET28 plasmid DNA was incubated with or without 1 μM purified wild-type and mutated CPS-6 proteins in 50 mM Tris–HCl pH7.4, 200 mM NaCl, 2 mM MgCl2 and 2.5 mM DTT in 37°C for 25 min. For the DNase activity assays shown in Figure 5C, a quantity of 25 ng of pET28 plasmid DNA was incubated with 0.6 μM CPS-6 WT, K131D/F132N and P207E in 50 mM Tris–HCl pH7.4, 200 mM NaCl, 2 mM MgCl2 and 2.5 mM DTT in 37°C for 40 min. For the DNase activity assays shown in Figure 5G, purified wild-type protein (0–2 μM) was incubated with 48-nucleotide single-stranded 5′-end 32P-labeled or 3′-end 32P-labeled DNA (5′-ACGCTGCCGAATTCTGGCGTTAGGAGATACCGATAAGCTTCGGCTTAA-3′) at a concentration of 60 nM in the buffers containing 10 mM HEPES, pH7.0, 100 mM NaCl, 2 mM MgCl2, and 2 mM DTT for 3 h. For the DNase activity assays shown in Figure 5H, purified wild-type protein (0–1.5 μM) was incubated with 15-nucleotide single-stranded 5′-end or 3′-end FAM-labeled DNA (5′-AATGTTGGGAGGAAA-3′) at a concentration of 100 nM in the buffers containing 10 mM HEPES, pH 7.0, 100 mM NaCl, 2 mM MgCl2, and 2 mM DTT for 3 h. The digested samples were resolved in a 20% TBE and urea gel and visualized using the Typhoon FLA-9000 (GE Healthcare Life Sciences) imaging system.
Crystallization and structural determination
Crystals of CPS-6 H148A/F122A were grown by the hanging-drop vapor diffusion method at 4°C. The crystallization drop was made by mixing 0.3 μl of protein solution and 0.3 μl of reservoir solution. Protein (18 mg/ml in the buffer: 50 mM Tris–HCl, pH 7.4, 200 mM NaCl and 2.5 mM DTT) was crystallized using a reservoir solution containing 0.1 M sodium citrate tribasic dihydrate (pH 5.5) and 22% PEG1000. Crystals of CPS-6 H148A/F122A bound with DNA (5′-TTTTTTGT-3′) were grown by the hanging-drop vapor diffusion method at 4°C using a reservoir solution containing 0.1 M HEPES (pH 7.5) and 30% PEG1000. Diffraction data were collected at the BL15A beamline at NSRRC, Hsinchu (Taiwan) and were processed and scaled by HKL2000. All the diffraction statistics are listed in Table 1. Structures were solved by the molecular replacement method using the crystal structure of CPS-6 H148A (PDB ID: 3S5B) as the search model by program Phenix. The models were built by Coot and refined by Phenix.
Table 1. Crystallographic statistics for CPS-6 H148A/F122A and CPS6-H148A/F122A bound with ssDNA.
| Data collection and processing | CPS-6 H148A/F122A | CPS-6 H148A/F122A-ssDNA |
|---|---|---|
| Wavelength (Å) | 1.0 | 1.0 |
| Space group | P21 | P212121 |
| Cell dimensions, a, b, c (Å) | 70.6, 45.6, 81.0 | 56.7, 72.6, 127.9 |
| β (°) | 103.2 | 90.0 |
| Resolution (Å) | 1.9–27.5 | 2.3–20.0 |
| (last shell) | (1.89-1.92)a | (2.27-2.35)a |
| Observed/unique reflections | 145 855/40 272 | 155 880/25 101 |
| Data redundancy | 3.6 (3.4) | 6.2 (3.7) |
| Completeness (%) | 99.7 (99.6) | 99.8 (98.5) |
| Rsym (%) | 7.0 (30.8) | 7.2 (50.9) |
| I/σ(I) | 16.8 (4.5) | 23.1 (2.2) |
| Refinement statistics | ||
| Resolution range | 1.9–27.5 | 2.3–19.8 |
| Reflections (work/test) | 36 960/2029 | 24 707/2042 |
| R-factor/R-free (%) | 19.8/24.9 | 18.9/25.0 |
| Number of non-hydrogen atoms- | ||
| Protein/metal/solvent/DNA | 3856/2/401/0 | 3829/2/159/151 |
| Model quality | ||
| RMSD in bond length (Å) | 0.007 | 0.008 |
| Bond angle (o) | 1.152 | 1.134 |
| Average B-factor- | ||
| protein/metal/solvent/DNA (Å2) | 16.9/13.9/21.4/– | 26.9/24.9/24.6/45.0 |
| Ramachandran plot- | ||
| Most favored | 97.9 | 96.8 |
| Additionally allowed | 2.1 | 2.8 |
| Generally allowed | 0 | 0.4 |
| Disallowed | 0 | 0 |
aValues in parentheses refer to the highest resolution shell.
Differential scanning fluorimetry
The differential scanning fluorimetry experiments were performed by the LightCycler® 480 system (Roche). The concentration was optimized for CPS-6 H148A, H148A/P207E and H148A/K131D/F132N at 8, 2 and 1 μM, respectively, in 50 mM Tris–HCl, pH7.4, 200 mM NaCl and 2.5 mM DTT. The SYPRO Orange (Invitrogen) dye was added to the protein samples to make a 5x dye working concentration. The protein and dye preparations were loaded into a multiwell plate (MultiWell Plate 96 White) and sealed with LightCycler® 480 Sealing Foil (Roche). The temperature was increased from 20 to 90°C at a heating rate of 0.012°C/s and the emitted fluorescence at 568 nm (excited at 483 nm) was recorded every 0.02°C. The melting temperature (Tm) was calculated by the LightCycler® Protein Melting Analysis.
Circular dichroism (CD) spectrometry
The CD spectra of the recombinant CPS-6 proteins with protein concentration at 10 μM were recorded at 25°C on an AVIV 62DS CD spectrometer (Lakewood, NJ, USA). The CD spectra (Figure 5E) were measured in the range of 190–250 nm using a quartz cell with 1-mm light path. The spectra were recorded as the average of three scans for each recombinant protein sample after correction for the baseline contribution of the buffer. The data were reported as the mean residue molar ellipticities ([θ]).
Filter binding assay
The 48-nuclotide single-stranded DNA (the same one used for nuclease activity assays) 5′-end labeled with [γ-32P]ATP by T4 polynucleotide kinase was used for the filter binding assays. The 32P-labeled DNA (14 fmol) was incubated with a serial dilution of the CPS-6 H148A, H148A/K131D/F132N or H148A/P207E proteins (Figure 5E) in a binding buffer containing 10 mM HEPES, pH 7.0, 100 mM NaCl, 2 mM DTT and 2 mM EDTA for 60 min at room temperature. The reaction mixtures were then passed through the filter binding assay apparatus (Bio-Dot SF microfiltration apparatus, Bio-Rad). After extensive washing, the protein–DNA complex-bound nitrocellulose membrane and free DNA-bound nylon membrane were air-dried and exposed to a phosphorimaging plate. The intensities of the nitrocellulose-bound protein-DNA complex and nylon membrane-bound free DNA were quantified by the program AlphaImager IS-2200 (Alpha Innotech, USA). The apparent Kd values were estimated by fitting to a one-site binding curve using GraphPad Prism 4.
RESULTS
CPS-6 H148A/F122A mutant only has residual endonuclease activity
To understand how CPS-6 binds and digests DNA, we first expressed the N-terminal-His-tagged CPS-6 (residues 63–305) deprived of the mitochondrial targeting sequence from Escherichia coli. The purified recombinant wild-type CPS-6 (referred to as CPS-6 WT) was incubated with a 3′-end P32-labeled single-stranded 14-nucleotide DNA, which was degraded gradually into small fragments (<7 nucleotides) in the CPS-6 concentration-course experiments (Figure 1A). Previous studies suggest that His148 in CPS-6 functions as the general base in DNA hydrolysis, whereas Phe122 is involved in DNA binding (8). To obtain inactive CPS-6 mutants that could not digest DNA during co-crystallization experiments, we further expressed and purified three CPS-6 mutants—H148A, F122A and H148A/F122A—as described previously (8). Both the H148A and H148A/F122A mutants were eluted as homodimers in size exclusion chromatography, suggesting that they were assembled in a similar way as wild-type CPS-6 (Figure 1B). The endonuclease activities of wild-type CPS-6 and the H148A, F122A and H148A/F122A mutants were analyzed by plasmid nicking assays. These assays showed that most of the supercoiled DNA (87%) was degraded by wild-type CPS-6, but less supercoiled DNA was degraded by the mutants H148A (18%) and F122A (32%), and the least DNA was digested by the double mutant H148A/F122A (8%) (Figure 1C). Taken together, these results show that CPS-6 functions as a homodimer to nick DNA into small fragments and the double mutant H148A/F122A only has residual endonuclease activity.
Crystal structure of CPS-6 H148A/F122A bound to single-stranded DNA
We used the double mutant CPS-6 H148A/F122A with a largely reduced nuclease activity for co-crystallization with DNA. The CPS-6 H148A/F122A mutant was crystallized alone and in complex with single-stranded DNA by the hanging drop vapor diffusion method. The CPS-6 H148A/F122A mutant alone was crystallized in the space group P21, with one dimer per asymmetric unit and diffracted X-ray up to a resolution of 1.9 Å. The crystal structure of the CPS-6 H148A/F122A mutant was refined using the CPS-6 H148A structure (PDB entry 3S5B) as the starting model since these two mutants crystallized in the same unit cell. The crystal structure was refined to an R-factor of 19.8% for 36,960 reflections and an R-free of 24.9% for 2029 reflections at a resolution of 1.9 Å (Table 1). The average rmsd value between CPS-6 H148A (PDB: 3S5B) and CPS-6 H148A/F122A (this study, PDB: 5GKC) is 0.33 Å for 408 superimposed Cα atoms, suggesting that no major structural change was induced when F122 was mutated to alanine.
CPS-6 H148A/F122A was further co-crystallized with single-stranded DNA (5′-TTTTTTGT-3′) by the hanging drop vapor diffusion method in the space group P212121, with one CPS-6 dimer per asymmetric unit. X-ray diffraction data up to a resolution of 2.3 Å were collected and the crystal structure was determined by molecular replacement using the crystal structure of CPS-6 H148A (PDB ID: 3S5B) as the searching model. After location of the two CPS-6 protomers, five oligonucleotides (T1-ZT5 in each protomer) were built into the extra electron density in the Fourier maps. The CPS–6/DNA complex structure was refined to an R-factor of 18.9% for 24 707 reflections and an R-free of 25.0% for 2042 reflections at a resolution of 2.3 Å (Table 1).
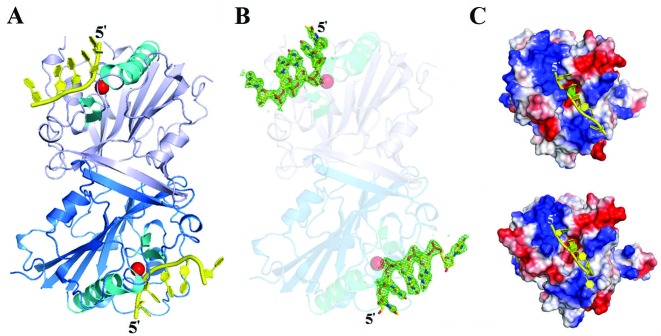
The crystal structure of dimeric CPS-6 H148A/F122A (chains A and B) bound to two single-stranded DNA molecules (chains C and D) in one asymmetric unit is illustrated in Figure 2A. The CPS-6 structure in the complex is similar to that of the free-form CPS-6 H148A/F122A, with an average rmsd of 0.31 Å for 408 superimposed Cα atoms, suggesting that DNA binding did not induce any significant protein conformational change. Only five out of the eight nucleotides (5′-TTTTTTGT-3′) were visible in the omitted Fourier (2Fo-Fc) maps (Figure 2B), and they were all assigned as thymidine (T1–T5) based on the shape of each base. These five nucleotides could represent a digested 5-nt DNA molecule or an intact 8-nt DNA that has disordered ends. The electrostatic surface potential for the DNA-binding region is consistently positive in both protomers, especially in the 5′-end (Figure 2C). A Mg2+ ion is modeled in the active site of both protomers, which is located close to the DNA backbone. The two DNA molecules are distantly located from each other and facing opposite directions. Therefore, the CPS-6/DNA complex structure supports the hypothesis that CPS-6 binds and degrades two separate DNA substrates.
Figure 2.
Crystal structure of CPS-6 H148A/F122A mutant bound with single-stranded DNA. (A) Overall crystal structure of the dimeric CPS-6 in complex with single-stranded DNA at 2.3 Å resolution. The two protomers are displayed in marine blue and light blue, respectively. The ββα-metal motifs are displayed in cyan with an Mg2+ ion (red sphere) bound at the active site. (B) Omit (2Fo −Fc) map illustration for the DNA substrates that were bound to two CPS-6 protomers (contoured at 3.0σ). Only five out of the eight nucleotides (5′-TTTTTTGT-3′) are visible in the omitted Fourier maps. (C) The electrostatic surface potential of CPS-6 reveals a positively-charged surface (in blue) near the active site that matches well with the DNA binding region. The Mg2+ ion in the active site is displayed as a green sphere.
How CPS-6 binds and degrades DNA at atomic level
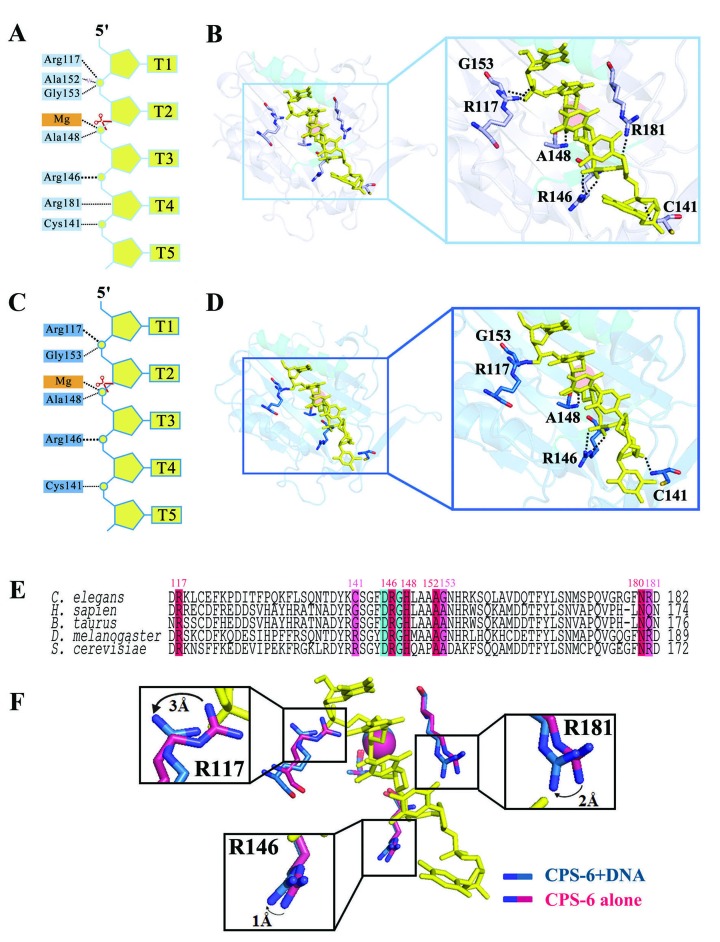
A close look at the CPS-6-DNA complex structure reveals that CPS-6 binds DNA mainly at the phosphate backbones via hydrogen-bonded interactions (Figure 3A and B for protomer A, Figure 3C and D for protomer B). The DNA bases are pointed away from the protein surface and in no way interact with CPS-6, thereby avoiding sequence-specific interactions. The hydrogen-bond networks between CPS-6 and DNA (summarized in Figure 3) show a total of six to eight hydrogen bonds between CPS-6 and the DNA. The Mg2+ ion is bound in the ββα-metal motif and bridges the CPS-6 and DNA molecules in each protomer. The protein-DNA interactions are slightly different in the two protomers, likely due to different crystal packing environments as the 3′ end of one DNA molecule (chain C) made contacts with the neighboring protein. However, in both of the protomers, Arg117, Cys141, Arg146, His148 (mutated to Ala), Gly153 and Arg181 are preserved for DNA binding. These DNA-binding residues are conserved in human, bovine, fruit fly and yeast EndoG (Figure 3E). The interactions between CPS-6 and DNA reveal the structural impact of the amino acid residues involved in DNA binding, including the side chain of Arg117, Arg181 and Arg146 that all shift up to 3 Å (Figure 3F). We conclude that Arg117, Arg146 and Arg181 play concerted and critical roles in the binding of nucleic acids.
Figure 3.
Interactions between CPS-6 and DNA at the atomic level. (A, C) Schematic diagram showing the hydrogen bond network between CPS-6 and DNA for protomers A and B, respectively. An Mg2+ ion is bound in the ββα-metal motif and bridges CPS-6 and DNA in each protomer. (B, D) Close-up of the hydrogen bonding at the CPS-6-DNA interface for protomers A and B, respectively. (E) The DNA-binding residues of human, bovine, fruit fly and yeast EndoG are conserved. (F) Interactions between CPS-6 and DNA have a structural impact on the DNA-binding residues, Arg117, Arg146 and Arg181.
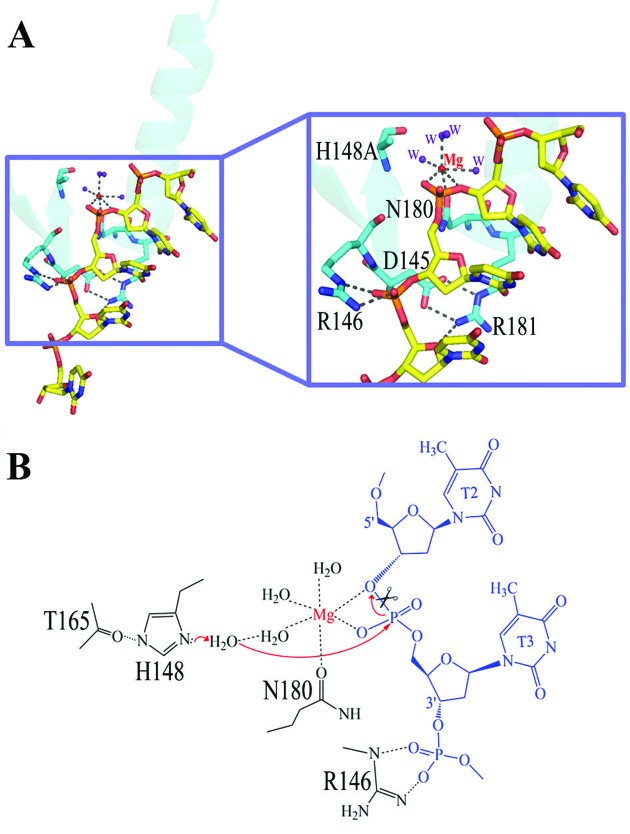
How is the bound DNA cleaved by CPS-6? The Mg2+ located within the ββα-metal motif is coordinated by six ligands in an octahedral geometry: Asn180, two scissile phosphate oxygen atoms, and three water molecules (Figure 4). Asp145 forms two salt bridges with the side chain of Arg181 to stabilize the conformation of the ββα-metal motif. The general base His148 is mutated to Ala in the crystal structure and its side-chain is likely polarized by the hydrogen bond with the carbonyl group of Thr165. We propose a hydrolysis mechanism for CPS-6 whereby His148 functions as the general base to polarize a water molecule (which is missing in the crystal structure due to H148A mutation), for nucleophilic attack on a scissile phosphate (Figure 4B). The Mg-bound water functions as the general acid to provide a proton to the cleaved DNA product. The bound DNA substrate is thereby hydrolyzed in a sequence-independent manner to produce cleaved products with a 5′-end phosphate and a 3′-end OH group. Moreover, we superimposed the ββα-metal motif in CPS-6-DNA to the one in Vvn–DNA complex (PDB entry: 1OUP) and found that the ssDNA bound to CPS-6 matches to one strand of the duplex DNA in Vvn–DNA complex. This result suggests that CPS-6 likely binds double-stranded DNA in a way similar to Vvn and CPS-6 likely nicks one strand of the duplex DNA upon binding.
Figure 4.
The DNA hydrolysis mechanism by EndoG/CPS-6. (A) Close-up of the ββα-metal motifs of CPS-6 bound to DNA. (B) Schematic diagram of the proposed DNA hydrolysis mechanism by CPS-6. The Mg2+ located within the ββα-metal motif is coordinated by six ligands in an octahedral geometry: Asn180, two scissile phosphate oxygens, and three water molecules. Asp145 forms two salt bridges with the side chain of Arg181 to stabilize the conformation of the ββα-metal motif. His148 was mutated to Ala in the crystal structure and its side-chain is likely polarized by Thr165. His148 acts as a general base to activate a water molecule, which in turn makes an in-line attack on the scissile phosphate. The Mg-bound water functions as the general acid to provide a proton to the cleaved DNA product.
Dimeric conformation of CPS-6 is critical for its optimal endonuclease activity
We were intrigued as to why CPS-6 has to function as a homodimer, since each protomer binds and degrades separate DNA substrates. To reveal the molecular basis for its dimeric conformation, we prepared two obligatory CPS-6 monomeric mutants for biochemical assays. One of the mutants, CPS-6 H148A/P207E, had been constructed previously to mimic oxidized monomeric CPS-6, with the interfacial Pro207 being oxidized during oxidation treatments (see Figure 5A) (24). We found that CPS-6 H148A/P207E was indeed eluted as a monomer in the size exclusion chromatography (Superdex 75), as compared to CPS-6 H148A that was eluted as a homodimer (Figure 5B). We further constructed another obligatory monomeric CPS-6 mutant, H148A/K131D/F132N, by altering two key interfacial residues (K131 and F132) predicted to be important for CPS-6 dimer formation based on the crystal structure of the CPS-6 H148A mutant (PDB entry: 3S5B) (8). CPS-6 H148A/K131D/F132N mutant protein also eluted as a monomer at a position identical to that of H148A/P207E (Figure 5B). We further assayed the folding stability of CPS-6 mutants by differential scanning fluorimetry. Unexpectedly, the monomeric mutants were more stable than the dimeric one with a melting point of 58°C for H148A/P207E and 61°C for H148A/K131D/F132N, as compared to 46°C for CPS-6 H148A (Figure 5D), suggesting that the monomeric CPS-6 mutants were well folded proteins. Moreover, we analyzed the secondary structures of these CPS-6 proteins by circular dichroism (CD). The CD spectrum of wild-type CPS-6 was similar but not identical to those of H148A/P207E and H148A/K131D/F132N mutants (Figure 5E), suggesting possible conformational changes in the monomeric mutants. Taken together these results suggest that the CPS-6 mutants, H148A/P207E and H148A/K131D/F132N, shift to monomers with a slightly varied secondary structures as compared to the dimeric wild-type CPS-6.
To determine the endonuclease activity of the CPS-6 mutants, we further expressed and purified CPS-6 P207E and CPS-6 K131D/F132N mutants, both of which contain the functional general base residue His148 for DNA hydrolysis. We then performed plasmid-nicking assays using pET28 as the DNA substrates. About 90% of supercoiled DNA was degraded by wild-type CPS-6, but only 27% and 13% of supercoiled DNA were degraded by the K131D/F132N and P207E mutants, respectively (Figure 5C). These results show that the monomeric CPS-6 proteins have significantly lower endonuclease activity than the dimeric wild-type enzyme. To further assay the molecular basis for the reduced endonuclease activity of the monomeric CPS-6, we measured the DNA-binding activity of these mutants by filter binding assays. CPS-6 proteins were incubated with 5′-end P32-labeled single-stranded 48-nucleotide DNA with a concentration ranging from 0 to 2 μM. Dissociation constants (Kd) were estimated at 159 nM for CPS-6 H148A, 659 nM for H148A/K131D/F132N and 804 nM for H148A/P207E (Figure 5F). These results suggest that the monomeric CPS-6 mutants bind DNA with about 4-to-5-fold lower affinities than wild-type CPS-6. Thus, we conclude that monomeric CPS-6 mutants bind DNA with poorer affinity and, therefore, have diminished endonuclease activities compared to the wild-type dimeric CPS-6.
To further determine if enzymatic activity is affected by the dimer-to-monomer conformational change, we incubated CPS-6 proteins (0–2 μM) with a single-stranded 48-nucleotide DNA that was P32-labeled at either the 5′ or 3′ end as substrates. We used RecJ, a 5′-to-3′ exonuclease, as a control to generate a 5′-end P32-labeled mononucleotide in Figure 5G (25). In the experiment using 5′-end P32-labeled DNA as the substrate, the nuclease activity of the wild-type enzyme was markedly greater than that of the P207E mutant protein (Figure 5G, left panel). Interestingly, in the experiment using 3′-end P32-labeled DNA as the substrate, the P207E mutant appeared to have increased 3′-to-5′ exonuclease activity, as a single 3′-end P32-labeled nucleotide was preferentially generated (Figure 5G, right panel). This DNA digestion pattern is distinct from that of the wild-type CPS-6, suggesting that the CPS-6 P207E mutant functions predominantly as an exonuclease with 3′ to 5′ directionality. Hence, we suggest that the dimer-to-monomer conformational change not only diminishes the endonuclease activity of CPS-6, but also changes its enzyme activity from an endonuclease to an exonuclease.
To corroborate this finding, we further incubated CPS-6 (0–1.5 μM) with a single-stranded 15-nucleotide DNA that had a different length and sequence to the 48-nucleotide DNA used in Figure 5G. The 15-nt DNA were labeled with the Fluorescein AMidite (FAM) at either 5′ or 3′ end. In the experiment using 5′-end FAM-labeled DNA as the substrate, distinct endo- and exo-nucleolytic digestion patterns were observed for the wild-type CPS-6 and P207E mutant, respectively (Figure 5H, left panel). In the experiment using 3′-end FAM-labeled DNA as the substrate, an immediate appearance of a 3′-end FAM-labeled mononucleotide confirmed the 3′-to-5′ exonuclease activity of CPS-6 P207E mutant (Figure 5H, right panel). Taken together these results, we suggest that CPS-6 P207E mutation may induce not only a dimer-to-monomer conformational change, but also a conformational change within the monomer that binds and degrades DNA with enhanced 3′-to-5′ exonuclease activity.
DISCUSSION
In this study, CPS-6 H148A/F122A dimer was successfully co-crystallized with two single-stranded DNA molecules, each bound at the ββα-metal motif (Figure 2). Further analysis demonstrated that R146 in the conserved 145DRGH148 motif forms hydrogen bonds with phosphate groups on the DNA substrate to stabilize the protein-DNA interface (Figure 3), while Asp145 forms salt bridges with Arg181 to maintain the structure of the ββα-metal motif (Figure 4). In addition, His148 (mutated to Ala) is suggested to function as the general base activating the water molecule that attacks the scissile phosphate for DNA hydrolysis (Figure 4B). These results provide the molecular basis for why these three residues—Asp145, Arg146 and His148—are highly conserved in the 145DRGH148 motif of EndoG proteins (26).
The dimeric conformation of CPS-6 is crucial for maintenance of its optimal endonuclease activity, given that the monomeric CPS-6 mutants P207E and F131D/F132N have 3-fold and 7-fold lower DNA degradation activity, respectively, compared to wild-type enzyme. Close cooperation between the two protomers of the CPS-6 dimer is thus essential to maintain the structural conformation that plays a regulatory role in the enzymatic function of CPS-6. The change of nucleic acid-binding pattern from endo- (as a dimer) to exo- (as a monomer) might have accounted for the marked decrease of the CPS-6 binding affinity to the same DNA substrate (Figure 5E). The predominant 3′-to-5′ exonuclease activity observed for the P207E mutant (Figure 5) provides further evidence for a possible endo-to-exonuclease functional switch controlled by conformational alternations between dimer and monomer. A previous study on RNase J reveals a similar endonuclease-to-exonuclease switch upon monomer-to-dimer conformational change (27). Different from EndoG whose dimer-to-monomer conformational change was induced by protein oxidation, the monomer-to-dimer switch of RNase J is induced by divalent cations, which further stimulate the 5′-to-3′ exonuclease activity of RNase J. Hence, the interconversion between endonuclease and exonuclease activity mediated by protein dimerization appears to be an evolved strategy for the regulation of the nuclease activity by environmental changes.
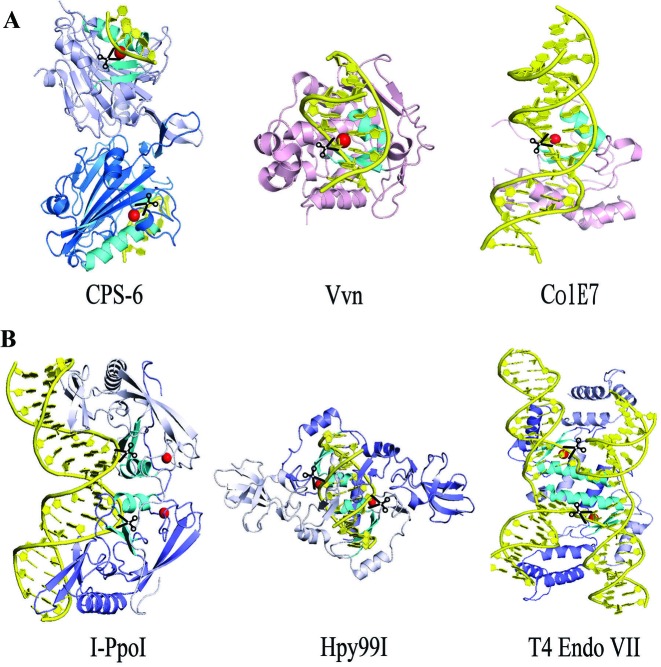
Most sequence- or structure-specific nucleases are homodimeric enzymes so that they can cleave two strands of nucleic acids in duplex or structured DNA, such as I-PpoI (28), Hpy99I (29) and T4 Endo VII (30) (see Figure 6). I-PpoI is a His-Cys box homing endonuclease, whereas Hpy99I is a type II restriction enzyme. Both I-PpoI and Hpy99I are dimeric proteins containing a pair of endonuclease active sites comprising ββα-metal motifs. The distances and orientations of the two ββα-metal motifs in I-PpoI and Hpy99I make them capable of cleaving two strands of B-form duplex DNA to generate cleaved products with 4-nucleotide and 5-nucleotide staggered ends, respectively. Another example of a ββα-metal endonuclease, T4 EndoVII, is also a dimeric enzyme, which recognizes the structured DNA of Holliday junctions. The crystal structure of T4 Endo VII reveals a different relative orientation and distance between the two ββα-metal motifs that bind and cleave two DNA strands in the Holliday junction (see Figure 6B). These examples show that sequence- or structure-specific endonucleases form a homodimer because they need to cleave two strands of DNA in duplex or structured DNA substrates. The two ββα-metal motifs in these enzymes are located close to each other as they bind and cleave the same DNA substrate.
Figure 6.
Structural comparison of non-specific and specific endonucleases that contain ββα-metal motifs. (A) Overall conformations of three non-specific endonucleases: CPS-6 (this study, PDB entry: 5GKP), Vvn (PDB entry: 1OUP) and ColE7 (PDB entry: 2IVH). (B) Overall conformations of three sequence- or structure-specific endonucleases: I-PpoI (PDB entry: 1A74), Hpy99I (PDB entry: 3GOX) and T4 endo VII (PDB entry: 2QNC). The ββα-metal motifs are displayed in cyan. The endonuclease cleavage sites are marked by scissors.
In contrast, most sequence non-specific endonucleases are usually monomers, such as DNase I (31), ColE7 (32) and Vvn (33), which randomly nick DNA substrates and do not need to digest two strands of DNA at a time. However, EndoG/CPS-6 is a dimeric non-specific endonuclease, with the two ββα-metal motifs of the CPS-6 dimer located far apart from each other and pointing in different directions. Therefore, EndoG/CPS-6 is a unique non-specific endonuclease in that it is a dimer with two protomers that bind and cleave two separate DNA substrates. Our results show that monomeric CPS-6 not only has significantly diminished endonuclease activity but also has increased exonuclease activity, suggesting that the dimeric conformation has a critical regulatory role for the endonuclease activity of CPS-6. Our previous results suggest that the dimeric conformation of EndoG/CPS-6 is regulated by the redox potential, and that EndoG/CPS-6 shifts to monomers under oxidative conditions due to oxidation of the dimer interfacial residues (24). The redox potential thereby modulates the endonuclease activity of EndoG/CPS-6 by changing its conformation from dimers to monomers. Our crystal structure of CPS-6 bound to two separate DNA substrates explains how CPS-6 degrades DNA without sequence specificity and why this unique non-specific endonuclease fully functions as a dimer. In conclusion, our study explains how the non-specific endonuclease CPS-6 functions as a homodimer, and provides evidence to show that its endonuclease activity can be regulated by dimer-to-monomer conformational changes. The structural information presented here might provide a molecular basis for development of new strategies to stabilize EndoG dimer conformation and activity as a means to boost mitochondrial function under conditions of oxidative stress.
Acknowledgments
Portions of this research were carried out at the National Synchrotron Radiation Research Center, a national user facility supported by the National Science Council of Taiwan. The Synchrotron Radiation Protein Crystallography Facility is supported by the National Core Facility Program for Biotechnology.
ACCESSION NUMBERS
5GKC and 5GKP.
FUNDING
Academia Sinica; Ministry of Science and Technology, Taiwan, ROC. Funding for open access charge: Academia Sinica, Taiwan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Li L.Y., Luo X., Wang X. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature. 2001;412:95–99. doi: 10.1038/35083620. [DOI] [PubMed] [Google Scholar]
- 2.Parrish J., Li L., Klotz K., Ledwich D., Wang X., Xue D. Mitochondrial endonuclease G is important for apoptosis in C. elegans. Nature. 2001;412:90–94. doi: 10.1038/35083608. [DOI] [PubMed] [Google Scholar]
- 3.Zassenhaus H.P., Denniger G. Analysis of the role of the NUC1 endo/exonuclease in yeast mitochondrial DNA recombination. Curr. Genet. 1994;25:142–149. doi: 10.1007/BF00309540. [DOI] [PubMed] [Google Scholar]
- 4.Curtis P.J., Burdon M.G., Smellie R.M.S. The purification from rat liver of a nuclease hydrolyzing ribonucleic acid and deoxyribonucleic acid. Biochem. J. 1966;98:813–817. doi: 10.1042/bj0980813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Linn S., Lehman I.R. An endonuclease from mitochondria of Neurospora crassa. J. Biol. Chem. 1966;241:2694–2699. [PubMed] [Google Scholar]
- 6.Low R.L. Mitochondrial endonuclease G function in apoptosis and mtDNA metabolism: a historical perspective. Mitochondrion. 2003;2:225–236. doi: 10.1016/S1567-7249(02)00104-6. [DOI] [PubMed] [Google Scholar]
- 7.Ruiz-Carrillo A., Renaud J. Endonuclease G: a (dG)n-(dC)n-specific DNase from higher eukaryotes. EMBO J. 1987;6:401–407. doi: 10.1002/j.1460-2075.1987.tb04769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin J.L.J., Nakagawa A., Lin C.L., Hsiao Y.-Y., Yang W.-Z., Wang Y.-T., Doudeva L.G., Skeen-Gaar R.R., Xui D., Yuan H.S. Structural insights into apoptotic DNA degradation by CED-3 Protease Suppressor-6 (CPS-6) from Caenorhabditis elegans. J. Biol. Chem. 2012;287:7110–7120. doi: 10.1074/jbc.M111.316075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Widlak P., Li L.Y., Wang X., Garrard W.T. Action of recombinant human apoptotic endonuclease G on naked DNA and chromatin substrates: cooperation with exonuclease and DNase I. J. Biol. Chem. 2001;276:48404–48409. doi: 10.1074/jbc.M108461200. [DOI] [PubMed] [Google Scholar]
- 10.Cote J., Ruiz-Carrillo A. Primers for mitochondrial DNA replication generated by Endonuclease G. Science. 1993;261:765–769. doi: 10.1126/science.7688144. [DOI] [PubMed] [Google Scholar]
- 11.Ikeda S., Ozaki K. Action of mitochondrial endonuclease G on DNA damaged by L-ascorbic acid, peplomycin, and cis-diamminedichloroplatinum (II) Biochem. Biophys. Res. Commun. 1997;235:291–294. doi: 10.1006/bbrc.1997.6786. [DOI] [PubMed] [Google Scholar]
- 12.Ikeda S., Hasegawa H., Kaminaka S. A 55-kDa endonuclease of mammalian mitochondria: comparison of its subcellular localization and endonucleolytic properties with those of endonuclease G. Acta Med. Okayama. 1997;51:55–62. doi: 10.18926/AMO/30782. [DOI] [PubMed] [Google Scholar]
- 13.Huang K.-J., Ku C.-C., Lehman I.R. Endonuclease G: A role for the enzyme in recombination and cellular proliferation. Proc. Natl. Acad. Sci. U.S.A. 2006;103:8995–9000. doi: 10.1073/pnas.0603445103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zan H., Zhang J., Al-Qahtani A., Pone E.J., White C.A., Lee D., Yel L., Mai T., Casali P. Endonuclease G plays a role in immunoglobulin class switch DNA recombination by introducing double-strand breaks in switch regions. Mol. Immun. 2011;48:610–622. doi: 10.1016/j.molimm.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buttner S., Eisenberg T., Carmona-Gutierrez D., Ruli D., Knauer H., Ruckenstuhl C., Sigrist C., Wissing S., Kollroser M., Frohlich K.-U., et al. Endonuclease G regulates budding yeast life and death. Mol. Cell. 2007;25:233–246. doi: 10.1016/j.molcel.2006.12.021. [DOI] [PubMed] [Google Scholar]
- 16.Susin S.A., Lorenzo H.K., Zamzami N., Marzo I., Snow B.E., Brothers G.M., Mangion J., Jacotot E., Costantini P., Loeffler M., et al. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature. 1999;397:441–446. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- 17.Kalinowska M., Garncarz W., Pietrowska M., Garrard W.T., Widlak P. Regulation of the human apoptotic DNase/RNase Endonuclease G: involvement of Hsp70 and ATP. Apoptosis. 2005;10:821–830. doi: 10.1007/s10495-005-0410-9. [DOI] [PubMed] [Google Scholar]
- 18.Parrish J.Z., Xue D. Cuts can kill: the roles of apoptotic nucleases in cell death and animal development. Chromosoma. 2006;115:89–97. doi: 10.1007/s00412-005-0038-0. [DOI] [PubMed] [Google Scholar]
- 19.Parrish J.Z., Xue D. Functional genomic analysis of apoptotic DNA degradation in C. elegans. Mol. Cell. 2003;11:987–996. doi: 10.1016/s1097-2765(03)00095-9. [DOI] [PubMed] [Google Scholar]
- 20.Wang X., Yang C., Chai J., Shi Y., Xue D. Mechanisms of AIF-mediated apoptotic DNA degradation in Caenorhabditis elegans. Science. 2002;298:1587–1592. doi: 10.1126/science.1076194. [DOI] [PubMed] [Google Scholar]
- 21.Wang X., Wang J., Gengyo-Ando K., Gu L., Sun C.-L., Yang C., Shi Y., Kobayashi T., Shi Y., Mitani S., et al. C. elegans mitochondrial factor WAH-1 promotes phosphatidylserine externalization in apoptotic cells through phospholipid scramblase SCRM-1. Nat. Cell Biol. 2007;9:541–549. doi: 10.1038/ncb1574. [DOI] [PubMed] [Google Scholar]
- 22.Zhou Q., Li H., Li H., Nakagawa A., Lin J., Lee E., Harry B., Skeen-Gaar R., Suehiro Y., William D., et al. Mitochondrial endonuclease G mediates breakdown of paternal mitochondria upon fertilization. Science. 2016;353:394–399. doi: 10.1126/science.aaf4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loll B., Gebhardt M., Wahle E., Meinhart A. Crystal structure of the EndoG/EndoGI complex: mechanism of EndoG inhibition. Nucleic Acids Res. 2009;37:7312–7320. doi: 10.1093/nar/gkp770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin J.L.J., Nakagawa A., Skeen-Gaar R., Yang W.-Z., Zhao P., Zhang Z., Ge X., Mitani S., Xue D., Yuan H.S. Oxidative stress impairs cell death by repressing the nuclease activity of mitochondrial Endonuclease G. Cell Rep. 2016;16:279–287. doi: 10.1016/j.celrep.2016.05.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han E.S., Cooper D.L., Persky N.S., Sutera V.A., Jr, Whitaker R.D., Montello M.L., Lovett S.T. RecJ exonuclease: substrates, products and interaction with SSB. Nucleic Acids Res. 2006;34:1084–1091. doi: 10.1093/nar/gkj503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schafer P., Scholz S.R., Gimadutdinow O., Cymerman I.A., Bujnicki J.M., Ruiz-Carrillo A., Pingoud A., Meiss G. Structural and functional characterization of mitochondrial EndoG, a sugar non-specific nuclease which plays an important role during apoptosis. J. Mol. Biol. 2004;338:217–228. doi: 10.1016/j.jmb.2004.02.069. [DOI] [PubMed] [Google Scholar]
- 27.Zhao Y., Lu M., Zhang H., Hu J., Zhou C., Xu Q., Shah A.M.U.H., Xu H., Wang L., Hua Y. Structural insights into catalysis and dimerization enhanced exonuclease activity of RNase J. Nucleic Acids Res. 2015;43:5550–5559. doi: 10.1093/nar/gkv444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galburt E.A., Chevalier B., Tang W., Jurica M.S., Flick K.E., Raymond J., Monnat J., Stoddard B.L. A novel endonuclease mechanism directly visualized for I-PpoI. Nat. Struct. Biol. 1999;6:1096–1099. doi: 10.1038/70027. [DOI] [PubMed] [Google Scholar]
- 29.Sokolowska M., Czapinska H., Bochtler M. Crystal structure of the ββα-Me type II restriction endonuclease Hpy99I with target DNA. Nucleic Acids Res. 2009;37:3799–3810. doi: 10.1093/nar/gkp228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biertümpfel C., Yang W., Suck D. Crystal structure of T4 endonuclease VII resolving a Holliday junction. Nature. 2007;449:616–620. doi: 10.1038/nature06152. [DOI] [PubMed] [Google Scholar]
- 31.Weston S.A., Lahm A., Suck D. X-ray structure of the DNase I-d(GGTATACC)2 complex at 2.3 A resolution. J. Mol. Biol. 1992;226:1237–1256. doi: 10.1016/0022-2836(92)91064-v. [DOI] [PubMed] [Google Scholar]
- 32.Hsia K.-C., Chak K.-F., Liang P.-H., Cheng Y.-S., Ku W.-Y., Yuan H.S. DNA binding and degradation by the HNH protein ColE7. Structure. 2004;12:205–214. doi: 10.1016/j.str.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 33.Li C.-L., Hor L.-I., Chang Z.-F., Tsai L.-C., Yang W.-Z., Yuan H.S. DNA binding and cleavage by the periplasmic nuclease Vvn: a novel structure with a known active site. EMBO J. 2003;22:4014–4025. doi: 10.1093/emboj/cdg377. [DOI] [PMC free article] [PubMed] [Google Scholar]