Abstract
We have previously shown that clusters of guanine quadruplex (G4) structures can form in the human herpes simplex-1 (HSV-1) genome. Here we used immunofluorescence and immune-electron microscopy with a G4-specific monoclonal antibody to visualize G4 structures in HSV-1 infected cells. We found that G4 formation and localization within the cells was virus cycle dependent: viral G4s peaked at the time of viral DNA replication in the cell nucleus, moved to the nuclear membrane at the time of virus nuclear egress and were later found in HSV-1 immature virions released from the cell nucleus. Colocalization of G4s with ICP8, a viral DNA processing protein, was observed in viral replication compartments. G4s were lost upon treatment with DNAse and inhibitors of HSV-1 DNA replication. The notable increase in G4s upon HSV-1 infection suggests a key role of these structures in the HSV-1 biology and indicates new targets to control both the lytic and latent infection.
INTRODUCTION
Guanine-rich DNA sequences can form stable four-stranded guanine quadruplex (G4) structures based on the formation of G-quartets, which are stabilized by Hoogsteen-type hydrogen bonds between guanines and monovalent cations between the G-quartets (1).
In eukaryotes, G4s have been shown to occur in functionally important regions of the genome: in telomeres, G-rich micro- and mini-satellites, within promoters, and in ribosomal DNA (rDNA) repeat arrays (2–4). Human DNA G4 motifs have been reported to be associated with recombination prone regions (5) and to show mutational patterns that preserved the potential to form G4 structures (4,6).
The presence of G4 DNA structures in human cells has recently been supported by specific antibodies derived from phage display selection (7) and hybridoma technology (8).
Besides eukaryotes, G4 relevance and presence have recently emerged also in prokaryotes (9,10) and viruses (11). The presence of functionally significant G4 DNA motifs in the human immunodeficiency virus (HIV) has been reported by us and others both in the promoter (12–14) and Nef coding regions (15). The herpes simplex virus-1 (HSV-1) genome has a very high GC content (68%) which peaks at 84.7% GC in simple sequence repeats (SSRs) (16). Recently we provided evidence for the presence of very stable G4-forming regions located in the HSV-1 inverted repeats (17). In particular, multiple conserved and extended clusters of G4 forming sequences were observed, covering about 2,000 bp of the 152,000 bp-viral genome.
HSV-1 first lytic infection occurs within mucosal epithelial cells, where the expression of viral genes proceeds in a regulated cascade in which three classes of viral genes are temporally expressed: immediate-early (IE), early (E) and late (L) (18). The virus next enters sensory neurons where latency is established; it can later reactivate resulting in the generation of new virions that cause recurrent disease (19).
Both in the case of HSV-1 and HIV-1, treatment of infected cells with G4 ligands greatly impaired viral infectivity (13,15,17); in particular, treatment with BRACO-19 stabilized G4s in the HSV-1 genome and inhibited viral replication (17).
Given the extraordinary extension of G4 forming regions in the HSV-1 genome, we here aimed at visualizing G4s in eukaryotic cells infected with HSV-1. By employing the anti-G4 monoclonal antibody 1H6 (8), we were able to show strong enrichment of G4 structures in cells upon infection. G4 formation depended on the viral cycle, with the highest G4 signal observed at the time of viral replication. The observed G4s mainly localized in viral replication compartments (RCs) and treatment with viral DNA polymerase inhibitors greatly decreased the G4 antibody signal.
MATERIALS AND METHODS
Cells and viruses
Vero cells (Sigma Aldrich, Milan, Italy) and TZM-bl reporter cell line (obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID,NIH, from Dr J.C. Kappes, Dr X. Wu and Tranzyme Inc.) were grown in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum (FBS) and PenStrep 1× (Life Technologies, Monza, Italy). Wild-type (wt) HSV-1 strain F was a kind gift from Bernard Roizman (University of Chicago, IL, USA), recombinant HSV-1 expressing VP16-GFP (HSV-1 v41) was kindly provided by Peter O'Hare (Imperial College London, UK) (20). For virus infection, wt or mutant viruses were incubated with cells at different multiplicities of infection (MOI) in serum-free medium. After 1 h of incubation at 37°C, the inoculum was replaced with complete medium. Mock-infected cells were treated in the exact same way except that serum-free medium was added in place of the virus. HIV-1NL4-3 stock was prepared transfecting HEK293T with the proviral genome (NIH AIDS Reagent Program, Division of AIDS, NIAID,NIH, from Dr Malcolm Martin). For HIV-1 infection, TZM-bl cells were infected with wt virus at different MOIs. After 2 h, cells were washed with phosphate buffered saline (PBS) 1× and grown in complete medium.
Antibodies and immunofluorescence
The mouse monoclonal 1H6 antibody has been previously reported (8,21). The rabbit 3–83 antiserum specific for ICP8 was a kind gift from David M. Knipe (Harvard Medical School, Boston, USA) (22) anti-HSV-1 ICP8 monoclonal antibody conjugated with FITC was obtained from Santa Cruz Biotechnology Inc.
For immunofluorescence studies, Vero or TZM-bl cells were seeded at 5 × 104 cells/well in 8-well plates (8-well cultureslide, BD Falcon™) or at 0.5-1 × 105 cells/well on glass 12-mm-diameter coverslips in 24-well plates and grown overnight at 37°C. Following infection at multiple MOIs (from 0.5 to 8) and incubation for an appropriate time (from 2 to 20 h.p.i. or 24 h.p.i. for HIV-1 infection), cells were fixed in 4% paraformaldehyde (PFA, Sigma Aldrich) in PBS 1× for 20 min at room temperature (rt) and permeabilized with 0.5% Tween-20 (Sigma Aldrich) in PBS 1×. After blocking in PBS containing 5% of FBS, cells were incubated with 1H6 antibody at 1 μg/ml for 2 h at rt, rinsed with PBS-T (0.2% Tween-20 in PBS 1×) and incubated with Alexa Fluor-546 goat anti-mouse IgG at 1:500 (Molecular Probes, Life Technologies) for 1 h at rt and, where needed, followed by 1 h incubation at rt with FITC-conjugated anti-HSV-1 ICP8 at 1:200 or anti-ICP8 serum at 1:1000 dilution and Alexa Fuor-488 goat anti-rabbit IgG at 1:500 (Molecular Probes, Life Technologies) for 30 min. Nuclei were stained with far-red fluorescent DNA dye (DRAQ5®, 1:1000, Cell Signaling Technology, USA) for 5 min at rt. For enzyme treatment, Vero cells were incubated with 50 μg/ml RNase-A (Thermo Scientific) in PBS 1× or 0.12 U/μl DNAse I (Life Technologies) in saline buffer 1× (100 mM Tris pH = 7.5, 25 mM MgCl2, 5 mM CaCl2) for 1 h at 37°C, then blocked with FBS and stained with antibodies as described above. Mock-infected cells were treated in the exact same way as infected cells in each type of experiment and staining, except that serum-free medium was added in place of the virus. For viral replication inhibition, cells were treated with 400 μg/ml phosphonoacetic acid (PAA) (Sigma Aldrich) or acyclovir (ACV) (100 μM) from 1 h before infection up to fixation. For fluorescence quantification experiments, images were acquired using a Nikon A1Rsi+ Laser Scanning confocal microscope equipped with NIS-Elements Advanced Research software (Nikon Instruments Inc., Melville, USA), with 20× and 60× ocular objectives. Laser excitation and emission filters were: blue laser at 488 nm, emission filter 500–550 nm; yellow-green laser at 561 nm, emission filter 570–620 nm; red laser at 640 nm, emission filter 663–738 nm. Excitation and emission wavelength at the maximum peak and emission color of the fluorophores used in this work were: DAPI 358–461 nm—blue; FITC 490–525 nm—green; Alexa Fluor®488 496–519 nm—green; GFP 488–509 nm—green; Alexa Fluor®546 556–573 nm—orange; DRAQ5® 646–681/697 nm—red. To achieve constant quantification circumstances and to maintain high reproducibility of the data, FocalCheck™ Fluorescent Microsphere Standards (Life Technologies, Italy) were used to set the Confocal Laser Scanning Microscopy (CLSM) unit calibration parameters. For quantification, overall fluorescence intensity detected by the microscope software at 20× (Figure 2C and Supplementary Figure S2) was normalized for the total number of G4-positive cell nuclei. All samples were further normalized for the total number of cells and divided by the mock sample signal, which was set to 100%. Cells were counted using ImageJ software; at least three pictures per condition (200–500 cells/picture) were considered. Colocalization analysis from 3D acquisitions was acquired using the NIS-Elements Advanced Research software. Final images included in this work are representative of multiple experiments. For each repetition, mock-infected cells and at least one reference time point of viral replication were included for comparison. All viral conditions were assayed at least three times.
Figure 2.
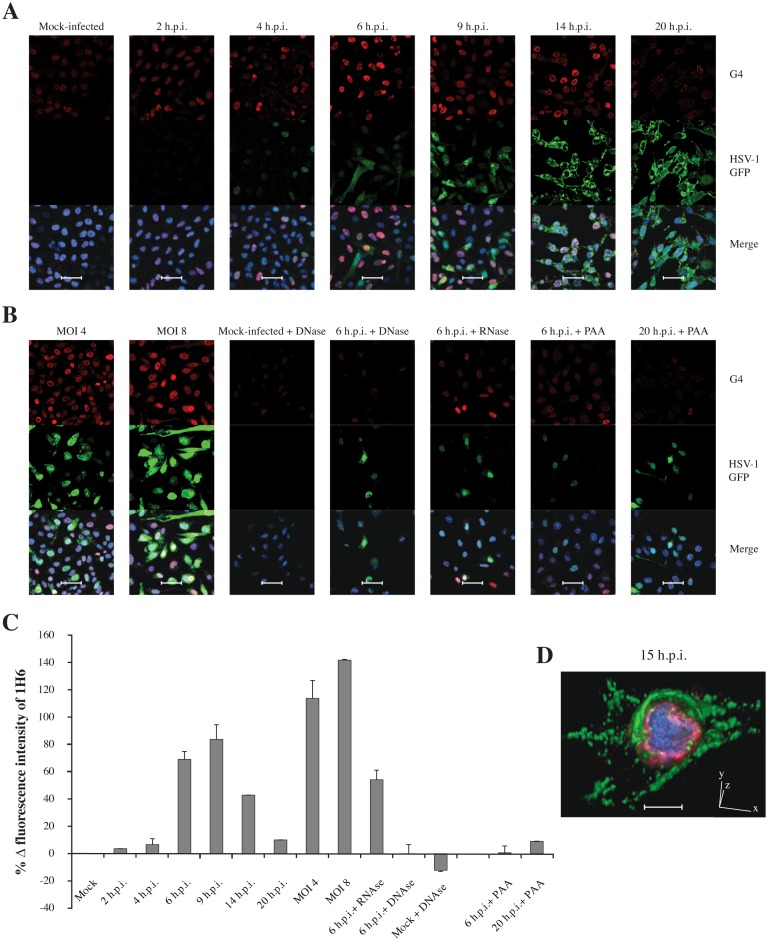
Immunofluorescence confocal microscopy of HSV-1 v41-infected mammalian cells at 60× magnification. (A) Infected cells visualized at different h.p.i., corresponding to a single cycle of replication. (B) Cells infected with increasing MOIs of HSV-1 v41 visualized at 6 h.p.i. and infected cell treated with DNase, RNAse and PAA, an inhibitor of the viral DNA polymerase, visualized either at 6 h.p.i. or 20 h.p.i., as indicated. In all images: the red signal indicates G4s, detected with the 1H6 primary Ab and Alexa Fluor-546 secondary Ab; the green signal derives from the viral recombinant L protein VP16-GFP and indicates the presence of the virus in cells; the blue signal is obtained staining the DNA with DRAQ5®. In the merge images the blue signal is shown overlapped with the red and green fluorescence. Mock-infected cells were treated in the same conditions. Shown are representative images at 60× magnification. Each condition was tested at least three times, always including mock-infected and infected cells as reference. Scale bars: 50 μm. (C) Quantification of the G4 (1H6) signal. Data refer to intensity values supplied by the instrument for the non-saturated signal in the 20× images (Supplementary Figure S2), opportunely normalized to the number of cell nuclei and to mock-infected cell fluorescence intensity. (D) Three-dimension (3D) confocal microscopy of HSV-1 v41 infected cell at 15 h.p.i.. Red fluorescence indicates G4s (1H6), which are mainly localized at the internal layer of the nuclear membrane; the green fluorescence shows the late viral protein VP16-GFP, mainly accumulated in the cytoplasm at this time p.i.; the blue fluorescence shows the DNA dispersed in the cell nucleus. Scale bar: 10 μm.
Surface plasmon resonance (SPR) analysis
Surface plasmon resonance (SPR) was performed on the Biacore T100 platform (GE Healthcare, Life Science, Milan, Italy). The following oligonucleotides were used (Sigma-Aldrich, Milan, Italy): gp054a 5′-GGGGTTGGGGCTGGGGTTGGGG-3′; un2 5′-GGGGGCGAGGGGCGGGAGGGGGCGAGGGG-3′; un3 5′-GGGAGGAGCGGGGGGAGGAGCGGG-3′; Oxy2 5′-TTTTGGGGTTTTGGGG-3′; OxyTel 5′-GGGGTTTTGGGGTTTTGGGGTTTTGGGG-3′; hTel21 5′-GGGTTAGGGTTAGGGTTAGGG-3′; hTel22 5′-AGGGTTAGGGTTAGGGTTAGGG-3′; hTel54 5′-TCGAGGGTTAGGGTTAGGGTTAGGGTTAGGGTTAGGGTTAGGGTTAGGGTTAC-3′; bcl-2 5′-GGGCGCGGGAGGAAGGGGGCGGG-3′; c-myc 5′-TGGGGAGGGTGGGGAGGGTGGGGAAGG-3′; c-kit1 5′-AGGGAGGGCGCTGGGAGGAGGG-3′; c-kit2 5′-CGGGCGGGCGCGAGGGAGGGT-3′;
hTelScra 5′-GGATGTGAGTGTGAGTGTGAGG-3′; LTRScra 5′-TTTTTGGAGCGTGTGTGCGCGAGAGCGTGCGCGTGGCGAGCGTTGAGTGGTTTTT-3′; rnd 5′-AAAAACTACTGCACGCTCGCTACGACGACACTGTCGCGCATACAAGCTGCAAAAA-3′ (Random in (23)); T9 5′-TTTTTTTTT-3′; T30 5′-TTTTTTTTTTTTTTTTTTTTTTTTTTTTTT-3′; hp 5′-CGCAGCGTGGCTTTGTTTGCCACGCTGCG-3′; dsDNA: hTelScra+complementary sequence; NRAS RNA 5′-GGGAGGGGCGGGUCUGGG-3′; U3-III HIV RNA 5′-GGGAGGCGUGGCCUGGGCGGGACUGGGG-3′; U3-IV HIV RNA 5′-GGGCGGGACUGGGGAGUGG-3′; hTel2 RNA 5′-UAGGGUUAGGGU-3′; TAR HIV RNA 5′-GGCAGAUCUGAGCCUGGGAGCUCUCUGCC-3′; rnd RNA 5′-GAGCGUGCGCGUGCGAGCGUGAGUGAGCGUGGG-3′. To allow the correct orientation of 1H6 binding site on the chip, 1H6 was incubated for 1 h at rt in the presence of the G4-folded un3. The Mouse Antibody Capture kit (GE Healthcare, Life Science, Milan, Italy) was next used to capture 1H6-un3 complex on Serie S sensor chip CM5. Immobilization of the anti-mouse IgG (GE Healthcare, Life Science, Milan, Italy) was performed by amine coupling in HEPES-NaCl running buffer (HEPES pH 7.4 10 m M, NaCl 150 mM, ethylenediaminetetraacetic acid (EDTA) 3 mM), while 1H6-un3 capturing was performed in HEPES-KCl buffer (HEPES 10 mM, KCl 200 mM, EDTA 3 mM). Un3 was dissociated with a flow of HEPES-KCl buffer followed by injection of KCl 1M. Flow cell 1 was blank immobilized with anti-mouse IgG to permit reference subtraction. Folding of oligonucleotides into G4s was performed in HEPES-KCl buffer after heat denaturation at 95°C for 5 min and gradual cooling at room temperature. Binding analysis of each oligonucleotide was performed at a flow rate of 20 μl/min, with contact time of 340 sec and dissociation time of 240 sec in HEPES-KCl buffer. Sensorgrams were obtained in the concentration range of 0.25-8 μM. After each oligonucleotide injection, the chip surface was regenerated with KCl 1M solution. All sensorgrams were corrected by reference subtraction of blank flow cell response and buffer injection response. Data were fitted to a global 1:1 binding model using BIAevaluation software (GE Healthcare). The stability of 1H6 binding was evaluated by measuring RU response 20 sec before end of the dissociation phase.
Flow cytometry analysis
For FACS analysis, Vero cells were seeded at 2 × 106 cells/dish in 10-cm-dishes (BD Falcon™), grown overnight at 37°C and infected with HSV-1 v41 at a MOI of 0.5 as described above. At 8 h.p.i., cells were washed with PBS 1× and fixed in IC Fixation Buffer (eBiosciences, CA, USA). HSV-1 infected cells were incubated with 1H6 antibody in permeabilization buffer 1× (eBiosciences) for 1 h, followed by incubation with secondary antibody Alexa Fluor-633 goat anti-mouse at 1:500 (Molecular Probes, Life Technologies). All incubations were carried out at rt, according to the manufacturer's instructions. Mock-infected Vero cells were treated as infected cells and used as negative control. A total of 30,000 events/sample were acquired with a LRS 2 instrument using FACS DIVA Software (BD Bioscience, San Jose, CA, USA) and analyzed with FlowJo (Tree Star, OR, USA).
Immuno-electron microscopy (Immuno-EM)
Vero cells were seeded at 1 × 105 cells/well in 12-well plates and grown overnight at 37°C. Immuno-EM samples were prepared as previously described (21). At 8 and 15 h.p.i., cells infected with wt HSV-1 at MOI 5 were washed with PBS 1× and fixed in 2.5% glutaraldehyde buffered in 0.1 M cacodylate buffer (Sigma Aldrich) for 1 h at 4°C. Cells were washed with 0.1 M cacodylate buffer, post-fixed in 1% OsO4/1.5% K3Fe(CN6) for 30–60 min, dehydrated and embedded in epoxy resin according to standard protocols. Sections were collected on nickel grids, treated with 1% periodic acid (Sigma Aldrich) for 20 min at rt, rinsed with distilled water and blocked in blocking buffer (Tris-buffered saline (TBS) containing 1% bovine serum albumin, 0.1% glycine, 0.1% cold water fish skin gelatin and 5% FBS). Grids were incubated overnight with 1H6 and rabbit 3–83 anti-ICP8 serum, followed by incubation for 2 h at rt with anti-mouse secondary antibody conjugated to 5-nm gold (Sigma Aldrich) at 1:100 or anti-rabbit secondary antibody conjugated to 10-nm gold (Sigma Aldrich) at 1:100, respectively. Samples were rinsed with distilled water and contrasted with uranyl acetate and lead citrate. Images were collected with a FEI Tecnai G12. For statistical analysis, the number of G4 and ICP8 dots in 35–45 images per condition was counted with the aid of ImageJ software. The number of recorded dots was calculated on the total images area (mm2) ± SEM. The frequency of G4s in virions was calculated as the number of G4 dots in virions on the number of virions in samples treated with 1H6 (41 images) and compared to the number of dots in virions/number of virions in the negative control treated only with the secondary Ab (38 images). The Student's t-test was used to calculate the statistical significance.
RESULTS
The monoclonal antibody 1H6 binds with high efficiency to HSV-1 G4-forming sequences
We have previously shown that at least four G-rich repeated sequences in the HSV-1 genome can fold into G4 structures in vitro (17). Three sequences feature four GGGG-tracts, while one sequence has four GGG-tracts, displaying the ability to form multiple monomolecular 4- or 3-tetrad stacked G4, respectively (Table 1).
Table 1. HSV-1 G4-forming sequences described in (17).
| HSV-1 sequences | Sequence (5′ → 3′) |
|---|---|
| gp054 | GGGGXTGGGGXTGGGGXTGGGG |
| un1 | GGGGGAGAGGGGAGAGGGGGGGAGAGGGG |
| un2 | GGGGGCGAGGGGCGGGAGGGGGCGAGGGG |
| un3 | GGGAGGAGCGGGGGGAGGAGCGGG |
X indicates T or C. G-tracts are underlined.
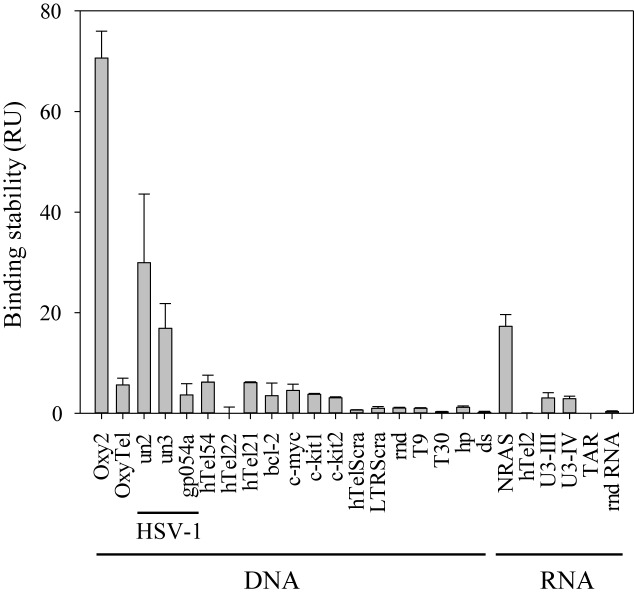
Since the anti-G4 1H6 antibody (Ab) has been reported to have broad specificity for many but not all G4 structures (8), we initially tested its affinity for the selected HSV-1 G4-forming sequences in vitro by SPR. Three HSV-1 sequences were tested: un2 (antiparallel), un3 (parallel) and gp054a (mixed-type); un1 could not be tested because unstable in solution (17). A series of control sequences was alongside tested: the tetramolecular Oxytrichia telomeric G4, which 1H6 was developed against (Oxy2), the monomolecular Oxytrichia G4 (OxyTel), three different-length human telomeric G4s (hTel21, hTel22 and hTel54), four G4s of oncogene promoters (bcl-2, c-myc, c-kit1, c-kit2) (24–27), two G-rich oligonucleotides corresponding to the scrambled sequences of the human telomeric and HIV-1 LTR promoter G4s (hTelScra and LTRScra) (13,15,17), one random ssDNA (rnd), two polyTs (T9 and T30), one hairpin oligonucleotide (hp) and a dsDNA. RNA sequences were also tested, in particular: two cellular G4s found in the 5′-UTR of the transcript of the human NRAS proto-oncogene (NRAS) (28) and in telomeric transcripts (hTel2) (29), two G4s that form in the HIV-1 RNA genome (U3-III and U3-IV) (30); one hairpin RNA from the HIV transcript (TAR) (31) and one unstructured random RNA (rndR). The absolute binding affinity at the thermodynamic equilibrium could be obtained only for few of these sequences. In particular, Oxy2 displayed the best affinity (KD 62.0 ± 4.2 nM); hTel54 had a 10-fold lower affinity (KD 654.5 ± 70.7 nM), slightly lower than un2, un3 and NRAS RNA (KD 535.3 ± 20.5, 479.7 ± 14.1 nM and 539.2 ± 16.2, respectively); gp054 was three times a lesser good binder than un2 (KD 1.67 ± 0.46 μM). The other sequences displayed either no or unspecific interaction with 1H6 (Supplementary Figure S1). KD values represent the binding affinity at the equilibrium. However, other kinetic parameters can be obtained by SPR, such as the binding stability (32). To compare all tested sequences, we measured the stability of 1H6/oligonucleotide right before the end of the dissociation phase, which can be roughly compared to the final wash step in an ELISA assay (8). In these conditions, for the DNA sequences, the best stabilization was again obtained with Oxy2, un2 and un3 sequences (Figure 1); gp054, hTel54, hTel21, OxyTel, bcl-2, c-myc, c-kit1 and c-kit2 (all G4 forming sequences) also showed stabilization though to a lower extent; hTel22 and all non-G4-forming oligonucleotides displayed no stabilization (Figure 1). For the RNA sequences, NRAS G4 displayed stabilization similar to that of un3; all other sequences displayed very low or non-significant stabilization (Figure 1). These data indicate that two of the three tested HSV-1 G4s are bound at least 30 times better than ssDNA. Interestingly, un2 and un3 are the most abundant repeats in all HSV-1 strains (17). The SPR data also indicate the excellent G4 selectivity of 1H6, which recognizes G4s with different efficiency, depending on the conformation, and does not bind unstructured oligonucleotides.
Figure 1.
SPR analysis of 1H6 antibody binding stability to HSV-1 G4-forming oligonucleotides and control sequences. HSV-1 G4-forming oligonucleotides (un2, un3, gp054a) (Table 1) and the controls (0.25-8 μM) were injected on chip-immobilized 1H6. The binding stability values at 4 μM oligonucleotides are shown. Values were measured in the late dissociation phase. For DNA sequences: Oxy2 forms a tetramolecular G4 against which 1H6 was initially developed in animals; OxyTel is the corresponding monomolecular G4. HTel21, hTel22 and hTel54 are different-length G4-forming telomeric repeats; bcl-2, c-myc, c-kit1 and c-kit2 are oncogene promoter G4s; hTelScra and LTRScra are G-rich non-G4-forming oligonucleotides; rnd is a random sequence; T9 and T30 are polyT oligonucleotides; hp is a hairpin forming sequence; dsDNA is double stranded DNA. For RNA sequences: NRAS and hTel2 are two cellular G4s; U3-III and U3-IV are two HIV-1 G4s; TAR is a hairpin RNA; rndR is a unstructured random RNA. Results are shown as RU (response units) values ± SEM (n = 3).
G4 formation in infected cells depends on the HSV-1 cycle and peaks during viral replication
Based on the above encouraging data, showing that 1H6 Ab binds with high efficiency HSV-1 G4s versus most other cellular G4s and non-structured sequences possibly present in a mammalian cell, we proceeded using 1H6 to visualize G4s in HSV-1 infected cells. To this end, HSV-1 susceptible cells (Vero) were infected with HSV-1 v41, a recombinant virus expressing the viral protein VP16 fused to the green fluorescent protein (GFP). This mutant virus is characterized by normal replication kinetics and yields (20). VP16 is an essential and abundant L tegument protein that regulates IE gene transcription, assembly and viral egress. VP16-GFP is normally incorporated into the virion, resulting in fluorescent particles, which allow monitoring of viral steps in infected cells (20,33). In general, no significant VP16-GFP signal is detectable at the earliest stages of infection (up to 3 h.p.i.), whereas between 3 and 5–6 h.p.i. newly synthesized VP16-GFP becomes visible in punctate foci diffused in the nucleus. At 7 to 12 h.p.i., VP16-GFP is also detected in the cytoplasm. After 12 h.p.i., VP16-GFP accumulates in vesicular-like foci mainly in the cytoplasm. At late stages, accumulation of high signal intensity VP16-GFP occurs at the boundaries between cells (20,33). G4s, VP16-GFP and DNA were visualized at different times post-infection (p.i.), including pre-replication (2-4 h.p.i), replication (6-9 h.p.i.) and post-replication events (14-20 h.p.i.), to fully comprise a single cycle of replication (34).
The VP16-GFP signal steadily increased from 2–5 h.p.i., when it was just above background levels and localized in the nucleus, to 6–20 h.p.i. when it sharply increased and localized entirely in the cytoplasm (Figure 2A and Supplementary Figure S2), fully matching its previously reported behavior (20) and representing a typical HSV-1 infection.
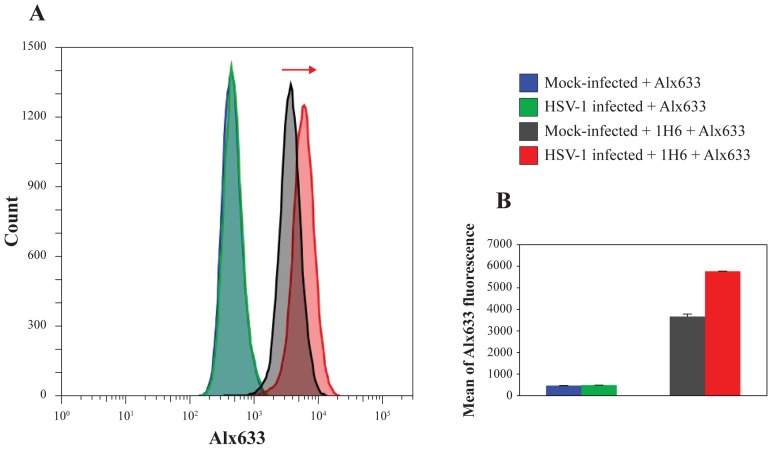
In these conditions, intensity of the G4 signal was barely above that of mock-infected cells at 2–4 h.p.i. (4–10%), it remarkably increased at 6–9 h.p.i. (50–80%), progressively decreased at 14 h.p.i. (30–40%) and at 20 h.p.i. reverted to signal just above that of mock-infected cells (10%) (Figure 2A, C and Supplementary Figure S2a). Negative controls, i.e. infected and mock-infected cells lacking treatment with primary antibodies and treated with secondary antibodies, showed no unspecific background in these conditions (Supplementary Figure S3). Quantitative flow cytometry analysis confirmed that the G4 signal in 1H6-stained cells at 8 h.p.i. increased by 57% compared to mock-infected cells (Figure 3).
Figure 3.
Quantification of G4s in HSV-1 infected cells by flow cytometry. (A) Analysis of the increase of 1H6 fluorescence in HSV-1 infected cells (in red) in comparison with mock-infected cells (in black) as indicated by the red arrow. No effect on fluorescence was observed in the presence of the Alexa Fluor-633 secondary antibody only (Alx633) in neither mock-infected nor infected cells (see histogram on the left, curves are indicated in blue and green respectively). Figures are representative of two independent experiments, each made in triplicate. (B) Mean of 1H6 fluorescence in mock-infected and infected cells: a 57% increase, in terms of mean of 1H6 fluorescence was observed in infected cells compared to mock-infected cells within the replication time range. The mean of Alx633 in mock-infected and infected cells resulted superimposable and no increase was observed.
As for G4 localization, at 2–9 h.p.i. the G4 signal was almost uniformly localized in the cell nucleus, with visible bright punctate foci. Differently, at 14–20 h.p.i. the G4 signal gradually clustered to the nuclear membrane (Figure 2A and corresponding magnification in Supplementary Figure S4). In particular, high-resolution 3D and 2D analysis (Figure 2D and Supplementary Figure S5, respectively) showed that at 15 h.p.i. G4s (red signal) were clustered at the internal layer of the nuclear membrane, the DNA (blue signal) was uniformly distributed within the cell nucleus, with portions of it close to the nuclear membrane, and the viral protein (green signal) was dispersed in the cytoplasm. The nuclear membrane localization of viral G4s is in line with the reported lateral diffusion of the HSV-1 machinery, which allows budding of the virus from the nucleus after viral DNA replication has occurred (35,36). Cells infected with increasing virus amounts (MOIs 1, 4, 8) displayed increased G4 signal (Figure 2B, C and Supplementary Figure S2b), indicating that G4 formation was strictly dependent on the amount of virus in the cell. Since 1H6 recognized one RNA G4 in our SPR testing, to detect the nature of the visualized G4s, infected cells were treated with DNase I or RNase A: DNase-treated samples drastically reduced G4s to levels below those of mock–infected cells, whereas treatment with RNAse induced only a minor (20%) decrease in G4s (Figure 2B, C and Supplementary Figure S2c), which may account for viral/cellular RNA G4s. As expected, VP16-GFP was not affected by these treatments. These data indicate that G4s visualized in infected cells are mainly DNA G4s.
Cells treated with PAA or ACV, two specific inhibitors of the viral DNA polymerase, which replicates the viral DNA at around 6–8 h.p.i. (17,37,38), displayed G4 signal similar to that of mock-infected cells when tested at 6–8 and 20 h.p.i. (Figure 2B and Supplementary Figure S2c). VP16-GFP was poorly affected by PAA and ACV at 6–8 h.p.i.; at 20 h.p.i. it localized prevalently in the nucleus, due to the reported compound interference with L viral proteins (39) (Figure 2B and Supplementary Figure S2c).
To check if G4s formed within the cell as a response to the viral infection, cells were infected with an unrelated virus, i.e. the human immunodeficiency virus HIV-1, and visualized for G4s with 1H6 Ab. HIV-1 has a much shorter genome compared to HSV-1 (around 9,000 versus 150,000 nt) and it has been shown to form only few G4s in key regions of the genome (six confirmed G4s in HIV-1 versus 67–70 G4 repeats in HSV-1) (13,15,17), even if more putative G4 sequences are present in both viruses. In addition, only few copies of the HIV-1 DNA genome integrate into each host cell chromosome (40). Therefore, HIV-1 while imposing to the cell a stress typical of virus infection, does not display G4s and DNA replication rate as high as HSV-1. Infection of cells at different MOIs of HIV-1 showed no virus-dependent increase in G4s (Supplementary Figure S6): a statistically significant decrease was observed only at MOI 2, effect likely due to the virus-dependent cytotoxicity at this MOI which largely decreased the number of cells (Supplementary Figure S6).
These data altogether suggest that HSV-1 G4s are mainly responsible for the increased G4 signal in HSV-1 infected cells during viral replication.
G4s localize in virus replication compartments in infected cells
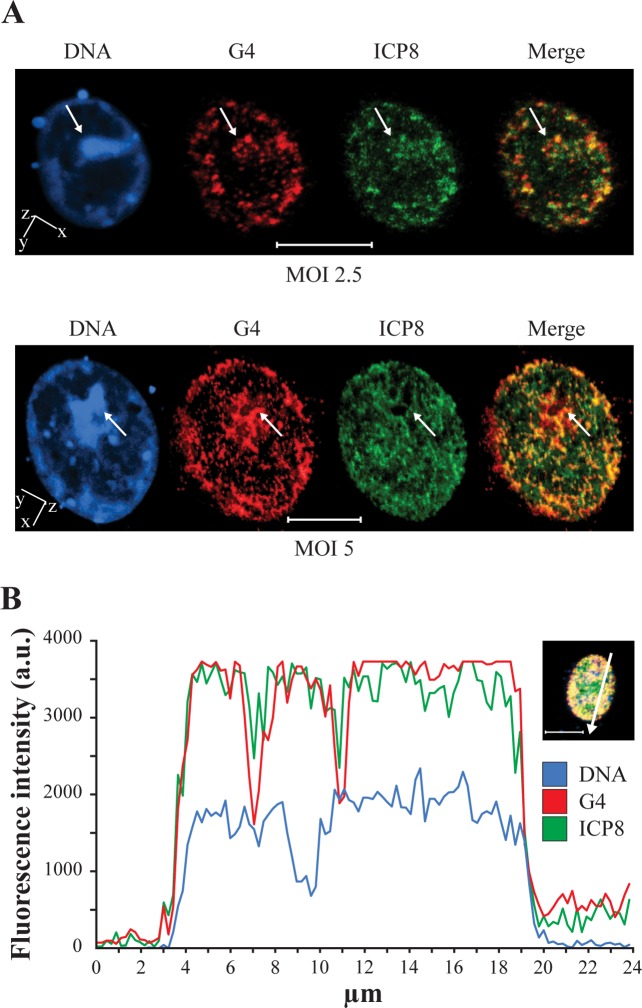
To detect if the G4s observed in infected cells localized in virus-related compartments within the cell, an anti-ICP8 antibody was used. ICP8 is a DNA binding viral protein required for formation of viral pre-replication and replication compartments (RCs) (41–44). Cells infected with wt HSV-1 were co-stained at 6–8 h.p.i. with anti-ICP8 and anti-G4 (1H6) antibodies: the ICP8 protein localized in the cell nucleus, as previously reported (22) and an extended G4/RC colocalization was appreciable in the 2D images (Supplementary Figure S7). Accurate 3D analysis of several cells confirmed the presence of multiple overlapping foci of G4s and RCs at the internal layer of the nuclear membrane and distributed in the cell nucleus (yellow/orange signal, Figure 4A). In contrast, the most intense regions stained by the DNA probe DRAQ5®, reasonably corresponding to the cell chromosome DNA, were particularly poor in G4s and RCs (white arrows, Figure 4A). The observed degree of colocalization was further confirmed by the intensity profiles acquired in the 3D single-cell along an ideal arrow entirely sectioning the cell nucleus: the G4 and RC signals displayed overlapping profiles (red and green lines, respectively, Figure 4B), whereas they were distinct from the cell DNA profile (blue line, Figure 4B). The Pearson correlation coefficient (PCC) (45), which is 1 in an ideal 100% overlapping, was 0.89. As negative control, the same analysis was performed in the nucleus of a cell stained with VP16-GFP and 1H6, the signals of which were not expected to overlap. Indeed, intensity profiles did not overlap and PCC corresponded to 0.14 (Supplementary Figure S8).
Figure 4.
Colocalization of G4s and the viral protein ICP8 by 3D confocal microscopy. ICP8 is a marker for HSV-1 replication compartments (RCs) (41). (A) Cells were infected with different amounts of wt HSV-1 (strain F), MOI 2.5 (upper panel) and 5 (lower panel). At 6–8 h.p.i. cells were stained with the anti-G4 (1H6) and anti-ICP8-FITC Abs. Blue, red and green indicate DNA, G4s and ICP8-dependent viral RCs, respectively. The images on the right (merge) show G4 (red) and ICP8 (green) overlapping as a yellow/orange signal. Spatial orientation is indicated. (B) Intensity profiles of DNA (blue), G4s (red) and ICP8-dependent RCs (green) obtained with NIS-Elements Advanced Research software, along an ideal 24 μm-long straight line (white) crossing the nucleus of a representative infected cell (right inset). The DNA profile defines the nuclear area where the G4 and ICP8 profiles are clearly also enclosed. G4 and ICP8 exhibit very similar fluorescence profile. The scale bar is set to 10 μm.
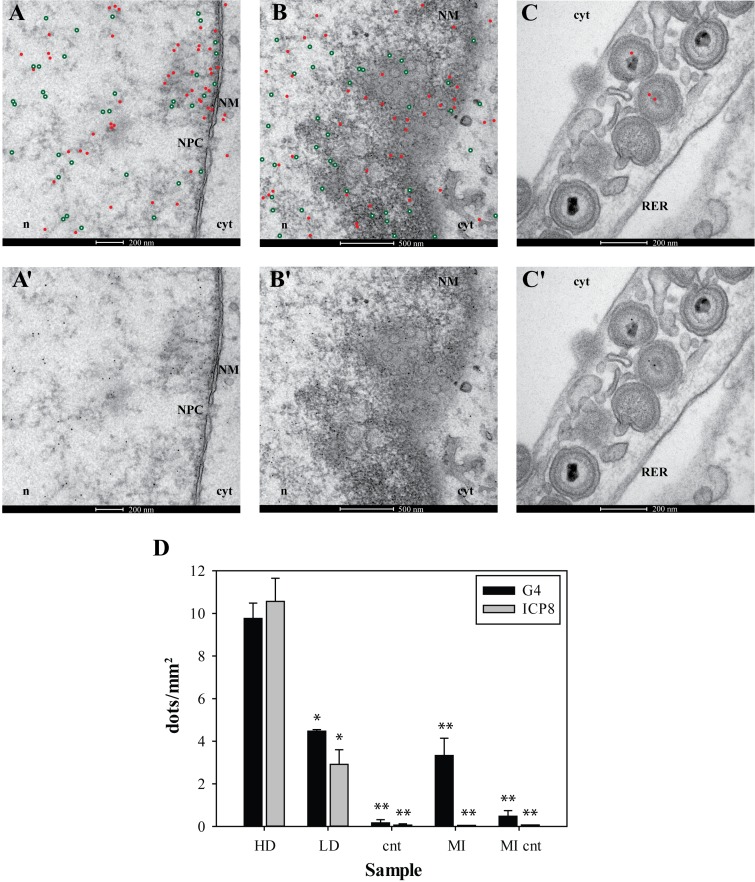
To further assess G4 localization, immune-EM was performed on cells infected with wt HSV-1, fixed at 8 and 15 h.p.i. and incubated with anti-G4 and anti-ICP8 serum, conjugated to 5- and 10-nm gold particles, respectively (Figure 5A–C’). At both times p.i., G4s and ICP8 were mostly found in close proximity; samples obtained at 15 h.p.i. were the most informative since at this time p.i. premature capsids ready for budding from the nuclear membrane begin to be visible. Indeed, G4s and ICP8 were detected in clusters close to the nuclear membrane, in which nuclear pores (Figure 5A–A’) were observed. Indeed nuclear egress has been reported to follow diverse pathways: budding of capsids at the inner nuclear membrane into the perinuclear space or via rough endoplasmic reticulum (RER) into Golgi cisternae, or direct access from the nucleus to the cytoplasm via nuclear pores (46,47). G4s and ICP8 were clearly assembled also in particularly crowded viral RCs producing many capsids in proximity to the nuclear membrane (Figure 5B–B’) (35,36,48). In several cases G4s were observed within the core of immature capsids, which enclose viral DNA. In particular, capsids in the nucleoplasm, before and after fusion at the nuclear membrane and in the perinuclear RER (Figure 5B–C’ and Supplementary Figure S9) were found to contain G4s. The amount and distribution of G4s in mock-infected cells was distinctly different from those in HSV-1 infected cells: a lower number of G4s were detected in the cell nucleus and mostly not close to the nuclear membrane (Supplementary Figure S10, panels a–d). The presence of G4s was negligible in HSV-1 infected cells lacking treatment with anti-G4 and anti-ICP8 Abs (Supplementary Figure S10, panels e and f). These data were statistically analyzed by quantification of G4s and ICP8, performed by counting the number of 5 nm (red, G4) and 10 nm (green, ICP8) dots in all samples (Figure 5D). About 35–45 images per sample were considered and data were reported as the number of dots per mm2. In nuclear areas close to the nuclear membrane, the highest concentration of both G4s and ICP8 was observed (9.8 and 10.5 dots/mm2, respectively). Other nuclear areas that displayed a lower density of G4 dots (≤5 dots/mm2) were calculated separately. In this case a less marked correlation between G4s and ICP8 was observed (4.5 and 3 dots/mm2, respectively). To note that the lower density areas represented only 14.7% of all tested areas. The mock-infected samples displayed 3.3 G4 dots/mm2 and no ICP8 was detected; infected and mock-infected samples lacking treated with the primary Abs showed very low amount of unspecific labeling (Figure 5D). The number of G4s in capsids (Figure 5B–C’ and Supplementary Figure S9) was low but statistically significant (0.23/capsid, P < 0.01), whereas no specific signal in virions was found in infected cells treated with only the secondary Ab. These data altogether indicate that G4s visualized in infected cells are present in virus-related compartments (RCs and newly formed virions) and thus strongly support the presence of G4s in the viral genome during the HSV-1 infective cycle.
Figure 5.
Immuno-EM of HSV-1 infected cells, fixed at 15 h.p.i. and incubated with anti-G4 Ab (1H6) and 3–83 anti-ICP8 serum. Primary 1H6 and anti-ICP8 Abs were detected with 5- and 10-nm gold particles, respectively. To improve image clarity, gold particles were highlighted with red dots (indicating G4s) and green encircled white dots (indicating ICP8) in A, B, C. The original images are provided in the corresponding panels A', B', C'. (A-A’) G4s and ICP8 are gathered in close proximity to the nuclear membrane (NM) where a nuclear pore complex (NPC) is present. Nuclear egress via NCP is one of the pathways used by HSV-1 capsids to move from the nucleus (n) to the cytoplasm (cyt) (46,47); (B-B’) G4s and ICP8 clustered close to the NM, where many newly formed virions are budding. (C-C’) After nuclear egress, enveloped virions, some of which show the presence of G4s in their core, are transported into RER. ‘n’ indicates the nucleoplasm, cyt the cytoplasm. Bars length is indicated in each panel. Panel (D): statistical analysis of G4 and ICP8 distribution in cells. G4 and ICP8 dots were calculated in 35–45 images per each condition and represented as dot/mm2. Cell areas with ≤5 dots/mm2 were considered low-density (LD) areas and calculated separately with respect to high-density (HD) areas. Cnt are infected cells treated only with secondary Abs. MI are mock-infected cells treated with both the primary and secondary Abs; MI cnt are mock-infected cells treated only with the secondary Abs. In all data sets: mean ± sem, Student's t-test, *P < 0.05, **P < 0.01.
DISCUSSION
We showed here that HSV-1 infected cells were highly enriched in G4s. In particular, the amount of G4s depended on the virus amount (MOI) and on the viral step, being more intense around the time of viral DNA replication. This finding is in line with the reported presence of G4s in the duplex DNA in the eukaryotic genome during events that promote dissociation of the G-rich strand from its complementary strand (49), i.e. during transcription (50), replication (51), DNA repair (52) or under molecular crowded conditions (53).
Besides G4s forming in the viral genome, the enrichment of G4s during HSV-1 infection might also derive from virus-induced manipulation of host cell factors (54). Indeed, a number of DNA viruses, which require cellular DNA replication proteins for viral replication (i.e. adenovirus, simian virus 40 and human papillomavirus), encode functions to promote cellular S phase. However, this is not the case of HSV-1, which encodes a large number of genes associated with its DNA synthesis (55,56); in fact, HSV-1 infection results in the activation of only a limited number of cellular promoters contained in the cellular genome (57) and it is rather linked to inhibition of cellular DNA synthesis associated with cell cycle arrest in G1/S (44). In addition, we have shown that HIV-1 does not modify the number of G4s in the infected cell.
Confocal microscopy colocalization analysis in our study showed an almost complete overlapping between G4s induced during the viral infection and RCs where ICP8, an essential component of the HSV-1 DNA replication machinery implicated in the assembly of viral pre-replication and RCs (41–44), localized. This evidence supports formation of viral G4s during viral replication. ICP8 has been recently shown to co-localize with telomeric foci (58) in the cell nucleus, however, no G4 dependence has been reported: it is thus unlikely that colocalization signals obtained in our experiments derive from telomeric G4s/ICP8 colocalization. Moreover, G4s in infected cells were much more abundant than those in mock-infected cells and their localization within the nucleus at different times p.i. moved in a pattern compatible with the processing of viral DNA during the virus cycle. In particular, G4s were distributed in foci corresponding to RCs all over the nucleus during viral DNA replication (6 h.p.i.) (43,44,59–61), while they clustered at the nuclear membrane at later times (14 h.p.i.), when late maturation of viral RCs (i.e. replication, late-gene transcription and encapsidation) takes place (36,38).
Interestingly, immune-EM analysis allowed to detect G4s in immature virions before and after budding from the nuclear membrane (Figure 5). In the virions the viral genome is linear and double-stranded and no replication events occur. The presence of G4s at this level is compatible with the reported induction of G4s under molecular crowded conditions (53) and with the recently provided evidence that G4s are also present in post-mitotic DNA (21).
Attempts to characterize the DNA enriched for 1H6 binding sites have thus far been unsuccessful (8,21), however, since (i) the HSV-1 genome is extremely rich in G-rich regions that fold in G4 in vitro (17); (ii) a G4 ligand was able to suppress viral DNA replication and infectivity (17); (iii) ACV and PAA, compounds that selectively inhibit the viral DNA polymerase, decrease the amount of G4s in the infected cell (shown here); (iv) G4s in infected cells show striking virus cycle- and virus amount-dependence (shown here); (v) G4s in infected cells associate with viral RCs and are present in immature virions before and after budding at the nuclear membrane (shown here) convincingly indicate that the observed G4s are viral structures.
In latently infected neurons in vivo, an average of 20–50 genome copies, with peaks up to 900 genome copies per cell, have been measured (62). During the in vivo outburst of the lytic infection, almost 10,000 HSV genome copies per cell have been estimated to be produced before cells were eliminated by CD8+ T cell-mediated response (63). In vitro, the number of new genome copies depends on the initial amount of the infecting virus (MOI), as shown in Figure 2B, C and Supplementary Figure S2b; given that no limitation from the immune system of the host is present in our cell system in vitro, an even higher amount of HSV-1 genomes per cell may be expected. In each HSV-1 genome we have confirmed 67–70 G4 repeats, however, other isolated G4s are also present (17). With these figures, the number of HSV G4s that form in a limited range of time in vitro may reach very high levels per cell, numbers that justify the extensive G4 signal that we observed.
The viral G4 massive presence during the viral cycle suggests a key epigenetic role played by these structures during the viral replication cycle and points to possible new actors in the viral biology. For instance, one G4 repeat is embedded in the promoter of one key protein responsible for in vivo HSV-1 neurovirulence and induction of autophagy (17,64); almost all G4 repeats are in the terminal repeats of the HSV-1 genome, the functions of which are not yet fully understood. It is possible that the interaction of G4s with key factors in the initial steps of virus replication regulates the virus cycle itself. These new mechanistic insights may support the design of innovative anti-HSV-1 drugs. While ACV has been the antiviral drug of choice for the treatment of HSV-1 infections (65) since its first introduction in the 1980s, the emergence of resistance to ACV has created an obstacle for the treatment of HSV-1 (66). Because of the inherent different mechanism of action, G4 ligands could be envisaged as therapeutic options against HSV-1 strains resistant to current anti-herpetic drugs. In this direction, we have recently shown the availability of small molecules that target the HIV-1 G4s with a degree of selectivity vs the cellular telomeric G4s, resulting in very good antiviral properties (67). These data indicate the possibility to identify ligands selective for HSV-1 G4s. In addition, the overwhelming presence of viral G4s during replication, as indicated in this work, could also be exploited to develop antiviral compounds even when they do not display outstanding selective recognition properties against HSV-1 G4s. Indeed, we have already reported that BRACO-19, a G4 ligand lacking selectivity against viral G4s, displays good anti-HSV-1 activity (17). Moreover, the HSV-1 genome is responsible for both the lytic and latent infection, the latter lasting for the entire life of the human host: the availability of targets in the viral genome may also pave the way for the development of drugs able to eradicate the virus.
Supplementary Material
Acknowledgments
We thank Prof. P. M. Lansdorp for kindly providing the 1H6 Antibody and for helpful discussion; Prof. D. M. Knipe for providing 3–83 anti-ICP8 serum and Prof. A. Calistri for HSV-1 v41. We thank Prof. M. Palumbo for helpful discussion and Dr M. Rossetto for advise in the SPR analysis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Bill and Melinda Gates Foundation [GCE grant numbers OPP1035881, OPP1097238]; European Research Council [ERC Consolidator grant 615879]; Italian Ministry of University and Research [FIRB-Ideas RBID082ATK to S.N.R.]. Funding for open access charge: Bill and Melinda Gates Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Huppert J.L. Four-stranded nucleic acids: structure, function and targeting of G-quadruplexes. Chem. Soc. Rev. 2008;37:1375–1384. doi: 10.1039/b702491f. [DOI] [PubMed] [Google Scholar]
- 2.Eddy J., Maizels N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006;34:3887–3896. doi: 10.1093/nar/gkl529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huppert J.L., Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakken S., Rognes T., Hovig E. The disruptive positions in human G-quadruplex motifs are less polymorphic and more conserved than their neutral counterparts. Nucleic Acids Res. 2009;37:5749–5756. doi: 10.1093/nar/gkp590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mani P., Yadav V.K., Das S.K., Chowdhury S. Genome-wide analyses of recombination prone regions predict role of DNA structural motif in recombination. PLoS One. 2009;4:e4399. doi: 10.1371/journal.pone.0004399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rhodes D., Lipps H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015;43:8627–8637. doi: 10.1093/nar/gkv862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biffi G., Tannahill D., McCafferty J., Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henderson A., Wu Y., Huang Y.C., Chavez E.A., Platt J., Johnson F.B., Brosh R.M., Jr, Sen D., Lansdorp P.M. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 2014;42:860–869. doi: 10.1093/nar/gkt957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beaume N., Pathak R., Yadav V.K., Kota S., Misra H.S., Gautam H.K., Chowdhury S. Genome-wide study predicts promoter-G4 DNA motifs regulate selective functions in bacteria: radioresistance of D. radiodurans involves G4 DNA-mediated regulation. Nucleic Acids Res. 2013;41:76–89. doi: 10.1093/nar/gks1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du X., Wojtowicz D., Bowers A.A., Levens D., Benham C.J., Przytycka T.M. The genome-wide distribution of non-B DNA motifs is shaped by operon structure and suggests the transcriptional importance of non-B DNA structures in Escherichia coli. Nucleic Acids Res. 2013;41:5965–5977. doi: 10.1093/nar/gkt308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Metifiot M., Amrane S., Litvak S., Andreola M.L. G-quadruplexes in viruses: function and potential therapeutic applications. Nucleic Acids Res. 2014;42:12352–12366. doi: 10.1093/nar/gku999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amrane S., Kerkour A., Bedrat A., Vialet B., Andreola M.L., Mergny J.L. Topology of a DNA G-quadruplex structure formed in the HIV-1 promoter: a potential target for anti-HIV drug development. J. Am. Chem. Soc. 2014;136:5249–5252. doi: 10.1021/ja501500c. [DOI] [PubMed] [Google Scholar]
- 13.Perrone R., Nadai M., Frasson I., Poe J.A., Butovskaya E., Smithgall T.E., Palumbo M., Palu G., Richter S.N. A dynamic G-quadruplex region regulates the HIV-1 long terminal repeat promoter. J. Med. Chem. 2013;56:6521–6530. doi: 10.1021/jm400914r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piekna-Przybylska D., Sullivan M.A., Sharma G., Bambara R.A. U3 region in the HIV-1 genome adopts a G-quadruplex structure in its RNA and DNA sequence. Biochemistry. 2014;53:2581–2593. doi: 10.1021/bi4016692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perrone R., Nadai M., Poe J.A., Frasson I., Palumbo M., Palu G., Smithgall T.E., Richter S.N. Formation of a unique cluster of G-quadruplex structures in the HIV-1 Nef coding region: implications for antiviral activity. PLoS One. 2013;8:e73121. doi: 10.1371/journal.pone.0073121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ouyang Q., Zhao X., Feng H., Tian Y., Li D., Li M., Tan Z. High GC content of simple sequence repeats in Herpes simplex virus type 1 genome. Gene. 2012;499:37–40. doi: 10.1016/j.gene.2012.02.049. [DOI] [PubMed] [Google Scholar]
- 17.Artusi S., Nadai M., Perrone R., Biasolo M.A., Palu G., Flamand L., Calistri A., Richter S.N. The Herpes Simplex Virus-1 genome contains multiple clusters of repeated G-quadruplex: implications for the antiviral activity of a G-quadruplex ligand. Antiviral Res. 2015;118:123–131. doi: 10.1016/j.antiviral.2015.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Honess R.W., Roizman B. Regulation of herpesvirus macromolecular synthesis: sequential transition of polypeptide synthesis requires functional viral polypeptides. Proc. Natl. Acad. Sci. U.S.A. 1975;72:1276–1280. doi: 10.1073/pnas.72.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roizman B., Knipe D.M., Whitley R.J. Herpes Simplex Viruses. 6th edn. Philadelphia, PA: Lippincott Williams & Wilkins; 2013. [Google Scholar]
- 20.La Boissiere S., Izeta A., Malcomber S., O'Hare P. Compartmentalization of VP16 in cells infected with recombinant herpes simplex virus expressing VP16-green fluorescent protein fusion proteins. J. Virol. 2004;78:8002–8014. doi: 10.1128/JVI.78.15.8002-8014.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffmann R.F., Moshkin Y.M., Mouton S., Grzeschik N.A., Kalicharan R.D., Kuipers J., Wolters A.H., Nishida K., Romashchenko A.V., Postberg J., et al. Guanine quadruplex structures localize to heterochromatin. Nucleic Acids Res. 2016;44:152–163. doi: 10.1093/nar/gkv900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knipe D.M., Senechek D., Rice S.A., Smith J.L. Stages in the nuclear association of the herpes simplex virus transcriptional activator protein ICP4. J. Virol. 1987;61:276–284. doi: 10.1128/jvi.61.2.276-284.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tosoni E., Frasson I., Scalabrin M., Perrone R., Butovskaya E., Nadai M., Palu G., Fabris D., Richter S.N. Nucleolin stabilizes G-quadruplex structures folded by the LTR promoter and silences HIV-1 viral transcription. Nucleic Acids Res. 2015;43:8884–8897. doi: 10.1093/nar/gkv897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dai J., Chen D., Jones R.A., Hurley L.H., Yang D. NMR solution structure of the major G-quadruplex structure formed in the human BCL2 promoter region. Nucleic Acids Res. 2006;34:5133–5144. doi: 10.1093/nar/gkl610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuryavyi V., Phan A.T., Patel D.J. Solution structures of all parallel-stranded monomeric and dimeric G-quadruplex scaffolds of the human c-kit2 promoter. Nucleic Acids Res. 2010;38:6757–6773. doi: 10.1093/nar/gkq558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mathad R.I., Hatzakis E., Dai J., Yang D. c-MYC promoter G-quadruplex formed at the 5′-end of NHE III1 element: insights into biological relevance and parallel-stranded G-quadruplex stability. Nucleic Acids Res. 2011;39:9023–9033. doi: 10.1093/nar/gkr612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rankin S., Reszka A.P., Huppert J., Zloh M., Parkinson G.N., Todd A.K., Ladame S., Balasubramanian S., Neidle S. Putative DNA quadruplex formation within the human c-kit oncogene. J. Am. Chem. Soc. 2005;127:10584–10589. doi: 10.1021/ja050823u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumari S., Bugaut A., Huppert J.L., Balasubramanian S. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martadinata H., Phan A.T. Structure of propeller-type parallel-stranded RNA G-quadruplexes, formed by human telomeric RNA sequences in K+ solution. J. Am. Chem. Soc. 2009;131:2570–2578. doi: 10.1021/ja806592z. [DOI] [PubMed] [Google Scholar]
- 30.Perrone R., Butovskaya E., Daelemans D., Palu G., Pannecouque C., Richter S.N. Anti-HIV-1 activity of the G-quadruplex ligand BRACO-19. J. Antimicrob. Chemother. 2014;69:3248–3258. doi: 10.1093/jac/dku280. [DOI] [PubMed] [Google Scholar]
- 31.Richter S., Ping Y.H., Rana T.M. TAR RNA loop: a scaffold for the assembly of a regulatory switch in HIV replication. Proc. Natl. Acad. Sci. U.S.A. 2002;99:7928–7933. doi: 10.1073/pnas.122119999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lynch H.E., Stewart S.M., Kepler T.B., Sempowski G.D., Alam S.M. Surface plasmon resonance measurements of plasma antibody avidity during primary and secondary responses to anthrax protective antigen. J. Immunol. Methods. 2014;404:1–12. doi: 10.1016/j.jim.2013.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knipe D.M., Cliffe A. Chromatin control of herpes simplex virus lytic and latent infection. Nat. Rev. Microbiol. 2008;6:211–221. doi: 10.1038/nrmicro1794. [DOI] [PubMed] [Google Scholar]
- 34.Lehman I.R., Boehmer P.E. Replication of herpes simplex virus DNA. J. Biol. Chem. 1999;274:28059–28062. doi: 10.1074/jbc.274.40.28059. [DOI] [PubMed] [Google Scholar]
- 35.Scott E.S., O'Hare P. Fate of the inner nuclear membrane protein lamin B receptor and nuclear lamins in herpes simplex virus type 1 infection. J. Virol. 2001;75:8818–8830. doi: 10.1128/JVI.75.18.8818-8830.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simpson-Holley M., Baines J., Roller R., Knipe D.M. Herpes simplex virus 1 U(L)31 and U(L)34 gene products promote the late maturation of viral replication compartments to the nuclear periphery. J. Virol. 2004;78:5591–5600. doi: 10.1128/JVI.78.11.5591-5600.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.James S.H., Prichard M.N. Current and future therapies for herpes simplex virus infections: mechanism of action and drug resistance. Curr. Opin. Virol. 2014;8C:54–61. doi: 10.1016/j.coviro.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 38.Weller S.K., Coen D.M. Herpes simplex viruses: mechanisms of DNA replication. Cold Spring Harb. Perspect. Biol. 2012;4:a013011. doi: 10.1101/cshperspect.a013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Honess R.W., Watson D.H. Herpes simplex virus resistance and sensitivity to phosphonoacetic acid. J. Virol. 1977;21:584–600. doi: 10.1128/jvi.21.2.584-600.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brady T., Agosto L.M., Malani N., Berry C.C., O'Doherty U., Bushman F. HIV integration site distributions in resting and activated CD4+ T cells infected in culture. Aids. 2009;23:1461–1471. doi: 10.1097/QAD.0b013e32832caf28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liptak L.M., Uprichard S.L., Knipe D.M. Functional order of assembly of herpes simplex virus DNA replication proteins into prereplicative site structures. J. Virol. 1996;70:1759–1767. doi: 10.1128/jvi.70.3.1759-1767.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quinlan M.P., Chen L.B., Knipe D.M. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell. 1984;36:857–868. doi: 10.1016/0092-8674(84)90035-7. [DOI] [PubMed] [Google Scholar]
- 43.Taylor T.J., McNamee E.E., Day C., Knipe D.M. Herpes simplex virus replication compartments can form by coalescence of smaller compartments. Virology. 2003;309:232–247. doi: 10.1016/s0042-6822(03)00107-7. [DOI] [PubMed] [Google Scholar]
- 44.de Bruyn Kops A, Knipe D.M. Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA binding protein. Cell. 1988;55:857–868. doi: 10.1016/0092-8674(88)90141-9. [DOI] [PubMed] [Google Scholar]
- 45.Adler J., Parmryd I. Quantifying colocalization by correlation: the Pearson correlation coefficient is superior to the Mander's overlap coefficient. Cytometry A. 2010;77:733–742. doi: 10.1002/cyto.a.20896. [DOI] [PubMed] [Google Scholar]
- 46.Leuzinger H., Ziegler U., Schraner E.M., Fraefel C., Glauser D.L., Heid I., Ackermann M., Mueller M., Wild P. Herpes simplex virus 1 envelopment follows two diverse pathways. J. Virol. 2005;79:13047–13059. doi: 10.1128/JVI.79.20.13047-13059.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wild P., Leisinger S., de Oliveira A.P., Schraner E.M., Kaech A., Ackermann M., Tobler K. Herpes simplex virus 1 Us3 deletion mutant is infective despite impaired capsid translocation to the cytoplasm. Viruses. 2015;7:52–71. doi: 10.3390/v7010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Myllys M., Ruokolainen V., Aho V., Smith E.A., Hakanen S., Peri P., Salvetti A., Timonen J., Hukkanen V., Larabell C.A., et al. Herpes simplex virus 1 induces egress channels through marginalized host chromatin. Sci. Rep. 2016;6:28844. doi: 10.1038/srep28844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat. Struct. Mol. Biol. 2006;13:1055–1059. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- 50.Duquette M.L., Handa P., Vincent J.A., Taylor A.F., Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheung I., Schertzer M., Rose A., Lansdorp P.M. Disruption of dog-1 in Caenorhabditis elegans triggers deletions upstream of guanine-rich DNA. Nat. Genet. 2002;31:405–409. doi: 10.1038/ng928. [DOI] [PubMed] [Google Scholar]
- 52.van Kregten M., Tijsterman M. The repair of G-quadruplex-induced DNA damage. Exp. Cell Res. 2014;329:178–183. doi: 10.1016/j.yexcr.2014.08.038. [DOI] [PubMed] [Google Scholar]
- 53.Miyoshi D., Matsumura S., Nakano S., Sugimoto N. Duplex dissociation of telomere DNAs induced by molecular crowding. J. Am. Chem. Soc. 2004;126:165–169. doi: 10.1021/ja036721q. [DOI] [PubMed] [Google Scholar]
- 54.Hobbs W.E., 2nd, DeLuca N.A. Perturbation of cell cycle progression and cellular gene expression as a function of herpes simplex virus ICP0. J. Virol. 1999;73:8245–8255. doi: 10.1128/jvi.73.10.8245-8255.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ebert S.N., Subramanian D., Shtrom S.S., Chung I.K., Parris D.S., Muller M.T. Association between the p170 form of human topoisomerase II and progeny viral DNA in cells infected with herpes simplex virus type 1. J. Virol. 1994;68:1010–1020. doi: 10.1128/jvi.68.2.1010-1020.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hammarsten O., Yao X., Elias P. Inhibition of topoisomerase II by ICRF-193 prevents efficient replication of herpes simplex virus type 1. J. Virol. 1996;70:4523–4529. doi: 10.1128/jvi.70.7.4523-4529.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Everett R.D. Activation of cellular promoters during herpes virus infection of biochemically transformed cells. EMBO J. 1985;4:1973–1980. doi: 10.1002/j.1460-2075.1985.tb03880.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deng Z., Kim E.T., Vladimirova O., Dheekollu J., Wang Z., Newhart A., Liu D., Myers J.L., Hensley S.E., Moffat J., et al. HSV-1 remodels host telomeres to facilitate viral replication. Cell Rep. 2014;9:2263–2278. doi: 10.1016/j.celrep.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bush M., Yager D.R., Gao M., Weisshart K., Marcy A.I., Coen D.M., Knipe D.M. Correct intranuclear localization of herpes simplex virus DNA polymerase requires the viral ICP8 DNA-binding protein. J. Virol. 1991;65:1082–1089. doi: 10.1128/jvi.65.3.1082-1089.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Bruyn Kops A, Knipe D.M. Preexisting nuclear architecture defines the intranuclear location of herpesvirus DNA replication structures. J. Virol. 1994;68:3512–3526. doi: 10.1128/jvi.68.6.3512-3526.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.de Bruyn Kops A, Uprichard S.L., Chen M., Knipe D.M. Comparison of the intranuclear distributions of herpes simplex virus proteins involved in various viral functions. Virology. 1998;252:162–178. doi: 10.1006/viro.1998.9450. [DOI] [PubMed] [Google Scholar]
- 62.Sawtell N.M., Poon D.K., Tansky C.S., Thompson R.L. The latent herpes simplex virus type 1 genome copy number in individual neurons is virus strain specific and correlates with reactivation. J. Virol. 1998;72:5343–5350. doi: 10.1128/jvi.72.7.5343-5350.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schiffer J.T., Abu-Raddad L., Mark K.E., Zhu J., Selke S., Koelle D.M., Wald A., Corey L. Mucosal host immune response predicts the severity and duration of herpes simplex virus-2 genital tract shedding episodes. Proc. Natl. Acad. Sci. U.S.A. 2010;107:18973–18978. doi: 10.1073/pnas.1006614107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Orvedahl A., Alexander D., Talloczy Z., Sun Q., Wei Y., Zhang W., Burns D., Leib D.A., Levine B. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 65.Vere Hodge R.A., Field H.J. Antiviral agents for herpes simplex virus. Adv. Pharmacol. 2013;67:1–38. doi: 10.1016/B978-0-12-405880-4.00001-9. [DOI] [PubMed] [Google Scholar]
- 66.Bacon T.H., Levin M.J., Leary J.J., Sarisky R.T., Sutton D. Herpes simplex virus resistance to acyclovir and penciclovir after two decades of antiviral therapy. Clin. Microbiol. Rev. 2003;16:114–128. doi: 10.1128/CMR.16.1.114-128.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Perrone R., Doria F., Butovskaya E., Frasson I., Botti S., Scalabrin M., Lago S., Grande V., Nadai M., Freccero M., et al. Synthesis, binding and antiviral properties of potent core-extended naphthalene diimides targeting the HIV-1 long terminal repeat promoter G-quadruplexes. J. Med. Chem. 2015;58:9639–9652. doi: 10.1021/acs.jmedchem.5b01283. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.