The structure of the A. gambiae serine proteinase inhibitor (serpin) SRPN18 was determined, revealing an unusually short and constricted reactive-center loop.
Keywords: serpins, serine proteases, insect immunity, enzyme inhibitors, mosquito, Anopheles gambiae, malaria vector
Abstract
Serine protease inhibitors (serpins) in insects function within development, wound healing and immunity. The genome of the African malaria vector, Anopheles gambiae, encodes 23 distinct serpin proteins, several of which are implicated in disease-relevant physiological responses. A. gambiae serpin 18 (SRPN18) was previously categorized as non-inhibitory based on the sequence of its reactive-center loop (RCL), a region responsible for targeting and initiating protease inhibition. The crystal structure of A. gambiae SRPN18 was determined to a resolution of 1.45 Å, including nearly the entire RCL in one of the two molecules in the asymmetric unit. The structure reveals that the SRPN18 RCL is extremely short and constricted, a feature associated with noncanonical inhibitors or non-inhibitory serpin superfamily members. Furthermore, the SRPN18 RCL does not contain a suitable protease target site and contains a large number of prolines. The SRPN18 structure therefore reveals a unique RCL architecture among the highly conserved serpin fold.
1. Introduction
Serpins (serine protease inhibitors) represent the largest family of protease inhibitors and are found in all higher eukaryotes and some bacteria, archaea and viruses (Olson & Gettins, 2011 ▸; Silverman et al., 2010 ▸). Proteolytic events are integral to a wide variety of signaling pathways and govern diverse physiological functions, such as development (Hashimoto et al., 2003 ▸; Ligoxygakis et al., 2003 ▸; Francis et al., 2012 ▸), coagulation (Huntington, 2013 ▸), cell migration (Ravenhill et al., 2010 ▸; Declerck & Gils, 2013 ▸; Yamamoto et al., 2013 ▸; Huasong et al., 2015 ▸), tumor suppression (Mahajan et al., 2013 ▸), fibrinolysis (Rau et al., 2007 ▸; Al-Horani, 2014 ▸) and immunity (Ashton-Rickardt, 2013 ▸; Silverman et al., 2010 ▸; Gatto et al., 2013 ▸). In general, the proteases that comprise these signaling pathways are expressed as zymogens, becoming activated upon proteolytic cleavage in order to elicit a rapid, controlled physiological response. Serpins function to silence these responses in order to avoid negative physiological consequences stemming from uncontrolled activation (Olson & Gettins, 2011 ▸). Recent studies in insects have implicated serpins in a variety of essential physiological events, including development, mating, anticoagulation and immunity (Silverman et al., 2010 ▸; Gubb et al., 2010 ▸; Meekins et al., 2016 ▸).
Serpins typically range in size from 350 to 400 residues and contain a highly conserved fold consisting of three β-sheets (A, B and C) surrounded by up to nine α-helices (A–I) (Huntington, 2011 ▸). The central motif governing serpin activity is the reactive-center loop (RCL), a solvent-exposed loop that acts as a bait for the target protease. Despite the conservation of the overall serpin fold, there is a high degree of diversity among the RCLs of different serpins (Irving et al., 2000 ▸). The specificity and regulation of serpin–protease interactions largely stems from the primary sequence and conformation of the RCL (Irving et al., 2000 ▸; Ye et al., 2001 ▸). An essential component of the RCL is the scissile bond, which is defined as the peptide bond between two amino-acid residues denoted P1 and P1′ and cleaved by the target protease, forming an acyl-enzyme intermediate (Loebermann et al., 1984 ▸). After formation of the intermediate complex, the serpin undergoes a dramatic conformational transformation whereby the RCL inserts into β-sheet A, forming an additional β-strand, and the protease is translocated to the opposing end of the serpin (Stratikos & Gettins, 1999 ▸; Huntington, McCoy et al., 2000 ▸; Dunstone & Whisstock, 2011 ▸). The biophysical basis of this inhibitory transformation stems from the higher stability of the resultant complex compared with the native, metastable structure of the serpin (Whisstock & Bottomley, 2006 ▸; Huntington, 2006 ▸). As a result of the conformational change, both the serpin and protease are unable to perform further reactions and remain in an SDS-stable complex until degraded by cellular processes (Huntington, Read et al., 2000 ▸). Thus, the canonical serpin mechanism for inactivating proteases is designated suicide inhibition.
Interestingly, many serpins do not function as protease inhibitors via the canonical inhibitory mechanism despite containing the conserved serpin fold (Stein et al., 1989 ▸). Although many of the non-inhibitory serpins are susceptible to proteolytic cleavage, they invariably contain structural alterations, mostly within the RCL, that are unfavorable for rapid formation of the inhibitory complex. These features include α-helical secondary structure in the RCL, bulky residues in the RCL hinge region, a highly constrained RCL, complete hydrogen bonding in the breach region of β-sheet A, proline residues preceding the scissile bond or the presence of a glycosylation site within the RCL (Hopkins et al., 1993 ▸; Huntington et al., 1997 ▸; Simonovic et al., 2001 ▸; Al-Ayyoubi et al., 2004 ▸; Chaillan-Huntington et al., 1997 ▸; Widmer et al., 2012 ▸; Stein et al., 1991 ▸; Hood et al., 1994 ▸; McCarthy & Worrall, 1997 ▸). These non-inhibitory serpins invariably contain distinct structural features associated with protein or ligand binding that form the basis of their physiological function (Simonovic et al., 2001 ▸; Al-Ayyoubi et al., 2004 ▸; Widmer et al., 2012 ▸; Zhou et al., 2006 ▸; McGowan et al., 2006 ▸; Klieber et al., 2007 ▸). In addition, several serpins have been discovered that inhibit cysteine proteases via a noncanonical mechanism whereby the protease becomes trapped in a complex with the RCL until both the protease and the serpin are degraded by proteolytic processes (Guo et al., 2015 ▸; Fish & Bjork, 1979 ▸; Mast et al., 1992 ▸; Zhou et al., 1997 ▸; Annand et al., 1999 ▸; Schick et al., 1998 ▸). Despite their noncanonical properties, non-inhibitory and cysteine protease-inhibiting serpins are critical to a variety of essential biological processes.
Serpins in the African malaria vector mosquito Anopheles gambiae (designated SRPNs) have gained attention as key regulators of immunity and potential targets for vector control (Gulley et al., 2013 ▸; Suwanchaichinda & Kanost, 2009 ▸). 18 SRPN genes encoding 23 proteins have been identified in A. gambiae, but only a limited number have been characterized (Gulley et al., 2013 ▸; Christophides et al., 2002 ▸, 2004 ▸). The only known targets of inhibitory serpins in A. gambiae are clip domain-containing serine proteases (CLIPs; An, Budd et al., 2011 ▸). Catalytically active CLIPs circulate as zymogens, becoming active upon cleavage between the CLIP and serine protease (SP) domains, thus initiating a proteolytic cascade that culminates in a specific physiological response (Barillas-Mury, 2007 ▸). A. gambiae CLIPB9 has been shown to convert pro-phenoloxidase (PPO) to phenoloxidase (PO), which results in a melanization immune response (An, Budd et al., 2011 ▸). SRPN2 inhibits CLIPB9 in A. gambiae, thus regulating melanization and avoiding the negative-fitness consequences of an uncontrolled immune response (An, Budd et al., 2011 ▸; Michel et al., 2005 ▸). Furthermore, SRPN6 and SRPN10 have been shown to be upregulated in response to parasite or bacterial infection, and SRPN6 has been shown to have antiparasitic effects (Danielli et al., 2003 ▸; Abraham et al., 2005 ▸). Despite these insights into the importance of mosquito serpins, our understanding of the protease–serpin networks that regulate mosquito physiology is in its infancy and information regarding the uncharacterized A. gambiae serpins is limited. Insights into these uncharacterized A. gambiae serpins is important to further understand the physiology and vector competence of this medically important mosquito species.
SRPN18 (AGAP007691; XP_003435746) is among the sparsely characterized serpins in A. gambiae. Previous studies indicate that A. gambiae SRPN18 is expressed throughout all life stages in multiple tissues and the hemolymph, and it is predicted to be secreted based on the presence of a signal peptide (Suwanchaichinda & Kanost, 2009 ▸). SRPN18 expression doubles within 3 h of a blood meal and returns to pre-blood-meal levels within 24 h post-blood feeding (Marinotti et al., 2006 ▸). However, its role in A. gambiae physiology is entirely unknown. The SRPN18 gene clusters tightly with SRPN7 and SRPN14 on chromosomal arm 2L, close to the SRPN2 cluster (Suwanchaichinda & Kanost, 2009 ▸). Microarray data indicate that AgSRPN18 is repressed upon infection with Wolbachia bacteria, which have been shown to protect mosquitoes from Plasmodium infection, although the significance of these data is unknown (Kambris et al., 2010 ▸; Hughes et al., 2011 ▸). Previous examination of the primary sequence of the SRPN18 RCL resulted in the prediction that SRPN18 is non-inhibitory (Gulley et al., 2013 ▸; Suwanchaichinda & Kanost, 2009 ▸). This is based on the absence of an RCL hinge region, a conserved span of four residues that provides RCL flexibility and is integral for inhibitory complex formation.
Here, we present the structure of A. gambiae SRPN18 to a resolution of 1.45 Å. The high-resolution crystal structure of SRPN18 was determined, including nearly complete resolution of the RCL. These data provide additional insights into mosquito serpins and may provide a basis for identifying the physiological function of SRPN18 in A. gambiae.
2. Materials and methods
2.1. Macromolecule production
2.1.1. SRPN18 cloning and recombinant SRPN18 protein expression
cDNA fragments encoding full-length mature proteins were amplified using the gene-specific primers SRPN18-1F, 5′-TCATCACGGCGATCCTACGACAG-3′, and SRPN18-1R, 5′-TTGAATTCTCAAAACTGTTCATCGG-3′. The reverse primer contained an EcoRI restriction site (bold). A second round of PCR was performed with the forward primer SRPN18-2F containing codons for a His6 tag (underlined) as well as an NcoI site (bold), 5′-ATCCATGGGCCATCATCATCATCATCATCACGGC-3′, with SRPN18-1R as the reverse primer. The PCR products were digested with NcoI and EcoRI, and then inserted into the pET-28a vector (Novagen) using the same restriction sites. The resulting plasmid (pET28a-SRPN18) was transformed into BL21 (DE3) competent Escherichia coli cells and stored at −80°C (Table 1 ▸).
Table 1. Macromolecule-production information.
| Source organism | A. gambiae |
| DNA source | Adult female A. gambiae G3 strain |
| Forward primer | 5′-TCATCACGGCGATCCTACGACAG-3′ |
| Reverse primer | 5′-TTGAATTCTCAAAACTGTTCATCGG-3′ |
| Cloning vector | Not applicable |
| Expression vector | pET-28a |
| Expression host | E. coli BL21 (DE3) |
| Complete amino-acid sequence of the construct produced | MGHHHHHHGDPTTDDAIVAANNKFTLEYFKACYDEKCNCAVSPYHVRLALSMFYPLAGAAVQEDFQVAFGLPEDVHAAIEQQQRLAQQLHDGQHLKALSFVLVEETLRLDSEFERLFHRTFQTTVEPVDLTDDIPSALAVNSFYQRANTEIEDFIGEGDVFSLPPCHKLMLFSGVSVLTPLAIRFNPADTALELFQFINAPTQRVSTMHTTAFVRRCLHNELRCKVVDMPFDAASGLSMLVLLPYDGTELRQIVNSITPAHLAQIDERLQSCWTDLKLPKFFVREKTDPKQTLGKLGYGGVFEIDDLHVFHDSGRTRLNGFIQHCYLAVSESGSGIPAPPDTPSEFEFHANRPFMFLIRRTMDGNVLQVGNFSKYIDPDEQF |
Recombinant SRPN18 protein was produced using an E. coli expression system (Table 1 ▸). The full coding region, minus the predicted signal peptide, was amplified using gene-specific primers. SRPN18 protein was expressed using BL21 (DE3) competent E. coli cells. Cells containing the plasmid were grown overnight at 37°C from bacterial stocks on LB agar plates containing 50 µg ml−1 kanamycin. A single colony was inoculated into a 250 ml flask containing 50 ml LB with 50 µg ml−1 kanamycin and then shaken overnight at 37°C and 150 rev min−1. 15 ml of the overnight culture was used to inoculate two 2 l flasks of 500 ml LB with 50 µg ml−1 kanamycin. The inoculated culture was incubated at 37°C with shaking at 225 rev min−1 for approximately 2 h to an OD600 of between 0.6 and 0.8. Protein expression was induced using 0.1 mM IPTG with incubation for at least 8 h at 20°C and 150 rev min−1. The culture was centrifuged at 4000 rev min−1 for 20 min and the pellet was stored at −80°C.
2.1.2. Recombinant SRPN18 purification
Recombinant SRPN18 purification was performed as described previously, with the following modifications (Zhang et al., 2015 ▸). Cell pellets from expression were resuspended in 50 ml buffer A (50 mM NaCl, 20 mM Tris–HCl pH 8.0) supplemented with protease-inhibitor cocktail (Roche). Cells were lysed by sonication (Vibra Cell High Intensity Ultrasonic Processor 750 W model), and soluble and insoluble fractions were separated by centrifugation at 10 000g for 30 min at 4°C. Soluble portions were retained and purified by nickel-affinity, ion-exchange and size-exclusion chromatography using an ÄKTAxpress purification system (GE Healthcare) at 4°C. The clarified lysate was loaded onto a 5 ml HisTrap HP column (GE Healthcare) at 1 ml min−1. Bound proteins were washed with 25 ml buffer A. Nonspecifically bound proteins were then eluted using a gradient of buffer B (500 mM imidazole, 50 mM NaCl, 20 mM Tris–HCl pH 8.0). Elution of SRPN18 was carried out with a linear gradient from 10 to 100% buffer B over eight column volumes, and all elution peaks were collected and analyzed by SDS–PAGE. SRPN18-containing fractions were pooled and loaded onto a 5 ml HiTrap Q HP anion-exchange column (GE Healthcare) equilibrated with buffer A. Elution was carried out with a linear gradient from 0 to 100% buffer C (500 mM NaCl, 20 mM Tris pH 8.0) over 20 column volumes, and the purity of SRPN18 was analyzed by SDS–PAGE. SRPN18-containing fractions were pooled again and concentrated to 1.0 ml in a Vivaspin 20 10 kDa molecular-weight cutoff concentrator (GE Healthcare). The concentrated protein was loaded onto a Superdex 75 10 300 GL size-exclusion column (GE Healthcare) and eluted in buffer D (400 mM NaCl, 20 mM Tris pH 8.0) at 0.2 ml min−1. Protein fractions were analyzed by SDS–PAGE and protein concentrations were determined by the Bradford assay using Coomassie Plus Protein Assay Reagent (Pierce) and bovine serum albumin as a standard (Sigma). SRPN18 fractions were pooled and concentrated for crystallization screening to 12.8 mg ml−1 in buffer D via a Vivaspin 20 10 kDa molecular weight cutoff concentrator (GE Healthcare).
2.2. Crystallization
Crystallization screening was conducted in high-throughput Compact 300 (Rigaku Reagents) sitting-drop vapor-diffusion plates at 20°C using 0.5 µl protein solution and 0.5 µl crystallization solution equilibrated against 100 µl of the latter (Table 2 ▸). Prismatic crystals were obtained in 3–4 d from Index HT screen (Hampton Research) condition F11 [25%(w/v) PEG 3350, 100 mM bis-tris pH 6.5, 200 mM NaCl]. Crystals were transferred into a solution consisting of crystallization solution supplemented with 5% PEG 400 and equilibrated for 1 min. The crystals were then transferred into solutions containing increasing concentrations of PEG 400 from 5% to 25%(w/v) PEG 3350, 100 mM bis-tris pH 6.5, 200 mM NaCl, 20% PEG 400 before cooling and storing them in liquid nitrogen.
Table 2. Crystallization.
| Method | Vapor diffusion, sitting drop |
| Plate type | Sitting drop |
| Temperature (K) | 293 |
| Protein concentration | 12.8 |
| Buffer composition of protein solution | 400 mM NaCl, 20 mM Tris pH 8.0 |
| Composition of reservoir solution | 25%(w/v) PEG 3350, 100 mM bis-tris, 200 mM NaCl, 20% PEG 400 |
| Volume and ratio of drop | 1 µl (1:1) |
| Volume of reservoir (µl) | 75 |
2.3. Data collection and processing
Initial X-ray diffraction data were collected at 93 K in the University of Kansas Protein Structure Laboratory using a Rigaku RU-H3R rotating-anode generator (Cu Kα) equipped with Osmic Blue focusing mirrors and a Rigaku R-AXIS IV++ image-plate detector. Higher resolution data were collected at the Advanced Photon Source beamline 17-ID using a Dectris PILATUS 6M pixel-array detector.
2.4. Structure solution and refinement
Intensities were integrated using XDS (Kabsch, 2010a ▸,b ▸) and the Laue class check and data scaling were performed with AIMLESS (Evans, 2011 ▸). The highest probability Laue class was mmm and space group P212121. The Matthews coefficient (V M; Matthews, 1968 ▸) and solvent content were estimated to be V M = 4.0 Å3 Da−1 with 69.2% solvent content and V M = 2.0 Å3 Da−1 with 38.5% solvent content for one and two molecules in the asymmetric unit, respectively. A homology model for molecular replacement was created with CHAINSAW (Stein, 2008 ▸) using the previously determined SRPN2 structure (PDB entry 3pzf), the amino-acid sequence of which is 37.9% similar to that of SRPN18 (An, Lovell et al., 2011 ▸). Molecular-replacement searches for two molecules in the asymmetric unit were conducted using in-house diffraction data with Phaser (McCoy et al., 2007 ▸) via the PHENIX (Adams et al., 2010 ▸) interface in all possible space groups with point symmetry 222. The top solution was found in space group P212121 and consisted of a noncrystallographic dimer related by the spherical polar coordinates ω = 91.057°, φ = −177.973°, χ = 176.861° with the NCS axis is nearly parallel to the crystallographic a axis. Following initial refinement with PHENIX, the R factors converged at R = 40% and R free = 44%. The model was improved using automated model building with ARP/wARP (Langer et al., 2008 ▸) and the final model, refined against the synchrotron diffraction data, was obtained by iterative rounds of refinement and manual model building with PHENIX and Coot (Emsley et al., 2010 ▸), respectively. Structure validation was conducted with MolProbity (Chen et al., 2010 ▸). TLS refinement (Painter & Merritt, 2006 ▸; Winn et al., 2001 ▸) was incorporated into the later stages of refinement to model anisotropic atomic displacement parameters. Residues Ser343–Ser353 of chain A and Ser341–Gly344 of chain B, which are part of the RCL, were disordered and could not be modeled. Disordered side-chain atoms were truncated to the point where electron density could be observed. Figures were prepared using PyMOL (Schrödinger). Solvent-accessible solvent area was determined using AREAIMOL (Lee & Richards, 1971 ▸; Saff & Kuijlaars, 1997 ▸) from the CCP4 program suite (Winn et al., 2011 ▸). Relevant crystallographic data are provided in Table 3 ▸. Coordinates and structure factors were deposited in the Worldwide Protein Data Bank with accession code 5c98.
Table 3. Crystallographic data for SRPN18 refined to 1.45 Å resolution.
Values in parentheses are for the highest resolution shell.
| Data collection | |
| Unit-cell parameters (Å) | a = 40.42, b = 87.49, c = 194.79 |
| Space group | P212121 |
| Resolution (Å) | 48.70–1.45 (1.47–1.45) |
| Wavelength (Å) | 1.0000 |
| Temperature (K) | 100 |
| Observed reflections | 808528 |
| Unique reflections | 122919 |
| 〈I/σ(I)〉 | 20.1 (2.5) |
| Completeness (%) | 99.6 (98.9) |
| Multiplicity | 6.6 (6.7) |
| R merge † (%) | 4.3 (84.1) |
| R meas ‡ (%) | 4.7 (90.3) |
| R p.i.m. ‡ (%) | 1.8 (34.6) |
| CC1/2 § | 0.999 (0.746) |
| Refinement | |
| Resolution range (Å) | 36.69–1.45 |
| Reflections (working/test) | 116670/6160 |
| R/R free ¶ (%) | 16.5/18.5 |
| No. of atoms (protein/water) | 5773/537 |
| Model quality | |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 0.974 |
| Average B factors (Å2) | |
| All atoms | 25.0 |
| Protein | 24.3 |
| Water | 32.0 |
| Coordinate error (maximum likelihood) (Å) | 0.14 |
| Ramachandran plot | |
| Most favored (%) | 98.0 |
| Additionally allowed (%) | 2.0 |
R
merge = 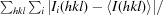
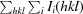 , where Ii(hkl) is the intensity measured for the ith reflection and 〈I(hkl)〉 is the average intensity of all reflections with indices hkl.
, where Ii(hkl) is the intensity measured for the ith reflection and 〈I(hkl)〉 is the average intensity of all reflections with indices hkl.
R meas is the redundancy-independent (multiplicity-weighted) R merge (Evans, 2006 ▸, 2011 ▸). R p.i.m. is the precision-indicating (multiplicity-weighted) R merge (Diederichs & Karplus, 1997 ▸; Weiss, 2001 ▸).
CC1/2 is the correlation coefficient of the mean intensities between two random half-sets of data (Karplus & Diederichs, 2012 ▸; Evans, 2012 ▸).
R = 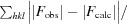
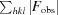 ; R
free is calculated in an identical manner using a randomly selected 5% of reflections that were not included in the refinement.
; R
free is calculated in an identical manner using a randomly selected 5% of reflections that were not included in the refinement.
3. Results and discussion
3.1. Overall SRPN18 structure
We determined the crystal structure of A. gambiae SRPN18 (residues 23–391) to a resolution of 1.45 Å (Table 3 ▸). SRPN18 contains a conserved serpin fold with three β-sheets (A, B and C) surrounded by 13 α-helices (Fig. 1 ▸ a). An analysis of the SRPN18 structure against the DALI database revealed a high degree of homology to a wide range of serpins owing to the conservation of the serpin fold, with a root-mean-square deviation (r.m.s.d.) between Cα atoms of SRPN18 and its closest structural homologs of approximately 2.3 Å (Supplementary Table S1).
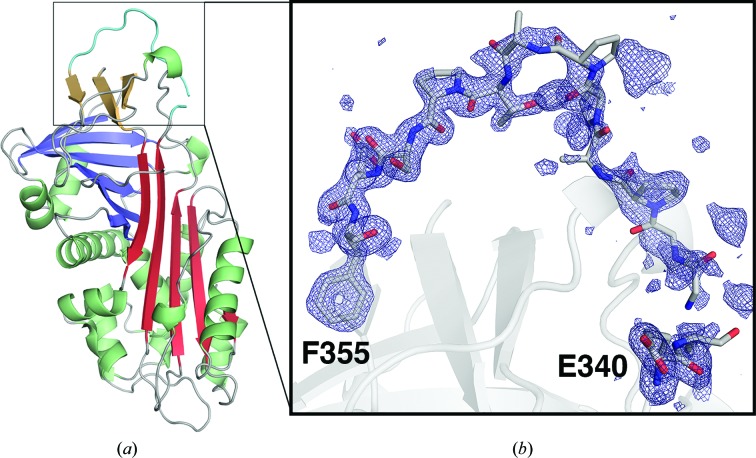
Figure 1.
Crystal structure of SRPN18 including the reactive-center loop (RCL). (a) Ribbon diagram of the SRPN18 crystal structure determined to a resolution of 1.45 Å. β-Sheet A, red; β-sheet B, blue; β-sheet C, yellow; α-helices, green; RCL, cyan. (b) F o − F c OMIT electron-density map (3σ) of the SRPN18 RCL (residues Glu340–Phe355). Map clipping is expanded to 5 Å from the RCL atoms.
SRPN18 crystallized as an NCS dimer with two molecules in the asymmetric unit (Supplementary Fig. S1). The two molecules are identical, with the exception of the observable electron density for the RCL (Glu340–Pro362). In chain A residues Gly344–Pro352 were disordered and could not be modeled, as is commonly found in serpin structures. However, the RCL of SRPN18 chain B was resolved completely, with the exception of two residues, Gly342 and Ser343 (Fig. 1 ▸ b). The SRPN18 RCL in chain B forms a continuous loop and is located directly above β-sheet C. The RCL interacts directly with β-sheet C via multiple hydrogen bonds, which are also present between residues within the RCL itself.
3.2. Comparative structural analysis of the SRPN18 RCL
The RCL is integral to the canonical inhibitory activity of serpins. Therefore, the resolution of the SRPN18 RCL in chain B permitted investigation into the architecture of the RCL in order to gain insight into its potential role in SRPN18 inactivity. We performed a comparative structural analysis of the RCL of chain B in the SRPN18 structure and the RCLs of previously published serpin structures. This comparative analysis was performed with 21 serpin structures, all of which contain an RCL that is completely, or nearly completely, resolved (Table 4 ▸). The structures represent 17 distinct serpins: 13 are inhibitory against serine proteinases [protein Z-dependent protease inhibitory protein (ZPI), heparin cofactor II (HCII), protease nexin-1 (PN1), plasminogen activator inhibitor-1 (PAI-1), 1-antitrypsin (A1AT), antithrombin (AT), AtSerpin1, alaserpin, antichymotrypsin (ACH), tengpin, squamous cell carcinoma antigen 1 (SCCA1), serpin 1K and protein C inhibitor (PCI)], one is inhibitory against cysteine proteinases (Bombyx mori serpin 18) and four are non-inhibitory [ovalbumin, heat-shock protein 47 (Hsp47), maspin and myeloid and erythroid nuclear termination stage-specific antigen (MENT)]. These structures provide a diverse and comprehensive basis to analyze the functional implications of the SRPN18 RCL structure.
Table 4. Serpin structures used for comparative structural analysis of the RCL with accessible surface area (ASA).
| Serpin | Protease complex | Species | PDB code | Function | RCL residues | ASA (Å2) | Reference |
|---|---|---|---|---|---|---|---|
| SRPN18 | Anopheles gambiae | 5c98 | 16 | 1095† | This work | ||
| Serpin 18 | Bombyx mori | 4r9i | Cysteine protease inhibitor, silk production | 17 | 1273 | Guo et al. (2015 ▸) | |
| Ovalbumin | Gallus gallus | 1ova | Non-inhibitory, storage protein | 22 | 1318 | Stein et al. (1991 ▸) | |
| Hsp47 | Canis lupus familiaris | 3zha | Non-inhibitory, collagen chaperone | 19 | 1473 | Widmer et al. (2012 ▸) | |
| Maspin | Homo sapiens | 1xqg | Non-inhibitory, tumor suppression | 20 | 1522 | Al-Ayyoubi et al. (2004 ▸) | |
| HCII | Homo sapiens | 1jmj | Inhibitory, anti-coagulation | 22 | 1524 | Baglin et al. (2002 ▸) | |
| ZPI | Protein Z | Homo sapiens | 3h5c | Inhibitory, anti-coagulation | 22 | 1663 | Huang et al. (2010 ▸) |
| β-AT | Homo sapiens | 1e04 | Inhibitory, anti-coagulation | 25 | 1714 | McCoy et al. (2003 ▸) | |
| Tengpin | Thermoanaerobacter tengcongensis | 2pee | Inhibitory, unknown | 24 | 1732 | Zhang et al. (2007 ▸) | |
| α-AT | Homo sapiens | 1e03 | Inhibitory, anti-coagulation | 24 | 1800 | McCoy et al. (2003 ▸) | |
| PAI-1 | Homo sapiens | 1b3k | Inhibitory, anti-coagulation | 21 | 1870 | Sharp et al. (1999 ▸) | |
| PN1 | Thrombin | Homo sapiens | 4dy7 | Inhibitory, anti-coagulation | 22 | 1883 | Li & Huntington (2012 ▸) |
| SCCA1 | Homo sapiens | 2zv6 | Inhibitory, anti-apoptotic | 24 | 1983 | Zheng et al. (2009 ▸) | |
| PCI | Homo sapiens | 2ol2 | Inhibitory, anti-coagulation | 24 | 2024 | Li et al. (2007 ▸) | |
| Alaserpin | Trypsin | Manduca sexta | 1k9o | Inhibitory, development | 24 | 2026 | Ye et al. (2001 ▸) |
| MENT | Gallus gallus | 2dut | Non-inhibitory, chromatin condensation | 25 | 2039† | McGowan et al. (2006 ▸) | |
| ACH | Mus musculus | 1yxa | Inhibitory, anti-inflammation | 25 | 2040 | Horvath et al. (2005 ▸) | |
| A1AT | Homo sapiens | 1hp7 | Inhibitory, anti-inflammation | 22 | 2113 | Kim et al. (2001 ▸) | |
| PAI-1 | Danio rerio | 4dte | Inhibitory, anti-coagulation | 22 | 2124 | Bager et al. (2013 ▸) | |
| Serpin 1K | Manduca sexta | 1sek | Inhibitory, immunity | 23 | 2281 | Li et al. (1999 ▸) | |
| α-AT | Factor IXa | Homo sapiens | 3kcg | Inhibitory, anti-coagulation | 24 | 2337 | Johnson et al. (2010 ▸) |
| At-Serpin1 | Arabidopsis thaliana | 3le2 | Inhibitory, anti-apoptotic | 25 | 2357 | Lampl et al. (2010 ▸) |
To determine the accessible surface area of the entire RCL in SRPN18 and MENT, unresolved residues (SRPN18, Gly342–Ser343; MENT, Ile375–Asn376) were modeled into the structure before calculation using Coot (Emsley et al., 2010 ▸).
The most conspicuous feature of the SRPN18 RCL is its short length compared with those of other serpins, spanning a total of 16 residues (Table 4 ▸). RCL length is a crucial factor for canonical serpin inhibition, as a sufficient RCL length is required for full insertion into β-sheet A (Huntington, 2011 ▸). The next shortest RCLs in our analysis belong to the cysteine proteinase inhibitor B. mori serpin 18 (which uses a distinct inhibitory mechanism) and the non-inhibitory Hsp47, which contain 17 and 19 residues, respectively. The shortest RCL of an inhibitory serpin against serine proteinases is that of PAI-1, containing 21 residues. Among the inhibitory serpins in our analysis, the RCL length averages 23 residues and ranges from 21 to 25. The structures of AT, ZPI, PCI and PAI-1 have been determined in their cleaved state, with the RCL inserted into β-sheet A, and reveal a necessity for 16 RCL residues prior to the P1–P1′ scissile bond for full insertion (Schreuder et al., 1994 ▸; Huang et al., 2010 ▸; Li & Huntington, 2008 ▸; Jensen & Gettins, 2008 ▸). Thus, cleavage at even the most C-terminal residue in the SRPN18 RCL would result in an RCL segment that is too short for complete insertion, providing credence to previous assertions that it is non-inhibitory.
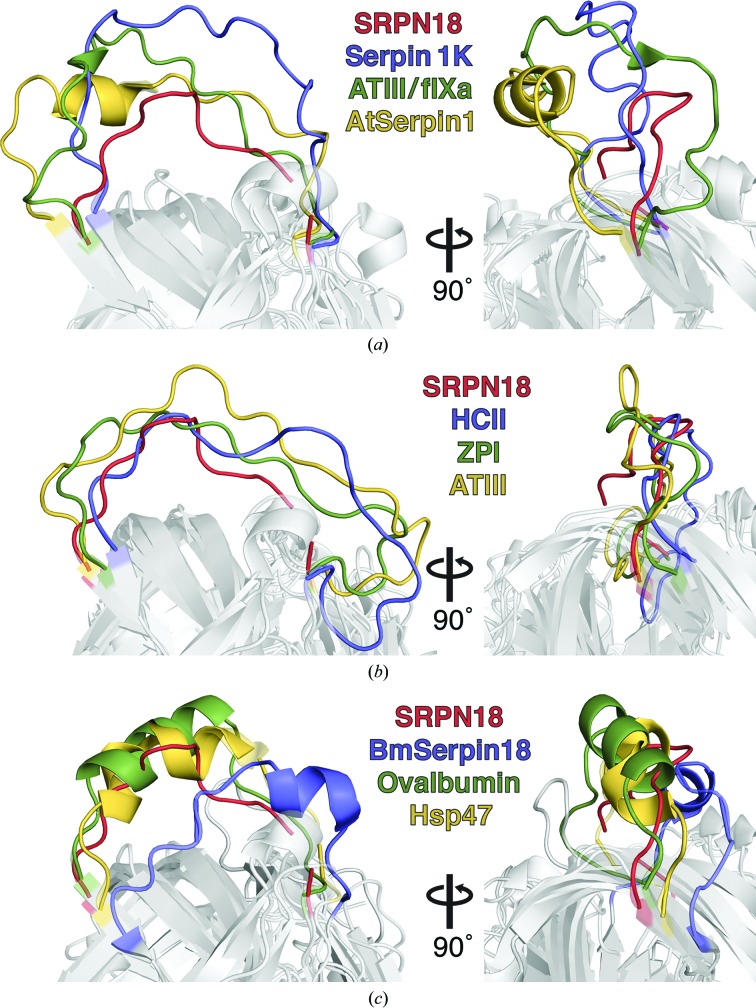
The length of the RCL also has implications for the accessibility of the RCL to the target protease (Johnson et al., 2010 ▸; Jin et al., 1997 ▸; Baglin et al., 2002 ▸). In general, increased accessibility correlates with increased inhibition to the extent that RCL flexibility may be controlled allosterically to regulate the level of serpin activity. To determine the level of constriction of the SRPN18 RCL, accessible surface area (ASA) was quantified in all of the serpins included in our comparative analyses (Table 4 ▸). Indeed, the RCL of SRPN18 is extremely constricted in comparison to other serpins, with an ASA of 1095 Å2, which was the lowest of any of the serpin structures in our analysis. The next lowest ASA was found for the cysteine proteinase inhibitor B. mori serpin 18 (1273 Å2). Structural alignment of SRPN18 and inhibitory serpins with high ASAs [serpin 1K, 2281 Å2; AT (in complex with factor IXa), 2337 Å2; AtSerpin1, 2357 Å2] highlights the level of constriction and the overall lack of flexibility inherent to the SRPN18 RCL (Fig. 2 ▸ a). Inhibitory serpins that contain lower ASAs, closer to that of SRPN18 (HCII, 1524 Å2; ZPI, 1663 Å2; AT, 1714 Å2), are found in structures containing partial RCL hinge-loop insertions, which are expelled to increase the accessibility of the RCL as a mechanism of allosteric regulation (Fig. 2 ▸ b). The RCLs that most closely resemble SRPN18 are either cysteine proteinase inhibitors or non-inhibitory serpins (B. mori serpin 18; ovalbumin, 1318 Å2; Hsp47, 1473 Å2; Fig. 2 ▸ c). It is interesting to note that all three of these RCLs contain α-helices and therefore do not directly resemble SRPN18. Together, these data suggest that the SRPN18 RCL is unprecedented with respect to its constriction and is likely to represent a minimum length that can be accommodated within the serpin fold.
Figure 2.
Structural alignment of the SRPN18 RCL with those of other serpins. The panel on the right is rotated 90° around the y axis. (a) Structural alignment of the SRPN18 RCL (red) with inhibitory serpins whose RCLs contain high accessible surface areas. Serpin 1K, PDB entry 1sek, blue (Li et al., 1999 ▸); AT (in complex with factor IXa; ATIII/fIXa), PDB entry 3kcg, green (Johnson et al., 2010 ▸); AtSerpin1, PDB entry 3le2, orange (Lampl et al., 2010 ▸). (b) Structural alignment of the SRPN18 RCL (red) with inhibitory serpins whose RCLs contain low accessible surface areas. HCII, PDB entry 1jmj, blue (Baglin et al., 2002 ▸); ZPI, PDB entry 3h5c, green (Huang et al., 2010 ▸); AT, PDB entry 1e04, orange (McCoy et al., 2003 ▸). (c) Structural alignment of the SRPN18 RCL with non-inhibitory or cysteine proteinase inhibitory (B. mori serpin 18; BmSerpin18) serpins with low accessibility scores. B. mori serpin 18, PDB entry 4r9i, blue (Guo et al., 2015 ▸); ovalbumin, PDB entry 1ova, green (Stein et al., 1991 ▸); Hsp47, PDB entry 3zha, orange (Widmer et al., 2012 ▸).
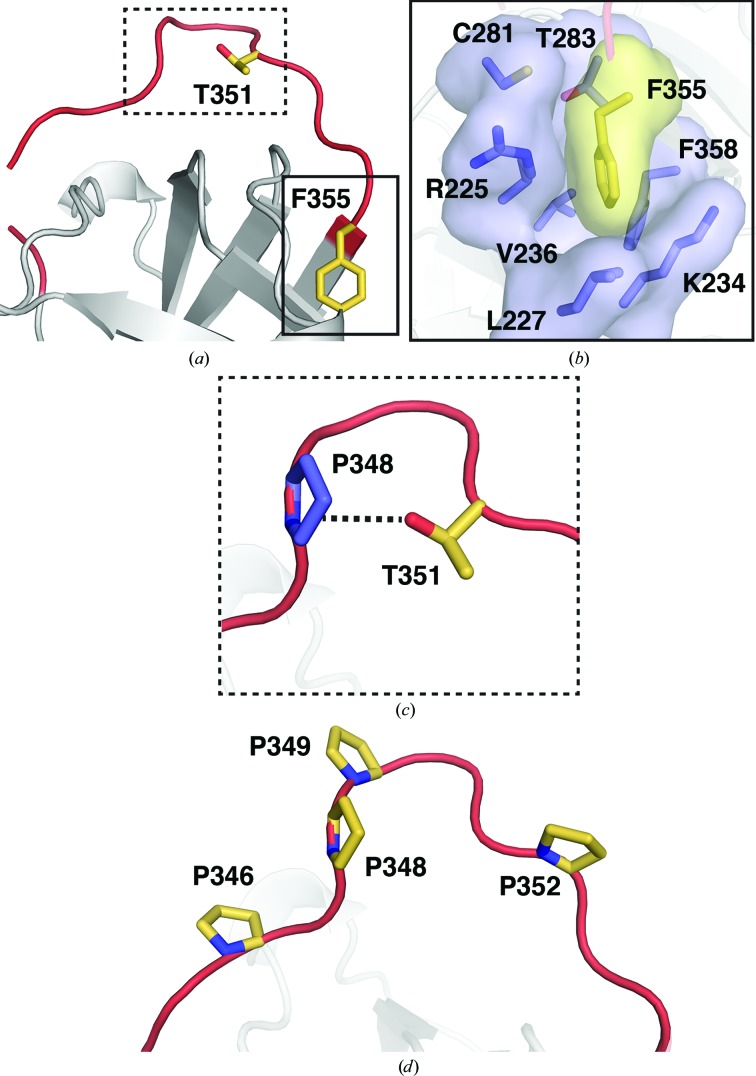
In addition to the constrained overall accessibility of the SRPN18 RCL, the structure reveals the absence of an accessible P1 residue that can function as a bait for proteolytic attack. Previous studies tenuously predicted the most likely P1 residue for SRPN18 to be Phe355 based on sequence alignment (Gulley et al., 2013 ▸; Suwanchaichinda & Kanost, 2009 ▸). However, the SRPN18 structure indicated that Phe355 is located at the C-terminus of the RCL and is buried within a pocket composed of Arg225, Leu227, Lys234, Val236, Phe358, Cys281 and Thr283 (Figs. 3 ▸ a and 3 ▸ b). Thus, Phe355 is not exposed to the solvent and is therefore inaccessible to a target protease. P1 residues in other serpins are invariably located towards the most solvent-exposed apex of the RCL and are commonly situated slightly towards the C-terminus as opposed to the hinge region (Huntington, 2011 ▸). In SRPN18 this region is occupied by Thr351, making it a potential candidate for a P1 residue (Fig. 3 ▸ a). However, the structure revealed that Thr351 is directed away from the surface and that the hydroxyl group forms a hydrogen bond to Pro348 (Fig. 3 ▸ c). As such, it appears to be incapable of acting as an effective P1 residue despite its position within the RCL. Thus, SRPN18 seems to lack a suitable P1 residue, which is critical for initiating the inhibitory mechanism. In addition, the SRPN18 RCL contains four proline residues (Pro346, Pro348, Pro349 and Pro352), three of which are found in the hinge region (Fig. 3 ▸ d). Previous studies have shown that prolines located N-terminal to the P2 residue result in a breakdown of inhibitory function (Hopkins et al., 1993 ▸; Hopkins & Stone, 1995 ▸; Gettins, 2002 ▸). This is because prolines disrupt β-strands, resulting in an inability of the RCL to insert completely into β-sheet A. Not surprisingly, proline residues prior to the P2 position in the RCL are almost exclusively found amongst non-inhibitory serpins, including Hsp47, maspin, thyroxine-binding globulin (TBG), corticosteroid-binding globulin (CBG) and pigment epithelium-derived factor (PEDF). In addition, the four prolines found in the SRPN18 RCL represent the extreme end of the spectrum. Analysis of 347 serpin primary sequences revealed only one instance (Mesocricetus auratus CBG, UniProtKB Q60543.1) of another serpin with four prolines in the RCL prior to the P2 residue, and only three instances (Mus musculus CBG, Q06770.1; M. musculus PEDF, P97298.2; Homo sapiens PEDF, P36955.4) of three prolines in this region of the RCL.
Figure 3.
Features of the SRPN18 RCL that are not conducive towards inhibition. (a) The SRPN18 RCL (red) showing the side chains of Phe355 and Thr351 (yellow). (b) The area highlighted with a solid box in (a) showing Phe355 (yellow) buried within a pocket formed by residues found within β-sheet C (blue). (c) The area highlighted with a dotted box in (a) showing Thr351, which forms interactions with Pro348 (blue). (d) Location of the four proline residues in the SRPN18 RCL.
Overall, the SRPN18 RCL sequence and structure reveal several features associated with serpins that do not function via the canonical serine proteinase inhibitory mechanism: (i) minimal length, (ii) a constricted conformation, (iii) the absence of a suitable P1 residue and (iv) a large number of proline residues. Nevertheless, the expression pattern of SRPN18 and its maintenance through the evolutionary history of A. gambiae suggest that it serves a specific function (Gulley et al., 2013 ▸; Suwanchaichinda & Kanost, 2009 ▸). An ortholog of SRPN18 in Culex quinquefasciatus (Bartholomay et al., 2010 ▸), which contains a short RCL length and RCL prolines, suggests that SRPN18 was present in the common ancestor of both anopheline and culicine mosquitoes and has been maintained by evolution for at least 160 million years. Orthologs of SRPN18 are found in automatically annotated gene sets of the 16 additional recently sequenced anopheline genomes (Neafsey et al., 2015 ▸). Their future refinement will provide additional information of possible conservation of key functional elements. Additional investigations will be necessary to determine the precise role of SRPN18 in A. gambiae and to determine how its unique structure operates to fulfill its role in the organism.
Supplementary Material
PDB reference: SRPN18 from Anopheles gambiae, 5c98
Supporting Information: Supplementary Figures S1 and S2.. DOI: 10.1107/S2053230X16017854/tt5090sup1.pdf
Acknowledgments
We thank Dr Fei Philip Gao for help with the SRPN18 purification. Use of the IMCA-CAT beamline 17-ID at the Advanced Photon Source was supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with Hauptman–Woodward Medical Research Institute. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-06CH11357. The research reported in this publication was supported by the Institute of Allergy and Infectious Diseases of the National Institutes of Health under award No. R01AI095842 (to KM). The use of the KU COBRE Protein Structure Laboratory was supported by NIH Grant No. P30 GM110761 from the National Institute of General Medical Sciences. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the Center of Biomedical Research Excellence in Protein Structure and Function or the National Institutes of Health. This is contribution 17-119-J from the Kansas Agricultural Experiment Station.
References
- Abraham, E. G., Pinto, S. B., Ghosh, A., Vanlandingham, D. L., Budd, A., Higgs, S., Kafatos, F. C., Jacobs-Lorena, M. & Michel, K. (2005). Proc. Natl Acad. Sci. USA, 102, 16327–16332. [DOI] [PMC free article] [PubMed]
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Al-Ayyoubi, M., Gettins, P. G. W. & Volz, K. (2004). J. Biol. Chem. 279, 55540–55544. [DOI] [PubMed]
- Al-Horani, R. A. (2014). Cardiovasc. Hematol. Agents Med. Chem. 12, 91–125. [DOI] [PubMed]
- An, C., Budd, A., Kanost, M. R. & Michel, K. (2011). Cell. Mol. Life Sci. 68, 1929–1939. [DOI] [PMC free article] [PubMed]
- An, C., Lovell, S., Kanost, M. R., Battaile, K. P. & Michel, K. (2011). Proteins, 79, 1999–2003. [DOI] [PMC free article] [PubMed]
- Annand, R. R., Dahlen, J. R., Sprecher, C. A., De Dreu, P., Foster, D. C., Mankovich, J. A., Talanian, R. V., Kisiel, W. & Giegel, D. A. (1999). Biochem. J. 342, 655–665. [PMC free article] [PubMed]
- Ashton-Rickardt, P. G. (2013). Immunol. Lett. 152, 65–76. [DOI] [PubMed]
- Bager, R., Johansen, J. S., Jensen, J. K., Stensballe, A., Jendroszek, A., Buxbom, L., Sørensen, H. P. & Andreasen, P. A. (2013). J. Mol. Biol. 425, 2867–2877. [DOI] [PubMed]
- Baglin, T. P., Carrell, R. W., Church, F. C., Esmon, C. T. & Huntington, J. A. (2002). Proc. Natl Acad. Sci. USA, 99, 11079–11084. [DOI] [PMC free article] [PubMed]
- Barillas-Mury, C. (2007). Trends Parasitol. 23, 297–299. [DOI] [PubMed]
- Bartholomay, L. C. et al. (2010). Science, 330, 88–90.
- Chaillan-Huntington, C. E., Gettins, P. G. W., Huntington, J. A. & Patston, P. A. (1997). Biochemistry, 36, 9562–9570. [DOI] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Christophides, G. K. et al. (2002). Science, 298, 159–165. [DOI] [PubMed]
- Christophides, G. K., Vlachou, D. & Kafatos, F. C. (2004). Immunol. Rev. 198, 127–148. [DOI] [PubMed]
- Danielli, A., Kafotos, F. C. & Loukeris, T. G. (2003). J. Biol. Chem. 278, 4184–4193. [DOI] [PubMed]
- Declerck, P. J. & Gils, A. (2013). Semin. Thromb. Hemost. 39, 356–364. [DOI] [PubMed]
- Diederichs, K. & Karplus, P. A. (1997). Nature Struct. Mol. Biol. 4, 269–275. [DOI] [PubMed]
- Dunstone, M. A. & Whisstock, J. C. (2011). Methods Enzymol. 501, 63–87. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Evans, P. (2011). Acta Cryst. D67, 282–292. [DOI] [PMC free article] [PubMed]
- Evans, P. (2012). Science, 336, 986–987. [DOI] [PubMed]
- Fish, W. W. & Bjork, I. (1979). Eur. J. Biochem. 101, 31–38. [DOI] [PubMed]
- Francis, S. E., Ersoy, R. A., Ahn, J.-W., Atwell, B. J. & Roberts, T. H. (2012). BMC Genomics, 13, 449. [DOI] [PMC free article] [PubMed]
- Gatto, M., Iaccarino, L., Ghirardello, A., Bassi, N., Pontisso, P., Punzi, L., Shoenfeld, Y. & Doria, A. (2013). Clin. Rev. Allerg. Immunol. 45, 267–280. [DOI] [PubMed]
- Gettins, P. G. W. (2002). Chem. Rev. 102, 4751–4804. [DOI] [PubMed]
- Gubb, D., Sanz-Parra, A., Barcena, L., Troxler, L. & Fullaondo, A. (2010). Biochimie, 92, 1749–1759. [DOI] [PubMed]
- Gulley, M. M., Zhang, X. & Michel, K. (2013). J. Insect Physiol. 59, 138–147. [DOI] [PMC free article] [PubMed]
- Guo, P.-C., Dong, Z., Zhao, P., Zhang, Y., He, H., Tan, X., Zhang, W. & Xia, Q. (2015). Sci. Rep. 5, 11863. [DOI] [PMC free article] [PubMed]
- Hashimoto, C., Kim, D. R., Weiss, L. A., Miller, J. W. & Morisato, D. (2003). Dev. Cell, 5, 945–950. [DOI] [PubMed]
- Hood, D. B., Huntington, J. A. & Gettins, P. G. W. (1994). Biochemistry, 33, 8538–8547. [DOI] [PubMed]
- Hopkins, P. C., Carrell, R. W. & Stone, S. R. (1993). Biochemistry, 32, 7650–7657. [DOI] [PubMed]
- Hopkins, P. C. & Stone, S. R. (1995). Biochemistry, 34, 15872–15879. [DOI] [PubMed]
- Horvath, A. J., Irving, J. A., Rossjohn, J., Law, R. H., Bottomley, S. P., Quinsey, N. S., Pike, R. N., Coughlin, P. B. & Whisstock, J. C. (2005). J. Biol. Chem. 280, 43168–43178. [DOI] [PubMed]
- Huang, X., Dementiev, A., Olson, S. T. & Gettins, P. G. W. (2010). J. Biol. Chem. 285, 20399–20409. [DOI] [PMC free article] [PubMed]
- Huasong, G., Zongmei, D., Jianfeng, H., Xiaojun, Q., Jun, G., Sun, G., Donglin, W. & Jianhong, Z. (2015). Brain Res. 1600, 59–69. [DOI] [PubMed]
- Hughes, G. L., Ren, X., Ramirez, J. L., Sakamoto, J. M., Bailey, J. A., Jedlicka, A. E. & Rasgon, J. L. (2011). PLoS Pathog. 7, e1001296. [DOI] [PMC free article] [PubMed]
- Huntington, J. A. (2006). Trends Biochem. Sci. 31, 427–435. [DOI] [PubMed]
- Huntington, J. A. (2011). J. Thromb. Haemost. 9, 26–34. [DOI] [PubMed]
- Huntington, J. A. (2013). J. Thromb. Haemost. 11, 254–264. [DOI] [PubMed]
- Huntington, J. A., Fan, B., Karlsson, K. E., Deinum, J., Lawrence, D. A. & Gettins, P. G. W. (1997). Biochemistry, 36, 5432–5440. [DOI] [PubMed]
- Huntington, J. A., McCoy, A., Belzar, K. J., Pei, X. Y., Gettins, P. G. W. & Carrell, R. W. (2000). J. Biol. Chem. 275, 15377–15383. [DOI] [PubMed]
- Huntington, J. A., Read, R. J. & Carrell, R. W. (2000). Nature (London), 407, 923–926. [DOI] [PubMed]
- Irving, J. A., Pike, R. N., Lesk, A. M. & Whisstock, J. C. (2000). Genome Res. 10, 1845–1864. [DOI] [PubMed]
- Jensen, J. K. & Gettins, P. G. W. (2008). Protein Sci. 17, 1844–1849. [DOI] [PMC free article] [PubMed]
- Jin, L., Abrahams, J. P., Skinner, R., Petitou, M., Pike, R. N. & Carrell, R. W. (1997). Proc. Natl Acad. Sci. USA, 94, 14683–14688. [DOI] [PMC free article] [PubMed]
- Johnson, D. J., Langdown, J. & Huntington, J. A. (2010). Proc. Natl Acad. Sci. USA, 107, 645–650. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010a). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010b). Acta Cryst. D66, 133–144. [DOI] [PMC free article] [PubMed]
- Kambris, Z., Blagborough, A. M., Pinto, S. B., Blagrove, M. S., Godfray, H. C., Sinden, R. E. & Sinkins, S. P. (2010). PLoS Pathog. 6, e1001143. [DOI] [PMC free article] [PubMed]
- Karplus, P. A. & Diederichs, K. (2012). Science, 336, 1030–1033. [DOI] [PMC free article] [PubMed]
- Kim, S.-J., Woo, J.-R., Seo, E. J., Yu, M.-H. & Ryu, S.-E. (2001). J. Mol. Biol. 306, 109–119. [DOI] [PubMed]
- Klieber, M. A., Underhill, C., Hammond, G. L. & Muller, Y. A. (2007). J. Biol. Chem. 282, 29594–29603. [DOI] [PubMed]
- Lampl, N., Budai-Hadrian, O., Davydov, O., Joss, T. V., Harrop, S. J., Curmi, P. M., Roberts, T. H. & Fluhr, R. (2010). J. Biol. Chem. 285, 13550–13560. [DOI] [PMC free article] [PubMed]
- Langer, G., Cohen, S. X., Lamzin, V. S. & Perrakis, A. (2008). Nature Protoc. 3, 1171–1179. [DOI] [PMC free article] [PubMed]
- Lee, B. & Richards, F. M. (1971). J. Mol. Biol. 55, 379–400. [DOI] [PubMed]
- Li, J., Wang, Z., Canagarajah, B., Jiang, H., Kanost, M. & Goldsmith, E. J. (1999). Structure, 7, 103–109. [DOI] [PubMed]
- Li, W., Adams, T. E., Kjellberg, M., Stenflo, J. & Huntington, J. A. (2007). J. Biol. Chem. 282, 13759–13768. [DOI] [PubMed]
- Li, W. & Huntington, J. A. (2008). J. Biol. Chem. 283, 36039–36045. [DOI] [PubMed]
- Li, W. & Huntington, J. A. (2012). Blood, 120, 459–467. [DOI] [PubMed]
- Ligoxygakis, P., Roth, S. & Reichhart, J. M. (2003). Curr. Biol. 13, 2097–2102. [DOI] [PubMed]
- Loebermann, H., Tokuoka, R., Deisenhofer, J. & Huber, R. (1984). J. Mol. Biol. 177, 531–557. [PubMed]
- Mahajan, N., Shi, H. Y., Lukas, T. J. & Zhang, M. (2013). J. Biol. Chem. 288, 11611–11620. [DOI] [PMC free article] [PubMed]
- Marinotti, O., Calvo, E., Nguyen, Q. K., Dissanayake, S., Ribeiro, J. M. & James, A. A. (2006). Insect Mol. Biol. 15, 1–12. [DOI] [PubMed]
- Mast, A. E., Enghild, J. J. & Salvesen, G. (1992). Biochemistry, 31, 2720–2728. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McCarthy, B. J. & Worrall, D. M. (1997). J. Mol. Biol. 267, 561–569. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Pei, X. Y., Skinner, R., Abrahams, J. P. & Carrell, R. W. (2003). J. Mol. Biol. 326, 823–833. [DOI] [PubMed]
- McGowan, S., Buckle, A. M., Irving, J. A., Ong, P. C., Bashtannyk-Puhalovich, T. A., Kan, W.-T., Henderson, K. N., Bulynko, Y. A., Popova, E. Y., Smith, A. I., Bottomley, S. P., Rossjohn, J., Grigoryev, S. A., Pike, R. N. & Whisstock, J. C. (2006). EMBO J. 25, 3144–3155. [DOI] [PMC free article] [PubMed]
- Meekins, D. A., Kanost, M. R. & Michel, K. (2016). Semin. Cell Dev. Biol. https//.org/10.1016/j.semcdb.2016.09.001. [DOI] [PMC free article] [PubMed]
- Michel, K., Budd, A., Pinto, S., Gibson, T. J. & Kafatos, F. C. (2005). EMBO Rep. 6, 891–897. [DOI] [PMC free article] [PubMed]
- Neafsey, D. E. et al. (2015). Science, 347, 1258522. [DOI] [PMC free article] [PubMed]
- Olson, S. T. & Gettins, P. G. W. (2011). Prog. Mol. Biol. Transl. Sci. 99, 185–240. [DOI] [PubMed]
- Painter, J. & Merritt, E. A. (2006). Acta Cryst. D62, 439–450. [DOI] [PubMed]
- Rau, J. C., Beaulieu, L. M., Huntington, J. A. & Church, F. C. (2007). J. Thromb. Haemost. 5, Suppl. 1, 102–115. [DOI] [PMC free article] [PubMed]
- Ravenhill, L., Wagstaff, L., Edwards, D. R., Ellis, V. & Bass, R. (2010). J. Biol. Chem. 285, 36285–36292. [DOI] [PMC free article] [PubMed]
- Saff, E. B. & Kuijlaars, A. B. J. (1997). Math. Intelligencer, 19, 5–11.
- Schick, C., Bromme, D., Bartuski, A. J., Uemura, Y., Schechter, N. M. & Silverman, G. A. (1998). Proc. Natl Acad. Sci. USA, 95, 13465–13470. [DOI] [PMC free article] [PubMed]
- Schreuder, H. A., de Boer, B., Dijkema, R., Mulders, J., Theunissen, H. J. M., Grootenhuis, P. D. & Hol, W. G. J. (1994). Nature Struct. Mol. Biol. 1, 48–54. [DOI] [PubMed]
- Sharp, A. M., Stein, P. E., Pannu, N. S., Carrell, R. W., Berkenpas, M. B., Ginsburg, D., Lawrence, D. A. & Read, R. J. (1999). Structure, 7, 111–118. [DOI] [PubMed]
- Silverman, G. A., Whisstock, J. C., Bottomley, S. P., Huntington, J. A., Kaiserman, D., Luke, C. J., Pak, S. C., Reichhart, J. M. & Bird, P. I. (2010). J. Biol. Chem. 285, 24299–24305. [DOI] [PMC free article] [PubMed]
- Simonovic, M., Gettins, P. G. W. & Volz, K. (2001). Proc. Natl Acad. Sci. USA, 98, 11131–11135. [DOI] [PMC free article] [PubMed]
- Stein, N. (2008). J. Appl. Cryst. 41, 641–643.
- Stein, P. E., Leslie, A. G. W., Finch, J. T. & Carrell, R. W. (1991). J. Mol. Biol. 221, 941–959. [DOI] [PubMed]
- Stein, P. E., Tewkesbury, D. A. & Carrell, R. W. (1989). Biochem. J. 262, 103–107. [DOI] [PMC free article] [PubMed]
- Stratikos, E. & Gettins, P. G. W. (1999). Proc. Natl Acad. Sci. USA, 96, 4808–4813. [DOI] [PMC free article] [PubMed]
- Suwanchaichinda, C. & Kanost, M. R. (2009). Gene, 442, 47–54. [DOI] [PMC free article] [PubMed]
- Weiss, M. S. (2001). J. Appl. Cryst. 34, 130–135.
- Whisstock, J. C. & Bottomley, S. P. (2006). Curr. Opin. Struct. Biol. 16, 761–768. [DOI] [PubMed]
- Widmer, C., Gebauer, J. M., Brunstein, E., Rosenbaum, S., Zaucke, F., Drogemuller, C., Leeb, T. & Baumann, U. (2012). Proc. Natl Acad. Sci. USA, 109, 13243–13247. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Winn, M. D., Isupov, M. N. & Murshudov, G. N. (2001). Acta Cryst. D57, 122–133. [DOI] [PubMed]
- Yamamoto, N., Kinoshita, T., Nohata, N., Yoshino, H., Itesako, T., Fujimura, L., Mitsuhashi, A., Usui, H., Enokida, H., Nakagawa, M., Shozu, M. & Seki, N. (2013). Int. J. Oncol. 43, 1855–1863. [DOI] [PMC free article] [PubMed]
- Ye, S., Cech, A. L., Belmares, R., Bergstrom, R. C., Tong, Y., Corey, D. R., Kanost, M. R. & Goldsmith, E. J. (2001). Nature Struct. Biol. 8, 979–983. [DOI] [PubMed]
- Zhang, Q., Buckle, A. M., Law, R. H., Pearce, M. C., Cabrita, L. D., Lloyd, G. J., Irving, J. A., Smith, A. I., Ruzyla, K., Rossjohn, J., Bottomley, S. P. & Whisstock, J. C. (2007). EMBO Rep. 8, 658–663. [DOI] [PMC free article] [PubMed]
- Zhang, X., Meekins, D. A., An, C., Zolkiewski, M., Battaile, K. P., Kanost, M. R., Lovell, S. & Michel, K. (2015). J. Biol. Chem. 290, 2946–2956. [DOI] [PMC free article] [PubMed]
- Zheng, B., Matoba, Y., Kumagai, T., Katagiri, C., Hibino, T. & Sugiyama, M. (2009). Biochem. Biophys. Res. Commun. 380, 143–147. [DOI] [PubMed]
- Zhou, Q., Snipas, S., Orth, K., Muzio, M., Dixit, V. M. & Salvesen, G. S. (1997). J. Biol. Chem. 272, 7797–7800. [DOI] [PubMed]
- Zhou, A., Wei, Z., Read, R. J. & Carrell, R. W. (2006). Proc. Natl Acad. Sci. USA, 103, 13321–13326. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: SRPN18 from Anopheles gambiae, 5c98
Supporting Information: Supplementary Figures S1 and S2.. DOI: 10.1107/S2053230X16017854/tt5090sup1.pdf