ABSTRACT
Circulating tumor cells (CTCs) have been proposed to be an active source of metastasis or recurrence of hepatocellular carcinoma (HCC). The enumeration and characterization of CTCs has important clinical significance in recurrence prediction and treatment monitoring in HCC patients. We previously developed a unique method to separate HCC CTCs based on the interaction of the asialoglycoprotein receptor (ASGPR) expressed on their membranes with its ligand. The current study applied the ligand-receptor binding assay to a CTC-chip in a microfluidic device. Efficient capture of HCC CTCs originates from the small dimensions of microfluidic channels and enhanced local topographic interactions between the microfluidic channel and extracellular extensions. With the optimized conditions, a capture yield reached > 85% for artificial CTC blood samples. Clinical utility of the system was further validated. CTCs were detected in all the examined 36 patients with HCC, with an average of 14 ± 10/2 mL. On the contrary, no CTCs were detected in healthy, benign liver disease or non-HCC cancer subjects. The current study also successfully demonstrated that the captured CTCs on our CTC-chip were readily released with ethylene diamine tetraacetic acid (EDTA); released CTCs remained alive and could be expanded to form a spheroid-like structure in a 3-dimensional cell culture assay; furthermore, sensitivity of released CTCs to chemotherapeutic agents (sorafenib or oxaliplatin) could be effectively tested utilizing this culture assay. In conclusion, the methodologies presented here offer great promise for accurate enumeration and easy release of captured CTCs, and released CTCs could be cultured for further functional studies.
KEYWORDS: Circulating tumor cells, culture, drug evaluation, hepatocellular carcinoma, ligand-receptor recognition, microfluidics
Abbreviations
- ASGPR
asialoglycoprotein receptor
- AO
acridine orange
- BSA
bovine serum albumin
- CTCs
Circulating tumor cells
- CPS1
carbamoyl phosphate synthetase 1
- DMEM
Dulbecco modified Eagle medium
- DMSO
dimethyl sulphoxide
- DAPI
4,6-diamidino-2-phenylindole
- EDTA
ethylene diamine tetraacetic acid
- EpCAM
epithelial cell adhesion molecule
- FBS
fetal bovine serum
- HCC
hepatocellular carcinoma
- PDMS
polydimethylsiloxane
- PBS
phosphate buffer saline
- P-CK
pan-cytokeratin
- PI
Propidium iodide
- TACE
transcatheter arterial chemoembolization
- 2-NBDG
2-[N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl) amino]-2-deoxy-d-glucose
- 3D
3-dimensional
- 3-MPTMS
3-mercaptopropyl trimethoxysilane
Introduction
Hepatocellular carcinoma (HCC) is the fifth most frequent cancer in the world, and is also one of the most aggressive cancers. If the tumor cannot be completely removed, the disease is ultimately fatal within 3–6 months.1-4 Surgical resection, liver transplantation, and local ablation are potentially curative for carefully selected HCC patients diagnosed at an early stage. However, these modalities are associated with higher postoperative recurrence rates, that jeopardize overall survival in these patients.5-10 It has been recognized that circulating tumor cells (CTCs) that are shed into the bloodstream from solid tumor origin are a potential continuous source of HCC to seed metastasis or recurrence. Therefore, the enumeration and characterization of CTCs has important clinical significance in recurrence prediction and treatment monitoring in HCC patients.11-13
Detection of CTCs is a technical challenge due to their extreme rarity and their low-recovery rate following traditional batch enrichment techniques.14,15 Multiple batch approaches based on one or more unique properties of tumor cells have been employed to detect CTCs for different cancer types, and numerous engineering efforts have focused on building macro-scale systems for CTC isolation.16-21 As an alternative to macro-scale systems, researchers have turned to emerging microfluidic technologies to build promising micro-scale CTC isolation systems since 2007 when a so-called CTC-chip was developed to capture rare CTCs.22 Microfluidic techniques allow us to manipulate and process cells on an individual basis and precisely control rare cells in mixtures with minimal damage to sensitive cell populations, and offer interesting perspectives for rare cell isolation, particularly when combined with specific antibodies.23 Among the macro-or micro-scale systems, the epithelial cell adhesion molecule (EpCAM) expressed on the surface of cells of epithelial origin has been used widely as a target for CTC capture.24,25 However, EpCAM expression patterns in the liver have been found to be different from that in other epithelial organs although the liver is an epithelial organ. In liver neoplasia, only small percentages (0–20%) of HCC cases were positive for EpCAM.26-29 Obviously, all of the EpCAM-based strategies are not appropriate for detection of HCC CTCs. In addition, captured CTCs by an antigen-antibody binding assay could not be easily released, allowing for their functional studies.30
We previously developed a unique method to magnetically separate CTCs in HCC patients, mediated by the interaction of the asialoglycoprotein receptor (ASGPR) with its ligand.31-34 As an abundant transmembrane receptor, ASGPR is exclusively expressed on the surface of hepatocytes and can recognize glycoproteins. Given the generally accepted absence of normal hepatocytes in circulation, blood cells labeled with the ASGPR ligand or antibody are thus considered to be circulating HCC cells. In the current study, the ligand-receptor binding assay was applied to a CTC-chip in a microfluidic device, providing an enhanced platform for isolation of CTCs from blood of patients with HCC; the captured CTCs on our CTC-chip were readily released with ethylene diamine tetraacetic acid (EDTA); released CTCs remained alive and could be expanded to form a spheroid-like structure in a 3-dimensional (3D) cell culture assay; furthermore, sensitivity of released CTCs to chemotherapeutic agents could be measured utilizing this assay.
Materials and methods
Patients and blood sample collection
The study enrolled 36 patients with advanced HCC. Peripheral blood samples from HCC patients were collected in BD Vacutainer Heparin Tubes (BD Biosciences, San Jose, CA, USA) and processed within 24 h. The study was approved by the Biomedical Ethics Committee of Eastern Hepatobiliary Surgery Hospital (Shanghai, CHN) and informed written consent was obtained from all patients.
Cell lines and culture
Human hepatoma cell line HepG2, human breast cancer cell line MCF-7, colon cancer cell line SW480, renal carcinoma cell line A498, gastric cancer cell line AGS, and lung cancer cell line A549 were purchased from American Type Culture Collection and cultured according to previously described methods.31-33
Spiking experiments with tumor cell lines
Artificial CTC blood samples were prepared by spiking human blood with cell lines. Briefly, 2 mL aliquots of peripheral blood from healthy adults were collected into BD Vacutainer Heparin Tubes, spiked with various numbers of HepG2 cells or MCF-7 cells, and then processed as described below.
Fabrication of a platform for HCC CTC capture
The new microfluidic chip was designed with mapping software L-Edit 8.30, including the patterned micropillar array layer made of polydimethylsiloxane (PDMS) (GE Silicones, RTV 615) and the glass substrate.35,36 The micropillar array layer was fabricated using a standard soft lithography method.37 The mold was made from negative photoresist (SU8-2025, MicroChem) on 3-inch silicon wafer. Holes were punched to introducing reagents and samples through Tygon tubings. The whole PDMS chip was then attached to glass slide and ready for further application.
Microfluidic device functionalization
After being rinsed by ethanol, the whole device became ready for subsequent treatment of surface modification. Streptavidin binding was accomplished by first rinsing 4% (v/v) solution of 3-mercaptopropyl trimethoxysilane (3-MPTMS) (Aladdin, LA, USA) in ethanol for 1 h at room temperature, followed by incubation in 0.02 μmol/mL N-y-mal-eimidobutyryloxysuccinimide ester (GMBS) (Sigma-Aldrich, St. Louis, USA) in ethanol for 30 min at room temperature. And then, the channels were filled with 20 μg/mL of streptavidin (Aladdin) in phosphate buffer saline (PBS) for 1 h to attach streptavidin to the GMBS.38 Asialofetuin (Sigma-Aldrich) was biotinylated using Sulfo-NHS-LC-Biotin (Thermo Fisher Scientific, MA, USA) according to the manufacturer's instructions. 100 μg/mL biotinylated asialofetuin solution in PBS containing 1% (w/v) bovine serum albumin (BSA) (Sigma-Aldrich) was introduced into the devices followed by PBS washes to remove unbound reagents.
Capture of HCC CTCs on the CTC-chip
One hour prior to experiment, the device was purged with 3% BSA and 0.05% Tween20. Samples were then loaded and the designated flow rate was controlled by a syringe pump. Afterwards, the microchip was washed with 100 μL PBS, fixed with 4% paraformaldehyde in ddH2O for 15 min, and then refilled with 0.1% Triton X-100 in PBS for cell permeabilization.
Multicolor immunofluorescence staining for captured CTCs on the CTC-chip
When captured HCC CTCs are used for enumeration, they were immunologically stained within the CTC-chip using triple immunofluorescence method with reference to the method previously described.32 A mouse monoclonal antibody cocktail against pan-cytokeratin (P-CK) (Miltenyi Biotec GmbH, Bergisch Gladbach, GER) and carbamoyl phosphate synthetase 1 (CPS1) (Abcam, MA, USA) and a rat anti-human CD45 monoclonal antibody (Santa Cruz, CA, USA) were used as primary antibodies. A Cy3-conjugated goat anti-rat IgG antibody (red color), an Alexa Fluor 488-conjugated rabbit anti-mouse IgG antibody (green color) (Beyotime, Shanghai, CHN) served as secondary antibodies, and cell nuclei were costained with 4,6-diamidino-2-phenylindole (DAPI, blue color) (Pierce, New Jersey, USA).
Identification and enumeration of CTCs on the CTC-chip
Stained CTC-chip was moved into fluorescence microscope observation (IX71; Olympus, Tokyo, Japan) and imaged. The objects that met preset criteria were carefully identified and counted. CTC counts were expressed as the number of cells per 2 mL of blood.
Release of captured CTCs from the CTC-chip
When captured CTCs are used for further culture, after cell capture inside the chip, a combination of EDTA solution (Sigma-Aldrich) and a higher flow rate of washing were applied for cell release. Briefly, 10 mmol/L EDTA was pumped into the chip device and incubated for 10 min at 37°C. Cell culture media was then introduced into the device at a flow rate of 5 mL/h. The flow rate was much higher than the cell capturing flow rate of 0.8 mL/h. Released cells were collected in a new sterile eppendorf tube with some culture media for further experiments. The experiment lasted for over 3 h (about 2.5 h for capturing and 0.5 h for releasing).
Released cell viability test
Propidium iodide (PI)-acridine orange (AO) (PtdIns-AO) assay was used to test the viability of released cells from spiking experiments with tumor cell lines according to the manufacturer's instructions. PI is a membrane-impermeant dye stain and induces dead cells with red fluorescence. AO is a membrane-permeable stain that binds to nucleic acids of all cells and induces green fluorescence. As for released CTCs in blood of HCC patients, due to their small numbers, their viability was observed via fluorescence imaging microscopy followed by direct incubation of them with a fluorescent d-glucose analog 2-[N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl) amino]-2-deoxy-d-glucose (2-NBDG) (Invitrogen, Carlsbad, CA). Living tumor cells have the ability of a fast 2-NBDG uptake and indicate green fluorescence.39,40
3D culture and drug-sensitivity testing of released CTCs
Released CTCs from the CTC-chip were cultured using a 3D cell culture assay.41 Briefly, released CTCs were resuspended in 150 μL Dulbecco modified Eagle medium (DMEM) (high glucose) (Gibco of Thermo Fisher Scientific, MA, USA). Matrigel (Becton, Dickinson, and Company, FranklinLakes, NJ, USA) was thawed and mixed equally with the CTC-containing DMEM. The prepared mixture was then incubated in a 24-well plate for 30 min at 37°C. Subsequently, 500 μL DMEM supplemented with 10% fetal bovine serum (FBS) were added to the plate, and the media was supplied every 2 d during culture. Spheroid formation was observed every day and counted on day 7. A spheroid was defined as 3D cell structure > 100 μm in diameter. As for drug-sensitivity testing, released CTCs were resuspended in DMEM containing sorafenib (Selleck Chemicals, Houston, TX, USA) pre-dissolved in dimethyl sulphoxide (DMSO) (Sigma-Aldrich) or containing oxaliplatin (R&D, MN, USA) pre-dissolved in ddH2O, and 500 μL of DMEM with sorafenib or oxaliplatin was added on top of the gel to give a final concentration of 10 μM sorafenib in 0.4% DMSO or a final concentration of 80 μM oxaliplatin.
Statistical analysis
Statistical analyses were performed using SPSS software (version 17.0; SPSS Inc., Chicago, IL, USA). Comparison of cell capture, release and viability rate with different assays was performed using Student's t test and a one-way analysis of variance (ANOVA). Comparison of categorical variables was performed using the χ2 test. Data are expressed as mean ± standard deviation or as a percentage. P < 0.05 was considered as statistically significant.
Results
Expression of ASGPR on the membrane of hepatoma cells
The results from flow cytometry showed that all 6 tested cancer cell lines derived from extra-hepatic origin displayed negative expression for ASGPR. On the contrary, the expression rate of ASGPR in human hepatoma cell line HepG2 was 95.8% (Fig. 1A). Indirect immunofluorescence laser confocal assay showed strong positive staining for ASGPR antibody on the cell surface of HepG2 (Fig. 1B), indicating that ASGPR is expressed and localized on the membrane of hepatoma cells.
Figure 1.
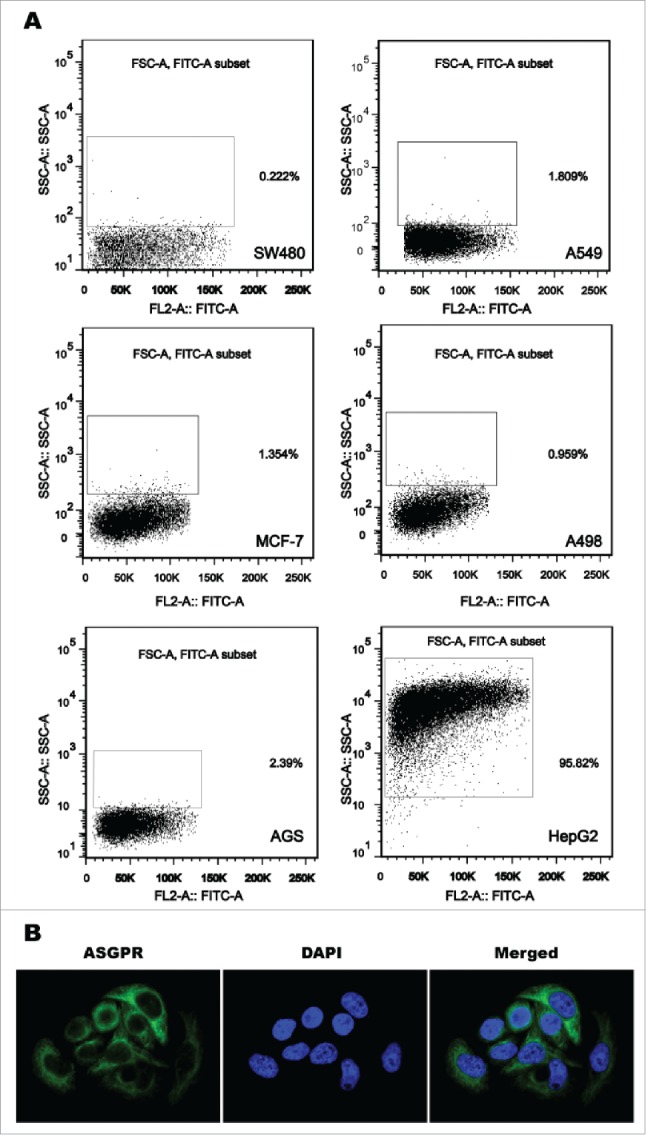
Expression of asialoglycoprotein receptor (ASGPR) on the membrane of hepatoma cells. (A) ASGPR was exclusively expressed in human hepatoma cell line HepG2 analyzed by flow cytometry, but not expressed in cancer cell lines derived from extrahepatic origin including breast cancer cell line MCF-7, colon cancer cell line SW480, renal carcinoma cell line A498, lung cancer cell line A549 and gastric cancer cell line AGS. (B) Strong positive staining for ASGPR antibody was localized on the outer cell membrane of HepG2 visualized by confocal laser scanning microscopy.
The unique design of microfluidic chips for CTC isolation
A syringe pump was used for sampling and introducing reagents (Fig. 2A). In order to significantly increase the number of interactions between target CTCs and the chip surface, our CTC-chip was fabricated using multilayer soft lithographyand consisting of an array of micropillars. In the system, HCC CTCs were captured by biotinylated asialofetuin (an ASGPR ligand), which was labeled on the streptavidin-coated surface of microfluidic chip channel (Fig. 2B, 2C and 2D).
Figure 2.
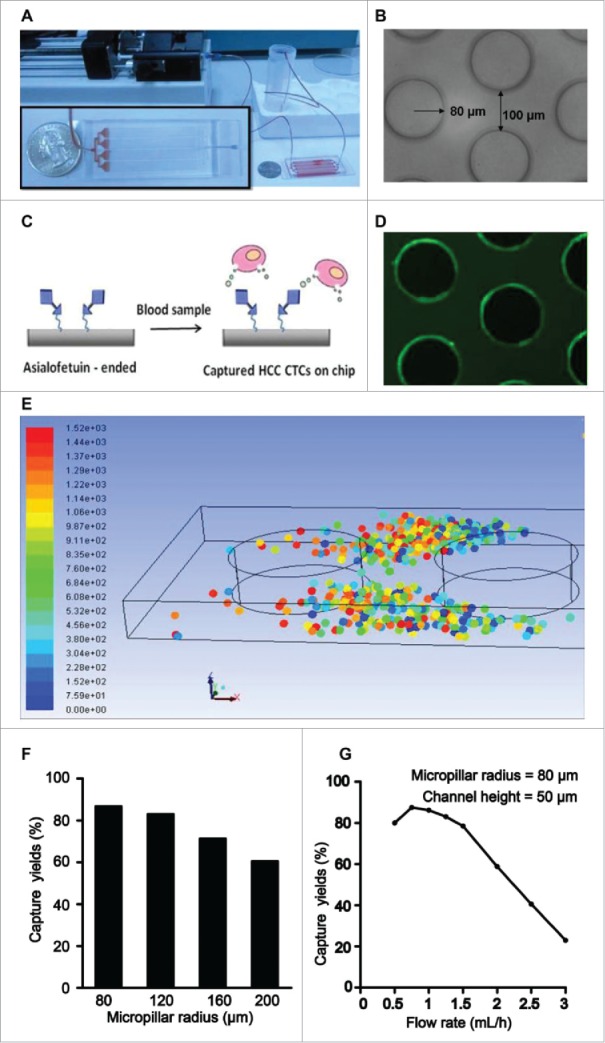
Representation of the configuration and operational mechanism of an integrated device for capturing circulating tumor cells (CTCs) of hepatocellular carcinoma (HCC). (A) Pictures of the microfluidic system for detection of HCC CTCs. (B) Optical micrograph of the patterned structures inside the chip. (C) Illustration of the HCC CTC detection on chip. (D) Fluoresce micrograph of the structures, which was coated with streptavidin and then introduced with biotin-FITC-IgG. (E) Simulation of the CTC capture inside the microchip. The dots of different color represent captured CTCs. (F) The relation between different micropillar sizes and the capture yieldat a same flow rate of 1 mL/h. (G) The relation between different flow rates and the capture yields with a same micropillar size of 80 µm.
A simple numerical simulation was conducted to investigate the relationship between the CTC capture efficiency and the micropillar structure (Fig. 2E). For the easy simulation, CTCs was assumed as zero mass, solid sphere structure at a diameter of 10 µm, the while blood cells were ignored. Affirmative capture was upon direct contact between the particle and inner channel wall. In the simulation process, 100 particles (different color dots in Fig. 2E) were loaded into the channel with different micropillar size (a radius of 80, 120, 160, 200 µm) at a same flow rate of 1 mL/h. As a result, the smaller radius gave a better capture efficiency (Fig. 2F). Similarly, a simulation for the flow effect was conducted to analyze the relationship of different flow rates at 0.5, 0.75, 1, 1.25, 1.5, 2, 2.5 and 3 mL/h with a same micropillar size of 80 µm. It was found that the lower flow rates had a better capture efficiency, and a flow rate of 0.8 mL/h gave the highest capture efficiency of 89% (Fig. 2G). However, when the flow rates were more than 1.5 mL/h, the capture efficiencies decreased gradually with an increasing flow rate (Fig. 2G). Overall, from above simple simulation, 80 µm pillar size of micropillar array columns at a flow rate of 0.8 mL/h gave the highest capture efficiency, and the captured CTCs were always positioned between the micropillars.
Microfluidic chip optimization for high-performance cell capture
Besides simulation, series of optimization experiements were performed by using the CTC-chip with the micropillar radius of 80 µm to obtain the best conditions. An artificial CTC blood sample (2 mL) was introduced onto a microfluidic chip. We first determined the minimum time required to capture target cells. Fig. 3A summarizes the correlation between different flow rates (0.5, 0.75, 1, 1.25 and 1.5 mL/h) and the cell capture efficiency. A maximal cell-capture yield of about 87% was achieved at a flow rate of 0.75 mL/h, which was consistent to the best capture results in the numerical simulation. The small difference of capture efficiency from that of the numerical simulation is possibly due to the strict postulations (zero mass, sphere structure, and positive capture upon contact and so on).
Figure 3.
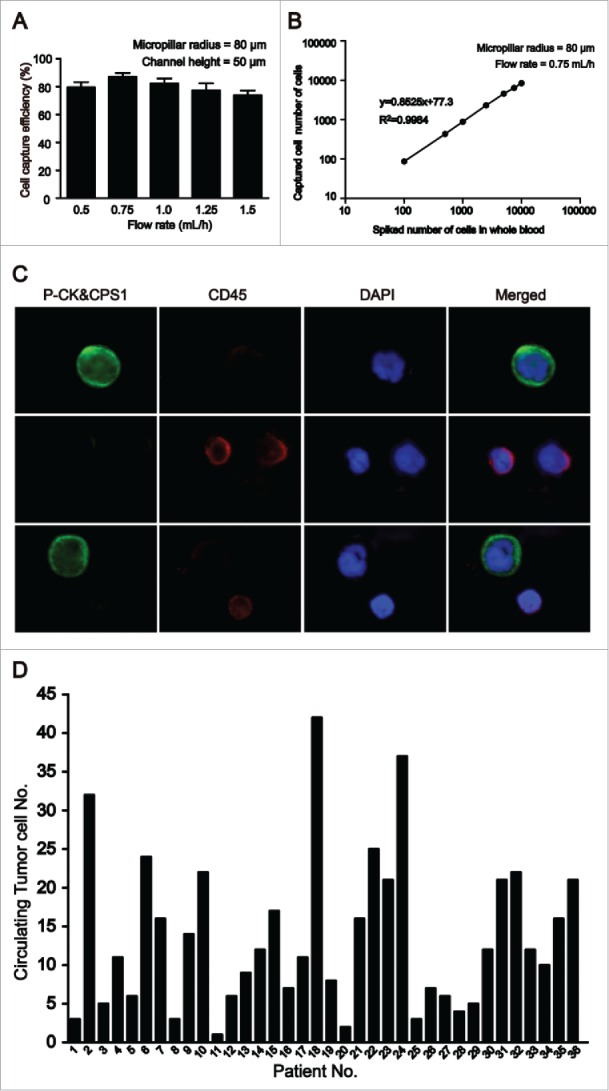
Efficient detection of circulating tumor cells (CTCs) from artificial CTC blood samples and clinical samples. (A) Target cell capture experiments with HepG2 cells-spiked blood samples at a series of flow rates. Error bars show standard deviations (n = 3). (B) Cell capture efficiencies at different numbers of target cells in HepG2-spiked blood samples. (C) CTCs detected in blood from patients with HCC (magnification, ×200). A large cells with a morphologically intact 4,6-diamidino-2-phenylindole (DAPI)-stained nucleus (blue), pan-cytokeratin (P-CK) and carbamoyl phosphate synthetase 1 (CPS1) (green) positive and CD45 (red) negative was considered CTCs. (D) Summary of CTC counts detected in 2 mL blood from 36 patients with HCC.
Efficient detection of CTCs from artificial CTC blood samples and clinical samples
After optimal cell-capture conditions were obtained, a series of artificial CTC blood samples at different densities (ranging from 102 to 104 HepG2 cells/mL) and clinical whole blood samples from HCC patients were examined using this CTC chip. As shown in Fig. 3B, the CTC chip can obtain a capture yield of up to 85% under separate experimental conditions, suggesting that asialofetuin-coated substrate can effectively capture the HCC cells at different concentrations. Simultaneously, no cancer cells were detected in any artificial CTC blood samples prepared withcell lines SW480 and MCF-7 (200 cells spiked), indicating specificity of asialofetuin-coated substrate to HCC CTCs and minimal nonspecific interactions between HCC CTCs and the CTC chip.
As previously described,32 the criteria for HCC CTC determination include a large cell with a morphologically intact DAPI-stained nucleus, P-CK and CPS1 positive and CD45 negative. A typical result of CTC detection is shown in Fig. 3C. CTCs were then detected in all the examined 36 HCC patients, with a range of 1–42/2 mL (Fig. 3D) and an average of 14 ± 10/2 mL. On the contrary, no CTCs were detected in healthy, benign liver disease including 3 with cirrhosis, 3 with chronic hepatitis B, 2 with chronic hepatitis C, 6 patients with benign intrahepatic space-occupying lesions (3 hepatic hemangioma and 3 liver cysts) or non-HCC cancer subjects. The correlations of CTC counts with clinical variables of HCC patients were summarized in Table 1.
Table 1.
Correlations between circulating tumor cell (CTC) counts and clinical variables of patients with hepatocellular carcinoma (HCC).
| Clinical variable | No. of patients | Mean ± SD | P |
|---|---|---|---|
| Age in years | 0.730 | ||
| ≤ 50 | 19 | 26 ± 21 | |
| > 50 | 17 | 28 ± 19 | |
| Sex | 0.879 | ||
| Male | 26 | 27 ± 19 | |
| Female | 10 | 26 ± 22 | |
| Tumor size in cm | 0.023 | ||
| ≤ 5 | 20 | 19 ± 18 | |
| > 5 | 16 | 36 ± 18 | |
| Portal vein tumor thrombus | 0.001 | ||
| With | 14 | 39 ± 17 | |
| Without | 22 | 21 ± 18 | |
| TNM staging* | < 0.001 | ||
| I-II | 21 | 17 ± 12 | |
| III-IV | 15 | 41 ± 20 |
Tumor-node-metastasis staging, Sixth Edition of International Union Against Cancer (UICC).
Efficient release of CTCs from CTC-chip and their viability
Following microfluidic capture, the release of the captured cells in microchannels was achieved by using a combination of EDTA treatment (receptor-ligand release) and a higher flow rate washing (high shear stress). Released cells were collected in a sterile eppendorf tube containing culture media. As shown in Fig. 4A, a higher flow washing alone yielded a release efficiency of about 25%, while a combination of treatment with 10 mmol/L EDTA and a higher flow rate washing increased an efficiency of release to > 85%. PtdIns-AO assay was used to test the viabilities of cultured cells (suspended in media), captured cells adherent to the chip surface, and released cells from the CTC-chip, and the results (Fig. 4B) showed that the cell viabilities was ∼89% for cultured cells, ∼82% for captured cells, and ∼80% for released cells, respectively. The difference was not statistically significant (P > 0.05), indicating that this release procedure caused minimum cell damage. In addition, the viability of released CTCs from clinical samples was observed under fluorescence spectroscopy after direct incubation of them with a fluorescent d-glucose analog 2-NBDG. As shown in Fig. 4C, after exposed to 2-NBDG for 20 min and then washed with PBS, released CTCs of HCC patients appeared to rapidly take up 2-NBDG as evidenced by a strong fluorescence, showing that these CTCs were alive.
Figure 4.
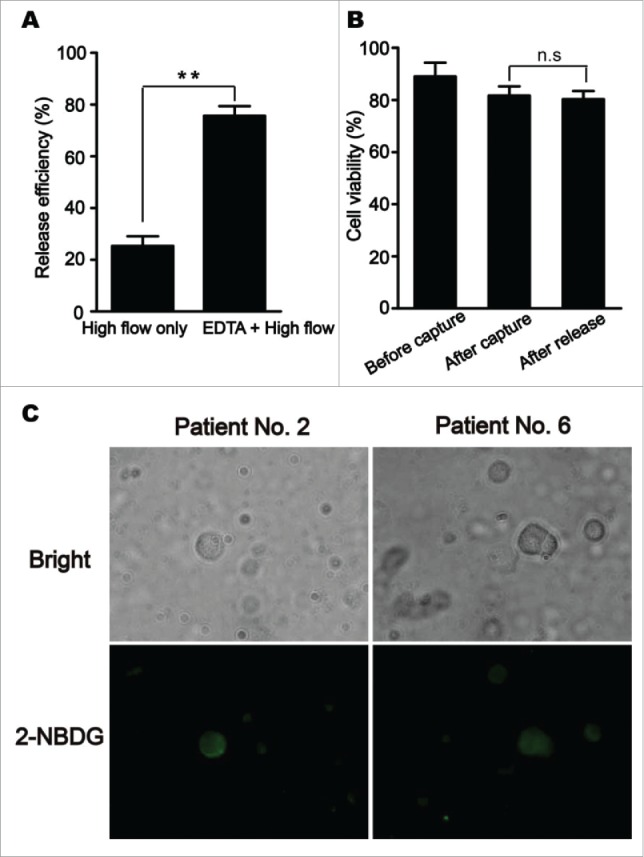
Efficient release of circulating tumor cells (CTCs) from CTC-chip and their viability. (A) A combination of treatment with ethylene diamine tetraacetic acid (EDTA) and a higher flow rate washing yielded a higher efficiency of CTC release from CTC-chip. (B) Comparison of cell viabilities among cultured cells (suspended in media), captured cells adherent to the chip surface, and released cells determined by propidium iodide-acridine orange (PI-AO) assay. Error bars represent range (n = 3), **P< 0.01. (C) Fluorescence imaging of released CTCs from clinical samples under fluorescence spectroscopy after direct incubation of them with a fluorescent d-glucose analog 2-[N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl) amino]-2-deoxy-d-glucose (2-NBDG).
Released CTCs allowing for in vitro expansion and drug sensitivity assay
Released cells were collected for a Matrigel-based induction of 3D tumor formation. Either released HepG2 cells or released CTCs of HCC patients formed spheroids, which increased in size over a period of 14 d (Fig. 5A). 31 patients had their released CTC successfully cultured into spheroids in all 36 advanced HCC patients. Before the drug sensitivity test in vitro, we identified the 3D cultured spheroids cells. The criteria for HCC CTC determination are considered to be a large spheroids cells with morphologically intact DAPI-stained nucleus, as well as, hepatoma cells express markers CPS1 and ASGPR positive (Fig. 5B). On the basis of this experiment, we tried to evaluate the sensitivity of released CTCs to chemotherapeutic drugs. Sorafenib and oxaliplatin are presented here as a representative anticancer drug for HCC. Blood samples containing more than 20 CTCs/2 mL that were measured with above-mentioned method were selected for drug evaluation. Culture of a same sample with drug solvent was as control. Two examples of sorafenib or oxaliplatin sensitivity tests are shown in Fig. 5C, where CTCs from HCC patients formed spheroids at day 7 with or without sorafenib or oxaliplatin. The results of all the 10 tested samples were summarized in Table 2, which showed a decline to varying degrees in the number of spheroids formed when sorafenib or oxaliplatin was added to the culture media. Compared to controls, CTCs treated sorafenib (5 samples) or oxaliplatin (5 samples) formed a significantly decreased number of spheroids (Fig. 5D, P < 0.01).
Figure 5.
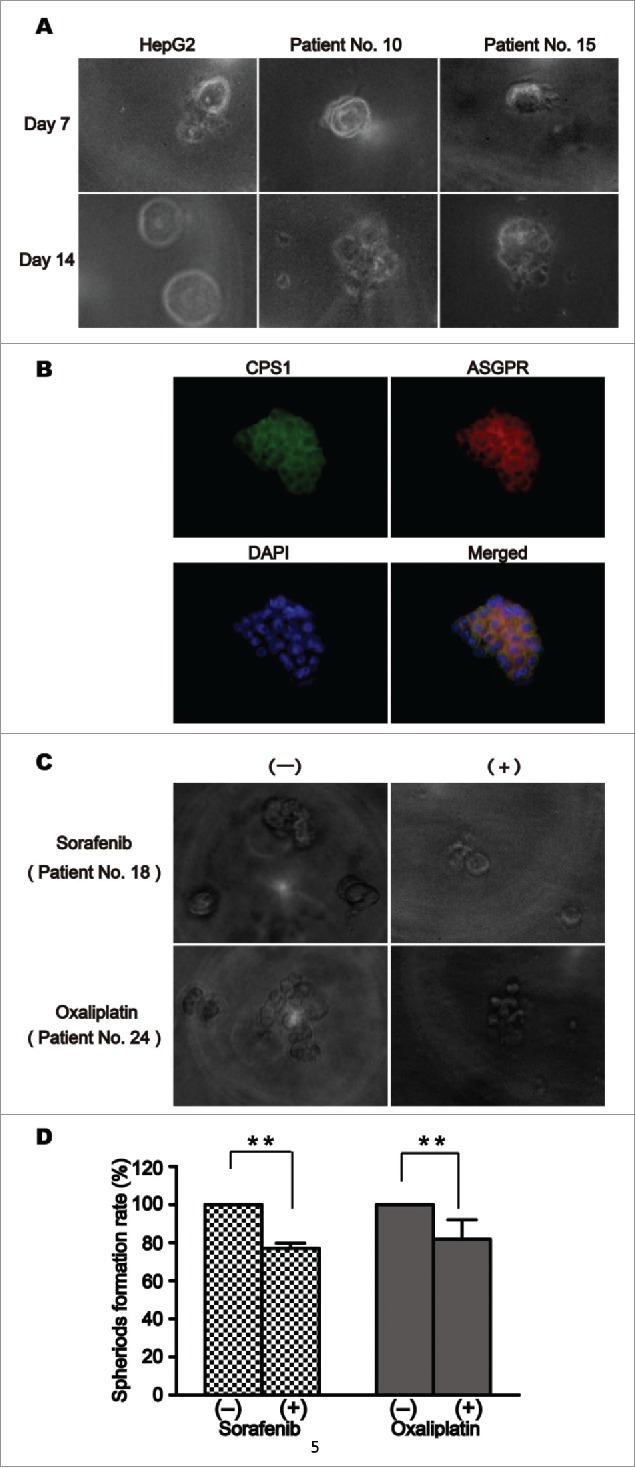
Culture and drug sensitivity assay of released circulating tumor cells (CTCs). (A) Released HepG2 cells and released CTCs of patients with hepatocellular carcinoma (HCC) from the CTC-chip formed spheroids at day 7 in a Matrigel-based induction of 3-dimensional tumor formation. (B)Triple immunofluorescence staining with antibodies against CPS1 (green), ASGPR (red), and DAPI (blue) in the 3D cultured spheroids cells. (C) Released CTCs from 2 patients with HCC formed spheroids at day 7 in culture with or without chemotherapeutic drugs. (D) Treatment of CTCs with sorafenib (5 samples) or oxaliplatin (5 samples) significantly decreased the number of spheroid formation (**P< 0.01).
Table 2.
Evaluating the sensitivity of released circulating tumor cells (CTCs) to chemotherapeutic drugs.
| Number of spheroids |
||||
|---|---|---|---|---|
| Patient no | CTC counts | Sorafenib (-) | Sorafenib (+) | Inhibition rate, % |
| 2 | 32 | 24 | 18 | 25.0 |
| 6 | 24 | 19 | 15 | 21.1 |
| 10 | 22 | 15 | 11 | 26.7 |
| 18 | 42 | 27 | 21 | 22.2 |
| 22 | 25 | 20 | 16 | 20.0 |
| Number of spheroids | ||||
| Patient no. |
CTC counts |
Oxaliplatin (-) |
Inhibition rate, %Oxaliplatin (+) |
|
| 23 | 21 | 17 | 11 | 35.3 |
| 24 | 37 | 32 | 29 | 9.4 |
| 31 | 21 | 16 | 13 | 18.8 |
| 32 | 22 | 19 | 16 | 15.8 |
| 36 | 21 | 17 | 15 | 11.8 |
Discussion
The uniqueness of this new cell-capture platform lies in the small dimensions of the use of microfluidic channels, which allows for enhanced local topographic interactions between the coated asialofetuin and the cellular surface, and results in a vastly improved cell-capture affinity compared to a previously described method 32 (Fig. 2). Efficient capture of HCC CTCs was upon direct contact between the particle and inner channel wall. Generally, at the appropriate flow rate, captured cells were close to the inner micro pillars wall. Efficient capture also likely originated from enhanced local topographic interactions between the microfluidic channel and extracellular extensions, based on the ligand-receptor recognition. The minimum micropillar radius required for optimal capture yield is believed to be compatible with the cell-protrusion lengths of tumor cells. Using this chip, HCC cells could be reliably captured by biotinylated asialofetuin, a ligand of ASGPR receptors on HCC cell surface. Captured HCC cells were identified by immunofluorescence staining of an antibody cocktail for P-CK and CPS1.32 Clinical utility of the system was then validated by the detection of CTCs in blood samples of HCC patients. Therefore, our platform provides a convenient and cost-efficient alternative for isolation and enumeration of CTCs in whole blood specimens from HCC patients.
Detection of fixed CTCs provides relatively little information about their functional capability and metastatic potential. The option to isolate viable CTCs in good condition without damage is a huge potential of CTCs since it is a necessary prerequisite for functional analysis. However, isolation of intact and viable CTCs represents a technical challenge. Most of current strategies for isolation of CTCs are based on the basic principle of antigen-antibody reaction, a well-known example being anti-EpCAM antibodies-based capture of CTCs. As we known, it is not easy for captured cells to release from immobilized antibodies without damage. Trypsin is usually used to dissociate antigen-antibody complex, but the dissociation of antibody-captured cells with trypsin is typically incomplete and may also damage the target cells.42 Over an antigen-antibody binding assay, the ligand-receptor binding assay has an advantage that captured CTCs by a specific ligand could be easily detached by a complexing agent without damageas the assay usually relies on a non-covalent and reversible interaction that requires calcium ions. Our results showed that captured CTCs on our CTC-chip were readily released with EDTA from immobilized ASGPR ligand, and the released CTCs remained alive.
After isolation of viable CTCs, their culture is required for further functional studies. In addition to a very limited number, CTCs are not relatively protected from cell death partly due to the harsh environment and shear stresses of the vascular circulation. Generally, it is more difficult to culture CTCs than primary tumor cells, and mimicry of the tumor microenvironment in vitro is particularly difficult to achieve. In this aspect, Lovitt et al. described a standardized and highly reproducible 3D cell culture model that better recreates the in vivo microenvironment and tumor biology compared to monolayer cell culture.41 In the present study, we tried to use this 3D cell culture assay to culture released CTCs from our CTC-chip. Fortunately, some of these CTCs could be expanded as 3D cell structures or spheroids for up to 14 d although it took a little longer for them to form a spheroid than the cell line HepG2.
Since this 3D human cell culture model has been demonstrated to be applicable for drug evaluation that mimics in vivo responses to drugs,43,44 it was used here to test the sensitivity of released CTCs to chemotherapeutic drugs. At present, sorafenib and oxaliplatin are 2 systemic therapy agents recommended for advanced HCC patients.45,46 Sorafenib is a multitargeted, small molecule tyrosine kinase inhibitor with multiple antitumor effects, including antiangiogenic, antiproliferative, and pro-apoptotic effects.45 Oxaliplatin is a novel platinum derivative analog that crosslinks DNA and induces apoptotic cell death.46 In addition to systemic therapy, oxaliplatin is more widely used as a chemotherapeutic agent in transcatheter arterial chemoembolization (TACE).47 As a minimally invasive procedure to restrict a tumor's blood supply, TACE is considered medically necessary as a primary treatment for surgically unresectable primary hepatocellular carcinoma. Small embolic particles mixed with chemotherapeutic agents are injected selectively into an artery directly supplying a tumor.48,49 In order to assess the usefulness of this method, we choose sorafenib and oxaliplatin as representative anticancer drugs and selected blood samples with a higher count of CTCs as tested samples. As expected, when cultured with media containing oxaliplatin or sorafenib, all the tested blood samples showed a decline to varying degrees in the number of spheroids formed.
In summary, this study presents a unique microfluidic chip to efficiently isolate CTCs in blood of patients with HCC, which allows for accurate enumeration of individually captured CTCs or for easy release of them. More importantly, we demonstrated that released CTCs remained alive and could be expanded to form a spheroid-like structure in a 3D cell culture assay. Furthermore, sensitivity of CTCs to chemotherapeutic agents can be measured utilizing this assay. It seems especially important for individual therapeutic decisions because CTCs can be serially obtained as a “liquid tumor biopsy” through a much less invasive and cost-effective approach. The 3D CTC culture assay described in this report has great potential to enable assessment of initial sensitivity and primary or secondary resistance to a specific drug, e.g. sorafenib or oxaliplatin. It could aid the physician in the selection of appropriate drug therapies in the hope of improving response outcomes.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This research was supported by China National Key Projects for Infectious Disease (No. 2012ZX10002012-010), and the National Natural Science Foundation of China (No.81172207, 21377026).
References
- 1.Shariff MI, Cox IJ, Gomaa AI, Khan SA, Gedroyc W, Taylor-Robinson SD. Hepatocellular carcinoma: current trends in worldwide epidemiology, risk factors, diagnosis and therapeutics. Expert Rev Gastroenterol Hepatol 2009; 3(4):353-67; PMID:19673623; http://dx.doi.org/ 10.1586/egh.09.35 [DOI] [PubMed] [Google Scholar]
- 2.Rahbari NN, Mehrabi A, Mollberg NM, Müller SA, Koch M, Büchler MW, Weitz J. Hepatocellular carcinoma: current management and perspectives for the future. Ann Surg 2011; 253(3):453-69; PMID:21263310; http://dx.doi.org/ 10.1097/SLA.0b013e31820d944f [DOI] [PubMed] [Google Scholar]
- 3.Bruix J, Sherman M. American association for the study of liver diseases. Management of hepatocellular carcinoma: an update. Hepatology 2011; 53(3):1020-2; PMID:21374666; http://dx.doi.org/ 10.1002/hep.24199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akoad ME, Pomfret EA. Surgical resection and liver transplantation for hepatocellular carcinoma. Clin Liver Dis 2015; 19(2):381-99; PMID:25921669; http://dx.doi.org/ 10.1016/j.cld.2015.01.007 [DOI] [PubMed] [Google Scholar]
- 5.Mazzaferro V, Llovet JM, Miceli R, Bhoori S, Schiavo M, Mariani L, Camerini T, Roayaie S, Schwartz ME, Grazi GL, et al.. Metroticket Investigator Study Group. Predicting survival after liver transplantation in patients with hepatocellular carcinoma beyond the Milan criteria: a retrospective, exploratory analysis. Lancet Oncol 2009; 10(1):35-43; PMID:19058754; http://dx.doi.org/ 10.1016/S1470-2045(08)70284-5 [DOI] [PubMed] [Google Scholar]
- 6.European Association for the Study of Liver . EASL clinical practical guidelines: management of alcoholic liver disease. J Hepatol 2012; 57(2):399-420; PMID:22633836; http://dx.doi.org/ 10.1016/j.jhep.2012.04.004 [DOI] [PubMed] [Google Scholar]
- 7.Bellissimo F, Pinzone MR, Cacopardo B, Nunnari G. Diagnostic and therapeutic management of hepatocellular carcinoma. World J Gastroenterol 2015; 21(42):12003-21; PMID:26576088; http://dx.doi.org/ 10.3748/wjg.v21.i42.12003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mazzola A, Costantino A, Petta S, Bartolotta TV, Raineri M, Sacco R, Brancatelli G, Cammà C, Cabibbo G. Recurrence of hepatocellular carcinoma after liver transplantation: an update. Future Oncol 2015; 11(21):2923-36; PMID:26414336; http://dx.doi.org/ 10.2217/fon.15.239 [DOI] [PubMed] [Google Scholar]
- 9.Schlachterman A, Craft WW Jr, Hilgenfeldt E, Mitra A, Cabrera R. Current and future treatments for hepatocellular carcinoma. World J Gastroenterol 2015; 21(28):8478-91; PMID:26229392; http://dx.doi.org/ 10.3748/wjg.v21.i28.8478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sacco R, Antonucci M, Bresci G, Corti A, Giacomelli L, Mismas V, Rainieri M, Romano A, Eggenhoffner R, Tumino E, Cabibbo G. Curative therapies for hepatocellular carcinoma: an update and perspectives. Expert Rev Anticancer Ther 2015; 14:1-7; PMID:26588992; http://dx.doi.org/ 10.1586/14737140.2016.1123625 [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Li J, Cao L, Xu W, Yin Z. Circulating tumor cells in hepatocellular carcinoma: detection techniques, clinical implications, and future perspectives. Semin Oncol 2012; 39(4):449-60; PMID:22846862; http://dx.doi.org/24415867 10.1053/j.seminoncol.2012.05.012 [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Shi ZL, Yang X, Yin ZF. Targeting of circulating hepatocellular carcinoma cells to prevent postoperative recurrence and metastasis. World J Gastroenterol 2014; 20(1):142-7; PMID:24415867; http://dx.doi.org/ 10.3748/wjg.v20.i1.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu H, Zhang X, Li J, Sun B, Qian H, Yin Z. The biological and clinical importance of epithelial-mesenchymal transition in circulating tumor cells. J Cancer Res Clin 2015; 141(2):189-201; PMID:24965746; http://dx.doi.org/26594792; 10.1007/s00432-014-1752-x [DOI] [PubMed] [Google Scholar]
- 14.Stoecklein NH, Fischer JC, Niederacher D, Terstappen LW. Challenges for CTC-based liquid biopsies: low CTC frequency and diagnostic leukapheresis as a potential solution. Expert Rev Mol Diagn 2015; 16:1-18. [Epub ahead of print]; PMID:26594792; http://dx.doi.org/ 10.1586/14737159.2016.1123095 [DOI] [PubMed] [Google Scholar]
- 15.Hyun KA, Kim J, Gwak H, Jung HI. Isolation and enrichment of circulating biomarkers for cancer screening, detection, and diagnostics. Analyst 2016; 141(2):382-92; PMID:26588824; http://dx.doi.org/ 10.1039/C5AN01762A [DOI] [PubMed] [Google Scholar]
- 16.Talasaz AH, Powell AA, Huber DE, Berbee JG, Roh KH, Yu W, Xiao W, Davis MM, Pease RF, Mindrinos MN, et al.. Isolating highly enriched populations of circulating epithelial cells and other rare cells from blood using a magnetic sweeper device. Proc Natl Acad Sci USA 2009; 106(10):3970-5; PMID:19234122; http://dx.doi.org/ 10.1073/pnas.0813188106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu T, Lu B, Tai YC, Goldkorn A. A cancer detection platform which measures telomerase activity from live circulating tumor cells captured on a microfilter. Cancer Res 2010; 70(16):6420-6; PMID:20663903; http://dx.doi.org/ 10.1158/0008-5472.CAN-10-0686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farace F, Massard C, Vimond N, Drusch F, Jacques N, Billiot F, Laplanche A, Chauchereau A, Lacroix L, Planchard D, et al.. A direct comparison of CellSearch and ISET for circulating tumour-celldetection in patients with metastatic carcinomas. Br J Cancer 2011; 105(6):847-53; PMID:21829190; http://dx.doi.org/ 10.1038/bjc.2011.294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saucedo-Zeni N, Mewes S, Niestroj R, Gasiorowski L, Murawa D, Nowaczyk P, Tomasi T, Weber E, Dworacki G, Morgenthaler NG, et al.. A novel method for the in vivo isolation of circulating tumor cells from peripheral blood of cancer patients using a functionalized and structured medical wire. Int J Oncol 2012; 41(4):1241-50; PMID:22825490; http://dx.doi.org/ 10.3892/ijo.2012.1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andreopoulou E, Yang LY, Rangel KM, Reuben JM, Hsu L, Krishnamurthy S, Valero V, Fritsche HA, Cristofanilli M. Comparison of assay methods for detection of circulating tumor cells in metastatic breast cancer: AdnaGen AdnaTest BreastCancer Select/Detect™ versus Veridex CellSearch™ system. Int J Cancer 2012; 130(7):1590-7; PMID:21469140; http://dx.doi.org/ 10.1002/ijc.26111 [DOI] [PubMed] [Google Scholar]
- 21.Mazzini C, Pinzani P, Salvianti F, Scatena C, Paglierani M, Ucci F, Pazzagli M, Massi D. Circulating tumor cells detection and counting in uveal melanomas by a filtration-based method. Cancers (Basel) 2014; 6(1):323-32; PMID:24514165; http://dx.doi.org/ 10.3390/cancers6010323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, et al.. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 2007; 450(7173):1235-9; PMID:18097410; http://dx.doi.org/ 10.1038/nature06385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qian W, Zhang Y, Chen W. Capturing cancer: Emerging microfluidic technologies for the capture and characterization of circulating tumor cells. Small 2015; 11(32):3850-72; PMID:25993898; http://dx.doi.org/ 10.1002/smll.201403658 [DOI] [PubMed] [Google Scholar]
- 24.Esmaeilsabzali H, Beischlag TV, Cox ME, Parameswaran AM, Park EJ. Detection and isolation of circulating tumor cells: principles and methods. Biotechnol Adv 2013; 31(7):1063-84; PMID:23999357; http://dx.doi.org/ 10.1016/j.biotechadv.2013.08.016 [DOI] [PubMed] [Google Scholar]
- 25.Li P, Stratton ZS, Dao M, Ritz J, Huang TJ. Probing circulating tumor cells in microfluidics. Lab Chip 2013; 13(4):602-9; PMID:23306378; http://dx.doi.org/10398165 10.1039/c2lc90148j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Boer CJ, van Krieken JH, Janssen-van Rhijn CM, Litvinov SV. Expression of EpCAM in normal, regenerating, metaplastic, and neoplastic liver. J Pathol 1999; 188(2):201-6; PMID:10398165; http://dx.doi.org/ 10.1002/(SICI)1096-9896(199906)188:2%3c201::AID-PATH339%3e3.0.CO;2-8 [DOI] [PubMed] [Google Scholar]
- 27.Proca DM, Niemann TH, Porcell AI, DeYoung BR. MOC31 immunoreactivity in primary and metastatic carcinoma of the liver. Report of findings and review of other utilized markers. Appl Immunohistochem Mol Morphol 2000; 8(2):120-5; PMID:10937059; http://dx.doi.org/10912937 10.1097/00022744-200006000-00006 [DOI] [PubMed] [Google Scholar]
- 28.Porcell AI, De Young BR, Proca DM, Frankel WL. Immunohistochemical analysis of hepatocellular and adenocarcinoma in the liver: MOC31 compares favorably with other putative markers. Mod Pathol 2000; 13(7):773-8; PMID:10912937; http://dx.doi.org/ 10.1038/modpathol.3880134 [DOI] [PubMed] [Google Scholar]
- 29.Went PT, Lugli A, Meier S, Bundi M, Mirlacher M, Sauter G, Dirnhofer S. Frequent EpCAM protein expression in human carcinomas. Hum Pathol 2004; 35(1):122-8; PMID:14745734; http://dx.doi.org/ 10.1016/j.humpath.2003.08.026 [DOI] [PubMed] [Google Scholar]
- 30.Lin M, Chen JF, Lu YT, Zhang Y, Song J, Hou S, Ke Z, Tseng HR. Nanostructure embedded microchips for detection, isolation, and characterization of circulating tumor cells. Acc Chem Res 2014; 47(10):2941-50; PMID:25111636; http://dx.doi.org/ 10.1021/ar5001617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu W, Cao L, Chen L, Li J, Zhang XF, Qian HH, Kang XY, Zhang Y, Liao J, Shi LH, et al.. Isolation of circulating tumor cells in patients with hepatocellular carcinoma using a novel cell separation strategy. Clin Cancer Res 2011; 17(11):3783-93; PMID:21527564; http://dx.doi.org/ 10.1158/1078-0432.CCR-10-0498 [DOI] [PubMed] [Google Scholar]
- 32.Li J, Chen L, Zhang X, Zhang Y, Liu H, Sun B, Zhao L, Ge N, Qian H, Yang Y, et al.. Detection of circulating tumor cells in hepatocellular carcinoma using antibodies against asialoglycoprotein receptor, carbamoyl phosphate synthetase 1 and pan-cytokeratin. PLoS One 2014; 9(4):e96185; PMID:24763545; http://dx.doi.org/ 10.1371/journal.pone.0096185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu H, Qian HH, Zhang XF, Li J, Yang X, Sun B, Ma JY, Chen L, Yin ZF. Improved method increases sensitivity for circulating hepatocellular carcinoma cells. World J Gastroenterol 2015; 21(10):2918-25; PMID:25780289; http://dx.doi.org/ 10.3748/wjg.v21.i10.2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J, Shi L, Zhang X, Sun B, Yang Y, Ge N, Liu H, Yang X, Chen L, Qian H, et al.. pERK/pAkt phenotyping in circulating tumor cells as a biomarker for sorafenib efficacy in patients with advanced hepatocellular carcinoma. Oncotarget 2016; 7(3):2646-59; PMID:26544731; http://dx.doi.org/11090344 10.18632/oncotarget.6104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quake SR, Scherer A. From micro- to nanofabrication with soft materials. Science 2000; 290(5496):1536-40; PMID:11090344; http://dx.doi.org/ 10.1126/science.290.5496.1536 [DOI] [PubMed] [Google Scholar]
- 36.Unger MA, Chou HP, Thorsen T, Scherer A, Quake SR. Monolithic microfabricated valves and pumps by multilayer soft lithography. Science 2000; 288(5463):113-6; PMID:10753110; http://dx.doi.org/ 10.1126/science.288.5463.113 [DOI] [PubMed] [Google Scholar]
- 37.Sia SK, Whitesides GM. Microfluidic devices fabricated in poly(dimethylsiloxane) for biological studies. Electrophoresis 2003; 24(21):3563-76; PMID:14613181; http://dx.doi.org/ 10.1002/elps.200305584 [DOI] [PubMed] [Google Scholar]
- 38.Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK, et al.. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci USA. 2010; 107(43):18392-7; PMID:20930119; http://dx.doi.org/16182371 10.1073/pnas.1012539107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zou C, Wang Y, Shen Z. 2-NBDG as a fluorescent indicator for direct glucose uptake measurement. J Biochem Biophys Methods 2005; 64(3):207-15; PMID:16182371; http://dx.doi.org/ 10.1016/j.jbbm.2005.08.001 [DOI] [PubMed] [Google Scholar]
- 40.Blodgett AB, Kothinti RK, Kamyshko I, Petering DH, Kumar S, Tabatabai NM. A fluorescence method for measurement of glucose transport in kidney cells. Diabetes Technol Ther 2011; 13(7):743-51; PMID:21510766; http://dx.doi.org/ 10.1089/dia.2011.0041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lovitt CJ, Shelper TB, Avery VM. Miniaturized three-dimensional cancer model for drug evaluation. Assay Drug Dev Technol 2013; 11(7):435-48; PMID:25310845; http://dx.doi.org/doi: 10.1089/adt.2012.483 [DOI] [PubMed] [Google Scholar]
- 42.Deng Y, Zhang Y, Sun S, Wang Z, Wang M, Yu B, Czajkowsky DM, Liu B, Li Y, Wei W, Shi Q. An integrated microfluidic chip system for single-cell secretion profiling of rare circulating tumor cells. Sci Rep 2014; 4:7499; PMID:25511131; http://dx.doi.org/25013076 10.1038/srep07499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu M, Bardia A, Aceto N, Bersani F, Madden MW, Donaldson MC, Desai R, Zhu H, Comaills V, Zheng Z, etal. Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science 2014; 345(6193):216-20; PMID:25013076; http://dx.doi.org/ 10.1126/science.1253533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Z, Shiratsuchi H, Lin J, Chen G, Reddy RM, Azizi E, Fouladdel S, Chang AC, Lin L, Jiang H, et al.. Expansion of CTCs from early stage lung cancer patients using a microfluidic co-culture model. Oncotarget 2014; 5(23):12383-97; PMID:25474037; http://dx.doi.org/ 10.18632/oncotarget.2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Llovet JM, Villanueva A, Lachenmayer A, Finn RS. Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat Rev Clin Oncol 2015; 12(7):408-24; PMID:26054909; http://dx.doi.org/ 10.1038/nrclinonc.2015.103 [DOI] [PubMed] [Google Scholar]
- 46.Qin S, Bai Y, Lim HY, Thongprasert S, Chao Y, Fan J, Yang TS, Bhudhisawasdi V, Kang WK, Zhou Y, et al.. Randomized, multicenter, open-label study of oxaliplatin plus fluorouracil/leucovorin versus doxorubicin as palliative chemotherapy in patients with advanced hepatocellular carcinoma from Asia. J Clin Oncol 2013; 31(28):3501-8; PMID:23980077; http://dx.doi.org/ 10.1200/JCO.2012.44.5643 [DOI] [PubMed] [Google Scholar]
- 47.Li JH, Xie XY, Zhang L, Le F, Ge NL, Li LX, Gan YH, Chen Y, Zhang JB, Xue TC, et al.. Oxaliplatin and 5-fluorouracil hepatic infusion with lipiodolized chemoembolization in large hepatocellular carcinoma. World J Gastroenterol 2015; 21(13):3970-7; PMID:25852283; http://dx.doi.org/ 10.3748/wjg.v21.i13.3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sacco R, Mismas V, Marceglia S, Romano A, Giacomelli L, Bertini M, Federici G, Metrangolo S, Parisi G, Tumino E, et al.. Transarterial radioembolization for hepatocellular carcinoma: An update and perspectives. World J Gastroenterol 2015; 1(21):6518-25; PMID:26074690; http://dx.doi.org/25038916 10.3748/wjg.v21.i21.6518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xie ZB, Ma L, Wang XB, Bai T, Ye JZ, Zhong JH, Li LQ. Transarterial embolization with or without chemotherapy for advanced hepatocellular carcinoma: a systematic review. Tumour Biol 2014; 35(9):8451-9; PMID:25038916; http://dx.doi.org/ 10.1007/s13277-014-2340-z [DOI] [PubMed] [Google Scholar]


