Abstract
We report an effective intracellular delivery strategy for proteins of high molecular weight using AuNP stabilized capsules. This strategy provides direct delivery to the cytosol, avoiding endosomal entrapment.
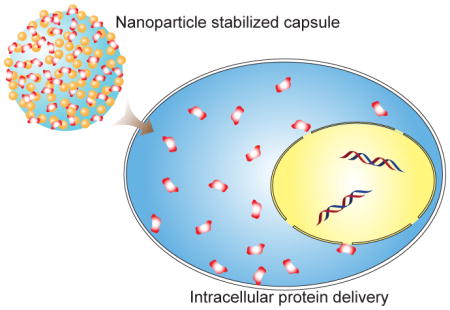
Graphical Abstract
Protein-based therapeutics have widespread applications in biomedical engineering, 1 cell engineering 2, 3 and regenerative medicine. 4, 5 An increasing number of proteins, including signalling proteins,6 antibodies7,8 and functional enzymes9,10 have been preclinically or clinically tested for the treatment of diseases. The vast majority of these studies, however, have focused on delivery to extracellular targets.
Intracellular delivery of proteins provides a transient and non-integrative means for the regulation of cellular protein functions, and it has recently attracted the interest of researchers and clinicians. However, despite significant advances in the development of intracellular protein delivery tools, major challenges still remain.11 A key aspect that remains unresolved is the release of the macromolecule in its active form into the cytosol, where the protein can serve a functional role. Unlike small molecules, proteins have sizes and surface properties that inhibit cytosolic access, typically through entrapment in the endosomal/lysosomal pathway.12 To address this problem, a number of delivery platforms have been developed. 13 For instance, endosomal escape agents, cell penetrating peptides, and endosomal lysis agents such as chloroquine14 have been used to facilitate protein delivery into the cytosol.15 Nevertheless, the efficiency of delivery of these traditional methods is still limited,16 in particular for proteins of large size.17,18
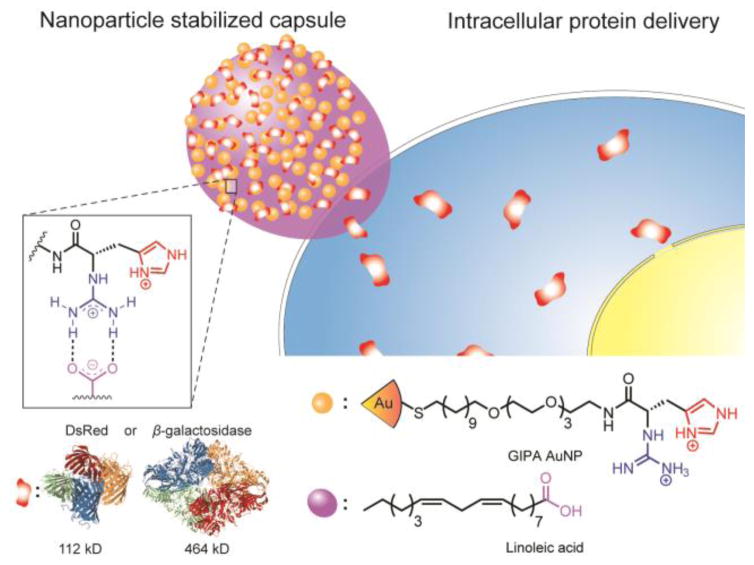
Membrane fusion is an alternative approach that conveys rapid release of cargo proteins into the cytosol by bypassing endosomal entrapment. 19 We have previously developed a nanoparticle-stabilized capsule (NPSC) platform for intracellular protein delivery through direct membrane fusion 20, 21 wherein the terminal functional group on the gold nanoparticles (AuNPs) is a tetrapeptide, His-Lys-Arg-Lys (HKRK). Efficient cytosolic release of proteins was observed as a result of dissociation from delivery carrier by live cell imaging. Despite their high efficiency, the tight binding of HKRK AuNPs to large proteins resulted in ineffective payload release into the cytosol (vide infra). We hypothesized that decreasing the overall charge of the terminal group on the AuNPs could reduce their interaction with proteins, thereby improving delivery efficiency for larger systems. We used 1-guanidino-2-(4-imidazole)propionic acid (GIPA, Fig. 1) as the terminal group of the AuNP ligand, providing effective cytosolic delivery of large proteins, including dsRed and β-galactosidase (β-gal), into the cytosol.
Fig. 1.
Schematic illustration of a new platform of intracellular protein (dsRed and β-galactosidase) delivery using GIPA AuNPs-stabilized capsule.
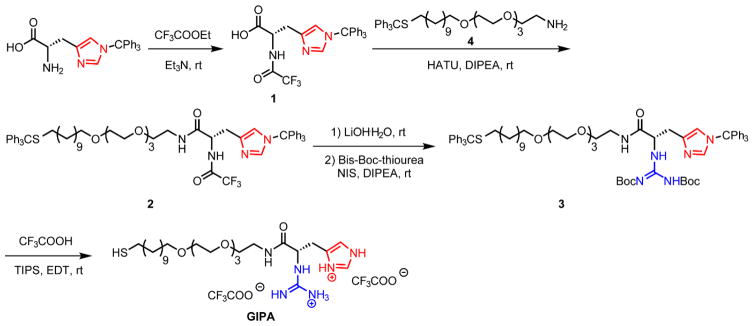
A key design parameters for NPSC formation is the presence of a guanidinium group to pin the particle to the fatty acid droplet through hydrogen bonding and electrostatic interactions. 22 The short peptide HKRK on the ligand terminal of our NPSC delivery platform also contains two lysine residues that increase positive charge density. As strong electrostatic interaction may result in inefficient release of proteins with high molecular weight, we truncated the structure of HKRK, leaving only imidazole and guanidine groups on the ligand. We synthesized the terminal based on a histidine derivative, adding the guanidine group to mimic a peptide terminal with reduced charge density (Fig. 2, ESI, Fig. S1–S8†). The imidazole residue on the histidine derivative provides a positive charge equally distributed between two nitrogen atoms at physiological pH. Moreover, it facilitates delivery due to a proton sponge function that promotes protein release if the payload is entrapped in endosomes.23 The guanidine group interacts with both the protein payload and the oil core to stabilize the NPSC structure.24
Fig. 2.
Synthesis of 1-guanidino-2-(4-imidazole)propionic acid-terminated (GIPA) ligand.
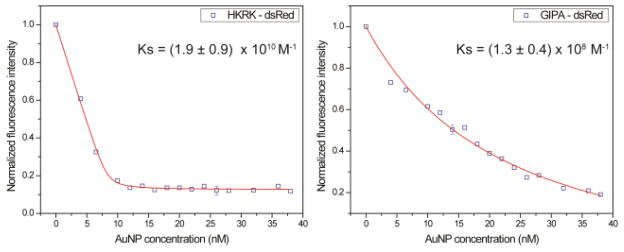
GIPA AuNPs were prepared using 2 nm AuNPs (ESI, Fig. S9, S10†) through a place-exchange reaction. From TEM results, there was no obvious difference in the core sizes before and after GIPA ligand exchange. The zeta potential of GIPA AuNPs was measured to determine the surface charge. As expected, although these AuNPs are positively charged (zeta potential: 15 ± 1 mV), their surface charge density was lower than HKRK AuNPs (zeta potential: 32 ± 1 mV).25 The reduced charge density of AuNPs would be expected to weaken the interaction of the NP with proteins. Since fluorescent proteins are typical models for easy tracking and demonstration of protein delivery without modification by fluorophores such as FITC, dsRed (a tetramer fluorescent protein with a molecular weight of 112 kD) 26 was titrated with GIPA AuNPs. When AuNPs bind with dsRed, the protein fluorescence is quenched 27 due to the energy transfer from the photo-excited fluorescent proteins to AuNPs.28,29 The titration30 results revealed that the binding constants of AuNPs to dsRed were different (Fig. 3). The binding constant (Ks) of HKRK AuNPs dsRed was (1.9 ± 0.9) × 1010 M−1, whereas the binding constant of GIPA AuNPs with dsRed was (1.3 ± 0.4) × 108 M−1.
Fig. 3.
Fluorescence titrations of AuNPs in the presence of fluorescent protein DS Red.
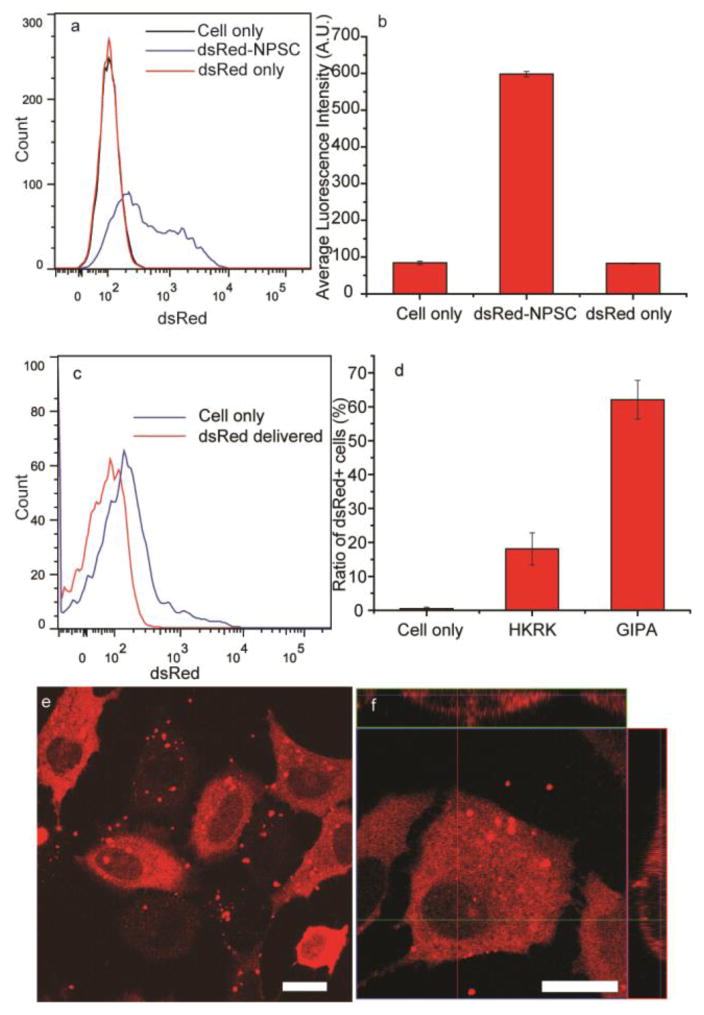
DsRed was likewise an advantageous protein to test the efficacy of GIPA NPSC-based protein delivery due to its strong fluorescence and large size. After the formation of dsRed-NPSCs, the overall capsule diameter was 130 ± 55 nm (ESI, Fig. S11a†), similar to previously reported values.20 After 24 hr, the capsule diameter was 150 ± 70 nm (ESI, Fig. S11b†), indicating no substantial change in size. The NPSCs were diluted with Dulbecco’s Modified Eagle Medium for cell culture experiments, and after 1 hr incubation, we measured the efficiency of dsRed delivery into cells. Flow cytometry results indicated that 65% cells were stained with dsRed (Fig. 4). The average fluorescence intensity was 7 times higher than control groups. In comparison, only 18% cells showed uptake of dsRed when delivered with HKRK NPSCs, indicating poor performance of this platform delivering proteins of large size. Notably, dsRed was observed evenly distributed throughout the cytosol, but not in nucleus (Fig. 4e). 3D image projections further confirmed this cytoplasmic distribution of dsRed (Fig. 4f), confirming the absence of punctate fluorescence indicative of endosomal entrapment. As proteins with molecular weight higher than 60 kD cannot diffuse passively into the nucleus,31 these results show that dsRed is in its native tetramer structure. In addition, after 24 hr culture following the delivery, no significant loss of cell viability was observed (ESI, Fig. S12†). Together these data demonstrate that GIPA AuNPs are able to efficiently deliver large proteins into the cytosol with retention of function, and presumably structure.
Fig. 4.
Delivery of dsRed to cytosol of HeLa cells. (a) Flow cytometry results of dsRed delivery by GIPA NPSCs. (b) Quantification of average fluorescence intensity of cells. (c) Flow cytometry results of dsRed delivery by HKRK NPSCs. (d) Quantification indicates GIPA NPSC has much higher efficiency for the delivery of dsRed. (e) LSCM image showing dsRed delivery into HeLa cells by GIPA NPSCs. f) Z-stack image of dsRed delivery. Scale bars: 20 μm.
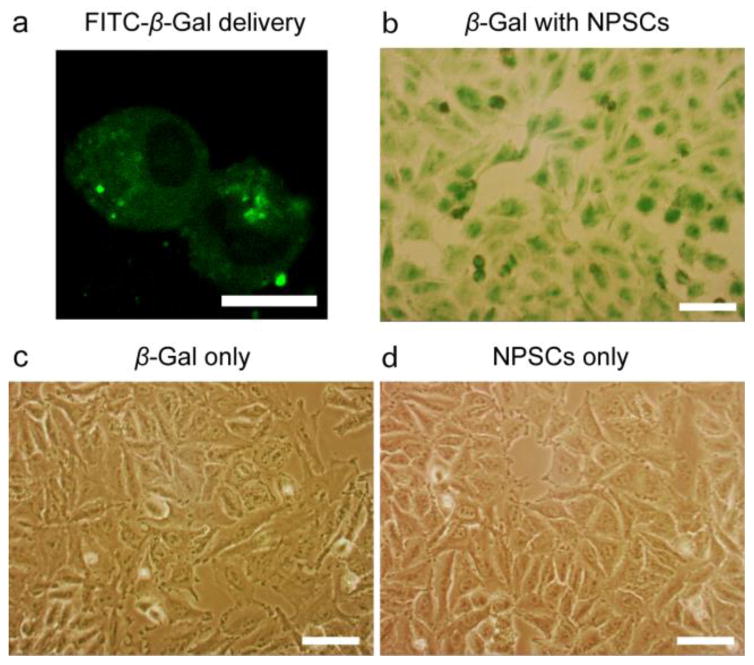
Delivery of enzymes into cells is a promising strategy for enzyme replacement and prodrug activation therapies. While we have demonstrated that caspase-3, an apoptotic enzyme of small size, can be rapidly delivered into the cytosol using a NPSC platform,20 the creation of delivery platform for enzymes of higher molecular weight would greatly expand the utility of this strategy. Rapid delivery of β-gal to the cytosol is a promising approach for efficient prodrug activation therapy in cancer cells,32,33 yet the large size of this enzyme (464 kD) in its tetrameric form is an obstacle for cytosolic access.18 We assessed whether the GIPA NPSC platform was capable of efficiently delivering β-gal into the cytosol in HeLa. The size of β-gal-GIPA NPSCs is 110 ± 50 nm when measured by dynamic light scattering (DLS; ESI, Fig. S13†). Delivery of fluorescein isothiocyanate labeled β-gal (FITC-β-gal) revealed the cytosolic but not nuclear distribution of the protein after 1 hr delivery (Fig. 5a), similar to that of dsRed. Due to its large size, β-gal cannot passively diffuse into nucleus. By delivering FITC-β-gal we were able to confirm that GIPA NPSCs efficiently deliver β-gal specifically into the cytosol. X-gal staining was then used to demonstrate retention of enzymatic activity of delivered β-gal. After delivery, the media was removed and cells were stained with X-gal for 4 hr. Notably, β-gal bound to GIPA NPSCs, but not β-gal alone, was efficiently delivered to the cell (Fig. 5 b–d). Thus, GIPA NPSC platforms are capable of delivering functional enzymes of large size into cells without hampering their function.
Fig. 5.
Distribution of β-gal in HeLa cells after delivery. (a) LSCM image showing FITC-β-gal delivery. Scale bar: 10 μm. (b) X-gal staining of delivered β-gal in HeLa cells. (c) X-gal staining of cells incubated with free β-gal alone. (d) X-gal staining of cells incubated with NPSCs alone. Scale bars: 100 μm.
Conclusions
We have developed an effective intracellular delivery strategy for proteins of high molecular weight using GIPA AuNPs stabilized capsules. By rational control of the interactions, payload proteins are rapidly delivered into the cytosol via a protein-GIPA NPSC complex. Both dsRed and β-galactosidase are effectively transduced into cells without hampering their functions. These studies demonstrate the use of supramolecular chemistry to tune the interaction between ligands of and proteins, providing a strategy for optimizing nanomaterial delivery vehicles.
Supplementary Material
Acknowledgments
This research was supported by NIH (GM077173).
Footnotes
Electronic Supplementary Information (ESI) available: Materials & Methods (ESI, Fig. S1–S13). See DOI: 10.1039/x0xx00000x
Notes and references
- 1.Kobsa S, Saltzman WM. Pediatr Res. 2008;63:513–519. doi: 10.1203/PDR.0b013e318165f14d. [DOI] [PubMed] [Google Scholar]
- 2.Zuris JA, Thompson DB, Shu Y, Guilinger JP, Bessen JL, Hu JH, Maeder ML, Joung JK, Chen ZY, Liu DR. Nat Biotechnol. 2015;33:73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peitz M, Pfannkuche K, Rajewsky K, Edenhofer F. Proc Natl Acad Sci USA. 2002;99:4489–4494. doi: 10.1073/pnas.032068699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou H, Wu S, Joo JY, Zhu S, Han DW, Lin T, Trauger S, Bien G, Yao S, Zhu Y, Siuzdak G, Schoeler HR, Duan L, Ding S. Cell Stem Cell. 2009;4:381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lorden ER, Levinson HM, Leong KW. Drug Deliv Transl Res. 2015;5:168–186. doi: 10.1007/s13346-013-0165-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrientos S, Brem H, Stojadinovic O, Tomic-Canic M. Wound Repair Regen. 2014;22:569–578. doi: 10.1111/wrr.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sassoon I, Blanc V. In: Antibody-Drug Conjugates. Ducry L, editor. Humana Press; Totowa, NJ: 2013. pp. 1–27. [DOI] [Google Scholar]
- 8.Peters C, Brown S. Biosci Rep. 2015;35:e00225. doi: 10.1042/BSR20150089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kan S-h, Aoyagi-Scharber M, Le SQ, Vincelette J, Ohmi K, Bullens S, Wendt DJ, Christianson TM, Tiger PMN, Brown JR, Lawrence R, Yip BK, Holtzinger J, Bagri A, Crippen-Harmon D, Vondrak KN, Chen Z, Hague CM, Woloszynek JC, Cheung DS, Webster KA, Adintori EG, Lo MJ, Wong W, Fitzpatrick PA, LeBowitz JH, Crawford BE, Bunting S, Dickson PI, Neufeld EF. Proc Natl Acad Sci USA. 2014;111:14870–14875. doi: 10.1073/pnas.1416660111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ardelt W, Shogen K, Darzynkiewicz Z. Curr Pharm Biotechnol. 2008;9:215–225. doi: 10.2174/138920108784567245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu Y, Sun W, Gu Z. J Controlled Release. 2014;194:1–19. doi: 10.1016/j.jconrel.2014.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu Z, Biswas A, Zhao M, Tang Y. Chem Soc Rev. 2011;40:3638–3655. doi: 10.1039/c0cs00227e. [DOI] [PubMed] [Google Scholar]
- 13.Sun W, Lu Y, Gu Z. Part Part Syst Charact. 2014;31:1204–1222. doi: 10.1002/ppsc.201400140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yeh YC, Tang R, Mout R, Jeong Y, Rotello VM. Angew Chem Int Ed. 2014;53:5137–5141. doi: 10.1002/anie.201400559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Räägel H, Säälik P, Pooga M. BBA-Biomembranes. 2010;1798:2240–2248. doi: 10.1016/j.bbamem.2010.02.013. [DOI] [PubMed] [Google Scholar]
- 16.Stewart MP, Lorenz A, Dahlman J, Sahay G. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2016;8:465–478. doi: 10.1002/wnan.1377. [DOI] [PubMed] [Google Scholar]
- 17.Erazo-Oliveras A, Muthukrishnan N, Baker R, Wang TY, Pellois JP. Pharmaceuticals. 2012;5:1177–1209. doi: 10.3390/ph5111177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brodin JD, Sprangers AJ, McMillan JR, Mirkin CA. J Am Chem Soc. 2015;137:14838–14841. doi: 10.1021/jacs.5b09711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watabe A, Yamaguchi T, Kawanishi T, Uchida E, Eguchi A, Mizuguchi H, Mayumi T, Nakanishi M, Hayakawa T. BBA-Biomembranes. 1999;1416:339–348. doi: 10.1016/s0005-2736(98)00238-7. [DOI] [PubMed] [Google Scholar]
- 20.Tang R, Kim CS, Solfiell DJ, Rana S, Mout R, Velazquez-Delgado EM, Chompoosor A, Jeong Y, Yan B, Zhu ZJ, Kim C, Hardy JA, Rotello VM. ACS Nano. 2013;7:6667–6673. doi: 10.1021/nn402753y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ray M, Tang R, Jiang Z, Rotello VM. Bioconjugate Chem. 2015;26:1004–1007. doi: 10.1021/acs.bioconjchem.5b00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wright AT, Griffin MJ, Zhong ZL, McCleskey SC, Anslyn EV, McDevitt JT. Angew Chem Int Ed. 2005;44:6375–6378. doi: 10.1002/anie.200501137. [DOI] [PubMed] [Google Scholar]
- 23.Midoux P, Pichon C, Yaouanc JJ, Jaffres PA. Br J Pharmacol. 2009;157:166–178. doi: 10.1111/j.1476-5381.2009.00288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang XC, Samanta B, Agasti SS, Jeong Y, Zhu ZJ, Rana S, Miranda OR, Rotello VM. Angew Chem Int Ed. 2011;50:477–481. doi: 10.1002/anie.201005662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghosh P, Yang X, Arvizo R, Zhu ZJ, Agasti SS, Mo Z, Rotello VM. J Am Chem Soc. 2010;132:2642–2645. doi: 10.1021/ja907887z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lounis B, Deich J, Rosell FI, Boxer SG, Moerner WE. J Phys Chem B. 2001;105:5048–5054. [Google Scholar]
- 27.Mout R, Tonga GY, Ray M, Moyano DF, Xing Y, Rotello VM. Nanoscale. 2014;6:8873–8877. doi: 10.1039/c4nr02097a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang Z, Le NDB, Gupta A, Rotello VM. Chem Soc Rev. 2015;44:4264–4274. doi: 10.1039/c4cs00387j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saraswat S, Desireddy A, Zheng D, Guo L, Lu HP, Bigioni TP, Isailovic D. J Phys Chem C. 2011;115:17587–17593. [Google Scholar]
- 30.You CC, De M, Han G, Rotello VM. J Am Chem Soc. 2005;127:12873–12881. doi: 10.1021/ja0512881. [DOI] [PubMed] [Google Scholar]
- 31.Dingwall C, Laskey RA. Annu Rev Cell Biol. 1986;2:367–390. doi: 10.1146/annurev.cb.02.110186.002055. [DOI] [PubMed] [Google Scholar]
- 32.Legigan T, Clarhaut J, Tranoy-Opalinski I, Monvoisin A, Renoux B, Thomas M, Le Pape A, Lerondel S, Papot S. Angew Chem Int Ed. 2012;51:11606–11610. doi: 10.1002/anie.201204935. [DOI] [PubMed] [Google Scholar]
- 33.Tietze LF, Krewer B. Chem Biol Drug Des. 2009;74:205–211. doi: 10.1111/j.1747-0285.2009.00856.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.