Significance
We used population genomic analyses to reconstruct the recent history and dispersal of a major clade of Mycobacterium tuberculosis in central Asia and beyond. Our results indicate that the fall of the Soviet Union and the ensuing collapse of public health systems led to a rise in M. tuberculosis drug resistance. We also show that armed conflict and population displacement is likely to have aided the export of this clade from central Asia to war-torn Afghanistan and beyond.
Keywords: Mycobacterium tuberculosis, evolution, antimicrobial resistance, phylodynamic analysis, tip-dating
Abstract
The “Beijing” Mycobacterium tuberculosis (Mtb) lineage 2 (L2) is spreading globally and has been associated with accelerated disease progression and increased antibiotic resistance. Here we performed a phylodynamic reconstruction of one of the L2 sublineages, the central Asian clade (CAC), which has recently spread to western Europe. We find that recent historical events have contributed to the evolution and dispersal of the CAC. Our timing estimates indicate that the clade was likely introduced to Afghanistan during the 1979–1989 Soviet–Afghan war and spread further after population displacement in the wake of the American invasion in 2001. We also find that drug resistance mutations accumulated on a massive scale in Mtb isolates from former Soviet republics after the fall of the Soviet Union, a pattern that was not observed in CAC isolates from Afghanistan. Our results underscore the detrimental effects of political instability and population displacement on tuberculosis control and demonstrate the power of phylodynamic methods in exploring bacterial evolution in space and time.
The Mycobacterium tuberculosis complex (MTBC) comprises seven main lineages. Of these, lineages 2, 3, and 4 are found across most of the globe, but their regional distribution varies and reflects historical and recent human population movements. L2 (“L2” and “Beijing lineage” are used interchangeably throughout the text) has a southeast Asian (1) or east Asian (2) origin and has received considerable attention as it is spreading globally (3), may be associated with accelerated progression of disease (4, 5), and is associated with increased antibiotic resistance (5). It also has been suggested that L2 exhibits an elevated mutation rate relative to other M. tuberculosis (Mtb) lineages, but studies have yielded differing results in this regard (6, 7).
There is no consensus in the literature regarding the age of the MTBC and its main lineages, and different studies have approached this question using distinct strategies. One such approach, the “out of Africa” hypothesis, is based on the assumption of codivergence of Mtb with its human host (1, 8), and suggests that the most recent common ancestor (MRCA) of extant Mtb existed roughly 40,000–70,000 y ago, with the bacillus subsequently spreading globally with human migrations out of Africa (9, 10). In contrast, the two studies that relied on genomic sequence data using ancient DNA (aDNA) analysis point to a 10-fold younger origin, around 6,000 y ago (11, 12). Even though calibration with aDNA is becoming the gold standard for dating evolutionary events, few noncontemporaneous MTBC genomes are available at present. One previous study relied on ∼1,000-y-old M. pinnipedii isolates, an animal-associated MTBC strain (11). A second study relied on Mtb sensu stricto genomes for calibration, but the isolates were only 200–250 y old (12). These two studies yielded similar rate estimates, despite including data from very different time periods. The substitution rate estimates of ∼5 × 10−8 substitutions/site/year (s/s/y) obtained in these aDNA studies are slightly lower than estimates from epidemiologic studies and other studies based on contemporaneous sampling, all of which produced rate estimates around 0.7 × 10−7 – 1.3 × 10−7 s/s/y, corresponding to 0.3–0.5 substitutions/genome/year (6, 13–18).
The origin and spread of the Beijing lineage have also been vigorously debated. According to a recent phylogeographic analysis of L2 genomes, the lineage emerged in Southeast Asia some 30,000 y ago and subsequently spread to northern China, where it experienced a massive population expansion, purportedly related to the Neolithic expansion of the Han Chinese population (1). The 30,000 y age was obtained by extrapolating from the aforementioned 70,000 y age of the MTBC. Another attempt to reconstruct the age and evolutionary history of L2 and its clonal complexes (CCs), based on a massive global collection of mycobacterial interspersed repetitive unit (MIRU) genotyping data complemented with genome sequencing, resulted in an age of approximately 6, 600 y for the whole lineage and 1,500–6,000 y for each of the CCs (2). However, this study also relied on strong assumptions, particularly concerning the underlying mutation model and mutation rate of the MIRU markers (2, 10).
Until recently, fine-scaled phylodynamic and phylogeograpic methods were applied mainly to rapidly evolving taxa, such as RNA viruses (19). The increased availability of whole-genome sequences has shifted the limits of measurably evolving pathogens to also encompass bacteria (20), including Mtb (13, 21), despite its relatively slow substitution rate compared with most other bacterial pathogens (22). Here we applied phylodynamic methods, calibrated with sampling dates (tip-dating), to a collection of Mtb isolates from Europe and southeast and central Asia. The isolates belong to a L2 clade that we term the central Asian clade (CAC). The CAC corresponds to the MIRU-defined CC1 (2) and includes the Russian clade A (23). The isolates included in the study cover a sampling period of 15 y, and even though we did not attempt to reconstruct the age of Mtb or L2 as a whole, our dated CAC phylogeny points to a nearly 100-fold younger origin of the lineage than was previously estimated.
We also show that the evolution and dispersal of the CAC in Eurasia have been shaped by recent historical events. Specifically, we find that being an ex-Soviet state is a major risk factor for high prevalence of multidrug-resistant tuberculosis (MDR-TB), and that this pattern also holds true within the CAC. We were able to trace the introduction of this clade to Afghanistan during the 1979–1989 Soviet–Afghan war and document its subsequent spread across Europe after migration events in the wake of recent armed conflicts. Our results highlight the detrimental effects of political instability and population displacement on global TB control and demonstrate the power of phylodynamic methods for understanding bacterial evolution in time and space.
Results and Discussion
Defining the CAC and the Afghan Strain Family.
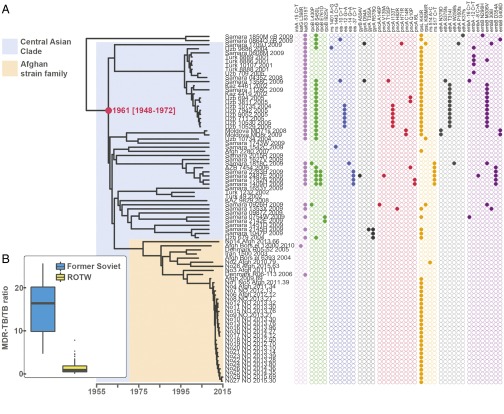
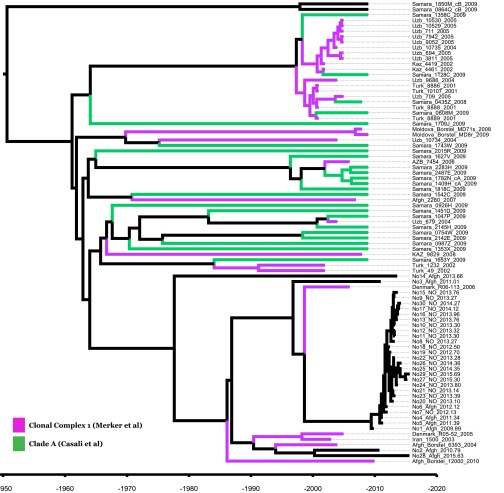
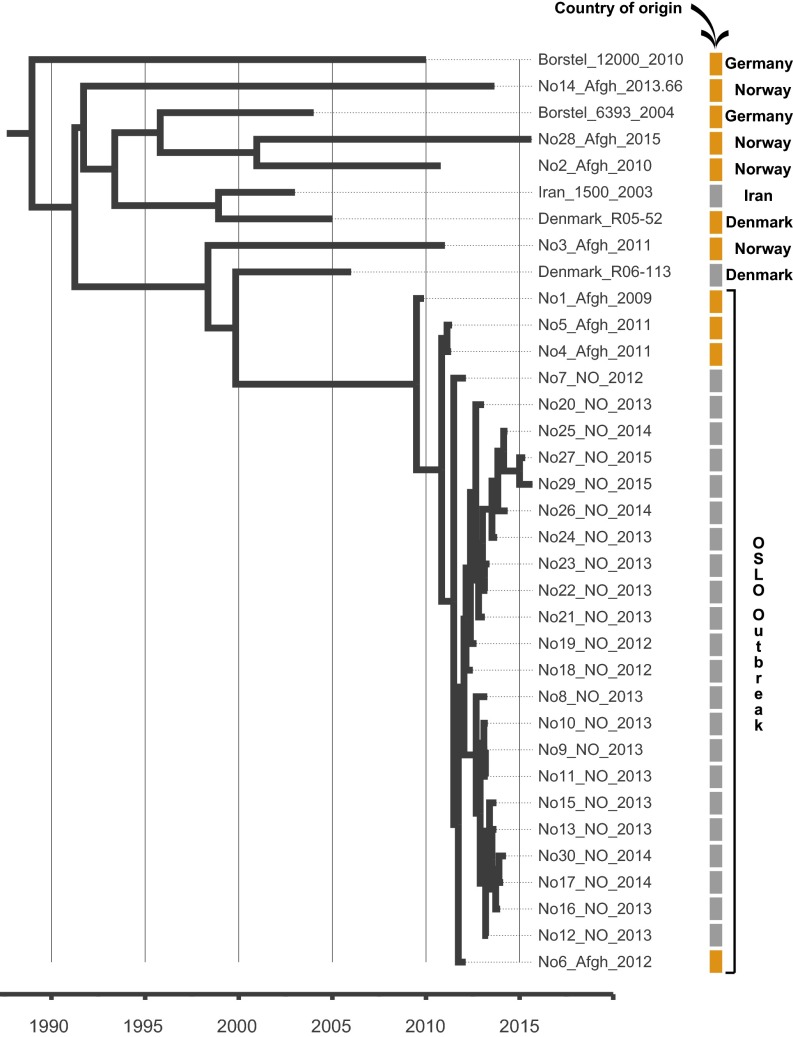
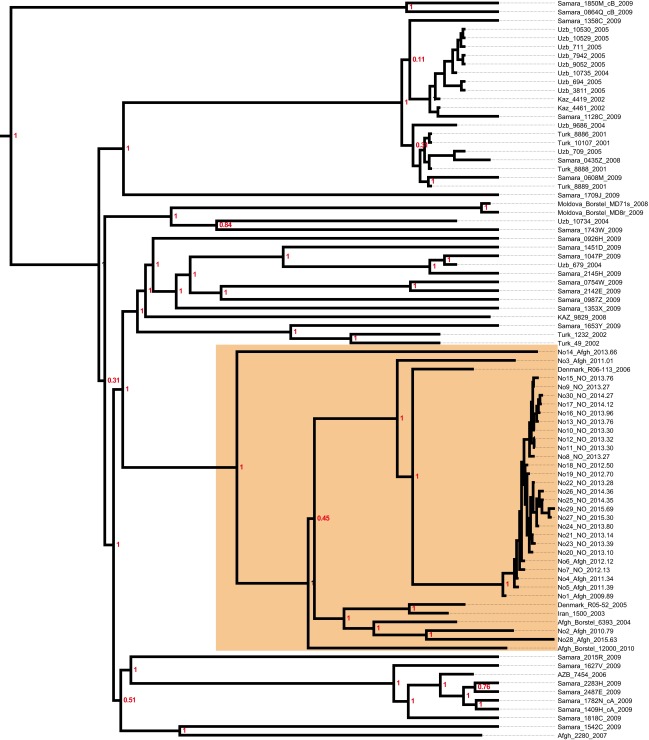
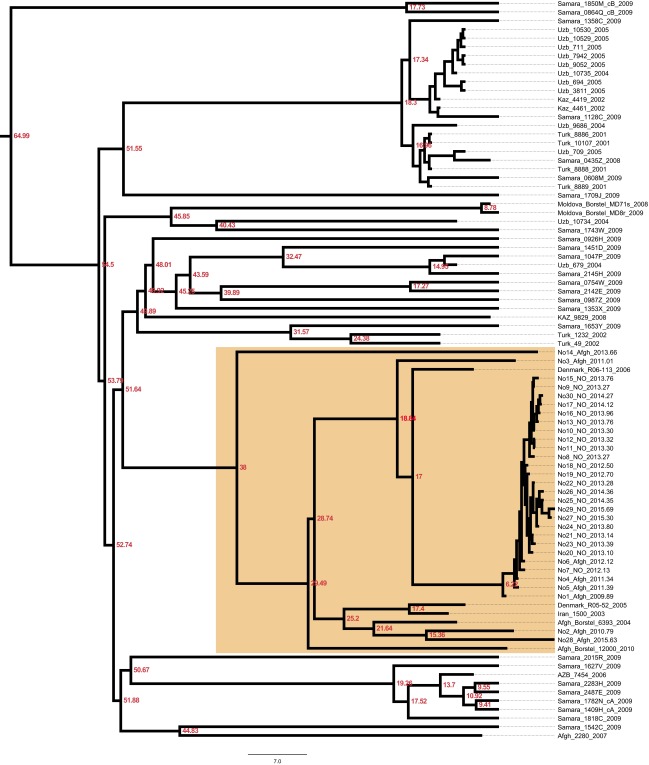
To investigate the recent history and spread of an Mtb L2 clade associated with Afghan refugees in Norway, Mtb genomes from a recent large TB outbreak affecting mainly Norwegian and Afghan nationals in Oslo, Norway were included in the study, along with related isolates from Norway, Denmark, Germany, and Moldova. Sequence data from two other relevant studies (2, 23) were included as well. A whole-genome SNP phylogeny was constructed as described in Materials and Methods. It was clear that the Oslo outbreak belongs to an Afghan strain family (ASF) (Fig. 1A, orange highlighting). This ASF belongs to a larger clade that includes the aforementioned clade A from Russia (23) and the CC1 isolates from a recent global study (2) (Fig. 1, blue highlighting). Interestingly, Casali et al. (23) noted that clade A isolates were consistently found at a greater frequency east of the Volga River, a natural border between Russian Europe to the west and Russian central Asia to the east, whereas the other dominant clade in Russia, clade B, was more frequent west of the river. Thus, we term this clade, which encompasses the previously defined clade A and CC1 isolates (2, 23), the CAC (Fig. 1A).
Fig. 1.
Phylogenetic placement and antibiotic resistance of Mtb isolates in the study. (A) Bayesian dated phylogeny of the CAC. The ASF and the CAC to which it belongs are highlighted in orange and blue, respectively. Filled dots indicate the presence of mutations colored by the compound to which they are known or predicted to confer resistance (magenta, isoniazid; purple, rifampicin; blue, kanamycin; green, fluoroquinolones; yellow, pyrazinamide; orange, streptomycin; red, ethionamide; gray, ethambutol). The TMRCA of the CAC lineage is indicated in red. Two clade B isolates (23) served as outgroups. (B) Relative prevalence of multidrug-resistant TB (MDR-TB) stratified by a history of Soviet Union allegiance (blue, ex-Soviet states; yellow, rest of the world).
The Fall of the Soviet Union and the Rise of MDR-TB.
Mapping of known and putative resistance mutations on the phylogeny revealed that isolates originating in central Asia were strongly enriched in resistance mutations relative to isolates of Afghan origin (Fig. 1A). The countries in central Asia were all part of the Soviet Union until its fall in 1991. To investigate geographic patterns of drug resistance in a wider context, we calculated relative MDR-TB prevalence (MDR-TB = Mtb resistant to first-line drugs isoniazid and rifampicin) in all countries of the world for which appropriate data were available. The countries were divided into two groups: ex-Soviet states and the rest of the world. Even though it is widely acknowledged that MDR-TB represents a particularly acute problem in many ex-Soviet countries, the strength of the association remains striking (W = 2,577; P < 0.001, Wilcoxon rank-sum test) (Fig. 1B).
To examine in more detail whether our CAC data support a role of the fall of the Soviet Union in the rise of resistance within the clade, we mapped individual resistance mutations to nodes in the dated phylogeny. From this phylogeny, it is clear that the vast majority of transmitted resistance mutations evolved in the years after the collapse of the Soviet Union (Fig. S1). Taken together, these findings support the notion that external factors, namely the fall of the Soviet Union and the ensuing breakdown of public health systems and drug stewardship, rather than features specific to the Beijing lineage, are to blame for high rates of drug resistance in former Soviet states.
Fig. S1.
Timed phylogeny with resistance mutations mapped to nodes. Only mutations present in at least two isolates were mapped. The colored dots at the bottom correspond the timing of individual events of resistance emergence in the phylogeny.
A Recent Origin of the CAC.
To investigate the temporal evolution and spread of the CAC and the ASF in detail, we performed Bayesian phylogenetic analyses using BEAST 1.8 (24) with tip-dates (sampling dates) for temporal calibration. We investigated root-to-tip distance as a function of sampling time and used tip-randomization to assess the strength of the temporal signal in the data (Materials and Methods). Both tests revealed a strong temporal signal in the data. Bayesian phylogenetic analyses using various clock and demographic models on various sample subsets resulted in similar ages of the MRCAs of both the CAC and the ASF (Table 1).
Table 1.
Estimated TMRCA for the CAC and ASF
| Sample | Substitution model | Demographic model | CAC TMRCA (95% HPD) | ASF TMRCA (95% HPD) | Substitution rate (×10−7) |
| ASF | HKY | Skyride | 1988 (1980–1996) | 3.13 (1.86–4.44) | |
| ASF | HKY | Exponential | 1987 (1977–1995) | 3.24 (2.03–4.48) | |
| ASF | HKY | Expansion | 1987 (1975–1995) | 3.26 (1.90–4.47) | |
| ASF | HKY | Logistic | 1987 (1977–1995) | 3.28 (2.06–4.56) | |
| ASF | HKY | Constant | 1986 (1975–1995) | 3.15 (1.88–4.44) | |
| CAC ÷ Samara | GTR | Skyride | 1957 (1935–1976) | ND | 2.42 (1.42–3.43) |
| CAC ÷ Samara | GTR | Constant | 1955 (1932–1974) | ND | 2.56 (1.57–3.61) |
| Representatives* | HKY | Constant | 1971 (1955–1984) | 1987 (1978–1994) | 3.78 (2.33–5.26) |
| Representatives* | HKY | Skyride | 1968 (1952–1983) | 1983 (1971–1993) | 3.34 (2.00–4.79) |
| CAC | GTR | Skyride | 1961 (1948–1972) | 1977 (1967–1986) | 2.79 (2.10–3.54) |
| CAC | GTR | Skyride (RC) | 1961 (1945–1975) | 1975 (1961–1987) | 2.80 (1.91–3-75) |
| CAC | HKY | Skyride | 1960 (1949–1972) | 1977 (1967–1986) | 2.78 (2.10–3.50) |
| CAC | GTR | Constant | 1950 (1932–1966) | 1973 (1960–1985) | 2.30 (1.63–3.01) |
| CAC | HKY | Constant | 1950 (1932–1966) | 1973 (1960–1984) | 2.31 (1.64–3.02) |
Estimates reported in the text are highlighted in bold type. Unless stated otherwise, a strict clock was used for all of the analyses. ND, not determined; RC, relaxed clock. Detailed information on the models and sample sets tested are provided in Materials and Methods.
Maximum of one isolate included per year per patient country of origin.
Our estimated time of the MRCA (TMRCA) of the CAC is 1961 CE [95% highest posterior density (HPD), 1948–1972 CE], which deviates considerably from a previous study based on MIRU data that estimated that the Beijing lineage CC1 is 4,415 y old (95% HPD, 2,569–7,509 y) (2). The CC1 isolates all fall within the CAC in our phylogeny (Fig. S2), and we thus expect the TMRCA of the CC1 to be less than or equal to the TMRCA of the CAC. The TMRCA estimates of CC1 were based on a mean MIRU mutation rate per year of 10−4 (2, 10).
Fig. S2.
Placement of CC1 and clade A isolates within the CAC phylogeny.
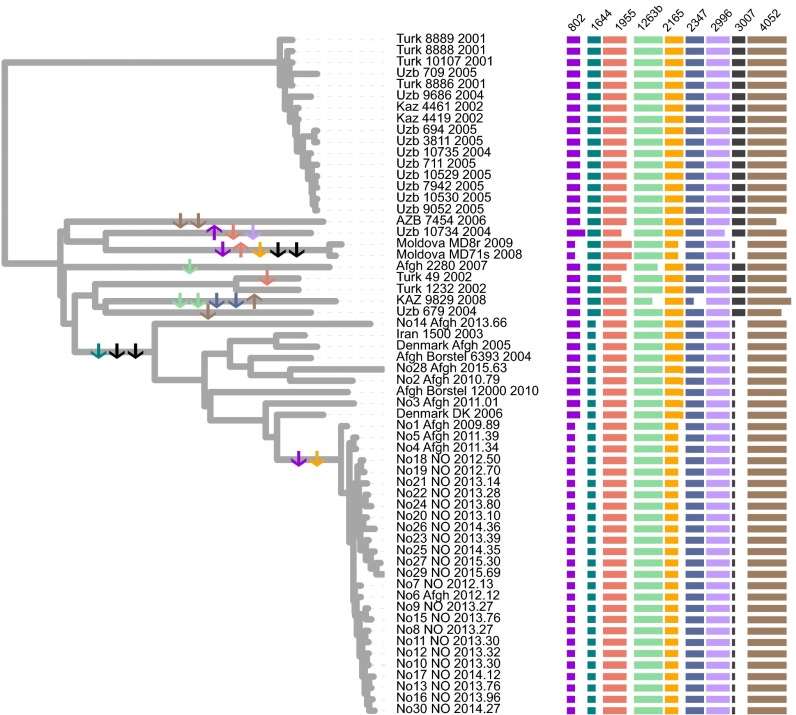
To investigate the mean MIRU evolutionary rate in our samples, we first constructed a tip-dated genome phylogeny including isolates for which MIRU data were available—that is, all isolates except those from Samara (23). We found that the total branch length of this phylogeny, corresponding to the total evolutionary time (y) elapsed, was 593 y [95% confidence interval (CI), 365–821 y]. Subsequently, we annotated and counted repeat expansion and contraction events (Fig. 2). Only 9 of the 24 MIRU loci had undergone any changes in repeat number among the sampled isolates. Across all 24 loci, we calculated a mean per-locus MIRU mutation rate of 1.62 × 10−3 (95% CI, 1.17 × 10−3 to 2.63 × 10−3 mutations/locus/y) (Dataset S3). This rate is ∼15-fold higher than the rate used in the previous study.
Fig. 2.
MIRU repeat changes mapped on whole-genome tip-dated phylogeny. Changes in repeat number relative to MtbC15-9 94–32 of nine variable MIRUs loci annotated on the right. Individual state change events are indicated by arrows in the phylogeny. The arrows are colored to match the color of individual MIRU loci, and the direction of the arrows indicates repeat expansion (up) or contraction (down). To the right, the lengths of the horizontal bars indicate repeat numbers for individual loci.
Nonetheless, this estimate for MIRU markers is in line with other recent rate estimates based on whole-genome sequencing of serial Mtb isolates from Macaque monkeys and model-based Bayesian estimates (25, 26). Also of note is the number of homoplasies in the MIRU data. Out of a total of 23 repeat gain/loss events, 7 occurred twice on independent occasions (i.e., on different branches) and thus are homoplasies; that is, 14 of the 23 events were homoplasic events. Furthermore, we observed five occasions of apparent simultaneous loss of two repeats, which are more parsimoniously explained by mutations involving two tandem repeats. Although the possibility of stepwise loss in unsampled strains cannot be ruled out, these findings suggest that MIRU evolution does not follow a strict stepwise mutation model as assumed previously (2), but might be better modeled applying a slipped-strand model that allows for the simultaneous insertion or deletion of multiple repeats (27). Taken together, our observations suggest that MIRU data might not be ideal for evolutionary inference over long time scales.
Spread of the CAC: The Role of Armed Conflict and Population Displacement.
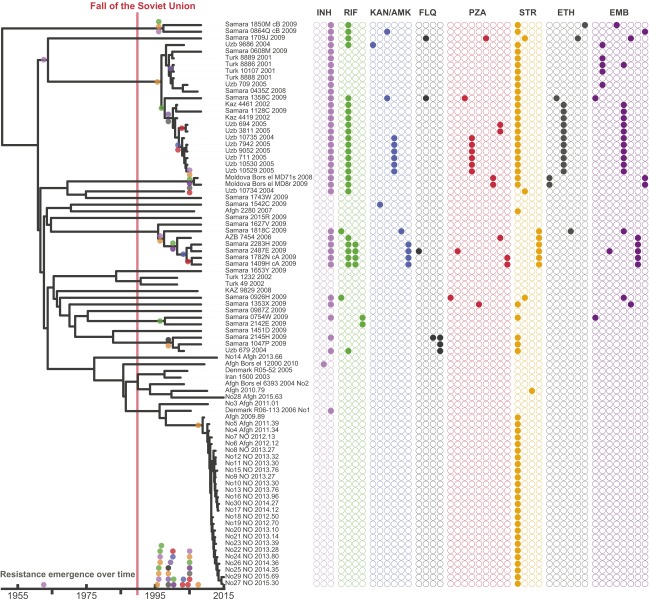
Our TMRCA estimates suggest that the CAC was introduced to Afghanistan from Soviet central Asia coincident with the 1979–1989 Soviet occupation of the country (Table 1). A dated phylogeny including only isolates belonging to the ASF revealed that, apart from the Oslo outbreak, individual isolates generally represented isolated TB cases among Afghan refugees in Europe. All cases had been diagnosed between 2003 and 2015, and (again excluding the Oslo outbreak) the isolates were situated on long terminal branches stretching 5–20 y back in time (Fig. 3). These observations suggest that these TB cases represent multiple individual introductions of the strain to Europe with Afghan refugees in the wake of the continued violent conflicts in the country. The long terminal branches are consistent with reactivation of latent disease in refugees, which in one case was followed by a local outbreak in Norway, identifiable by very short terminal branches (Fig. 3).
Fig. 3.
Bayesian evolutionary phylogeny of the ASF. Colored bars indicate country of origin of the patient: orange, Afghanistan; gray, other countries. The country of isolation is annotated on the right.
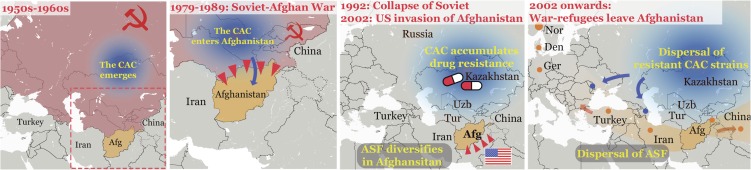
When interpreting our phylogenetic analyses in the light of historic events in the region, it appears that armed conflict has played a major role both in introducing the CAC to Afghanistan (Soviet invasion) and in the subsequent repeated export of the clade with Afghans fleeing the country in the wake of the American invasion in 2001. A hypothetical scenario for the spread of the CAC and the ASF in time and space is presented in Fig. 4.
Fig. 4.
A scenario for the spread of the CAC and ASF in time and space. Color shading and arrows indicate the emergence and spread of the CAC (blue) and ASF (orange). Dots represent cases or clusters of cases belonging to either the CAC or the ASF based on genome sequences, except the cases in Turkey, China, and Tajikistan, for which only MIRU data were available. Red shading of countries is used to indicate membership in the Soviet Union. Red triangles symbolize armed invasion. Afg, Afghanistan; Den, Denmark; Ger, Germany; Nor, Norway; Tur, Turkmenistan; Uzb, Uzbekistan.
Substitution Rates Over Time.
The origin and subsequent evolutionary history of Mtb have been subjects of much debate (1, 9, 11, 12). It has been suggested that a high degree of congruence between human and Mtb phylogenies supports a scenario of codivergence for the two organisms, and thus that the age of the MRCA of Mtb mirrors the timing of the migrations of anatomically modern humans out of Africa approximately 40,000–70,000 y ago (9). However, another study failed to identify such a congruence in phylogenies and did not find support for a codivergence scenario when using a host of formal tests (16). When it comes to the Beijing lineage, age estimates range from approximately 6,000 y (2, 9) to 30,000 y (1); however, the two studies using aDNA to calibrate MTBC phylogenies both estimated an age of approximately 6,000 y for the TMRCA of all extant Mtb (11, 12).
We estimated a substitution rate for the CAC of 2.79 × 10−7 s/s/y (95% HPD, 2.10 × 10−7−3.54 × 10−7), resulting in a TMRCA estimate of roughly 1961 (95% HPD, 1948–1972). The age of a CC corresponding to the CAC (CC1) was previously estimated as ∼4,000 y (2). The discrepancy between this estimate and the age of ∼55 y obtained here by tip-date calibration is striking; however, across all of the sampling subsets and clock and demographic models tested, the lowest bottom 95% HPD to the highest upper 95% HPD restricts the TMRCA of the CAC to the years 1932–1984. This time span also fits well with the estimated age of resistance mutations (Fig. S1), whereas these would likely predate the introduction of antibiotics if the CAC were thousands of years old.
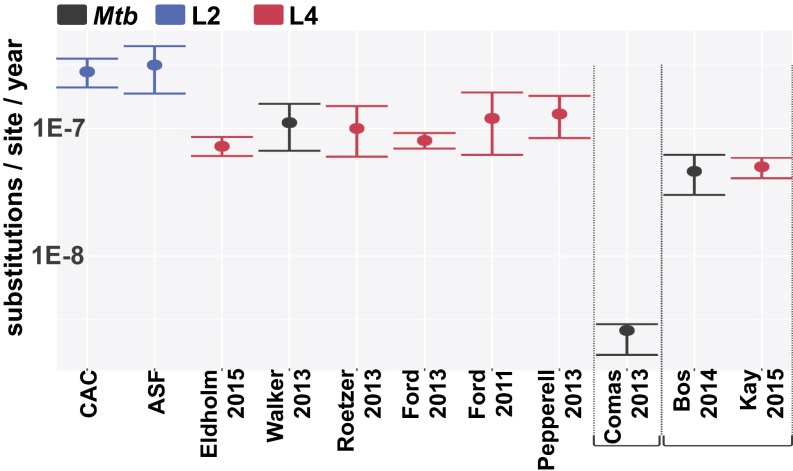
The substitution rate estimated for the CAC is slightly higher than previous rates estimated in studies of modern, heterochronous samples, but well within the margin of error for estimates obtained in comparable studies (Fig. 5). Interestingly, the other lineage-specific tip-dated rate estimates were all obtained for lineage 4 isolates, and thus it is possible that the higher rate obtained for the CAC (L2) in the present study might reflect an intrinsically higher mutation rate for L2 lineages (6). The similarity between rates from contemporaneous studies and the two studies using aDNA for temporal calibration is also striking, even if both Mtb aDNA studies point to slightly lower substitution rates. This difference may reflect time dependency in substitution rate estimates, owing to the fraction of slightly deleterious mutations eliminated over longer periods (28). A parallel observation of moderately decreased substitution rate estimates when older samples are included in the analysis also has been observed in mitochondrial genomes (29) and in the agent of the plague, Yersinia pestis (30).
Fig. 5.
Estimated Mtb substitution rates in the present study and previously published studies. Colors indicates the lineage to which the samples under study belong: blue, lineage 2; red, lineage 4; black, all). Studies using aDNA (11, 12) and human-Mtb codivergence (9) for calibration are annotated separately. The other studies used tip dating (6, 13, 14) or historical information (16), or counted mutations in paired isolates (15) or serial isolates (17).
That being said, although time dependency is a genuine and general phenomenon, the effect seems to be relatively subtle in Mtb (31), and seemingly incompatible with the extreme acceleration in substitution rates in recent times that would have to be invoked to reconcile these studies with 40,000–70,000 y age for Mtb generated under the ancient out of Africa scenarios (9). All current studies based both on ancient and modern samples in which substitution rates were directly inferred from the data support the notion that the MRCA of Mtb circulating today existed ∼6,000 y ago; however, this does not rule out the possibility that TB is a more ancient disease, as has been suggested by archeological studies (32, 33). Indeed, the MRCA of currently extant Mtb strains could be younger than that of TB as a result of a clonal replacement in the global Mtb population. It is also possible that the disease resembling TB in the archeological record was caused by an organism other than what is currently identified as Mtb.
Materials and Methods
Samples.
A total of 85 isolates were included in the study. Detailed information on the sampling scheme and samples is provided in SI Materials and Methods and in Datasets S1 and S2. Ethical approval was not required as the study was initiated within the legal mandate of the Norwegian Institute of Public Health (NIPH) to investigate and report on infectious disease outbreaks.
Calling SNPs.
Genomic DNA isolation and preparation of sequencing libraries was performed at the Norwegian Institute of Public Health following a published protocol (34), except using the Kapa HyperPlus Library Preparation Kit (Kapa Biosystems) rather than the Kapa High-Throughput Library Preparation Kit for DNA fragmentation and library preparation. All sequencing reads were paired end (read length 100–250 bp) and had been generated on the Illumina platform (NextSeq 500, HiSeq, or MiSeq). Reads were mapped against the M. tuberculosis H37rv genome (NC_000962.3) using SeqMan NGen (DNASTAR), resulting in a median alignment depth ranging from 25× to 719× for individual isolates. SNPs in or within 50 bp of regions annotated as PE/PPE genes, mobile elements, or repeat regions were excluded from all analyses. Heterozygous SNPs that were found at a frequency of 10–90% of reads in at least one isolate also were excluded. Finally, for inclusion of SNPs in our downstream analyses, a minimum depth of 10 reads in one strain and at least four reads in all strains was required.
Phylogenetic Evolutionary Inferences and Testing of Tip-Based Calibration.
A total of 1,212 concatenated genome-wide SNPs were used for phylogenetic analyses (Dataset S4). Based on Bayesian information criterion in jmodeltest2 (35), GTR was the best-fitting substitution model for the CAC and ASF datasets. Dated phylogenies, divergence times, and evolutionary rates were computed from the alignments using BEAST 1.8 (36). On observing that the BEAST chains (see below) failed to converge using the GTR model on the ASF dataset, we applied the HKY model (a simpler and the second-highest scoring model) for this subset and the GTR model for the other datasets. The XML-input file was modified to specify the number of invariant sites. SNPs were partitioned into three classes based on functional annotation: intergenic SNPs, synonymous SNPs, and nonsynonymous + noncoding RNA SNPs. Phylogenetic trees were visualized using Figtree v1.4.2 (tree.bio.ed.ac.uk/software/figtree) and ITOL v2 (37).
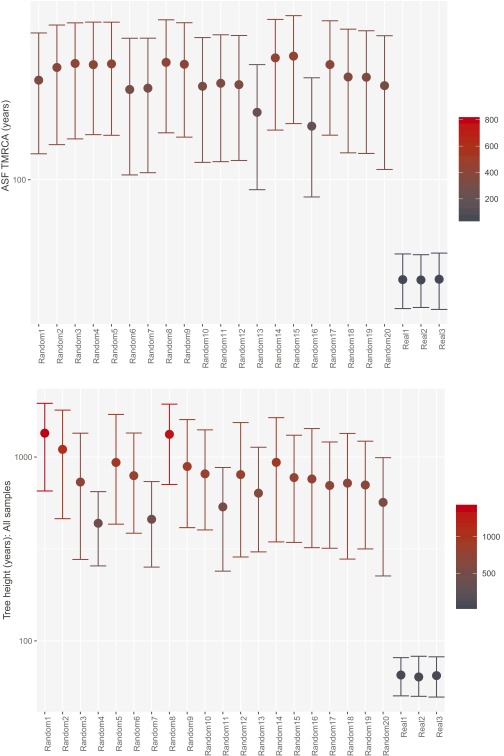
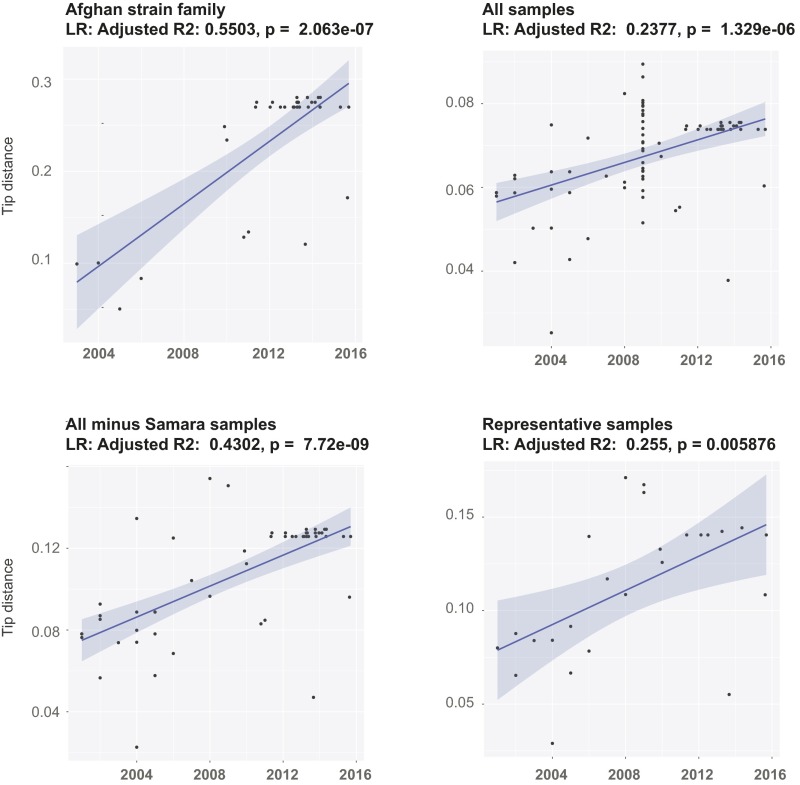
Maximum likelihood trees were computed in SeaView (38), and root-to-tip distances were extracted using Path-O-Gen (tree.bio.ed.ac.uk/software/pathogen/). As a complementary assessment of the temporal signal in the data, date randomization was performed on our datasets using a recently developed R package (39). Sampling dates of the genomes were randomly shuffled 20 times, and date-randomized datasets were analyzed with BEAST using the same parameters as for the original datasets (Fig. S3). For both datasets, there was no overlap between the TMRCA 95% HPD intervals between the real data and the randomized data (Fig. S3), suggesting that the data contain sufficient temporal structure and spread (40). Root-to-tip regression also revealed a clear temporal signal in the data (Fig. S4).
Fig. S3.
Calculated TMRCA of the ASF (Upper) and tree height including all isolates (Lower) after tip randomization. Error bars cover the 95% HPD.
Fig. S4.
Root-to-tip regression analyses on various sample sets.
To ensure that the estimates were not driven by any particular sample subset, we ran a root-to-tip regression on a subset of samples including a maximum of one sample per year per country of patient origin (the “representatives” sample subset). Estimated node ages and substitution rates were largely concordant between sample subsets, indicating that nonrandom sampling did not significantly affect the results overall (Table 1).
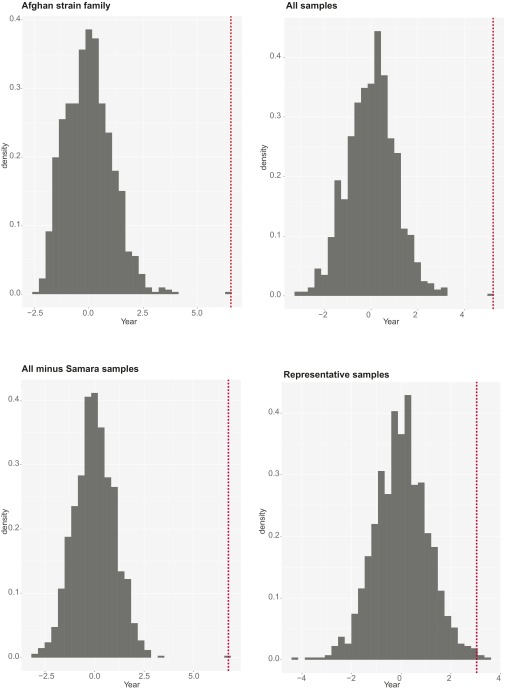
Finally, to assess the robustness of our root-to-tip regressions, we also randomized the tip distances 1,000 times, reran the regression analyses, and recorded the t statistics for the variable “year” for each iteration (Fig. S5). For each of the four sample sets, the estimate from observed data differed significantly from that using randomized data (P < 0.005 for all eight analyses).
Fig. S5.
Root-to-tip randomization test results. The x-axis denotes t values of the randomized tip distances. The red dotted lines indicate the original nonrandomized estimates.
We calibrated the trees using sampling dates spanning the years 2002–2015. We defined uniform prior distributions for the substitution rates (1 × 10−9 – 1 × 10−6 s/s/y), and assessed the performance of various clock and demographic models using stepping-stone sampling (Table S1). The GMRF Skyride demographic model (41) was found to fit both the ASF and CAC data best. A strict clock model was found to fit the ASF best, whereas an uncorrelated relaxed clock model scored highest on the CAC dataset. However, because the Markov chain Monte Carlo (MCMC) failed to converge properly over 100 million steps for the CAC dataset under a relaxed clock, and observing that the age and rate estimates were highly congruent between runs using either a strict or relaxed clock model (Table 1), we report the strict clock estimates for the CAC dataset as well.
Table S1.
Model evaluation based on stepping stone log-marginal likelihoods
| Rank | Clock | Substitution model | Demographic model | Stepping-stone log- marginal likelihood | Bayes factors (model relative to respective model) |
| ASF | |||||
| 1 | Strict | HKY | GMRF | −16,198,752.3 | |
| 2 | Strict | HKY | Exponential | −16,198,757.1 | 4.8 |
| 3 | Strict | HKY | Expansion | −16,198,759.1 | 6.8 |
| 4 | Strict | HKY | Constant | −16,198,764.0 | 11.7 |
| 5 | Strict | HKY | Logistic | −16,198,765.3 | 13.0 |
| 6 | Relaxed | HKY | GMRF | −16,199,610.5 | 858.3 |
| 7 | Relaxed | HKY | Constant | −16,199,617.0 | 864.7 |
| CAC | |||||
| 1 | Relaxed | GTR | GMRF | −16,214,587.8 | |
| 2 | Strict | GTR | GMRF | −16,215,969.4 | 1,381.6 |
| 3 | Strict | GTR | Constant | −16,215,993.4 | 1,405.6 |
| 4 | Strict | GTR | Exponential | −16,215,997.3 | 1,409.5 |
| 5 | Relaxed | GTR | Constant | −16,218,531.8 | 3,944.1 |
| 6 | Relaxed | GTR | Exponential | −16,218,534.9 | 3,947.1 |
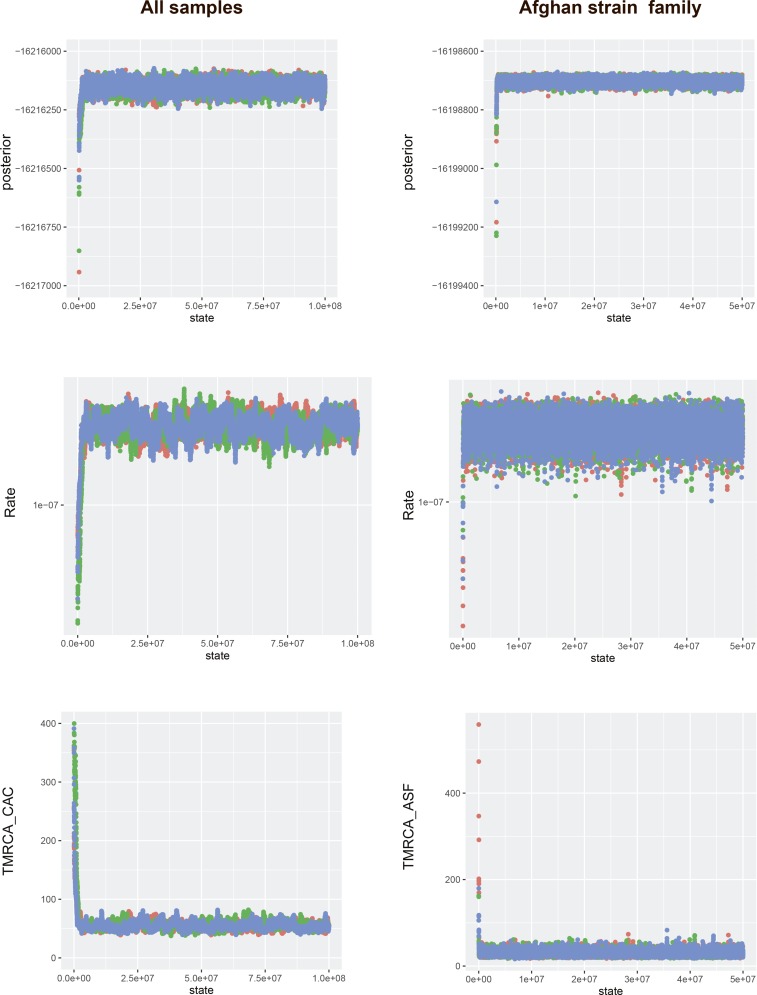
We estimated posterior distributions of parameters, including divergence times and substitution rates, using MCMC sampling. For each analysis, we ran three independent chains consisting of 30–300 million steps, the first 10% of which were discarded as a burn-in. Convergence to the stationary distribution was assessed within and between chains (Fig. S6), and sufficient sampling and mixing were checked by inspecting posterior samples (effective sample size >200). Parameter estimation was based on the samples combined from three different chains. The consensus tree was estimated from the combined samples using the maximum clade credibility method implemented in TreeAnnotator (beast.bio.ed.ac.uk/treeannotator). The Bayesian phylogenetic tree used to date the TMRCA of the CAC is shown annotated with posterior node probabilities in Fig. S7 and with individual node ages in Fig. S8. The results from the model testing are summarized in Table S1.
Fig. S6.
Assessing MCMC chain convergence. Panels show trace output for key parameters, posterior probabilities, substitution rates, and TMRCAs, for key nodes for the CAC and ASF datasets. Three independent chains were run for each dataset.
Fig. S7.
Tip date-calibrated Beast phylogeny including all isolates showing posterior probabilities of individual nodes.
Fig. S8.
Tip date-calibrated Beast phylogeny including all 85 isolates showing individual node ages.
Calculating MIRU Evolutionary Rates.
To calculate the yearly rate of MIRU evolution (contractions and expansions), we first constructed a BEAST phylogeny using a strict molecular clock and a GMRF skyride demographic model. We then extracted the total branch length of the phylogenetic tree using TreeStat (tree.bio.ed.ac.uk/software/treestat/). The sum of branch lengths corresponds to the evolutionary time of every branch from the sampled tips to the MRCA of all of the isolates. The number of repeats of each MIRU locus was annotated on the tree (Fig. 3). The total number of state changes over all 24 MIRU loci over the sum of years covered by the tree was then summed assuming a stepwise mode of MIRU evolution (Dataset S4).
Calculating Relative MDR-TB Prevalence.
TB and MDR-TB prevalence data were obtained from the World Health Organization (www.who.int/tb/country/data/download/en/). TB prevalence data were available for all countries for the year 2013, and point estimates of prevalence by 100,000 individuals were retrieved (e_prev_100k).
The data on MDR-TB prevalence were collected less systematically, relying on a mix of surveillance, surveys, and models. We used the estimated number of MDR-TB cases among all notified pulmonary TB cases (e_mdr_num), expressed as prevalence per 100,000 individuals by dividing by country population size estimates from the same source. We calculated the relative proportion of MDR-TB cases by dividing the prevalence of MDR-TB by the prevalence of TB and multiplying this number by 1,000.
SI Materials and Methods
We included samples from a TB outbreak detected at an Oslo educational institution for young adults in 2013, with the last cases belonging to the outbreak diagnosed in 2015. In addition, a search through an in-house database at the Norwegian Institute of Public Health revealed the presence of four Mtb isolates from Norway with an MIRU profile (Mtbc15-9 code 1047–189) with only two repeat differences from the larger outbreak (Mtbc15-9 code 10287–189). In total, 26 samples from 24 patients were available from the outbreak (all samples from culture-positive patients), along with the four isolates from the smaller cluster. The earliest cases in the outbreak, as well as the four cases in the smaller cluster, were all Afghan immigrants to Norway, indicating that these related MIRU types were representatives of a larger reservoir of strains circulating in Afghanistan.
To assess whether these two MIRU types were part of one or more larger groups of strains globally, we searched through the MIRU patterns published in a recent extensive global study of L2 isolates (4,987 isolates from 99 countries) (2). We included all sequenced isolates that differed at no more than two MIRU loci from either of the two types described above. Because this also included MIRU type 94–32, composing the majority of CC1, we included all sequenced CC1 isolates from the study reported by Merker et al. (2). It should be noted that the CC1 definition is based on MIRU typing. In the present study, the CC1 isolates were selected based on phylogenetic clustering at the genome level; for example, three of the isolates clustering with CC1 in the Merker et al. study (2) had been assigned to other CCs by MIRU typing. These three isolates were also included in our study and were found to form a monophyletic group together with the other CAC isolates.
An additional four isolates harboring the 1047–189 MIRU pattern and two isolates differing from the 10287–189 pattern at two loci were sequenced for the present study, including five from the global study (2) and one identified in an in-house database at Research Center Borstel, Germany. Finally, after initial analyses demonstrated that genomes belonging to the Russian clade A were closely related to CC1, we included a numerically matching sample of clade A genomes from a large genome study centered in Samara Oblast (23), Russia, as well as two clade B isolates to serve as an outgroup. The genomes included in the present study are available in the European Molecular Biology Laboratory (EMBL) database (accession nos. PRJEB12184, PRJEB9680, ERP006989, and ERP000192). Detailed information on samples included in the study is provided in Datasets S1 and S2. A total of 85 isolates were included in the study.
Supplementary Material
Acknowledgments
We thank Matthias Merker and Stefan Niemann (Research Center Borstel) for providing unpublished genome sequence data and information. The work of V.E. was partially funded by a postdoctoral fellowship from the Norwegian Research Council (Grant 221562). F.B. was supported by the European Research Council (Grant ERC260801–BIG_IDEA) and the National Institute for Health Research University College London Hospitals Biomedical Research Centre. C.S.P. was supported by the National Institutes of Health (Grant R01 AI113287).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences have been deposited in the European Molecular Biology Laboratory database, www.ebi.ac.uk/embl/index.htm (accession nos. PRJEB12184, PRJEB9680, ERP006989, and ERP000192).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1611283113/-/DCSupplemental.
References
- 1.Luo T, et al. Southern East Asian origin and coexpansion of Mycobacterium tuberculosis Beijing family with Han Chinese. Proc Natl Acad Sci USA. 2015;112(26):8136–8141. doi: 10.1073/pnas.1424063112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Merker M, et al. Evolutionary history and global spread of the Mycobacterium tuberculosis Beijing lineage. Nat Genet. 2015;47(3):242–249. doi: 10.1038/ng.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.European Concerted Action on New-Generation Genetic Markers and Techniques for the Epidemiology and Control of Tuberculosis Beijing/W genotype Mycobacterium tuberculosis and drug resistance. Emerg Infect Dis. 2006;12(5):736–743. doi: 10.3201/eid1205.050400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Jong BC, et al. Progression to active tuberculosis, but not transmission, varies by Mycobacterium tuberculosis lineage in The Gambia. J Infect Dis. 2008;198(7):1037–1043. doi: 10.1086/591504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drobniewski F, et al. Drug-resistant tuberculosis, clinical virulence, and the dominance of the Beijing strain family in Russia. JAMA. 2005;293(22):2726–2731. doi: 10.1001/jama.293.22.2726. [DOI] [PubMed] [Google Scholar]
- 6.Ford CB, et al. Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug-resistant tuberculosis. Nat Genet. 2013;45(7):784–790. doi: 10.1038/ng.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Werngren J, Hoffner SE. Drug-susceptible Mycobacterium tuberculosis Beijing genotype does not develop mutation-conferred resistance to rifampin at an elevated rate. J Clin Microbiol. 2003;41(4):1520–1524. doi: 10.1128/JCM.41.4.1520-1524.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Comas I, et al. Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat Genet. 2011;44(1):106–110. doi: 10.1038/ng.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Comas I, et al. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat Genet. 2013;45(10):1176–1182. doi: 10.1038/ng.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wirth T, et al. Origin, spread and demography of the Mycobacterium tuberculosis complex. PLoS Pathog. 2008;4(9):e1000160. doi: 10.1371/journal.ppat.1000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bos KI, et al. Pre-Columbian mycobacterial genomes reveal seals as a source of New World human tuberculosis. Nature. 2014;514(7523):494–497. doi: 10.1038/nature13591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kay GL, et al. Eighteenth-century genomes show that mixed infections were common at time of peak tuberculosis in Europe. Nat Commun. 2015;6:6717. doi: 10.1038/ncomms7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eldholm V, et al. Four decades of transmission of a multidrug-resistant Mycobacterium tuberculosis outbreak strain. Nat Commun. 2015;6:7119. doi: 10.1038/ncomms8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roetzer A, et al. Whole genome sequencing versus traditional genotyping for investigation of a Mycobacterium tuberculosis outbreak: A longitudinal molecular epidemiological study. PLoS Med. 2013;10(2):e1001387. doi: 10.1371/journal.pmed.1001387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walker TM, et al. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: A retrospective observational study. Lancet Infect Dis. 2013;13(2):137–146. doi: 10.1016/S1473-3099(12)70277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pepperell CS, et al. The role of selection in shaping diversity of natural M. tuberculosis populations. PLoS Pathog. 2013;9(8):e1003543. doi: 10.1371/journal.ppat.1003543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ford CB, et al. Use of whole genome sequencing to estimate the mutation rate of Mycobacterium tuberculosis during latent infection. Nat Genet. 2011;43(5):482–486. doi: 10.1038/ng.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lillebaek T, et al. Substantial molecular evolution and mutation rates in prolonged latent Mycobacterium tuberculosis infection in humans. Int J Med Microbiol. 2016;S1438-4221(16):30110–2. doi: 10.1016/j.ijmm.2016.05.017. [DOI] [PubMed] [Google Scholar]
- 19.Drummond A, Pybus O, Rambaut A, Forsberg R, Rodrigo A. Measurably evolving populations. Trends Ecol Evol. 2003;18(9):481–488. [Google Scholar]
- 20.Biek R, Pybus OG, Lloyd-Smith JO, Didelot X. Measurably evolving pathogens in the genomic era. Trends Ecol Evol. 2015;30(6):306–313. doi: 10.1016/j.tree.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Didelot X, Gardy J, Colijn C. Bayesian inference of infectious disease transmission from whole-genome sequence data. Mol Biol Evol. 2014;31(7):1869–1879. doi: 10.1093/molbev/msu121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eldholm V, Balloux F. Antimicrobial resistance in Mycobacterium tuberculosis: The odd one out. Trends Microbiol. 2016;24(8):637–648. doi: 10.1016/j.tim.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 23.Casali N, et al. Evolution and transmission of drug-resistant tuberculosis in a Russian population. Nat Genet. 2014;46(3):279–286. doi: 10.1038/ng.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29(8):1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aandahl RZ, Reyes JF, Sisson SA, Tanaka MM. A model-based Bayesian estimation of the rate of evolution of VNTR loci in Mycobacterium tuberculosis. PLOS Comput Biol. 2012;8(6):e1002573. doi: 10.1371/journal.pcbi.1002573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ragheb MN, et al. The mutation rate of mycobacterial repetitive unit loci in strains of M. tuberculosis from cynomolgus macaque infection. BMC Genomics. 2013;14:145. doi: 10.1186/1471-2164-14-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vogler AJ, et al. Effect of repeat copy number on variable-number tandem repeat mutations in Escherichia coli O157:H7. J Bacteriol. 2006;188(12):4253–4263. doi: 10.1128/JB.00001-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho SY, Phillips MJ, Cooper A, Drummond AJ. Time dependency of molecular rate estimates and systematic overestimation of recent divergence times. Mol Biol Evol. 2005;22(7):1561–1568. doi: 10.1093/molbev/msi145. [DOI] [PubMed] [Google Scholar]
- 29.Rieux A, et al. Improved calibration of the human mitochondrial clock using ancient genomes. Mol Biol Evol. 2014;31(10):2780–2792. doi: 10.1093/molbev/msu222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rasmussen S, et al. Early divergent strains of Yersinia pestis in Eurasia 5,000 years ago. Cell. 2015;163(3):571–582. doi: 10.1016/j.cell.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duchêne S, et al. Genome-scale rates of evolutionary change in bacteria. bioRxiv. 2016 doi: 10.1101/069492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baker O, et al. Human tuberculosis predates domestication in ancient Syria. Tuberculosis (Edinb) 2015;95(Suppl 1):S4–S12. doi: 10.1016/j.tube.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 33.Lee OY, et al. Mycobacterium tuberculosis complex lipid virulence factors preserved in the 17,000-year-old skeleton of an extinct bison, Bison antiquus. PLoS One. 2012;7(7):e41923. doi: 10.1371/journal.pone.0041923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eldholm V, et al. Evolution of extensively drug-resistant Mycobacterium tuberculosis from a susceptible ancestor in a single patient. Genome Biol. 2014;15(11):490. doi: 10.1186/s13059-014-0490-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: More models, new heuristics and parallel computing. Nat Methods. 2012;9(8):772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7(1):214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Letunic I, Bork P. Interactive Tree Of Life v2: Online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011;39(Web Server issue) suppl 2:W475-8. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gouy M, Guindon S, Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27(2):221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 39.Rieux A, Khatchikian C. 2015. TipDatingBeast: Using tip dates with phylogenetic trees in BEAST (R package) Available at https://cran.r-project.org/web/packages/TipDatingBeast/TipDatingBeast.pdf. Accessed October 25, 2016.
- 40.Firth C, et al. Using time-structured data to estimate evolutionary rates of double-stranded DNA viruses. Mol Biol Evol. 2010;27(9):2038–2051. doi: 10.1093/molbev/msq088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Minin VN, Bloomquist EW, Suchard MA. Smooth skyride through a rough skyline: Bayesian coalescent-based inference of population dynamics. Mol Biol Evol. 2008;25(7):1459–1471. doi: 10.1093/molbev/msn090. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.