Significance
The parasite Trypanosoma brucei causes African sleeping sickness. This disease, which lacks effective treatments, affects millions of humans and livestock in sub-Saharan Africa. This paper reveals a mechanism by which the parasite can evade our immune response. Indolepyruvate is a metabolite produced by the parasite and it manipulates the immune cells, called macrophages, preventing them from becoming fully active. The selective advantage for the parasite of excretion of indolepyruvate is possible modulation of host inflammatory responses to prolong host survival, thereby potentiating transmission of the parasite to the tsetse fly vector and ensuring completion of the life cycle. This discovery could lead to new drug targets and better treatments.
Keywords: immunometabolism, innate immunity, immune evasion, Trypanosoma brucei
Abstract
The parasite Trypanasoma brucei causes African trypanosomiasis, known as sleeping sickness in humans and nagana in domestic animals. These diseases are a major burden in the 36 sub-Saharan African countries where the tsetse fly vector is endemic. Untreated trypanosomiasis is fatal and the current treatments are stage-dependent and can be problematic during the meningoencephalitic stage, where no new therapies have been developed in recent years and the current drugs have a low therapeutic index. There is a need for more effective treatments and a better understanding of how these parasites evade the host immune response will help in this regard. The bloodstream form of T. brucei excretes significant amounts of aromatic ketoacids, including indolepyruvate, a transamination product of tryptophan. This study demonstrates that this process is essential in bloodstream forms, is mediated by a specialized isoform of cytoplasmic aminotransferase and, importantly, reveals an immunomodulatory role for indolepyruvate. Indolepyruvate prevents the LPS-induced glycolytic shift in macrophages. This effect is the result of an increase in the hydroxylation and degradation of the transcription factor hypoxia-inducible factor-1α (HIF-1α). The reduction in HIF-1α levels by indolepyruvate, following LPS or trypanosome activation, results in a decrease in production of the proinflammatory cytokine IL-1β. These data demonstrate an important role for indolepyruvate in immune evasion by T. brucei.
African trypanosomes, such as Trypanasoma brucei (T. brucei), are extracellular protozoan parasites of the mammalian hemolymphatic and CNSs that are transmitted by tsetse flies. These parasites cause persistent infections in humans (sleeping sickness) and animals (nagana) that are fatal unless treated (1). Because parasites are continuously exposed to the attention of their host’s immune response, but are reliant on an insect vector (tsetse flies) for transmission, trypanosomes must evade these responses while also prolonging host survival to ensure life cycle completion. The primary strategy of immune evasion by the trypanosomes involves the antigenic variation of the variant surface glycoprotein (VSG) coat on the surface of the parasite (2–4). This process gives rise to characteristic waves of parasitemia as the number of trypanosomes in circulation rises and falls, as the predominant VSG variants in the population expand and are subsequently cleared by antibody-mediated lysis. Before clearance, parasite loads at the peak of the parasitemia can be >108 cells per milliliter of blood (5). The innate immune response also plays a role in host defense against T. brucei. Exposure of macrophages to trypanosome-derived components, including released VSGs, results in an IFN-γ–dependent proinflammatory response that is required to control the initial parasitemia. However, macrophages from chronically infected animals appear to be impaired in their ability to produce proinflammatory cytokines (6–8). In the case of IL-1β levels, there have been conflicting reports following T. brucei experimental infections, with some studies reporting levels were elevated (9, 10), and other studies showing that production of IL-I was severely depressed in infected mice (11). Taken together, the data in the literature suggest that modulation of the innate immune response is extremely important in the progression of trypanosomiasis (8).
Trypanosomiasis is also associated with a profound disturbance in aromatic amino acid metabolism (12–14). Serum levels of aromatic amino acids (e.g., tryptophan) are significantly depressed in infected animals (15). This decrease is accompanied by the excretion of abnormal amounts of aromatic ketoacids, such as indolepyruvate, phenylpyruvate, and hydroxyphenylpyruvate at levels that correlate with the parasitemia (16–19). Indeed the presence and subsequent oxidation of these ketoacids explains the pungent odor and reddish brown color characteristic of the urine of camels infected with Trypanosoma evansi, a feature that has been used as a traditional diagnostic tool by camel herders to identify infected animals (20). These aromatic ketoacids are thought to be derived from the metabolic activities of the parasite rather than the host (19, 21, 22). The functional significance of these abnormal levels of aromatic ketoacids in the circulation remains obscure, although it has been suggested that they may contribute to the pathogenesis of trypanosomiasis, and specifically the neuropsychiatric symptoms, through alterations in the biogenic amine pool (15).
In this study we have examined whether these aromatic ketoacids might have a role in the modulation of the host innate response. Macrophage activation in innate immunity causes a shift in metabolism toward glycolysis (23, 24). This event is hypoxia-inducible factor-1α (HIF-1α)–dependent, with HIF-1α also being needed for induction of HIF-1α–dependent genes, including those encoding IL-1β. Here we demonstrate directly that aromatic ketoacids are transamination products constitutively and obligatorily excreted by bloodstream forms of T. brucei and that indolepyruvate specifically reduces HIF-1α levels, preventing the induction of HIF-1α–dependent genes, such as IL-1β, and metabolic reprogramming in macrophages. Our study suggests a potential role for indolepyruvate in the modulation of the host immune response by African trypanosomes.
Results
Aromatic Ketoacids Are Produced by Transamination Catalyzed by an Essential Cytoplasmic Aspartate Aminotransferase Activity in Bloodstream Forms of T. brucei.
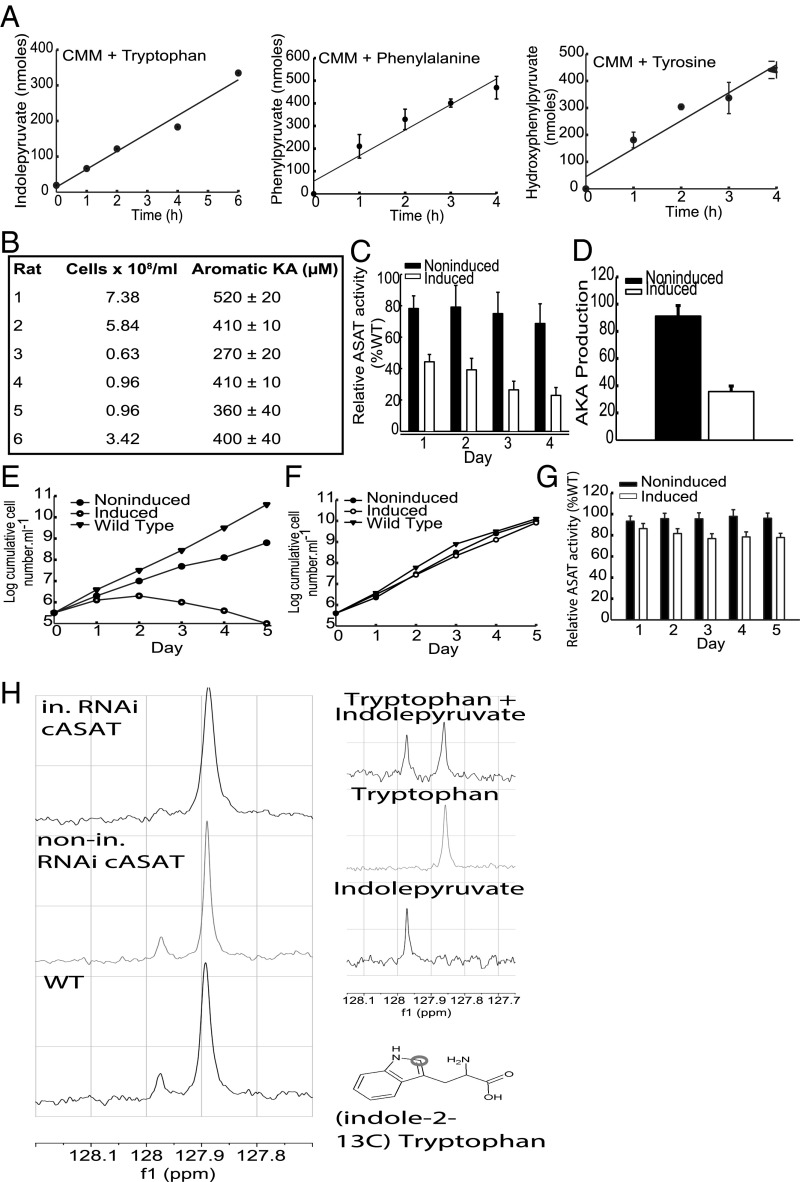
The high consumption of tryptophan and phenylalanine by bloodstream form trypanosomes (19, 21, 22, 25) suggested that the abnormal levels of aromatic ketoacids present in infected animals are transamination products generated by the parasites, but this has never been directly demonstrated. Therefore, aromatic ketoacid production by T. brucei incubated with tryptophan, tyrosine, or phenylalanine was measured directly using NAD-linked aromatic α-hydroxy acid dehydrogenase (26). Significant production rates of 50.2 ± 4.6, 77.3 ± 11.0, and 84.8 ± 12.7 nmols/5 × 107cells per hour were measured for indolepyruvate, hydroxyphenylpyruvate, and phenylpyruvate, respectively (Fig. 1A). Interestingly, cellular production of aromatic keto acids increased linearly with the concentration of the aromatic amino acid even above physiological levels (Fig. S1A). No significant aromatic ketoacid production was detected when aromatic amino acids were omitted from the medium. The same assay also demonstrated that aromatic ketoacids are produced in vivo in infected animals and were in the 0.2- to 0.5-mM range when the parasitemia was ∼108 parasites per milliliter (Fig. 1B). The levels of aromatic ketoacids produced did not correlate precisely with the number of parasites present in the rat. This variation may be because of differences in the ability of the rats to process indolepyruvate from the serum and the metabolic or cell-cycle status of parasites in the bloodstream. Nevertheless the data show that circulating levels of aromatic ketoacids are significant close to the peak of parasitemia. Production and excretion of aromatic ketoacids is a bloodstream-stage–specific phenomenon because there was no detectable production of aromatic ketoacids by the procyclic or insect gut form of the parasite (not shown). Thus, following uptake, aromatic amino acids are deaminated and the ketoacid product is excreted at sufficiently high rates that circulating levels of aromatic ketoacids in infected animals approach millimolar levels at the peak of parasitemia (i.e., >108 parasites per milliliter of blood).
Fig. 1.
Aromatic ketoacids production in bloodstream forms of T. brucei involves an essential cASAT activity. (A) Trypanosomes (5–6 × 107 cells/mL) were incubated in Creek’s Minimal Medium (CMM) supplemented with tryptophan, phenylalanine, or tyrosine (2 mM). At the indicated times, samples of the extracellular medium were assayed for aromatic ketoacid content using AHADH, as described in Materials and Methods. The results represent the mean ± SD of three determinations and the rates were estimated by linear regression analysis and expressed as nanomoles per 5 × 107cells per hour. (B) Serum was prepared from infected rat blood and concentration of aromatic ketoacids was determined using AHADH, as described in Materials and Methods. In each case the level of the parasitemia was also determined. (C) The total ASAT activity of noninduced and induced bloodstream TbcASAT RNAi cells was measured as described in Materials and Methods. The results were expressed as relative to the activity of the wild-type cells (46.7 ± 2.3 nmol⋅min−1⋅mg−1) and represent the mean ± SD of three determinations. (D) Total aromatic ketoacid production by noninduced and induced bloodstream TbcASAT RNAi cells was measured as described in Materials and Methods. The results are expressed as relative to wild-type production and represent the mean ± SD of three determinations. (E) The growth of the parental wild-type cells (▼) and a bloodstream form TbcASAT RNAi cell line cultured in the absence (●, noninduced) or presence (○, induced) of tetracycline was monitored and expressed as log-cumulative number of cells per milliliter. (F) The growth of the parental wild-type cells (▼) and a procyclic form TbcASAT RNAi cell line cultured in the absence (●, noninduced) or presence (○, induced) of tetracycline was monitored and expressed as log cumulative number of cells per milliliter. (G) The total ASAT activity of noninduced and induced procyclic TbcASAT RNAi cells were measured as described in Materials and Methods. The results were expressed as relative to the activity of the wild-type cells (203.3 ± 10.4 nmol⋅min−1⋅mg−1) and represent the mean ± SD of three determinations. (H) Bloodstream form wild-type (WT) MITat 1.1 and TbcASAT RNAi cells cultured in the presence (in. RNAicASAT) or absence (nonin. RNAicASAT) of tetracycline for 48 h were incubated in HMI-9 containing l-(indole-2-13C)-tryptophan for 5 h and the excreted end products were analyzed. The chemical shift of indolepyruvate (∼127.97 ppm) and l-tryptophan (∼127.86 ppm) are clearly distinguishable in a 13C NMR spectra. Molecular structure of (indole-2-13C) tryptophan, indicating the position of the 13C-labeled atom in the indole ring that was used for metabolite identification.
Fig. S1.
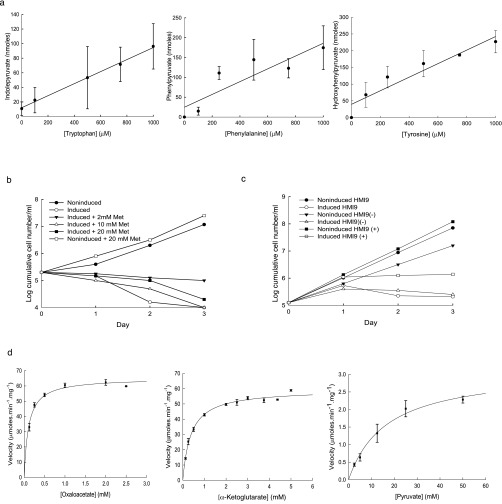
Aromatic ketoacids production in bloodstream forms of Trypanosoma brucei varies with the extracellular concentration of tryptophan, phenylalanine, or tyrosine and involves the essential activity of TbcASAT. Trypanosomes (2.5 × 107 cells/mL) were incubated (37 °C) in CMM containing dialyzed FCS and supplemented with various concentrations of tryptophan, phenylalanine, or tyrosine as indicated. After 4 h, samples of the extracellular medium were prepared by centrifugation and assayed for aromatic ketoacid content using AHADH as described in Materials and Methods. The results represent the mean ± SD of three determinations and the data were fitted by linear regression analysis. (B) The bloodstream form TbcASAT RNAi cell line cultured in HMI-9 medium supplemented with methionine (Met) from 2–20 mM and grown under induced (+tetracycline) or noninduced conditions. The growth of the cells was monitored and expressed as log cumulative number of cells per milliliter. (C) The bloodstream form TbcASAT RNAi cell line was cultured in HMI-9 medium, HMI-9 medium supplemented with 1 mM each of tryptophan, phenylalanine and tryosine [HMI(+)] or HMI-9 medium containing a fivefold lower concentration of each of the aromatic amino acids [HMI-9 (−)]. The cells were grown under induced (+tetracycline) or noninduced conditions. The growth of the cells was monitored and expressed as log-cumulative number of cells per milliliter. (D) The full-length ORF of TbcASAT (Tb10.70.3710,) was amplified from genomic DNA and inserted into pNIC28-Bsa4 using ligation independent cloning, for expression in BL21 (DE3) E. coli. Following IPTG-induced expression of the recombinant protein, the TbcASAT was purified via Ni2+ resin column. Protein concentration was ascertained via Bradford assay. The assays were performed in 100 mM Tris pH 7.5 containing 25 mM NaCl, 0.25 mM NADH, T. cruzi recombinant AHADH (20 units) as the coupling enzyme, 25 mM l-tryptophan, and various concentrations of oxaloacetate, α-ketoglutarate, or pyruvate as ketoacid acceptors. The assay was initiated by addition of 0.45 ng TbcASAT. Enzyme velocity was estimated from the decrease in absorbance at 340 nM and was expressed as μmoles⋅min−1⋅mg−1. The data were plotted and fitted to Michaelis–Menten curves using the Enzyme Kinetics Module (v1.3) operating in Sigma Plot (v11). The data represent the mean ± SE of triplicate determinations.
Several studies indicate that T. brucei cytoplasmic aspartate aminotransferase (TbcASAT) can efficiently catalyze transamination of aromatic amino acids (27, 28). Therefore, the functional role of TbcASAT in aromatic ketoacid production was investigated by conditional interference RNA (RNAi). Induction of the TbcASAT double-stranded RNA (dsRNA) resulted in a decrease of ∼70% in transcript levels within 24 h as demonstrated by quantitative RT-PCR (qRT-PCR), which gave a relative expression ratio (induced/noninduced) of 0.37 ± 0.05 (mean ± SD of three separate determinations). There was a similar reduction in cellular ASAT activity in the induced cells (Fig. 1C). Significantly, this decrease in activity was also accompanied by a proportionate decrease in aromatic ketoacid secretion by the induced cells (Fig. 1D). Knockdown of TbcASAT also had a rapid and deleterious effect on the growth of bloodstream forms (Fig. 1E). Cell division essentially ceased 24 h after induction of the TbcASAT dsRNA and was subsequently followed by cell death. A slight growth effect was also observed for the noninduced cells, which can be attributed to a small knockdown (∼20%) of ASAT activity in noninduced cells because of leaky expression of the dsRNA (Fig. 1C). Knockdown of TbcASAT had no effect on the growth of the procyclic form (Fig. 1F) despite specific knockdown of the TbcASAT transcript in procyclic forms, confirmed by qRT-PCR, which gave a relative expression ratio (induced/noninduced) of 0.35 ± 0.06 (mean ± SD of three separate determinations) 24 h after induction of the dsRNA. A significant ASAT activity (∼80%) remained, however, in the induced procyclic form (Fig. 1G), which can be attributed to the presence of the mitochondrial form (mASAT). This activity is found in procyclic but not bloodstream forms and is highly specific for aspartate (28).
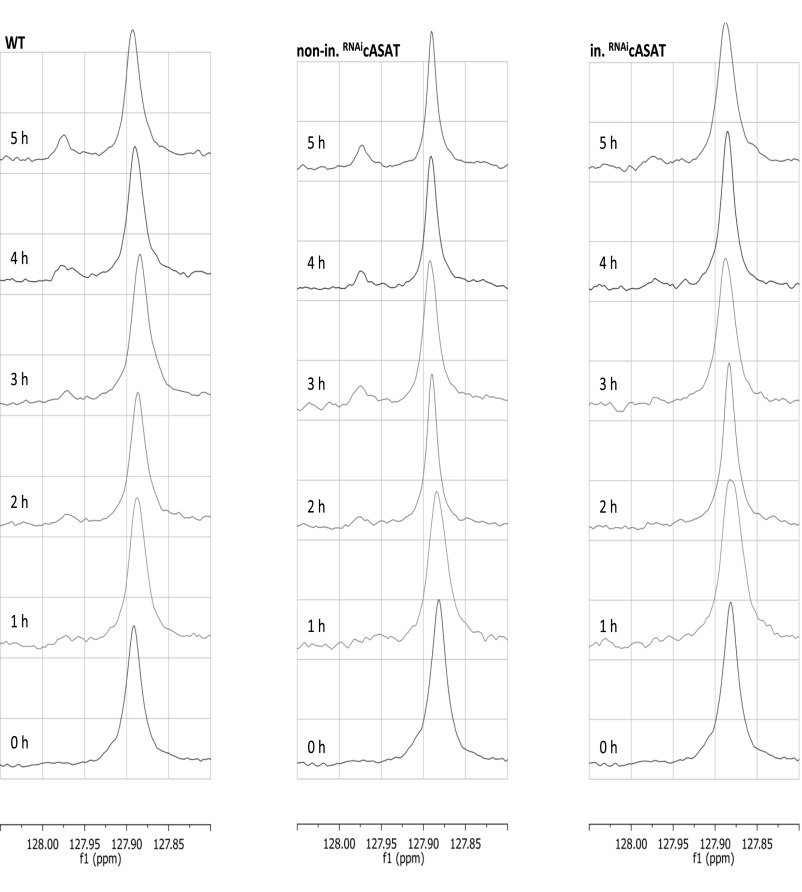
To demonstrate that aromatic ketoacids were produced by transamination, the metabolism of (indole-2-13C)-tryptophan by trypanosomes was investigated by solution-state NMR. The corresponding ketoacid, indolepyruvate (chemical shift ∼127.97 ppm), was easily distinguished from tryptophan (at ∼127.86 ppm) in a 13C spectra (Fig. 1H and Fig. S2). Indolepyruvate derived from tryptophan accumulated in the extracellular medium of wild-type and the noninduced TbcASAT RNAi cells over time, demonstrating that both wild-type and noninduced RNAi cells maintained similar continuous production rates of the ketoacid, whereas that of the induced RNAi cells was lower (Fig. S2). Importantly, indolepyruvate was the sole excreted metabolite produced from tryptophan under these conditions. Production of indolepyruvate from tryptophan decreased significantly following knockdown of TbcASAT [Fig. 1H, Top, compared with Middle (noninduced) and Bottom (wild-type)]. Taken together, these data demonstrated that bloodstream forms of T. brucei excrete significant amounts of aromatic ketoacids that are produced by transamination catalyzed by TbcASAT, and that this is an essential process during this stage of the life cycle.
Fig. S2.
Bloodstream T. brucei produce Indolepyruvate in the presence of tryptophan. Bloodstream from wild-type MITat 1.1 and TbcASAT RNAi cells cultured in the presence (in. RNAicASAT) or absence (nonin. RNAicASAT) of tetracycline for 48 h were incubated in HMI-9 containing l-(indole-2-13C)-tryptophan and the excreted end products were analyzed every hour for 5 h by 13C NMR, as described in Materials and Methods. The chemical shift of indolepyruvate (∼127.97 ppm) and l-tryptophan (∼127.86 ppm) are clearly distinguishable in a 13C NMR spectra.
Berger et al. (22, 27) have suggested that TbcASAT may play a key role in the transfer of amino groups from aromatic amino acids to ketothiobutyric acid as the final step in the recycling of methionine from methylthioadenosine, produced from S-adeosylmethionine during essential polyamine synthesis. However, methionine supplementation did not rescue the growth phenotype in cASAT knockdown cells (Fig. S1B), a result that is consistent with metabolomic data showing that a methionine recycling pathway does not appear to operate in bloodstream forms (29). Neither increasing nor decreasing extracellular levels of the aromatic amino acids ameliorated the growth defect in the knockdown cells, which indicated that the defect was not related to a detoxification role for TbcASAT (Fig. S1C). To investigate the acceptor ketoacid specificity of TbcASAT using tryptophan as the amino donor, the enzyme was expressed in Escherichia coli, purified to homogeneity, and subject to a kinetic analysis (Fig. S1D and Table S1). Significantly, TbcASAT catalyzed amino group transfer from tryptophan to oxaloacetate or α-ketoglutarate, whereas pyruvate could also act as a poor acceptor substrate. The highest affinity (lowest Km) was for oxaloacetate (0.1 mM), whereas the Vmax/Km ratio indicated that the catalytic efficiency was also greatest with the tryptophan/oxaloacetate pair (Table S1).
Table S1.
Kinetic constants for recombinant TbcASAT using various acceptor ketoacids with l-tryptophan as the amino group donor
| Ketoacid acceptor | Vmax (μmol⋅min−1⋅mg−1) | Km (mM) | Vmax/Km (μmol⋅min−1⋅mg−1⋅mM−1) |
| Oxaloacetate | 65.11 ± 1.29 | 0.10 ± 0.01 | 651.0 |
| α-Ketoglutarate | 59.42 ± 0.88 | 0.39 ± 0.03 | 152.3 |
| Pyruvate | 3.13 ± 0.37 | 16.65 ± 4.90 | 0.18 |
The kinetic constants (Km and Vmax) for the various ketoacid acceptors were estimated from the data presented in Fig. S1C by Michaelis–Menten analysis using the Enzyme Kinetics Module (v1.3) operating in Sigma Plot (v11). In all cases the concentration of the amino group donor l-tryptophan was 25 mM. The data are expressed are the mean ± SE of triplicate determinations.
Indolepyruvate Alters the Glycolytic State of the Cell Following LPS Stimulation.
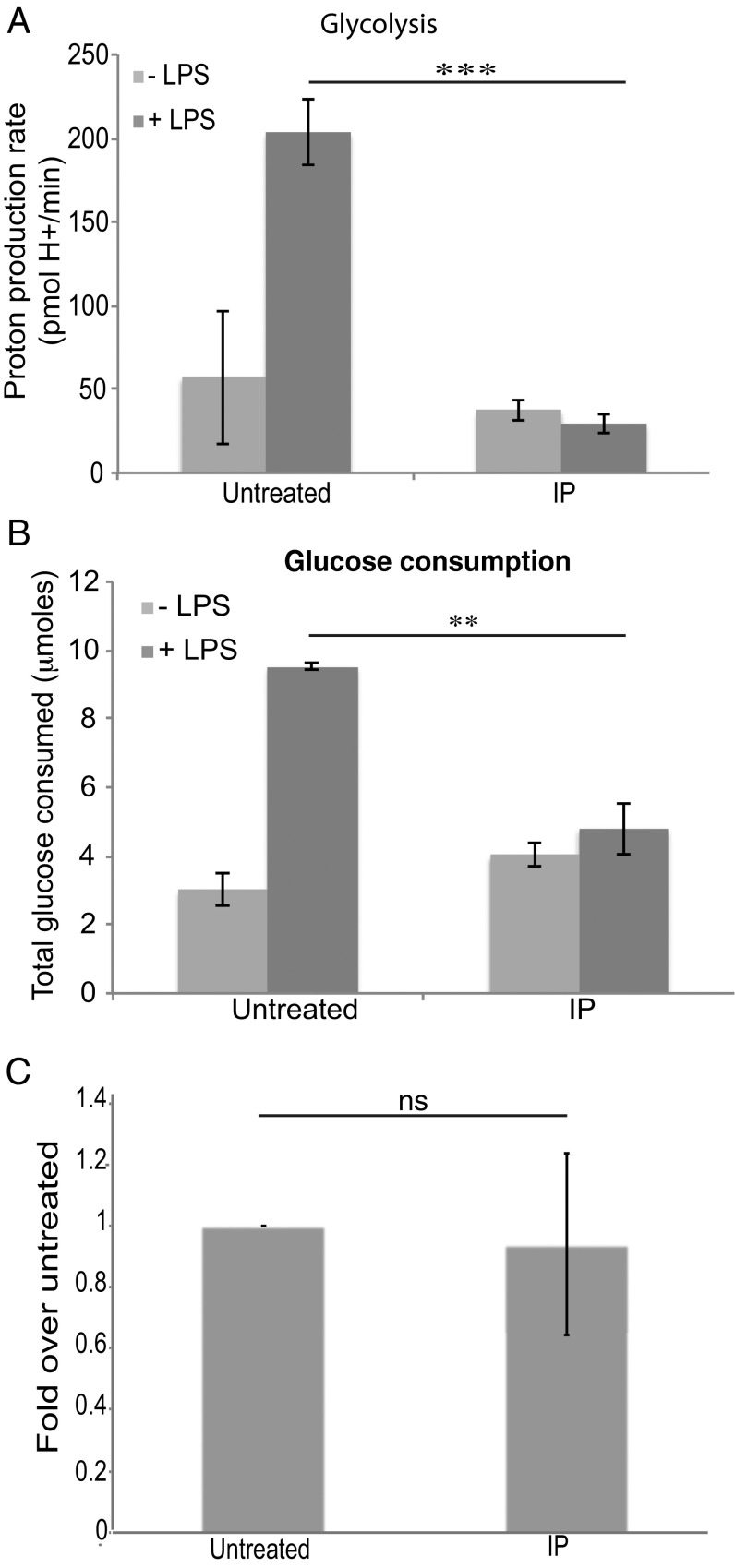
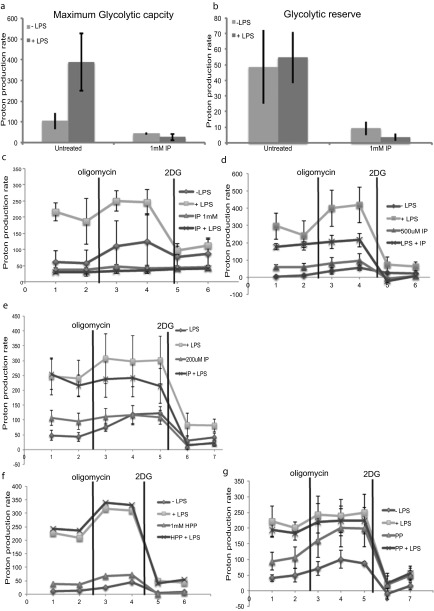
Because T. brucei clearly excrete aromatic ketoacids, we examined whether indolepyruvate, phenylpyruvate, or hydroxyphenylpyruvate would affect macrophage activation. We speculated that, like the ketoacid α-ketoglutarate, they might act to promote HIF-1α degradation via prolyl hydroxylase activation. We first used LPS to activate macrophages, because LPS is a potent inducer of proinflammatory cytokines. LPS is a Gram-negative bacterial product and the agonist for Toll-like receptor 4 (TLR4). LPS is also relevant during a natural T. brucei infection, because levels of circulating LPS are increased in both experimental and human African trypanosomiasis (29–31) and it is thought that this LPS contributes to pathology in trypanosomiasis by increasing the proinflammatory environment of the host (31, 32). The alteration in glycolytic rate (which is HIF-1α–dependent) in murine bone marrow-derived macrophages (BMDM), following LPS stimulation was examined. Circulating levels of aromatic ketoacids are likely to be in the millimolar range, so concentrations of up to 1 mM were used in these experiments. LPS increased glycolysis as predicted and indolepyruvate at a concentration of 1 mM inhibited this response (Fig. 2A). Indolepyruvate also reduced the maximum glycolytic capacity of the cells along with the glycolytic reserve (Fig. S3 A–C); 0.2 mM and 0.5 mM indolepyruvate didn’t appear to affect the glycolytic rate of these cells following LPS stimulation. However, the overall glycolytic state of the cell was altered as the maximum glycolytic capacity and the spare glycolytic capacity of the cells was reduced (Fig. S3 D and E). Phenylpyruvate and hydroxyphenylpyruvate had no effect on the glycolytic state of cells stimulated with LPS, even at a concentration of 1 mM (Fig. S3 F and G). We also examined the rate of glucose consumption from the media, an indicator of the level of glycolysis occurring within the cell. LPS increased the rate of glucose consumption and indolepyruvate again prevented this increase (Fig. 2B), thus confirming that indolepyruvate inhibits the ability of LPS to increase glycolysis. Indolepyruvate, at a concentration of 1 mM, was not toxic to the cells as assessed by LDH release (Fig. 2C).
Fig. 2.
Indolepyruvate inhibits the ability of LPS to induce glycolysis. BMDM were preincubated with indolepyruvate at a final concentration of 1 mM for 30 min before stimulation with LPS (100 ng/mL) for 24 h. (A) The proton production rate (PPR) was measured as described in Materials and Methods. ***P < 0.001. (B) The levels of glucose consumed from the supernatant was measured as described in Materials and Methods and (C) an LDH assay was performed to ensure indolepyruvate was not toxic at this concentration. **P < 0.01. One millimolar indolepyruvate was added to BMDM for 25 h and then the levels of cell death were measured as described in Materials and Methods. The level of cell death is presented as fold over untreated. These are representative of at least three independent experiments. IP, indolepyruvate; NS, not significant.
Fig. S3.
Indolepyruvate, but not phenylpyruvate or hydroxyphenylpyruvate, inhibits the ability of LPS to induce glycolysis. BMDM were preincubated with indolepyruvate at various concentrations, phenylpyruvate at 1 mM or hydroxyphenylpyruvate at 1 mM, for 30 min before stimulation with LPS (100 ng/mL) for 24 h. The proton production rate was measured as described in Materials and Methods. (A) The maximum glycolytic rate was measured following injection of oligomycin at a final concentration of 0.2 μM. (B) The glycolytic reserve was calculated by subtracting the rate of glycolysis from the maximum glycolytic rate. (C–G) The glycolytic profile of these BMDMs was measured using oligomycin at a final concentration of 0.2 μM and 2DG at a final concentration of 0.5 mM. These are representative of at least two independent experiments. HPP, hydroxyphenylpyruvate; IP, indolepyruvate; PP, phenylpyruvate.
Indolepyruvate Inhibits the LPS-Induced Signaling Pathway to HIF-1α but Has No Effect on the NF-κB, MAPK, or IFN-Stimulated Response Element Signaling Pathways.
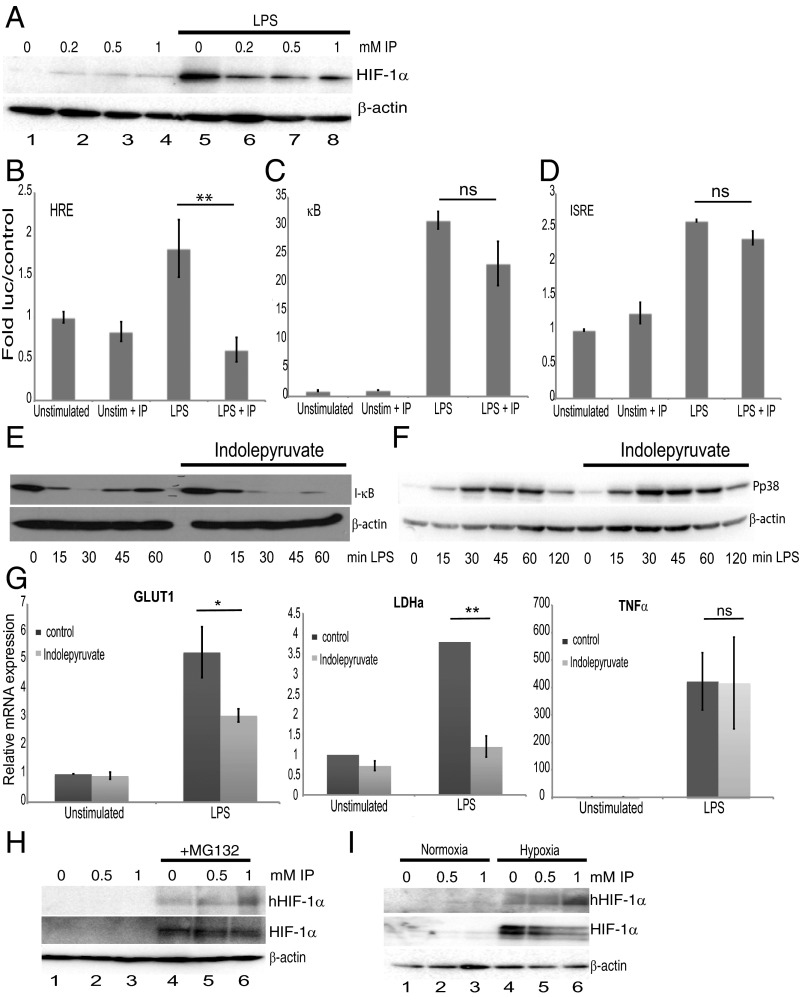
We next tested whether indolepyruvate affected HIF-1α activation. LPS increased HIF-1α protein levels and indolepyruvate inhibited this increase in a dose-dependent manner (Fig. 3A, compare lanes 6–8 to lane 5). Indolepyruvate also significantly inhibited the ability of LPS to induce a reporter gene, luciferase, under the control of a HIF-response element (HRE) (Fig. 3B) but not luciferase under the control of NF-κB (Fig. 3C) or the IFN response element (which binds IRF3) (Fig. 3D). Indolepyruvate also had no effect on LPS-induced I-κB degradation (Fig. 3E) or another LPS signal, p38 phosphorylation (Fig. 3F), even at the highest concentration of 1 mM. To confirm the role of HIF-1α in the mechanism of action of indolepyruvate, we examined two HIF-1α target genes, glucose transporter 1 (GLUT1) and lactate dehydrogenase A (LDHA) (33, 34). LPS induced the mRNA of both genes and this induction was inhibited in the presence of indolepyruvate (Fig. 3G). Indolepyruvate had no effect on the induction of the NF-κB–dependent gene TNF-α (Fig. 3G, Right). These results suggest that indolepyruvate specifically inhibits the signaling pathway leading to HIF-1α activation, while having little or no impact on the NF-κB, MAPK, or IFN-stimulated response element (ISRE) pathways.
Fig. 3.
Indolepyruvate inhibits the induction of HIF-1α by LPS but has little or no effect on the κB, ISRE, or MAPK pathways. (A) BMDM were incubated with various concentrations of indolepyruvate (as indicated) for 30 min before stimulation with LPS (100 ng/mL) for 24 h. The levels of HIF-1α were measured by Western blot. (B–D) 293-MTC cells were transfected with luciferase plasmids containing (B) HRE (**P < 0.01), (C) κB, or (D) ISRE promoters along with the TK Renilla control vector. Twenty-four hours later indolepyruvate, at a final concentration of 1 mM, was incubated on the cells for 30 min before stimulation with LPS (100 ng/mL) for 6 h. Results are normalized to Renilla luciferase activity and are presented relative to control cells containing empty vector. (E and F) BMDM were incubated with indolepyruvate at a final concentration of 1 mM for 30 min before stimulation with LPS (100 ng/mL) for varying times as indicated. The levels of (E) I-κB or (F) phosphorylated p38 (Pp38) were measured by Western blot. (G) GLUT1, LDHA, and TNF-α mRNA levels were measured by qPCR from mRNA isolated from BMDM incubated with 0.5 mM indolepyruvate for 30 min before stimulation with LPS (100 ng/mL) for 24 h. *P < 0.05; **P < 0.001. (H) BMDM were preincubated with MG132 for 30 min before addition of indolepyruvate at 0.5 and 1 mM for 6 h. The levels of hydroxylated HIF-1α and total HIF-1α were measured by Western blot. (I) Thirty minutes postincubation with indolepyruvate, BMDM were placed under normoxic or hypoxic conditions for 2 h. The levels of hydroxylated HIF-1α and total HIF-1α were measured by Western blot. (A–F, H, and I) are representative of at least three independent experiments. (G) Mean ± SEM, n = 3. IP, indolepyruvate.
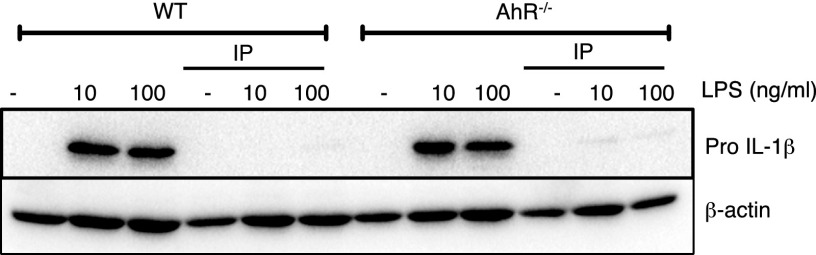
We next investigated the mechanism of inhibition of HIF-1α by indolepyruvate. Several studies suggest tryptophan catabolites, including breakdown products of indolepyruvate, act as endogenous ligands for the aryl hydrocarbon receptor (AhR), a ligand-activated transcription factor (35, 36). AhR was of particular interest because AhR dimerizes with aryl hydrocarbon receptor nuclear translocator (ARNT) (36, 37), the binding partner of HIF-1α; thus, an increase in AhR activation could cause a decrease in HIF-1α–ARNT complexes, reducing the production of HIF-1α–dependent target genes. Indolepyruvate was still able to inhibit LPS-induced pro–IL-1β production in BMDM from AhR knockout mice (Fig. S4). The AhR receptor is therefore unlikely to be required for the inhibition of LPS-induced pro–IL-1β by indolepyruvate.
Fig. S4.
Indolepyruvate inhibits LPS-induced IL-1β in both wild-type and AhR-deficient BMDM. BMDM were preincubated with indolepyruvate at 1 mM for 30 min before stimulation with LPS (10 ng/mL or 100 ng/mL, as indicated) for 24 h. The level of pro–IL-1β protein was measured by Western blot. This is representative of at least three independent experiments. IP, indolepyruvate.
Because indolepyruvate is a ketoacid, it might act in a similar way to αKG, which is a cofactor for the prolyl hydroxylases that hydroxylate HIF-1α and promote its degradation, so we next tested whether indolepyruvate could induce HIF-1α hydroxylation. The proteasomal inhibitor MG132 was used in these experiments as HIF-1α hydroxylation is difficult to detect under normal conditions because of the speed at which it is degraded once hydroxylated. The presence of indolepyruvate increased the levels of HIF-1α hydroxylation in BMDM (Fig. 3H, Upper, compare lanes 5 and 6 to lane 4). Under hypoxic conditions, which result in HIF-1α stabilization, the presence of indolepyruvate increased the levels of HIF-1α hydroxylation (Fig. 3I, Upper, compare lanes 5 and 6 to lane 4) and decreased the levels of HIF-1α protein (Fig. 3I, Middle, compare lanes 5 and 6 to lane 4). These results strongly suggest that indolepyruvate is promoting HIF-1α hydroxylation, leading to its degradation.
Indolepyruvate Inhibits the Ability of LPS to Induce Pro–IL-1β.
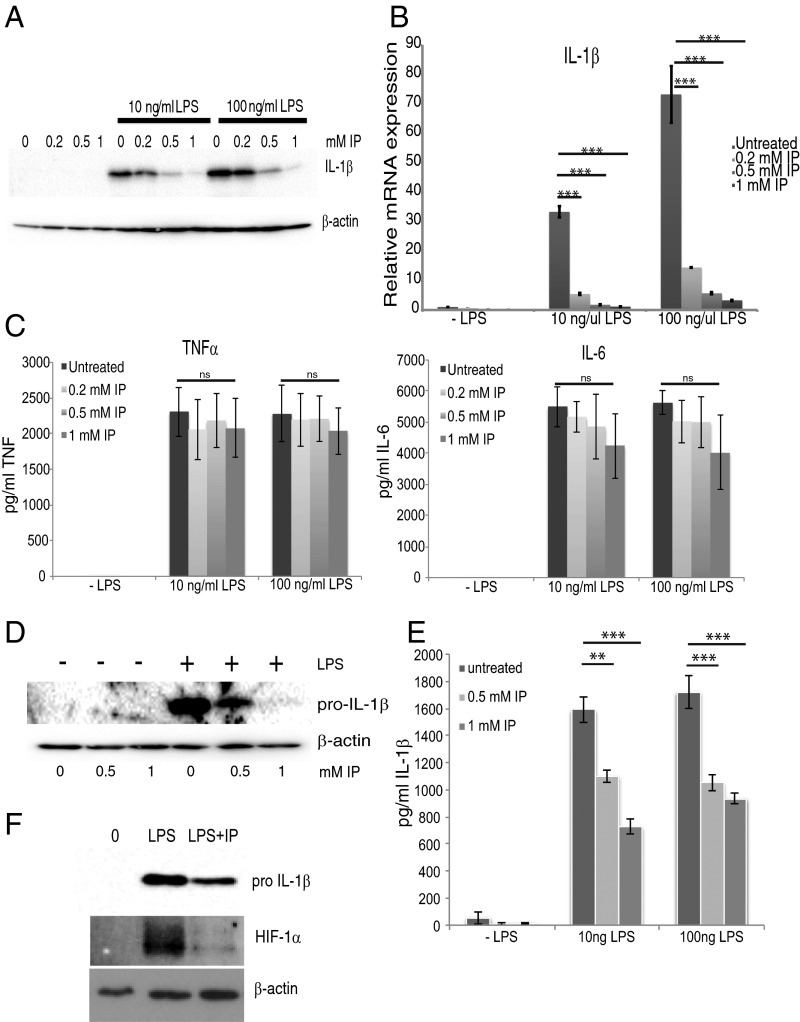
Previous studies have revealed that increased HIF-1α levels result in an induction of the critical proinflammatory cytokine pro–IL-1β (23). Preincubation of BMDM with indolepyruvate inhibited the ability of LPS to induce production of the cytokine IL-1β in a dose-dependent manner (Fig. 4A). Indolepyruvate also inhibits IL-1β mRNA production in a dose-dependent manner (Fig. 4B). Indolepyruvate had no effect on LPS-induced IL-6 or TNF production, which are HIF-1α–independent events (Fig. 4C). To determine if indolepyruvate has the same effect in human cells, we used peripheral blood mononuclear cells (PBMC). Indolepyruvate once again inhibited the ability of LPS to induce pro–IL-1β expression in these cells (Fig. 4D). Because LPS alone is sufficient to induce the production of mature IL-1β in PBMCs, an ELISA was used to measure mature IL-1β and, once again, indolepyruvate inhibited the ability of LPS to induce IL-1β production (Fig. 4E). We also examined the effect of indolepyruvate on the ability of LPS to induce HIF-1α and IL-1β in murine peritoneal macrophages (PECS). LPS induced HIF-1α and IL-1β in PECS and indolepyruvate inhibited this induction (Fig. 4F).
Fig. 4.
Indolepyruvate inhibits the induction of IL-1β by LPS. (A–C) BMDM were incubated with varying concentrations of indolepyruvate 30 min before stimulation with LPS (10 ng/mL or 100 ng/mL) for 24 h. (A) The levels of pro-IL-1β protein were measured by Western blot, whereas (B) IL-1β mRNA was measured by qPCR. ***P < 0.001. (C) Supernatants were examined by ELISA to determine the expression levels of TNF and IL-6. (D and E) PBMCs were incubated with varying concentrations of indolepyruvate 30 min before stimulation with LPS (100 ng/mL) for 24 h. (D) The levels of pro–IL-1β protein were measured by Western blot and (E) IL-1β mRNA was measured by qPCR. **P < 0.01; ***P < 0.001. (F) PECs were incubated with 1 mM indolepyruvate for 30 min before stimulation with LPS (100 ng/mL) for 24 h. The levels of pro–IL-1β and HIF-1α were measured by Western blot. A, D, and F are representative of three independent experiments; B, C, and E represent mean ± SEM, n = 3. IP, indolepyruvate.
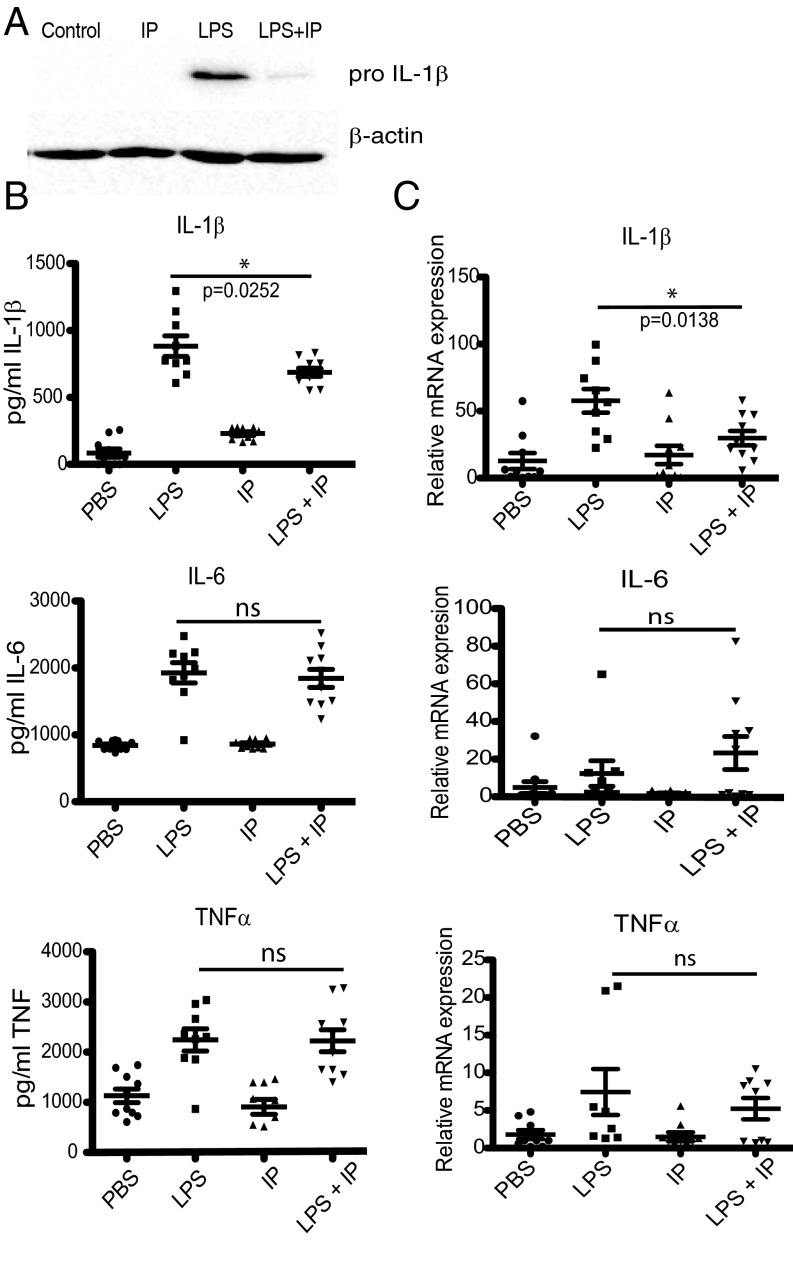
We next examined the effect of indolepyruvate on IL-1β induction in vivo. Mice injected with LPS into the peritoneum showed a significant increase in the production of pro–IL-1β protein in PECS isolated from the mice as assessed by Western blot (Fig. 5A), and IL-1β present in the blood of these mice as assessed by ELISA (Fig. 5B). Administering indolepyruvate to the mice significantly inhibited the production of IL-1β protein (Fig. 5 A and B). No effect was observed on IL-6 or TNF protein production (Fig. 5B). mRNA, isolated from the PECS of these mice, was also assessed and the induction of IL-1β mRNA was inhibited by indolepyruvate, whereas production of IL-6 or TNF mRNA was not (Fig. 5C). Phenylpyruvate was also tested to determine if it could inhibit IL-1β induction in vivo following LPS administration, but unlike indolepyruvate, phenylpyruvate showed no effect on the ability of LPS to induce IL-1β in vivo (Fig. S5). Therefore, indolepyruvate—but not phenylpyruvate—functions in vivo to modulate LPS-induced IL-1β production.
Fig. 5.
Indolepyruvate affects the ability of LPS to induce IL-1β and HIF-1α, but has no effect on TNF or IL-6 in vivo. Mice were injected intraperitoneally with indolepyruvate (50 mg/kg) or PBS for 30 min. LPS (15 mg/kg) or PBS were then injected intraperitoneally for 2 h. (A) Protein isolated from PECs was analyzed by Western blot for the presence of pro–IL-1β. (B) The levels of IL-1β, TNF, and IL-6 protein secreted into the serum were measured by ELISA. (C) The levels of IL-1β, TNF, and IL-6 mRNA were measured by qPCR using mRNA isolated from PECS. A is representative of three independent experiments. In B and C, n = 9 per group. IP, indolepyruvate.
Fig. S5.
Indolepyruvate affects the ability of LPS to induce IL-1β and HIF-1α, while having no effect on TNF or IL-6 in vivo. PECS and serum were isolated from mice following an in vivo experiment, as described in Materials and Methods. (A) mRNA isolated from PECs was analyzed by qPCR for IL-1β, TNF, and IL-6 qPCR. (B) The levels of IL-1β, TNF, and IL-6 protein secreted into the serum were measured by ELISA. n = 5 per group. IP, indolepyruvate; PP, phenylpyruvate.
Indolepyruvate Inhibits HIF-1α and IL-1β Induction in Response to Trypanosome Lysates.
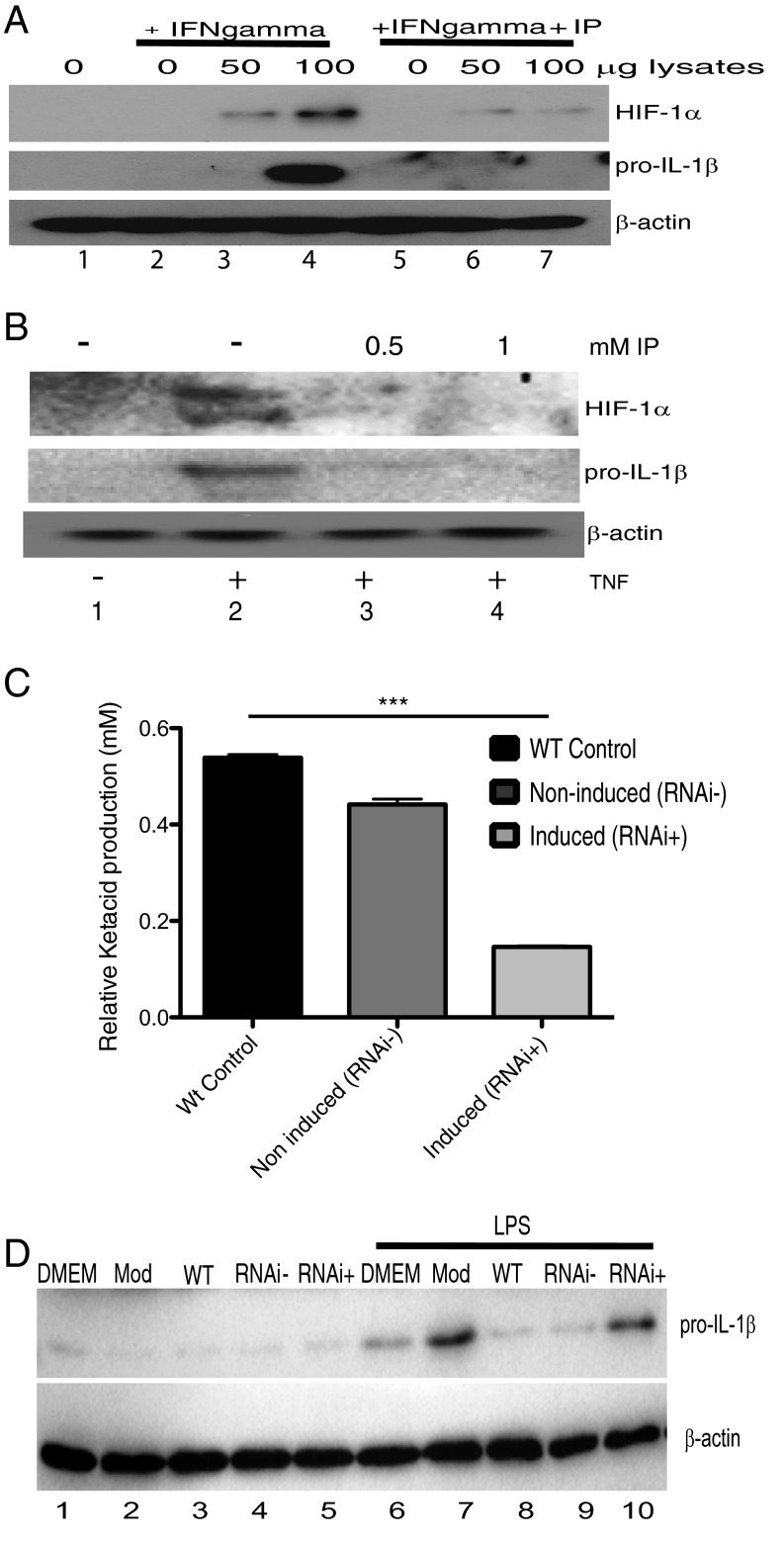
We next examined the effect of indolepyruvate on macrophage activation by trypanosome-derived components. We used total lysates from bloodstream T. brucei to stimulate BMDMs. Macrophages require preactivation by IFN-γ before they will respond to T. brucei derived proteins/components (6, 38), therefore we preincubated BMDM with IFN-γ before stimulation with the T. brucei lysates. HIF-1α and pro–IL-1β expression was detected in samples stimulated with increasing amounts of T. brucei lysates, and this production was inhibited in the presence of indolepyruvate (Fig. 6A, compare lanes 6 and 7 to lanes 3 and 4). TNF is produced following T. brucei infection and plays a key role in the regulation of parasitemia. As mentioned above, indolepyruvate has no effect on the induction of TNF expression by LPS (Fig. 3G). However, we wished to examine the effect of indolepyruvate on the ability of TNF itself to induce HIF-1α and pro–IL-1β expression in macrophages. Fig. 6B demonstrates that TNF can induce HIF-1α and pro–IL-1β expression, albeit at a lower level than LPS, and indolepyruvate again inhibited this response (Fig. 6B, compare lanes 3 and 4 to lane 2).
Fig. 6.
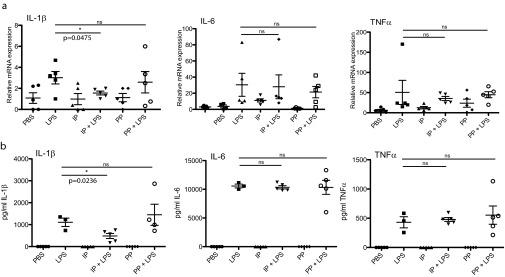
Indolepyruvate inhibits the ability of T. brucei lysates to induce pro–IL-1β expression. BMDM were incubated with 1 mM indolepyruvate for 30 min before stimulation with (A) T. brucei lysates (0, 50, 100 μg), prepared as described in Materials and Methods, alongside IFN-γ (100 ng/mL) for 8 h or (B) TNF-α (10 ng/mL) for 8 h. The levels of pro–IL-1β and HIF-1α protein were measured by Western blot. (C) Wild-type, noninduced, and induced bloodstream cASAT RNAi cells were grown in a modified DMEM (Mod). Supernatants were removed, centrifuged and filtered, and total ASAT activity was measured as described in Materials and Methods. ***P < 0.001. (D) BMDM were pretreated with supernatants (diluted 1:1) from C or control media for 1 h. Cells were then stimulated with 100 ng/mL LPS for 24 h. The levels of pro–IL-1β and HIF-1α protein were measured by Western blot. Mod: Modified DMEM; RNAi−, supernatants from noninduced TbcASAT RNAi cells; RNAi+, supernatants from induced TbcASAT RNAi cells; WT, supernatants from wild-type bloodstream cells. These experiments are representative of at least three independent experiments. IP, indolepyruvate.
Indolepyruvate Secreted from T. brucei Inhibits LPS-Induced pro–IL-1β.
Finally, to determine if the indolepyruvate secreted by T. brucei has an effect on the ability of LPS to induce pro–IL-1β, we incubated the wild-type bloodstream form of T. brucei or an induced cASAT RNAi cell line with media supplemented with tryptophan. The levels of indolepyruvate being produced were measured to confirm the reduction of indolepyruvate following knockdown of cASAT (Fig. 6C). BMDM were then cultured in this media and stimulated with LPS. LPS induced pro–IL-1β in control media but this induction was reduced in the presence of media from wild-type T. brucei (Fig. 6D, compare lane 8 to lane 7). When indolepyruvate was not present in the media (from induced cASAT RNAi T. brucei), there was no inhibition of the LPS-induced pro–IL-1β (Fig. 6D, compare lane 10 to lane 7). These results demonstrate that indolepyruvate produced by T. brucei can inhibit the ability of LPS to induce IL-1β. Taken together, these results identify indolepyruvate from T. brucei as an inhibitor of HIF-1α and IL-1β in macrophages, contributing to immune evasion by this parasite.
Discussion
African trypanosomiasis is characterized by alterations in plasma and urinary metabolic profiles, which display features of a diabetes-like state, such as glucose depletion, hyperketonemia, altered amino acid levels, and a marked α-keto aciduria that are pronounced at high parasitemia (14, 15, 39, 40). The extent to which these perturbations are a result of parasite or host metabolism, contribute to the pathogenesis of the disease, or if they have a function in the modulation of host responses is unclear. Of particular interest for this study are the effects on the aromatic amino acid pool in infected animals, which include a depletion of serum levels of such amino acids, especially tryptophan, which is accompanied by excretion of abnormal amounts of the corresponding aromatic α-ketoacid (15, 16, 18, 19). Here we show that these effects are clearly attributable to parasite metabolism. Significantly, a combination of biochemical, metabolic, and RNAi analysis demonstrated that these parasites secrete significant amounts of aromatic ketoacids, which are generated by the transamination of host aromatic amino acids catalyzed by TbcASAT. This process is obligatory in bloodstream forms of T. brucei and resultant levels of aromatic ketoacids can approach millimolar levels in serum.
Interestingly, aromatic ketoacid excretion increased as the extracelluar concentration of aromatic amino acid increased, which may indicate that amino acid uptake might be a rate-limiting step rather than transamination activity. Although the precise reason for the essential requirement for TbcASAT in bloodstream forms remains to be established, a number of results indicate that aromatic amino acids, especially tryptophan, act as amino acid donors for the transamination of oxaloacetate to generate essential intracellular aspartate. This view is consistent with metabolomic data, which show that intracellular aspartate is largely synthesized from oxaloacetate derived from glucose rather than by uptake of aspartate (41). This same study also noted that transport of aspartate is either absent or below detection in bloodstream forms. Moreover, PEPCK is also an essential enzyme in bloodstream forms and knockdown of this enzyme resulted in a significant decrease in labeling of asparate and pyrmidine pools (41). Analysis of the ketoacid acceptor specificity of TbcASAT in this study also showed that oxaloacetate is the preferred ketoacid acceptor when tryptophan is the amino donor, a substrate pair preference that agrees with the metabolomic data. Finally, although α-ketoglutarate can act as an acceptor in vitro, unlike oxaloacetate, there is no obvious metabolic route for the continuous net production of this ketoacid in bloodstream forms of T. brucei, which lack a functional TCA cycle and do not oxidize amino acids. Decreased intracellular levels of aspartate in the TbcASAT knockdown cells would readily account for the deleterious effect on cell growth for a number of reasons. These parasites are auxotropic for purines and production of adenine nucleotides from hypoxanthine, the purine source in the medium, requires aspartate during the synthesis of the adenylosuccinate precursor. Indeed the enzyme responsible, adenylosuccinate synthase, is also essential (42). It is noteworthy that the rate of succinate production from glucose (which can be taken as a measure of oxaoacetate production via PEPCK) by bloodstream forms, is 201 nmol⋅h−1⋅mg−1, which is equivalent to ∼56 nmols/h/5 × 107 cells (41), whereas the rate of excretion of indolepyruvate ranges from ∼20–50 nmols/h/5 × 107 cells depending on the extracellular concentration of tryptophan. Thus, production of oxaloacetate via PEPCK would be sufficient to meet the requirements of tryptophan transamination by TbcASAT. Additionally, although a biosynthethic pathway for pyrimidines is present in these parasites, aspartate is also essential for the production of uridine monophosphate and indeed, as noted previously, knockdown of PEPCK resulted in decrease labeling of the pyrimidine pool (41). So, at the very least, aspartate produced by TbASAT would have an essential role in nucleotide biosynthesis. A comparison of TbcASAT with the activity from Trypanosoma cruzi, a related but mainly intracellular parasite of mammals, suggests a selection for a preference for tryptophan in African trypanosomes. Kinetic studies have demonstrated that TbcASAT has a greater preference (lower Km) and a significantly higher Vmax/Km ratio for tryptophan as the amino group donor compared with tyrosine, phenylalanine, or aspartate, whereas the T. cruzi activity is less selective and transaminates aromatic amino acids and aspartate with equal efficiency (28).
Here we show that indolepyruvate, the transamination product of tryptophan, acts as a metabolic modulator of the innate immune response in vitro and in vivo. Indolepyruvate prevents the glycolytic shift in response to LPS, a critical response for macrophage function, by promoting the hydroxylation of the transcription factor HIF-1α, which targets HIF-1α for degradation. HIF-1α is known to bind to, and promote expression of multiple inflammatory genes, notably pro–IL-1β. This study demonstrates that indolepyruvate inhibits the ability of LPS to induce pro–IL-1β. Although indolepyruvate promotes HIF-1α degradation and IL-1β production at 0.2 mM, it shows more consistent inhibition at 0.5 and 1 mM, suggesting that the inhibition of the innate immune response by indolepyruvate is most important at the peak of the parasitemia, when levels of indolepyruvate in the blood are close to millimolar levels. At the peak of parasitemia, the levels of pathogen-associated molecular patterns and damage-associated molecular pattern molecules activating the innate immune response would be at their highest, leading to a potential overactivation of the innate immune response. This would be detrimental for the host and the well-being of the host is crucial for the survival of the parasite. Therefore, the ability of indolepyruvate to dampen down the immune response at the peak of the parasitemia would be very beneficial to the parasite.
Modulation of the innate immune response by T. brucei has previously been demonstrated. Garzón et al. (43) used LPS to stimulate an innate immune response and demonstrated that proteins secreted from T. brucei gambiense affected the host immune response. The secretome impaired LPS-induced maturation of murine dendritic cells as well as inhibiting the production of the cytokines TNF-α, IL-6, and IL-10. Mitchell et al. (11) demonstrated that LPS-induced IL-1 activity was profoundly depressed in cells isolated from both A/J and C57BL/6J mice following Trypanosoma congolense—another extracellular African trypanosome—infection. Day 7 of infection found no IL-1 activity in supernatants from either mouse strain. This effect may have been a result of indolepyruvate being present in the bloodstream of these mice. Aromatic ketoacids have also been shown to elicit an anti-inflammatory response. Aoki et al. (44) demonstrated that indolepyruvate, phenylpyruvate, and hydroxyphenylpyrvate, at concentrations similar to those used in this study, protected the skin of hairless mice from UVB-induced damage. Interestingly, indolepyruvate proved to be the most effective and topical application of indolepyruvate to the dorsal skin of hairless mice reduced the severity of UVB-induced skin lesions; it also suppressed the production of IL-1β and IL-6 in response to UVB radiation in these mice.
Macrophage activation occurs following infection with African trypanosomes but what causes this activation is poorly understood. It is likely that macrophages are exposed to several different activating agents. Some of these agents, such as VSG and trypanosome DNA, originate from the parasite; others will originate from the host, such as IFN-γ. The levels of circulating LPS also increase during African trypanosomiasis (30, 31, 29). This increase is likely because of an increase in gut permeability (45) and it is thought that this increase in LPS contributes to the pathology of the disease by increasing the proinflammatory environment of the host (30–32). Nyakundi et al. (9, 45) demonstrated that increases in cytokine production, including IL-1β, TNF-α, IL-6, and IFN-γ, following T. brucei infection, strongly correlated with the increase in LPS levels observed during infection. T. brucei-infected mice become hyperresponsive to LPS (6). The release of IFN-γ from several cell types, including T cells, is thought to prime macrophages making them more susceptible to LPS activation. Levels of LPS binding protein also increase in the plasma of infected mice, which would enhance the cellular response to LPS (46). Because of the increase in LPS present in the blood of infected patients, indolepyruvate may play a role in reducing the susceptibility of these patients to LPS or may be involved in the down-regulation of macrophage activation by all activating agents.
Indolepyruvate reduces HIF-1α protein levels. Because HIF-1α is a transcription factor, this reduction will lead to the reduction in several HIF-1α target genes, including GLUT1, VEGF, LDHA, and IL-1β. This study has concentrated on the impact of indolepyruvate on the proinflammatory cytokine, IL-1β. The exact role for IL-1β in T. brucei infection is not yet fully understood. IL-1β levels increase following T. brucei infection (9, 10) but Drennan et al. (38) demonstrated no significant difference between wild-type and IL-1R1−/− mice in T. brucei infection. A study by Quan et al. (47), however, demonstrated a role for the IL-1R1 in T. brucei infection. Intracerebral infusion of the IL-1R1 antagonist (IL-1ra) prevented the weight loss, or lack of weight gain, associated with infection. Infusion of IL-1ra and soluble type 1 TNF receptor (sTNFr1) reduced trypanosome-induced neurodegeneration, to a greater extent than TNFr1 on its own and reduced the expression of IL-1β and I-κB with no effect on TNF-α. Additional studies will be needed to determine if the impact of IL-1β during T. brucei infection would be far greater if the trypanosomes were not producing indolepyruvate. IL-1β is a potent pyrogen, and so inhibiting its production may indeed prevent fever and seizures during infection, but further studies will be needed to investigate this. Indolepyruvate may also enhance host survival by inhibiting endogenous trypanosome-induced gene expression, notably those targeted by HIF-1α, to lessen systemic pathologies associated with a persistent infection, such as anemia and cachexia, to increase the probability of parasite transmission. Although the physiological role of indolepyruvate during an infection requires further investigation, it is tempting to speculate that is has a function in regulating host inflammatory responses, particularly at the peak of the parasitemia when circulating levels of the ketoacid are likely to be highest. It is known that a proinflammatory response appears to be required to control the initial parasitemia (8) but indolepyruvate may limit the extent or systemic nature of this response during the clearance of parasites. Interestingly, the neurological symptoms associated with the late stage of the disease can occur in the absence of a CNS inflammatory response (48). Perhaps, as originally suggested by Newport et al. (15), disruption of the biogenic amine pool through metabolism of tryptophan and excretion of indolepyruvate contributes to the neurological effects, while simultaneously ameliorating a CNS inflammatory response.
Our results therefore confirm that indolepyruvate is produced by the bloodstream form T. brucei and can modulate the innate immune response. The levels of indolepyruvate present in a patient could therefore be an important diagnostic tool for the presence, and stage of disease, of T. brucei. Indolepyruvate might also have potential as an inhibitor of IL-1β in inflammatory diseases.
Materials and Methods
Reagents.
Reagents used were LPS (E. coli, serotype EH100, Alexis), CpG, Pam3Cys, R848 (Invitrogen), TNF (peprotech), and IFN-γ (R+D systems); in vivo LPS agonist (E. coli, serotype 055:B5); indolepyruvate and kynurenic acid (Sigma-Aldrich). ELISA kits used were IL-1β (DuoSet DY401), IL-6 (DuoSet DY406), TNF-α (Duoset DY410), and IL-1β (Quantikine, MLB00C). Antibodies used: β-actin (4267), IκB (44D4), phospho-p38 (9211), HydroxyHIF (3435) (Cell Signaling Technologies), anti–IL-1β (R&D, AF401-NA), anti–HIF-1α (Novus, NB100-449).
Mice and Cell Culture.
All BMDMs and PECs were isolated from C57BL/6 Harlan UK mice as previously described (23). The in vivo experiments were carried out in C57BL/6 Harlan UK mice. All experiments were carried out with prior ethical approval from the Trinity College Dublin Animal Research Ethics Committee. Each “n” represents BMDM/PECs from individual mice.
Trypanosome Cell Lines and Culture.
Trypanosomes were cultured in standard HMI-9 (bloodstream form) or SDM79 (procyclic form) medium containing 10% (vol/vol) FCS (49). The parental bloodstream (strain 13.90) and procyclic (29.13) conditional RNAi cell lines were maintained by selection in G418 and hygromycin, as described by Wirtz et al. (50). These cell lines express a T7 polymerase and tetracycline repressor. Monomorphic MITat 1.1 bloodstream cells were isolated from infected rats as described elsewhere (39).
NMR.
Bloodstream form wild-type and cASAT RNA1 cells were cultured in HMI-9 media for 24 h in the absence and presence of tetracycline (1 μg/mL). Cells were collected by centrifugation at 1,500 × g for 10 min, washed once in phosphate-buffered saline-glucose buffer (pH 7.4), centrifuged, and resuspended at 5 × 107 cells/mL in HMI-9 media supplemented with 150 μM single-labeled (indole-2-13C)-tryptophan for a 5-h incubation at 37 °C with 5% CO2. Cell viability was actively monitored via microscopy during incubation, and media samples were collected at 1-h intervals during the 5-h incubation period by removing cells using centrifugation at 14,000 × g for 5 min. Media samples were snap-frozen and stored at −80 °C until use for NMR. Next, 600-µL samples, consisting of 540 µL media and 60 µL D2O (lock), were run on an 800-MHz Agilent DD2/4.2 K premiumCOMPACT spectrometer. All 13C-NMR spectra were recorded at 25 °C, and samples were referenced to internal glucose at ∼98.64 ppm. Data were acquired with active proton decoupling and NOE, 7.35 s 90° pulse width, 1-s relaxation delay, spectral width −15.0 ppm to 235.2 ppm, with 3,200 scans performed. Postacquisition Line Broadening was 1-Hz exponential.
LDH Cytotoxicity Assay.
BMDM were seeded at 200,000 cells per well of a 96-well plate. Indolepyruvate was added to experimental wells at a final concentration of 1 mM for 25 h. Cytotox 96 nonradioactive cytotoxity assay (Promega) was carried out as per the manufacturer’s instructions.
RT-PCR.
Cells were lysed and total RNA was extracted using the RNAeasy kit (Qiagen) and was reverse-transcribed using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturers’ instructions. This cDNA served as a template for amplification of target genes by real-time PCR to determine the relative amounts of IL-1β (primers F: GGAAGCAGCCCTTCATCTTT, R: TGGCAACTGTTCCTGAACTC), LDHA (primers F: ATCTTGACCTACGTGGCTTGGA, R: CCATACAGGCACACTGGAATCTC), GLUT1 (primers F: GATCACTGCAGTTCGGCTATAA R: GTAGCGGTGGTTCCATGTT), TNF (primers F: GCCTCTTCTCATTCCTGCTT, R: TGGGAACTTCTCATCCCTTTG), or IL-6 (primers F: ACGATGATGCACTTGCAC R: ACTCCAGAAGACCAGAGGAA) mRNA. The cycling threshold method [2−(ΔΔCt)] was used for relative quantification by comparative method after normalization to RPS18 (primer F: GGATGTGAAGGATGGGAAGT, R: CCCTCTATGGGCTCGAATTT) or GAPDH expression (51).
Western Blotting.
Cells were lysed in SDS sample buffer [0.125 M Tris pH 6.8, 10% (vol/vol) glycerol, 0.02% SDS] and Western blot analysis was carried out as previously described (52). Western blots were developed using autoradiographic film or using a ChemiDoc MP gel imaging system (Bio-Rad).
ELISA.
For ELISA, 1 × 106 BMDMs per milliliter were pretreated for 30 min ± indolepyruvate, followed by 100 ng/mL LPS for 24 h. Supernatants were collected and cytokines quantified by ELISA according to manufacturers’ instructions (R&D Systems). Results are presented as mean ± SEM.
Reporter Gene Assays.
HEK293-MD2-TLR4-CD14 cells seeded at 200,000 cells per well into 96-well plates and transfected with expression vectors and luciferase reporter genes for NF-κB, ISRE, and HRE. phRL-TK reporter plasmid was cotransfected to allow normalization of data for transfection efficiency. After 18 h, cells were treated with indolepyruvate followed by LPS (100 ng/mL) and 6-h later reporter gene activity was measured. All reporter assays were done in triplicate and data are expressed as “relative stimulation” (mean SD) over the nonstimulated empty vector control, for a representative experiment, a total of three separate experiments being carried out.
Isolation of Human PBMC.
Human PBMC were isolated from human blood using Lymphoprep (Axis-Shield). Thirty milliliters of whole blood was layered on 20 mL lymphoprep and spun for 20 min at 2,000 rpm with no brake on. The PBMCs were then isolated from the middle layer and washed twice in PBS. PBMCs were maintained in RPMI supplemented with 10% (vol/vol) FCS, 2 mM l-glutamine, and 1% penicillin/streptomycin solution.
In Vivo Trial.
Mice were injected intraperitoneally with indolepyruvate (50 mg/kg), phenylpyruvate (50 mg/mg), or PBS for 30 min. LPS (15 mg/kg) or PBS were then injected intraperitoneally. Two-hours postinjection the mice were humanely euthanized and serum was isolated from whole blood and PECs were harvested.
cASAT RNAi.
A 446-bp fragment (from nuclotide 556–1002) from the cASAT (Tb927.10.3660) of T. brucei was amplified and cloned into the p2T7-177 RNAi vector. The construct was linearized with NotI and transformed into the RNAi parental cell lines using the Amaxa parasite nucleofection kit (51). The construct was targeted to the minichromosomes through homologous recombination within the 177-bp repeats. The cASAT-RNAi cell lines were selected by culturing in the presence of phleomycin and cloned by limiting dilution. Expression of the dsRNA was induced by addition of tetracycline (1 µg/mL). Total RNA was extracted using the Stratagene Absolutely RNA purification kit. The relative level of expression of the cASAT mRNA was estimated by qRT-PCR using the Brilliant SYBR Green qRT-PCR Master Mix Kit (Stratagene) in an MxPro 3000 instrument (Stratagene). The cycling threshold method [2-(ΔΔCt)] was used for relative quantification by comparative method after normalization to actin expression (51).
ASAT Assay.
Total ASAT specific activity was assayed at 340 nm in the direction of oxaloactate production using aspartate (12.5 mM) and α-ketoglutarate (10 mM) as substrates and malate dehydrogenase (5 units/mL) and NADH (0.2 mM) as a coupling enzyme/substrate as described in Bergmyer (53). At various times the trypanosomes were harvested by centrifugation and the ASAT specific activity (units per milligram) was determined in total cell lysates, prepared as described previously (54).
Aromatic Ketoacid Production.
The full ORF (TcCLB.506937.10) for NAD-linked aromatic α-hydroxy acid dehydrogenase (AHADH) was amplified and cloned into the pNIC28-Bsa4 expression vector. Following induction, recombinant AHADH was purified from the soluble fraction by Ni2+ affinity chromatography. Purified AHADH was used to measure the amount of total aromatic ketoacids in samples (up to 0.2 mL) using a standard assay (final volume of 1 mL) containing Tris buffer (25 mM, pH 7.4), NaCl (50 mM), NADH (0.25 mM), and AHADH (850 U). Total aromatic ketoacid present in the sample was determined by the decrease in absorbance at 340 nm. Appropriate blank controls were used to correct for sample volume and the assay was verified using standard solutions of phenlpyruvate.
In Vitro Trypanosome Supernatant Transfer Experiment.
Trypanosomes were grown in modified DMEM. Supernatants were harvested and used to pretreat BMDMs, followed by stimulation by LPS for 24 h. Cells were lysed in SDS sample buffer and analyzed by Western blot.
Glucose Consumption.
The levels of glucose present in BMDM supernatants were assayed enzymatically using a hexokinase/glucose-6-phosphate dehydrogenase assay. The assay (final volume 1 mL) was performed in assay buffer (Tris⋅Cl 50 mM, NaCl 50 mM, MgCl2 5 mM, pH 7.5) containing ATP (2.5 mM), NAD (2.5 mM), hexokinse (10 μ/mL) and glucose-6-phosphate dehydrogenase from Leuconostoc mesenteroides (10 μ/mL). The amount of glucose present was determined by the increase in absorbance at 340 nm following addition of the sample (typically 10 μL). The assay was verified using standard solutions of glucose.
Glycolysis Analysis Using Seahorse.
BMDMs were plated at 200,000 cells per well in XF24 plates overnight before 30-min pretreatment with varying concentrations of indolepyruvate, phenylpyruvate, or hydroxyphenylpyruvate, followed by stimulation with 100 ng/mL LPS for 24 h. XF24 Extracellular Flux analyzer (Seahorse Biosciences) was used to determine the proton production rate. Oligomycin (final concentration of 2 μM) and 2DG (final concentration of 50 mM) were used to determine the maximum glycolsis and gylcolytic reserve. Results were normalized to cell number and are represented as mean ± SD.
Hypoxia Experiment.
BMDMs were seeded in 24-well plates (1 × 106 cells/mL), incubated with indolepyruvate for 30 min. Half the cells were then maintained in normoxic conditions in a CO2 incubator, and half were placed in hypoxic conditions in a hypoxia chamber at 1% O2 (Coy Laboratories). The cells were then lysed in SDS sample buffer [0.125 M Tris pH 6.8, 10% (vol/vol) glycerol, 0.02% SDS] and a Western blot was performed.
In Vitro Trypanosome Supernatant Transfer Experiment.
For the in vitro trypanosome supernatant transfer experiment, 3 × 106 /mL MITat 1.1 wild-type and cASAT RNA1 cells were grown for 5 h in modified DMEM [DMEM + 10% (vol/vol) FCS, 0.05 mM bathocuprione disulphonic acid, 1.5 mM l-cysteine, 0.3 mM β-mercaptoethanol, 1 mM hypoxanthine, 3.0 g/L NaHCO3,, pH 7.5]. Cells were then centrifugued and the supernatants harvested. These supernatants were the used to pretreat BMDMs for 30 min, followed by stimulation by LPS at 100 ng/μL for 24 h. Cells were lysed in SDS sample loading buffer and analyzed by Western blot.
Statistical Analysis.
All statistical analysis was calculated using an unpaired two-tailed Student's t test unless otherwise specified.
SI Materials and Methods
cASAT Cloning.
The cASAT (Tb10.70.3710) ORF sequence was amplified from genomic DNA using PfuUltra II Fusion DNA polymerase and TACTTCCAATCCATGTCCAGGCCCTTTAAGGACTTAGCACCC (forward primer) and TATCCACCTTTACTGCTACTTGTTACGCACGTGTCGGACAAC (reverse primer), with the underlined region corresponding to the sequence of the ORF. The amplification product (∼1.2 kb) was inserted into pNIC28-Bsa4 using ligation independent cloning. Briefly BsaI-digested pNIC28-Bsa4 vector was treated with T4 DNA polymerase in the presence of dGTP at 22 °C for 30 min, then at 75 °C for 20 min to create single stranded overhangs, whereas the TbcASAT amplification product was also treated with T4 DNA Polymerase in the presence of dCTP at 22 °C for 30 min, then at 75 °C for 20 min to create the complementary single-stranded overhangs. The vector and insert were allowed to anneal and then used to transfect chemically competent Escherichia coli BL21 (DE3) cells. Selection was performed using kanamycin. Positive clones were subjected to small scale induction experiments and selected clones were subjected to full-length sequencing.
cASAT Recombinant Protein Expression and Activity Assays.
Expression of recombinant His-tagged TbcASAT was performed by incubation (500 mL) with isfopropyl-β-d-thiogalactopyranoside (IPTG; 0.2 mM) overnight at 18 °C. Following 18-h incubations, E. coli cells were collected by centrifugation and washed twice with cold PBS. The pellets were resuspended in 50 mL lysis buffer (300 mM NaCl, 20 mM Tris, 20 mM Imidazole, 1 mM 2-mercaptoethanol, pH 8) with 50 µg/mL Lysozyme at 4 °C. The cells were lysed using a French press homogenizer. Cell debris was removed by centrifugation at 14,000 × g for 30 min. The resulting supernatants were subjected to chromatography on Ni2+ agarose columns (5 mL) at 4 °C. The column was washed three times with lysis buffer, then the protein was eluted using lysis buffer with 250 mM imidazole added, and collected into fractions. The fractions were then combined, dialyzed against Tris⋅Cl (25 mM) and NaCl (100 mM) containing PLP (0.1 mM). Following dialysis active enzyme was concentrated using Amicon MW 3,500 centrifugal filters. The protein was stored in 20% (wt/vol) glyercol at −80 °C. The assays were performed in standard Tris buffer (100 mM, pH 7.5) NaCl (25 mM) and NADH (0.25 mM). Enzyme activity was determined by measuring the decrease in absorbance at 340 nM. In the case of aromatic amino acid substrates the coupling enzyme was Trypanosoma cruzi recombinant AHADH (20 units/mL); commercial malate dehydrogenase (20 units/mL) was used when asparate was the amino donor.
Acknowledgments
We thank Paul Voorheis for advice and comments, and Brigitta Stockinger (Medical Research Council National Institute for Medical Research) for access to bone marrow-derived macrophages from AhR−/− mice. This study was supported in part by the Science Foundation Ireland, the Wellcome Trust, and the European Research Council.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. E.J.P. is a Guest Editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1608221113/-/DCSupplemental.
References
- 1.Stich A, Abel PM, Krishna S. Human African trypanosomiasis. BMJ. 2002;325(7357):203–206. doi: 10.1136/bmj.325.7357.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cross GA. Cellular and genetic aspects of antigenic variation in trypanosomes. Annu Rev Immunol. 1990;8:83–110. doi: 10.1146/annurev.iy.08.040190.000503. [DOI] [PubMed] [Google Scholar]
- 3.Pays E, Vanhamme L, Berberof M. Genetic controls for the expression of surface antigens in African trypanosomes. Annu Rev Microbiol. 1994;48:25–52. doi: 10.1146/annurev.mi.48.100194.000325. [DOI] [PubMed] [Google Scholar]
- 4.Morrison LJ, Marcello L, McCulloch R. Antigenic variation in the African trypanosome: Molecular mechanisms and phenotypic complexity. Cell Microbiol. 2009;11(12):1724–1734. doi: 10.1111/j.1462-5822.2009.01383.x. [DOI] [PubMed] [Google Scholar]
- 5.Thuita JK, et al. Trypanosoma brucei rhodesiense transmitted by a single tsetse fly bite in vervet monkeys as a model of human African trypanosomiasis. PLoS Negl Trop Dis. 2008;2(5):e238. doi: 10.1371/journal.pntd.0000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Magez S, et al. The glycosyl-inositol-phosphate and dimyristoylglycerol moieties of the glycosylphosphatidylinositol anchor of the trypanosome variant-specific surface glycoprotein are distinct macrophage-activating factors. J Immunol. 1998;160(4):1949–1956. [PubMed] [Google Scholar]
- 7.Magez S, Radwanska M, Beschin A, Sekikawa K, De Baetselier P. Tumor necrosis factor alpha is a key mediator in the regulation of experimental Trypanosoma brucei infections. Infect Immun. 1999;67(6):3128–3132. doi: 10.1128/iai.67.6.3128-3132.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paulnock DM, Freeman BE, Mansfield JM. Modulation of innate immunity by African trypanosomes. Parasitology. 2010;137(14):2051–2063. doi: 10.1017/S0031182010001460. [DOI] [PubMed] [Google Scholar]
- 9.Nyakundi JN, Crawley B, Pentreath VW. The relationships between endotoxins, nitric oxide and inflammatory cytokines in blood and intestinal tissues in experimental Trypanosoma brucei brucei infections. Parasitology. 2002;124(Pt 6):597–604. doi: 10.1017/s0031182002001683. [DOI] [PubMed] [Google Scholar]
- 10.Sileghem M, Darji A, Hamers R, De Baetselier P. Modulation of IL-1 production and IL-1 release during experimental trypanosome infections. Immunology. 1989;68(1):137–139. [PMC free article] [PubMed] [Google Scholar]
- 11.Mitchell LA, Pearson TW, Gauldie J. Interleukin-1 and interleukin-2 production in resistant and susceptible inbred mice infected with Trypanosoma congolense. Immunology. 1986;57(2):291–296. [PMC free article] [PubMed] [Google Scholar]
- 12.Stibbs HH, Seed JR. Chromatographic evidence for the synthesis of possible sleep-mediators in Trypanosoma brucei gambiense. Experientia. 1973;29(12):1563–1565. doi: 10.1007/BF01943919. [DOI] [PubMed] [Google Scholar]
- 13.Stibbs HH, Seed JR. Short-term metabolism of (14-C) tryptophan in rats infected with Trypanosoma brucei gambiense. J Infect Dis. 1975;131(4):459–462. doi: 10.1093/infdis/131.4.459. [DOI] [PubMed] [Google Scholar]
- 14.Seed JR, Hall JE, Sechelski J. Phenylalanine metabolism in Microtus montanus chronically infected with Trypanosoma brucei gambiense. Comp Biochem Physiol B. 1982;71(2):209–215. doi: 10.1016/0305-0491(82)90242-5. [DOI] [PubMed] [Google Scholar]
- 15.Newport GR, Page CR, 3rd, Ashman PU, Stibbs HH, Seed JR. Alteration of free serum amino acids in voles infected with Trypanosoma brucei gambiense. J Parasitol. 1977;63(1):15–24. [PubMed] [Google Scholar]
- 16.Hall JE, Seed JR. Increased urinary excretion of aromatic amino acid catabolites by Microtus montanus chronically infected with Trypanosoma brucei gambiense. Comp Biochem Physiol B. 1984;77(4):755–760. doi: 10.1016/0305-0491(84)90309-2. [DOI] [PubMed] [Google Scholar]
- 17.Hall JE, Seed JR, Sechelski JB. Multiple alpha-keto aciduria in Microtus montanus chronically infected with Trypanosoma brucei gambiense. Comp Biochem Physiol B. 1985;82(1):73–78. doi: 10.1016/0305-0491(85)90130-0. [DOI] [PubMed] [Google Scholar]
- 18.el Sawalhy A, Seed JR, el Attar H, Hall JE. Catabolism of tryptophan by Trypanosoma evansi. J Eukaryot Microbiol. 1995;42(6):684–690. doi: 10.1111/j.1550-7408.1995.tb01616.x. [DOI] [PubMed] [Google Scholar]
- 19.El Sawalhy A, Seed JR, Hall JE, El Attar H. Increased excretion of aromatic amino acid catabolites in animals infected with Trypanosoma brucei evansi. J Parasitol. 1998;84(3):469–473. [PubMed] [Google Scholar]
- 20.Hunter AG. Urine odour in a camel suffering from surra (T. evansi infection) Trop Anim Health Prod. 1986;18(3):146–148. doi: 10.1007/BF02359524. [DOI] [PubMed] [Google Scholar]
- 21.Seed JR, Hall JE, Price CC. A physiological mechanism to explain pathogenesis in African trypanosomiasis. Contrib Microbiol Immunol. 1983;7:83–94. [PubMed] [Google Scholar]
- 22.Berger BJ, Dai WW, Wang H, Stark RE, Cerami A. Aromatic amino acid transamination and methionine recycling in trypanosomatids. Proc Natl Acad Sci USA. 1996;93(9):4126–4130. doi: 10.1073/pnas.93.9.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tannahill GM, et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature. 2013;496(7444):238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGettrick AF, O’Neill LA. How metabolism generates signals during innate immunity and inflammation. J Biol Chem. 2013;288(32):22893–22898. doi: 10.1074/jbc.R113.486464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Creek DJ, et al. Metabolomics guides rational development of a simplified cell culture medium for drug screening against Trypanosoma brucei. Antimicrob Agents Chemother. 2013;57(6):2768–2779. doi: 10.1128/AAC.00044-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cazzulo Franke MC, Vernal J, Cazzulo JJ, Nowicki C. The NAD-linked aromatic alpha-hydroxy acid dehydrogenase from Trypanosoma cruzi. A new member of the cytosolic malate dehydrogenases group without malate dehydrogenase activity. Eur J Biochem. 1999;266(3):903–910. doi: 10.1046/j.1432-1327.1999.00926.x. [DOI] [PubMed] [Google Scholar]
- 27.Berger LC, Wilson J, Wood P, Berger BJ. Methionine regeneration and aspartate aminotransferase in parasitic protozoa. J Bacteriol. 2001;183(15):4421–4434. doi: 10.1128/JB.183.15.4421-4434.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marciano D, et al. Biochemical characterization of stage-specific isoforms of aspartate aminotransferases from Trypanosoma cruzi and Trypanosoma brucei. Mol Biochem Parasitol. 2008;161(1):12–20. doi: 10.1016/j.molbiopara.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 29.Alafiatayo RA, Crawley B, Oppenheim BA, Pentreath VW. Endotoxins and the pathogenesis of Trypanosoma brucei brucei infection in mice. Parasitology. 1993;107(Pt 1):49–53. doi: 10.1017/s0031182000079397. [DOI] [PubMed] [Google Scholar]
- 30.Pentreath VW, et al. Endotoxin antibodies in African sleeping sickness. Parasitology. 1997;114(Pt 4):361–365. doi: 10.1017/s0031182096008530. [DOI] [PubMed] [Google Scholar]
- 31.Pentreath VW, Alafiatayo RA, Crawley B, Doua F, Oppenheim BA. Endotoxins in the blood and cerebrospinal fluid of patients with African sleeping sickness. Parasitology. 1996;112(Pt 1):67–73. doi: 10.1017/s0031182000065082. [DOI] [PubMed] [Google Scholar]
- 32.Greenwood BM. Possible role of a B-cell mitogen in hypergammaglobulinaemia in malaria and trypanosomiasis. Lancet. 1974;1(7855):435–436. doi: 10.1016/s0140-6736(74)92386-1. [DOI] [PubMed] [Google Scholar]
- 33.Chen C, Pore N, Behrooz A, Ismail-Beigi F, Maity A. Regulation of glut1 mRNA by hypoxia-inducible factor-1. Interaction between H-ras and hypoxia. J Biol Chem. 2001;276(12):9519–9525. doi: 10.1074/jbc.M010144200. [DOI] [PubMed] [Google Scholar]
- 34.Hayashi M, et al. Induction of glucose transporter 1 expression through hypoxia-inducible factor 1alpha under hypoxic conditions in trophoblast-derived cells. J Endocrinol. 2004;183(1):145–154. doi: 10.1677/joe.1.05599. [DOI] [PubMed] [Google Scholar]
- 35.Bittinger MA, Nguyen LP, Bradfield CA. Aspartate aminotransferase generates proagonists of the aryl hydrocarbon receptor. Mol Pharmacol. 2003;64(3):550–556. doi: 10.1124/mol.64.3.550. [DOI] [PubMed] [Google Scholar]
- 36.Nguyen LP, et al. D-amino acid oxidase generates agonists of the aryl hydrocarbon receptor from D-tryptophan. Chem Res Toxicol. 2009;22(12):1897–1904. doi: 10.1021/tx900043s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGuire J, Whitelaw ML, Pongratz I, Gustafsson JA, Poellinger L. A cellular factor stimulates ligand-dependent release of hsp90 from the basic helix-loop-helix dioxin receptor. Mol Cell Biol. 1994;14(4):2438–2446. doi: 10.1128/mcb.14.4.2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drennan MB, et al. The induction of a type 1 immune response following a Trypanosoma brucei infection is MyD88 dependent. J Immunol. 2005;175(4):2501–2509. doi: 10.4049/jimmunol.175.4.2501. [DOI] [PubMed] [Google Scholar]
- 39.Voorheis HP. The effect of T. brucei (S-42) on host carbohydrate metabolism: Liver production and peripheral tissue utilization of glucose. Trans R Soc Trop Med Hyg. 1969;63(1):122–123. doi: 10.1016/0035-9203(69)90088-1. [DOI] [PubMed] [Google Scholar]
- 40.Li JV, et al. Metabonomic investigation of single and multiple strain Trypanosoma brucei brucei infections. Am J Trop Med Hyg. 2011;84(1):91–98. doi: 10.4269/ajtmh.2011.10-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Creek DJ, et al. Probing the metabolic network in bloodstream-form Trypanosoma brucei using untargeted metabolomics with stable isotope labelled glucose. PLoS Pathog. 2015;11(3):e1004689. doi: 10.1371/journal.ppat.1004689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mony BM, et al. Genome-wide dissection of the quorum sensing signalling pathway in Trypanosoma brucei. Nature. 2014;505(7485):681–685. doi: 10.1038/nature12864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garzón E, et al. The Trypanosoma brucei gambiense secretome impairs lipopolysaccharide-induced maturation, cytokine production, and allostimulatory capacity of dendritic cells. Infect Immun. 2013;81(9):3300–3308. doi: 10.1128/IAI.00125-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aoki R, Aoki-Yoshida A, Suzuki C, Takayama Y. Protective effect of indole-3-pyruvate against ultraviolet b-induced damage to cultured HaCaT keratinocytes and the skin of hairless mice. PLoS One. 2014;9(5):e96804. doi: 10.1371/journal.pone.0096804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nyakundi JN, Crawley B, Smith RA, Pentreath VW. The relationships between intestinal damage and circulating endotoxins in experimental Trypanosoma brucei brucei infections. Parasitology. 2002;124(Pt 6):589–595. doi: 10.1017/s0031182002001701. [DOI] [PubMed] [Google Scholar]
- 46.Ngure RM, et al. Lipopolysaccharide binding protein in the acute phase response of experimental murine Trypanosoma brucei brucei infection. Res Vet Sci. 2009;86(3):394–398. doi: 10.1016/j.rvsc.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 47.Quan N, He L, Lai W. Intraventricular infusion of antagonists of IL-1 and TNF alpha attenuates neurodegeneration induced by the infection of Trypanosoma brucei. J Neuroimmunol. 2003;138(1-2):92–98. doi: 10.1016/s0165-5728(03)00122-x. [DOI] [PubMed] [Google Scholar]
- 48.MacLean L, Reiber H, Kennedy PG, Sternberg JM. Stage progression and neurological symptoms in Trypanosoma brucei rhodesiense sleeping sickness: Role of the CNS inflammatory response. PLoS Negl Trop Dis. 2012;6(10):e1857. doi: 10.1371/journal.pntd.0001857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hirumi H, Hirumi K. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J Parasitol. 1989;75(6):985–989. [PubMed] [Google Scholar]
- 50.Wirtz E, Leal S, Ochatt C, Cross GA. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol. 1999;99(1):89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- 51.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 52.Fitzgerald KA, et al. Mal (MyD88-adapter-like) is required for Toll-like receptor-4 signal transduction. Nature. 2001;413(6851):78–83. doi: 10.1038/35092578. [DOI] [PubMed] [Google Scholar]
- 53.Bergmeyer HU. Methods of Enzymatic Analysis. Vol 3 Chemie; Weinheim, Germany: 1983. [Google Scholar]
- 54.Spitznagel D, et al. Alanine aminotransferase of Trypanosoma brucei—A key role in proline metabolism in procyclic life forms. FEBS J. 2009;276(23):7187–7199. doi: 10.1111/j.1742-4658.2009.07432.x. [DOI] [PubMed] [Google Scholar]