Significance
Flower size can change rapidly in evolution; in particular, the frequent transition from animal-mediated out-crossing to self-pollination is often associated with a dramatic, yet rapid and specific, reduction in flower size. Here we demonstrate that the small petals of the selfing red Shepherd’s Purse (Capsella rubella) are because of a specific reduction in the activity of a general growth factor in petals. Different-strength versions of this growth gene were already present in the ancestral out-breeding population, and capture of a weak version from this pool can explain the rapid reduction of petal size in C. rubella. The additive effects of segregating small-effect mutations with low pleiotropy allowed specific modulation of petal size to enable adaptation to a new mode of reproduction.
Keywords: morphological evolution, growth control, standing variation, organ-specific evolution, intronic cis-regulatory element
Abstract
Mating system shifts recurrently drive specific changes in organ dimensions. The shift in mating system from out-breeding to selfing is one of the most frequent evolutionary transitions in flowering plants and is often associated with an organ-specific reduction in flower size. However, the evolutionary paths along which polygenic traits, such as size, evolve are poorly understood. In particular, it is unclear how natural selection can specifically modulate the size of one organ despite the pleiotropic action of most known growth regulators. Here, we demonstrate that allelic variation in the intron of a general growth regulator contributed to the specific reduction of petal size after the transition to selfing in the genus Capsella. Variation within this intron affects an organ-specific enhancer that regulates the level of STERILE APETALA (SAP) protein in the developing petals. The resulting decrease in SAP activity leads to a shortening of the cell proliferation period and reduced number of petal cells. The absence of private polymorphisms at the causal region in the selfing species suggests that the small-petal allele was captured from standing genetic variation in the ancestral out-crossing population. Petal-size variation in the current out-crossing population indicates that several small-effect mutations have contributed to reduce petal-size. These data demonstrate how tissue-specific regulatory elements in pleiotropic genes contribute to organ-specific evolution. In addition, they provide a plausible evolutionary explanation for the rapid evolution of flower size after the out-breeding-to-selfing transition based on additive effects of segregating alleles.
Mating system shifts toward self-fertilization occurred repeatedly during evolution, most likely to provide reproductive assurance and because of the transmission advantage of selfing mutations (1–3). In both plant and animal kingdoms this transition has been accompanied by a set of characteristic morphological changes in reproductive organs termed “the selfing syndrome” (4–7), implying that the mating system strongly constrains the evolution of reproductive-organ morphology. Still, it is unclear whether repeated evolution of these morphological changes is a result of positive selection, of the relaxation of purifying selection, or results from stronger genetic drift in selfing populations. In plants, the genetic basis underlying the reduction in flower size of selfing species is unclear. In particular, the observation that this reduction is often highly specific for floral organs contrasts with the pleiotropic activity of almost all known regulators of shoot-organ growth in both leaves and flowers, raising the question of how natural evolution has brought about organ-specific changes with a largely universal tool-kit. Different hypotheses have therefore been formulated to explain how such polygenic traits could be modified in a single organ (8, 9); these either assume mutations in an upstream gene affecting the regulation of a pleitropic gene in a given organ, or mutations affecting the activity of this pleitropic gene in an organ-specific manner. The latter would imply that such genes would have evolved organ-specific regulatory elements, allowing the regulation of their function independently in different organs (10).
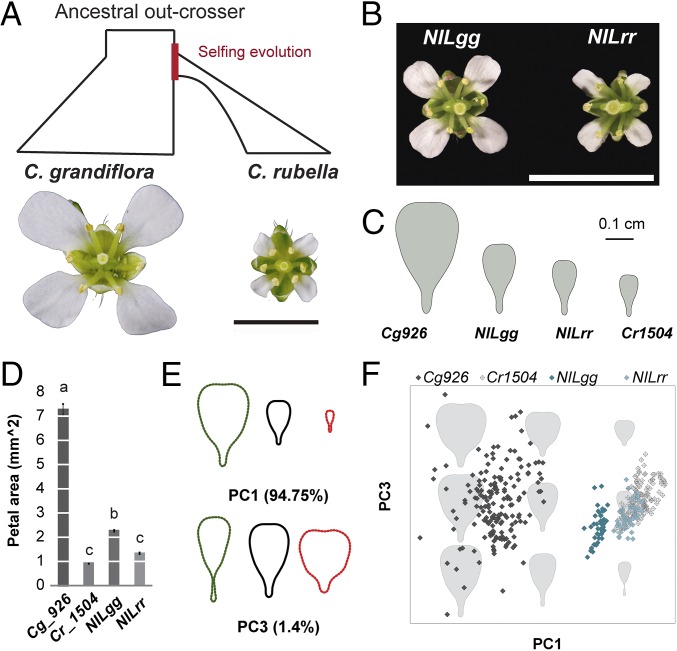
The genus Capsella provides a tractable model to study the genetics and evolution of the selfing syndrome (11–17). Within the last 200,000 y, breakdown of self-incompatibility in the out-breeding ancestor Capsella grandiflora gave rise to the self-fertilizing Capsella rubella (18). The latter has since undergone a severe reduction in effective population size and evolved the characteristic selfing syndrome (14) (Fig. 1A). One of the most prominent changes was a larger than fivefold reduction in flower size (Fig. 1 A and D) without altering overall plant size (14). Seven quantitative trait loci (QTLs) together explain ∼60% of the size difference between C. grandiflora and C. rubella petals (14) (SI Appendix, Fig. S1A). The responsible genes, causal polymorphisms, and evolutionary path underlying these QTLs are still unknown.
Fig. 1.
PAQTL_6 contributed to petal-size reduction after the transition to selfing. (A) The evolution of the selfing species C. rubella through the breakdown of the self-incompatibility system found in C. grandiflora, has been followed by a strong reduction of flower size. (B) Flowers of NILgg and NILrr plants differing at the petal-size QTL PAQTL_6. (Scale bars in A and B, 5 mm.) (C and D) The average petal shape and size of C. grandiflora (Cg926), C. rubella (Cr1504), NILgg, and NILrr (C) and the quantification of petal area (D) indicate that PAQTL_6 contributed to reducing flower size after the transition to selfing. Values are mean ± SEM from 11, 12, 18, and 18 individuals, respectively. Letters indicate significant differences as determined by Tukey’s honest significant difference test. (E and F) EFD-PCA of petal outlines demonstrates that PAQTL_6 affects the overall petal dimension. Effects of variation in PC1 and PC2, which together explain more than 95% of the phenotypic variance, are shown in E. Distribution of individual petals projected on the PC1/PC2 morphospace is shown in F. Note that swapping the genotype at PAQTL_6 from the homozygote for C. grandiflora allele (NILgg) to the homozygote for C. rubella allele (NILrr) brings the petal geometry closer to C. rubella.
Genetic evidence suggests that after the break-down of self-incompatibility in C. grandiflora, the selfing syndrome evolved relatively rapidly in the derived lineage C. rubella; crosses between geographically distant C. rubella accessions and comparative QTL mapping indicate a shared genetic basis for the selfing syndrome throughout most of C. rubella, suggesting that it evolved before the geographical spread of C. rubella (15, 14). Rapid changes in floral morphology and selfing efficiency have also been observed in artificial pollinator loss experiments in Mimulus guttatus (19). It is therefore conceivable that the sorting of standing variation from the founder population contributed to selecting mating system modifiers in the selfing lineage, improving autogamy and serving as an evolutionary path to the evolution of the selfing syndrome. However, testing this hypothesis has been difficult, because no genes/mutations underlying selfing-syndrome traits have been identified.
To address these issues, we herein identified the genetic basis and analyzed the evolutionary history of the QTL PAQTL_6, which was predicted to have the largest contribution to the evolution of petal size after the transition to selfing in the genus Capsella (14).
Results and Discussion
Organ-Specific Effects of Genetic Variation Contribute to the Selfing Syndrome in Capsella.
To confirm the effect of PAQTL_6 on petal size, we introgressed its C. grandiflora allele into C. rubella to generate a near isogenic line (NIL) (Fig. 1B). Petals of NIL homozygotes for the C. rubella allele (NILrr) were on average 35% smaller than those of NIL homozygotes for the C. grandiflora allele (NILgg), with this locus explaining up to 14.5% of the species difference (Fig. 1 B–D). Principal component analysis (PCA) on elliptic Fourier descriptors (EFD) of petal outlines showed that 94.75% of the total variance between the NILs and parental species could be captured with only one principal component (PC), PC1 (Fig. 1 E and F), representing variation mostly in petal area. PC1 clearly discriminated the parental species (Kruskal–Wallis test, P = 1.6 × 10−76), and NILgg versus NILrr petals (P = 1.4 × 10−53). PC3, which reflects variation in the length/width ratio could only moderately separate NILgg and NILrr petals (P = 0.0003). Thus, PAQTL_6 has contributed to reducing both dimensions of the petals with a slightly stronger effect on petal length. All flower organs are shorter in C. rubella than in C. grandiflora, with petals showing the strongest decrease (SI Appendix, Fig. S1D). In contrast, NILgg and NILrr plants differ exclusively in petal size (SI Appendix, Fig. S1 B–D). Allelic variation at the locus therefore specifically decreased petal size after the transition to selfing, suggesting that this QTL affects the function of a petal-specific growth regulator or modifies the activity of a general growth regulator specifically in petals.
Polymorphisms Within the Intron of a General Growth Regulator Underlie PAQTL_6.
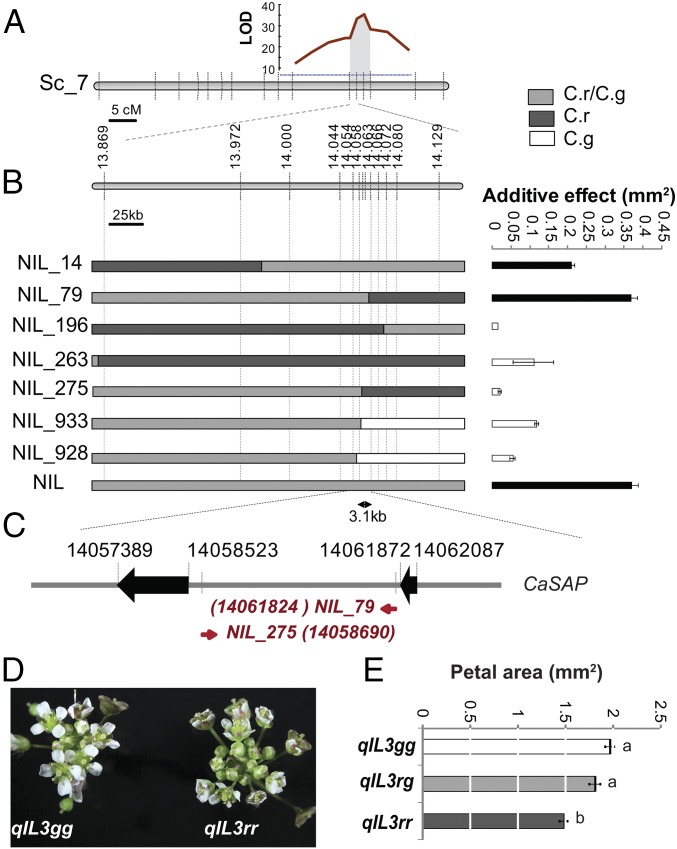
Genetic mapping on 300 progeny individuals of NIL plants heterozygous at PAQTL_6 (NILrg) refined the initial QTL position on chromosome 7 to an interval between 13.427 and 14.560 Mb (Fig. 2A). Screening over 3,000 progeny individuals of NILrg for recombinants in this interval and testing the petal-size segregation in their progenies narrowed the underlying polymorphisms to a 3.1-kb interval comprised of between 14,058,690 bp and 14,061,824 bp on chromosome 7 (Fig. 2 B and C). To confirm this location, we crossed the recombinants NIL_79 to NIL_275 and NIL_933 to NIL_139 to generate quasi-isogenic lines segregating for 3.1-kb (qIL3) and 70-kb (qIL70) intervals around PAQTL_6, respectively, but fixed for the flanking regions (Fig. 2 B, D, and E and SI Appendix, Fig. S2). In the progenies of both lines, homozygotes for the C. rubella allele displayed ∼25% smaller petals than homozygotes for the C. grandiflora allele, without differences in other organs (Fig. 2 D and E and SI Appendix, Fig. S2). Heterozygotes displayed an intermediate petal size, indicating an additive effect of the causal polymorphisms. Thus, segregating polymorphisms in a 3.1-kb interval on chromosome 7 underlie the organ-specific effect of PAQTL_6 on petal size.
Fig. 2.
Variation in the SAP intron underlies petal-size reduction. (A) Rough mapping localized PAQTL_6 between 13 and 16 Mb on Capsella scaffold 7. Logarithm of odds (LOD) score plot for petal size associated with the PAQTL_6 is shown. The gray area delimits the 2-LOD score interval; dashed horizontal blue line indicates 5% significance threshold. (B) Fine mapping of PAQTL_6. Additive phenotypic effects of allelic variation in the segregating regions (gray) on petal size in progenies of selected recombinants indicate that the causal polymorphisms are contained in a 3.1-kb region. Values are mean ± SEM from 12 to 16 individuals per genotype. Black filled bars indicate effects significantly different from 0 at P < 0.01 (Student’s t test). Diagrams (Left) represent the genotype of the selected recombinants. The color code corresponding to the different genotypes is indicated in the upper right corner. (C) The recombination breakpoints in the most informative recombinants identify the SAP intron as the region underlying PA_QTL6. Red numbers in brackets indicate the position of the first recombinant SNP. (D) qIL segregating only for the SAP intron vary in floral display. qIL3gg and qIL3rr indicates plants homozygous for the C. grandiflora and C. rubella allele respectively. (E) Average petal size of indicated genotypes. Values are mean ± SEM of 19, 22, and 24 individuals for qIL3gg, qIL3rg, and qIL3gg, respectively. Letters indicate significant differences as determined by Tukey’s honest significant difference test.
This interval contains part of the intron of the ortholog to the Arabidopsis thaliana STERILE APETALA (AtSAP) gene (Fig. 2C and SI Appendix, Fig. S3). AtSAP encodes an F-box protein acting as the specificity-determining component of an SCF-type E3 ubiquitin ligase; it is thought to act as a cadastral gene during flower development (20) and as a general growth promoter targeting the negative regulators of meristemoid proliferation, PEAPOD 1 and 2 for degradation (21). Indeed, three independent transfer DNA (T-DNA) insertions within the SAP locus in A. thaliana, cosegregated with a strong decrease in plant stature that affected the size of both vegetative and floral organs (SI Appendix, Fig. S4). Transforming the C. rubella SAP allele into the sap-1 loss-of-function mutant background rescued leaf and petal growth indicating that SAP function is conserved between Arabidopsis and Capsella. Transforming A. thaliana wild-type with a C. grandiflora SAP allele (SAPg_2) increased petal size by 30% compared with a 10% size increase when replacing the intron with the C. rubella allele (SAPr_2) to give SAPg_intron_r2 (SI Appendix, Fig. S5). Of note, these alleles were isolated from C. grandiflora and C. rubella BAC libraries and differ in several polymorphisms from the alleles segregating in our NILs. Thus, independent SAP genomic sequences are sufficient to reproduce the effect of PAQTL_6 in transgenic plants, validating SAP as the underlying gene and suggesting that functional differences in SAP are wide-spread between the two species.
Developmental Basis of Petal-Size Evolution.
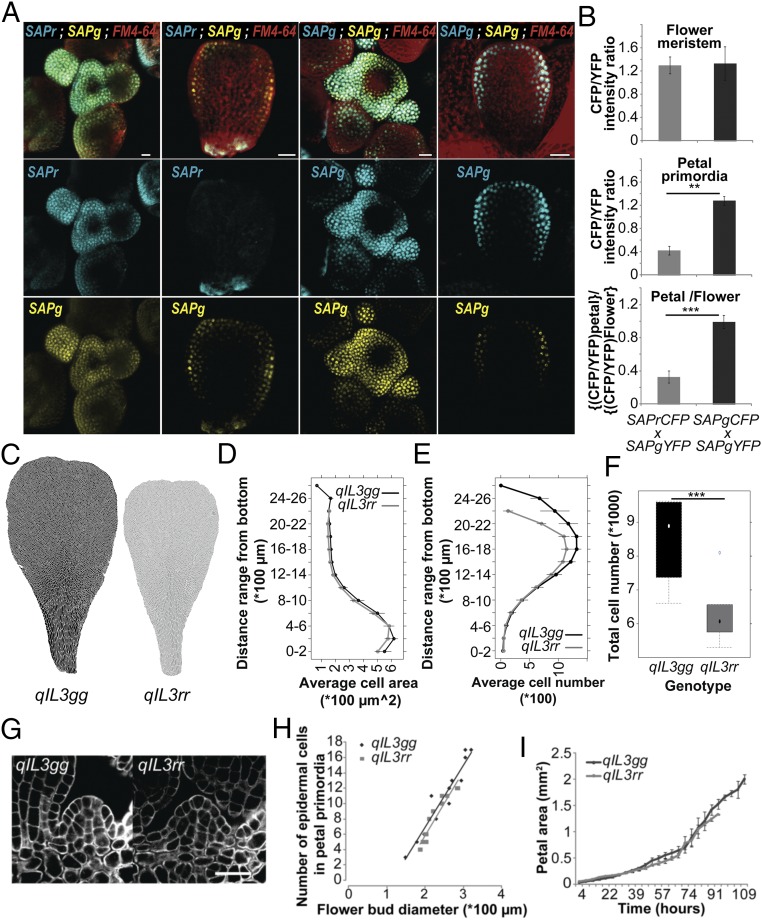
Although SAP has been recently shown to regulate meristemoid proliferation in leaves (21), it also affects the recruitment of cells into organ primordia as well as fertility and flower organ identity. To understand how decreased SAP activity affects petal size, we conducted a detailed cellular analysis of qIL3 and NIL petals (Fig. 3 C–F and SI Appendix, Fig. S7). Both qIL3rr and NILrr petals had fewer cells than qIL3gg and NILgg, respectively, whereas cell size was not affected (Fig. 3 C–F and SI Appendix, Fig. S7 G–J). This defect was particularly pronounced in the distal petal region, mirroring the sap-2 mutant phenotype in A. thaliana (Fig. 3 and SI Appendix, Fig. S7), and is consistent with predominant SAP expression in the distal region during petal development (Fig. 3A and SI Appendix, Fig. S6 J–L). The SAP polymorphism does not affect the number of cells recruited into the petal primordia or the rate of petal growth (Fig. 3 G–I). These results are consistent with the observations that C. rubella petals mainly differ from C. grandiflora petals by the total number of cells and that the petals of both parental species grow at the same rate (14). Therefore, this polymorphism reduces petal size by shortening the length of the cell proliferation period, leading to a reduced number of cells in the distal part of the petal.
Fig. 3.
Mutations in a petal-specific enhancer of SAP decrease petal cell number as a result of a shorter proliferation period. (A) Inflorescence meristems (Left) and young developing petals (Right) of A. thaliana plants expressing SAPr–CFP and SAPg–YFP (Left two columns) or SAPg–CFP and SAPg–YFP (Right two columns). Top row represent an overlay of the CFP, YFP, and FM4-64 channels, and the Middle and Bottom rows show the CFP and YFP channels, respectively. (Scale bars, 20 µm.) SAPg–CFP; SAPg–YFP plants were used to control for difference in tissue-specific accumulation between the CFP and YFP reporters. (B) Quantification of CFP and YFP fluorescence signal in the nucleus of flower-meristem and petal-primordia cells. Values are mean ± SEM from five nuclei in three F1 plants each. *P < 0.05, **P < 0.01, and ***P < 0.001, as determined with a Student’s t test. (C) Cell-outline segmentation of qIL3gg and qIL3rr petals. Segmented cells are shown in black for qIL3gg and gray for qIL3rr. (D) Average cell size in segments along the longitudinal petals axes is identical for qIL3gg and qIL3rr. Values are mean ± SEM from six petals per genotype. (E) qIL3gg plants develop more cells in the distal part of the petals than qIL3rr. Values are mean cell number ± SEM in segments along the longitudinal axes for six petals per genotype. (F) Total cell number in qIL3gg and qIL3rr petals (n = 6). (G) mPS-PI staining of petal primordia in qIL3gg and qIL3rr flower buds with a diameter of 250 µm. (Scale bar, 20 µm.) (H) Sizes of petal primordia expressed as the number of epidermal cells in the medial section do not differ between qIL3gg and qIL3rr plants when scaled by overall bud diameter. (I) Developmental series of petal growth in qIL3gg and qIL3rr plants indicates that petals in the two genotypes grow at an identical rate, but for a different period. Values are mean ± SEM from four individuals per genotype.
Polymorphisms Within an Organ-Specific Enhancer Reduce SAP Activity During Petal Growth.
Given the strong pleiotropic phenotypes of the sap T-DNA insertions in A. thaliana, it is unlikely that C. grandiflora/C. rubella polymorphisms globally reduce SAP function. Consistently, we did not observe any global changes in expression pattern or level of SAP or its presumed downstream target gene AGAMOUS (AG) (20) during qIL3 plant development (SI Appendix, Fig. S6N). There was no detectable difference in the splicing pattern between the two alleles (SI Appendix, Fig. S6O). A plausible explanation for the different allele effects is that they differ in expression specifically during petal development. To test this, we generated dual-reporter lines that allowed us to simultaneously monitor the expression of SAPg and SAPr during plant development (Fig. 3 A and B). Col-0 plants were transformed with either SAPr fused to CFP (SAPr–CFP) or SAPg fused to YFP (SAPg–YFP), using the SAP alleles from our NILs; both constructs reproduced the differential petal-growth promotion seen with the independent unmodified constructs, confirming their equivalent functionality (SI Appendix, Fig. S5). T1 plants were crossed and the expression of the two reporters was monitored in F1 individuals. In parallel, Col-0 plants were also transformed with SAPg fused to CFP (SAPg–CFP) and crossed to SAPg–YFP plants to control for differences in the behavior of the two reporter proteins. In these control plants, the CFP/YFP ratio was equivalent in the flower meristem and in cells of actively dividing petal primordia. In contrast, the CFP/YFP ratio decreases by 60% in the petal primordia of SAPr–CFP; SAPg–YFP plants compared with their flower meristems (Fig. 3B). Thus, the polymorphisms at PAQTL_6 reduce SAP expression specifically in petal primordia of C. rubella plants. Together with the fine-mapping, these findings argue that the causal polymorphisms modify the activity of an organ-specific enhancer in the highly conserved intron of the SAP locus (Fig. 4B) (22).
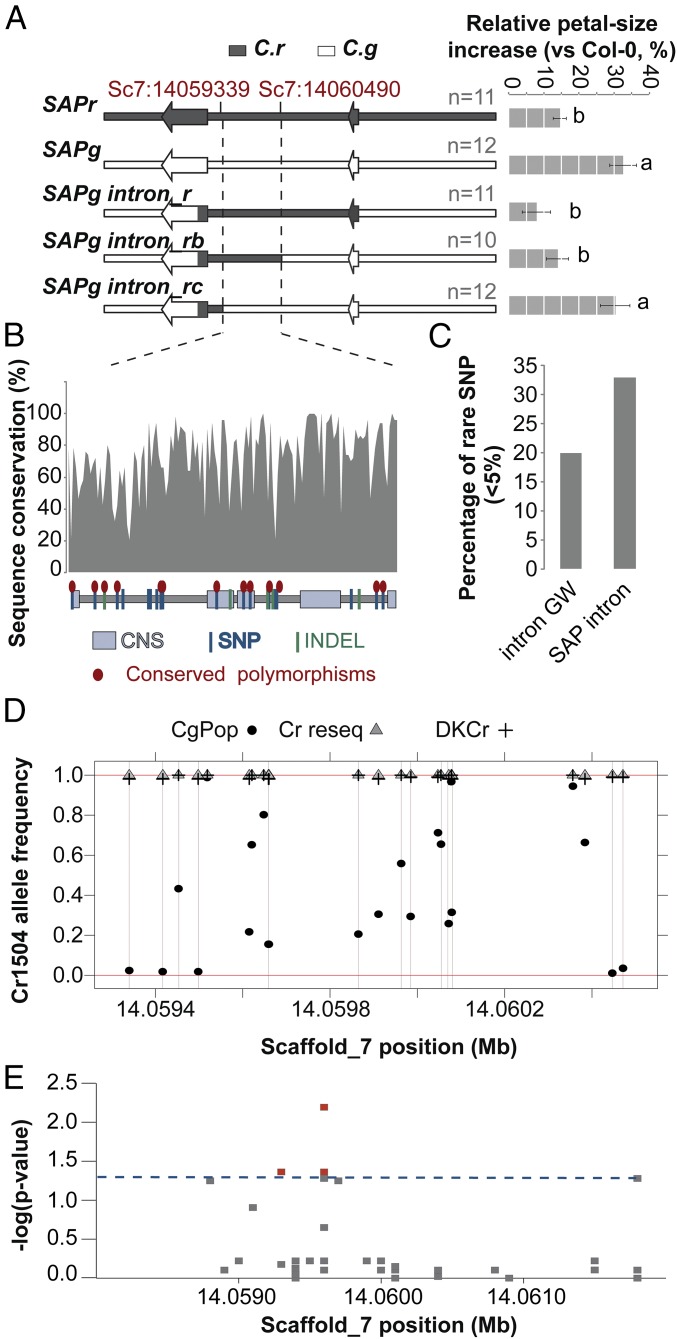
Fig. 4.
The SAP small-petal allele has been captured from C. grandiflora standing variation. (A) Effect of SAPg/SAPr chimeric genomic constructs on the size of Col-0 petals indicates that the causal polymorphisms are contained in a 1.1-kb region within SAP intron. The schemes of the constructs used are shown (Left); bar chart (Right) displays the percentage increase in petal size. Values are mean of the average petals size of each transgenic line normalized to the Col-0 petal-area average for each genotype. Error bars represent SEM. The number of independently transformed line used to obtain the mean values is indicated on the figure. Letters indicate significant differences as determined by Tukey’s honest significant difference test. (B) Sequence conservation within the SAP intron determined as the percentage identity between different Brassicaceae species within 10-bp windows. The scheme in the bottom represents the identified polymorphisms between Cr1504 and Cg926 as well as the conserved noncoding sequences (CNS) identified by ref. 22. Conserved polymorphisms shared between the two C. rubella versus the two C. grandiflora alleles used for transformation are indicated by red ellipses. (C) The SAP intron shows a higher percentage of rare SNPs within C. grandiflora compared with introns genome-wide (intron GW), suggesting purifying selection. (D) The analysis of Cr1504 allele frequency in C. grandiflora and C. rubella populations failed to identify fixed polymorphisms between the two species. All sites comprised in the 1.1-kb causal region underlying PAQTL_6, and polymorphic between Cr1504 and Cg926 were analyzed. Note that although the C. rubella alleles were at very low frequency in C. grandiflora at some positions, none of these frequencies were equal to 0. (E) Local association mapping identifies several polymorphisms within the SAP intron associated with petal-size variation in the C. grandiflora population. The y axis shows false-discovery rate adjusted negative log P values. Dashed blue line indicates 5% significance threshold.
Evolutionary History of the Small-Petal Allele.
We next investigated the evolutionary history of the small-petal SAPr allele. To this end, we first refined the position of the causal polymorphisms within the intron by transforming A. thaliana with a series of SAPr/SAPg chimeric constructs (Fig. 4A); this delineated a 1.1-kb interval at the 3′ end of the SAP intron as containing the causal polymorphisms. This region contained 23 polymorphic sites between the parental genotypes of the NILs, 14 of which were fixed between the two SAPr and the two SAPg alleles that recapitulate the PAQTL_6 effect in A. thaliana (Fig. 4 A and B and SI Appendix, Fig. S5B). We determined the allele frequencies at these 23 polymorphic sites in 180 resequenced C. grandiflora individuals from a single population, a species-wide sample of 13 C. grandiflora individuals, and a C. rubella species-wide sample of 73 individuals (see Materials and Methods for details). Besides the above transformation experiment using different C. grandiflora and C. rubella alleles of SAP, comparative QTL experiments also suggested that SAP underlies petal-size variation in different C. grandiflora × C. rubella crosses (SI Appendix, Fig. S8). This finding led us to expect that the causal polymorphisms are highly differentiated between the two species. Two different scenarios could explain the evolution of the “small-petal” allele: the selection of a new mutations in the selfing lineage or the capture and fixation of small-petal alleles already segregating in the ancestral out-crossing population. In the “new mutation” scenario, we would expect to find private polymorphisms in the Cr1504_SAP allele and other CrSAP alleles, whereas no species-specific polymorphism would be expected in the case of “capture and fixation.” None of the above 23 polymorphisms were fixed between the two species (Fig. 4 C and D), and there were no private polymorphisms in C. rubella, even though the C. rubella-like allele was present at very low frequency in C. grandiflora at five polymorphic sites. This lack of private polymorphisms could indicate that either the SAP small-petal allele has been captured from the standing variation in C. grandiflora, or that a new mutation in the selfing species has been introgressed into C. grandiflora because of more recent postdivergence hybridization. Recent hybridization would be expected to leave a genomic signature; in particular, because linkage disequilibrium is very low in C. grandiflora as a result of out-crossing, recent hybridization events should be detectable by the presence of long C. rubella-like haplotypes, not yet broken by recombination events, in the genome of C. grandiflora individuals. However, the C. rubella-like alleles were not present in long C. rubella-like haplotype blocks in the C. grandiflora individuals in question (highlighted in SI Appendix, Fig. S9B), arguing against recent hybridization reintroducing a derived C. rubella haplotype into C. grandiflora in these individuals. However, possible evidence for such recent hybridizations was seen in six C. grandiflora samples by counting the presence of C. rubella k-mers of increasing size in the data set used (SI Appendix, SI Text and Fig. S9A). Removing these individuals from the allele frequency calculation still did not reveal a C. rubella private polymorphism. Thus, the presence of C. rubella-like alleles on short (i.e., old) C. grandiflora haplotype blocks at all polymorphic sites indicates that the small-petal SAPr allele is likely to have been captured from standing variation in the ancestral out-crossing population.
SAP contains a large number of conserved noncoding sequences within its intron that suggest the existence of several functionally important elements (Fig. 4B). Consistently, we observed an excess of rare polymorphisms (minor allele < 5%) compared with average introns genome-wide in C. grandiflora (Fig. 4C), implying that purifying selection may be acting on these conserved sequences to prevent the fixation of deleterious mutations. Together with the observation that 5 of the 23 polymorphisms were rare in C. grandiflora, yet almost fixed in C. rubella, this finding suggests a scenario where less-efficient purifying selection in the selfing lineage as a result of stronger drift or positive selection led to the fixation of standing variation reducing petal size.
Several Polymorphisms Within the SAP Intron Associate with Petal-Size Variation in the Current Out-Crossing Population.
To determine whether the current out-crossing C. grandiflora population harbors functional standing variation in the SAP locus affecting petal size, we tested the association between the SNPs within the SAP intron and petal size in C. grandiflora. This approach revealed three segregating variants in the current C. grandiflora population that are significantly associated with petal size (Fig. 4E and SI Appendix, Fig. S10). One of these variants (SNP14059648) is also present in our QTL mapping population and NILs. Because some of the 23 candidate polymorphisms were filtered out from the initial genome-wide calling, we also performed a local variant calling for the given positions and used these improved genotypes to investigate whether any other polymorphisms beyond those included in the above analysis could be associated with petal size. A single-marker analysis comparing the effect of the parental C. rubella (Cr1504) allele from our NILs against all other alleles at each polymorphic site identified one additional polymorphism (SNP14059453) that was significantly associated with petal area (adjusted P value = 0.0037) (SI Appendix, Fig. S10A). The Cr1504 alleles are present at high frequency in C. grandiflora at both SNP14059453 and SNP14059648 (40% and 80%, respectively). The combined effect of these two SNPs on petal size is predicted to be 15% (SI Appendix, Fig. S10B). However, both SNPs differ between the two independent C. grandiflora alleles used for the above transformations, despite their very similar effects on petal size. Therefore, it appears that additional polymorphisms contribute to the functional difference between the C. grandiflora and C. rubella alleles from our NILs. The most plausible candidates for these are the five polymorphisms for which the Cr1504 allele is rare in C. grandiflora (Fig. 4D, and see above). One of these (SNP 14,059,340) is located within a conserved noncoding sequence and two others were present at highly conserved sites in close proximity to a conserved noncoding sequence (Fig. 4B) (SNP 14,060,447 and 14,060,471). It is therefore plausible that several of these sites are functional and may also affect petal size. Unfortunately, their low frequency prevented us from testing this hypothesis directly, because our association study did not provide sufficient statistical power to detect effects of such rare polymorphisms. Thus, testing the functional significance of these additional polymorphisms and determining whether their low frequency in C. grandiflora results from purifying selection acting to maintain large petals will require further study. However, the fact that at least two of the contributing polymorphisms, which are not in strong linkage disequilibrium and have an additive effect on petal size (SI Appendix, Fig. S10 B and C), were found to segregate at considerable frequencies in C. grandiflora (Fig. 4) indicates that the C. rubella small-petal SAP haplotype has combined several polymorphisms with individually small effects from the standing variation in C. grandiflora, rather than resulting from a single major-effect mutation.
Conclusions
These results demonstrate how standing variation in a tissue-specific regulatory module can contribute to rapid changes in morphology, and underscore the importance of a modular gene-regulatory architecture for enabling organ-specific morphological evolution (23–27). After the breakdown of the self-incompatibility system, the lineage leading to C. rubella has undergone a period of gene flow with the ancestral out-crossing species before experiencing a strong reduction in its effective population size (18). The fact that the SAP small-petal haplotype appears to have evolved from the ancestral standing genetic variation in the out-crossing species suggests that this period of gene flow has enabled the capturing of mating system modifiers that reduce flower size and improve autogamy. Indeed, previous studies have suggested that whereas flower size positively influences pollinator visitation (28; see also ref. 13 for extended discussion), the reduction of flower size in the selfing syndrome contributes to improving self-pollination (14). We have presented evidence that purifying selection acts on the SAP intron in the ancestral out-breeder. It is therefore plausible that selection maintains large petals for efficient pollinator attraction; in selfers, it is likely that this requirement was relieved, allowing the fixation of segregating variants that reduce flower size and improve selfing efficiency, and the evolution of the selfing syndrome over short evolutionary time scales. The fixation of haplotypes composed of several segregating small effect mutations with low pleiotropy can thus provide a fast track to organ-specific evolution. Extending such an evolutionary path to several loci within the genome would allow rapid changes in phenotypic means and explain the often complex genetic basis of selfing-syndrome traits (15, 14).
Materials and Methods
Additional methods are provided in SI Appendix.
Biological Materials.
The C. rubella and C. grandiflora populations and accessions used in this study have been described previously (14, 29, 30). The NIL segregating for PAQTL_6 was generated by introgressing the corresponding Cg926 allele into Cr1504 by four rounds of back-crossing followed by two additional rounds of selfing, while maintaining PAQTL_6 heterozygous. The crossing scheme used to generate the qIL is summarized in SI Appendix, Fig. S2. The sap-1 (N501593/SALK_001593) and sap-2 (N663979/SALK_129750) T-DNA mutants were obtained from the Nottingham Arabidopsis Stock Centre; sap-3 (FLAG_191C07) was obtained from the Institut National de la Recherche Agronomique.
Molecular Cloning and Plant Transformation.
Genomic chimeric constructs as well as reporter constructs were generated and subcloned into pBlueMLAPUCAP by ligation independent cloning using the In-Fusion HD Cloning Plus (Clontech) as indicated in SI Appendix, Table S1. The fragments were then transferred into the AscI site of the pBarMap vector, a derivate of pGPTVBAR (31). These genomic constructs were then used to transform A. thaliana Col-0 by floral dip (32). The sequences of the primers used are presented in SI Appendix, Table S2.
Morphological Measurements.
Size parameters were measured using ImageJ (https://imagej.nih.gov/ij/) from the digitalized images of the dissected organs. Morphometric analysis of petal outlines was performed using EDF for closed outlines, as described previously (33). To determine the size of the petal primordia, we performed a modified pseudo-Schiff propidium iodide (mPS-PI) staining on young flower buds of qIL3rr and qIL3gg as described in ref. 34. Petal cell size and cell number were determined from a dried-gel agarose print (35) of whole petals from fully open flowers.
Genetic Mapping.
To refine the position of the PAQTL_6, we screened about 3,300 NILrg progenies for plants having a recombination breakpoint between G09 and G09_20 (SI Appendix, Table S3). The selected recombinants were selfed and genotyped to identify between three and six plants homozygous for the C. grandiflora allele and three to six plants homozygous for the C. rubella allele in the remaining segregating region. We termed these plants “sister lines.” These plants were then selfed for another generation and the petal size of four replicates per progeny plant was measured as described above. The position of the recombination breakpoint for each of these recombinants was determined by genotyping the selected recombinants with additional markers in the focal region; these markers are presented in SI Appendix, Table S3.
Confocal Imaging and Analysis of the Dual Reporter Lines.
Reporter constructs were imaged using a scanning microscope Zeiss LSM710 using excitation wavelengths of 405, 488, or 561 nm and collecting CFP emission between 460 and 520 nm, FM6-64 signal between 630 and 760 nm, and YFP fluorescence between 510 and 570 nm. A maximum z projection of the 2-µm sections was used to quantify the CFP and YFP signal in five nuclei for each tissue type with imageJ (https://imagej.nih.gov/ij/). The CFP/YFP ratio was used to compare the expression of SAPr and SAPg in flower meristem and petal primordia.
Allele Frequencies and Population Genetic Analysis.
The population genetic analysis was performed on a dataset including 180 resequenced C. grandiflora individuals from a single population (36), a species-wide sample of 13 C. grandiflora individuals (30), and a C. rubella species-wide sample of 73 individuals. The latter include the sequencing data for 51 C. rubella individuals, which were downloaded from the European Nucleotide Archive (www.ebi.ac.uk/ena, data made publicly available by Daniel Koenig and Detlef Weigel, study number PRJEB6689) as well as SAP sequences resequenced on an Ion Torrent platform from 22 C. rubella accessions (SI Appendix, Table S4). Note that some of the sequences from the two C. rubella datasets may be redundant. We therefore treated the two datasets as independent samples in all our analyses. The dataset corresponding to the publicly available genomes was termed DKCr and the one including all resequenced SAP sequences was named CrReseq. Independently, the two datasets led to very similar results.
Haplotyes were reconstructed combining local assembly and multiple paired-end–based phasing approaches (SI Appendix, SI Materials and Methods). Local variant calling was done using SAMtools (37). Hierarchical clustering of Cr1504/Cg926 variants was done based on Euclidean distances. For this, Cr1504 nucleotides were coded as 0, heterozygous ones as 1, and Cg926 ones as 2.
We conducted a candidate gene-association mapping analysis using the C. grandiflora population genomics data from (36) and plink v1.07, where we tested all SNPs in the region with a minor allele frequency greater than 10%. Significance was assessed using the Benjamini–Hochberg false-discovery rate correction.
Supplementary Material
Acknowledgments
We thank Tanja Slotte and Barbara Neuffer for seeds; Doreen Mäker, Christiane Schmidt, and Monika Bischoff-Schäfer for plant care; Detlef Weigel and Daniel Koenig for having made publicly available Capsella rubella genome sequences; Emily Josephs and Niroshini Epitawalage for plant care and molecular laboratory assistance; and Isabel Bäurle and members of the M.L. group for discussion and comments on the manuscript. This work was supported by a Genome Canada and Genome Quebec Applied Bioproducts and Crops grant (to J.R.S. and S.I.W.); Deutsche Forschungsgemeinschaft Grant SI1967/1 within the framework of the research priority programme “Adaptomics” (to A.S.); and European Research Council Starting Grant 260455 (to M.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences of STERILE APETALA alleles from the recombinant inbred line population reported in this paper have been deposited in the GenBank database (accession nos. KX894525 and KX894526).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1613394113/-/DCSupplemental.
References
- 1.Jarne P, Auld JR. Animals mix it up too: The distribution of self-fertilization among hermaphroditic animals. Evolution. 2006;60(9):1816–1824. doi: 10.1554/06-246.1. [DOI] [PubMed] [Google Scholar]
- 2.Stebbins GL. Flowering Plants: Evolution Above the Species Level. Harvard Univ Press; Cambridge, MA: 1974. [Google Scholar]
- 3.Barrett SCH. The evolution of plant sexual diversity. Nat Rev Genet. 2002;3(4):274–284. doi: 10.1038/nrg776. [DOI] [PubMed] [Google Scholar]
- 4.Cutter AD. Reproductive evolution: Symptom of a selfing syndrome. Curr Biol. 2008;18(22):R1056–R1058. doi: 10.1016/j.cub.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 5.Darwin C. The Effects of Cross and Self Fertilisation in the Vegetable Kingdom. John Murray; London: 1876. [Google Scholar]
- 6.Ornduff R. Reproductive biology in relation to systematics. Taxon. 1969;18(2):121–133. [Google Scholar]
- 7.Thomas CG, Woodruff GC, Haag ES. Causes and consequences of the evolution of reproductive mode in Caenorhabditis nematodes. Trends Genet. 2012;28(5):213–220. doi: 10.1016/j.tig.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monteiro A, Podlaha O. Wings, horns, and butterfly eyespots: How do complex traits evolve? PLoS Biol. 2009;7(2):e37. doi: 10.1371/journal.pbio.1000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stern DL, Orgogozo V. The loci of evolution: How predictable is genetic evolution? Evolution. 2008;62(9):2155–2177. doi: 10.1111/j.1558-5646.2008.00450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaunt SJ, Paul Y-L. Changes in cis-regulatory elements during morphological evolution. Biology (Basel) 2012;1(3):557–574. doi: 10.3390/biology1030557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foxe JP, et al. Recent speciation associated with the evolution of selfing in Capsella. Proc Natl Acad Sci USA. 2009;106(13):5241–5245. doi: 10.1073/pnas.0807679106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo Y-L, et al. Recent speciation of Capsella rubella from Capsella grandiflora, associated with loss of self-incompatibility and an extreme bottleneck. Proc Natl Acad Sci USA. 2009;106(13):5246–5251. doi: 10.1073/pnas.0808012106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sicard A, Lenhard M. The selfing syndrome: A model for studying the genetic and evolutionary basis of morphological adaptation in plants. Ann Bot (Lond) 2011;107(9):1433–1443. doi: 10.1093/aob/mcr023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sicard A, et al. Genetics, evolution, and adaptive significance of the selfing syndrome in the genus Capsella. Plant Cell. 2011;23(9):3156–3171. doi: 10.1105/tpc.111.088237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slotte T, Hazzouri KM, Stern D, Andolfatto P, Wright SI. Genetic architecture and adaptive significance of the selfing syndrome in Capsella. Evolution. 2012;66(5):1360–1374. doi: 10.1111/j.1558-5646.2011.01540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koenig D, Weigel D. Beyond the thale: Comparative genomics and genetics of Arabidopsis relatives. Nat Rev Genet. 2015;16(5):285–298. doi: 10.1038/nrg3883. [DOI] [PubMed] [Google Scholar]
- 17.Sicard A, et al. Divergent sorting of a balanced ancestral polymorphism underlies the establishment of gene-flow barriers in Capsella. Nat Commun. 2015;6:7960. doi: 10.1038/ncomms8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brandvain Y, Slotte T, Hazzouri KM, Wright SI, Coop G. Genomic identification of founding haplotypes reveals the history of the selfing species Capsella rubella. PLoS Genet. 2013;9(9):e1003754. doi: 10.1371/journal.pgen.1003754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roels SA, Kelly JK. Rapid evolution caused by pollinator loss in Mimulus guttatus. Evolution. 2011;65(9):2541–2552. doi: 10.1111/j.1558-5646.2011.01326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Byzova MV, et al. Arabidopsis STERILE APETALA, a multifunctional gene regulating inflorescence, flower, and ovule development. Genes Dev. 1999;13(8):1002–1014. doi: 10.1101/gad.13.8.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z, et al. SCF(SAP) controls organ size by targeting PPD proteins for degradation in Arabidopsis thaliana. Nat Commun. 2016;7:11192. doi: 10.1038/ncomms11192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haudry A, et al. An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions. Nat Genet. 2013;45(8):891–898. doi: 10.1038/ng.2684. [DOI] [PubMed] [Google Scholar]
- 23.Indjeian VB, et al. Evolving new skeletal traits by cis-regulatory changes in bone morphogenetic proteins. Cell. 2016;164(1-2):45–56. doi: 10.1016/j.cell.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan YF, et al. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science. 2010;327(5963):302–305. doi: 10.1126/science.1182213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGregor AP, et al. Morphological evolution through multiple cis-regulatory mutations at a single gene. Nature. 2007;448(7153):587–590. doi: 10.1038/nature05988. [DOI] [PubMed] [Google Scholar]
- 26.Loehlin DW, Werren JH. Evolution of shape by multiple regulatory changes to a growth gene. Science. 2012;335(6071):943–947. doi: 10.1126/science.1215193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frankel N, et al. Morphological evolution caused by many subtle-effect substitutions in regulatory DNA. Nature. 2011;474(7353):598–603. doi: 10.1038/nature10200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glaettli M, Barrett SC. Pollinator responses to variation in floral display and flower size in dioecious Sagittaria latifolia (Alismataceae) New Phytol. 2008;174(4):1193–1201. doi: 10.1111/j.1469-8137.2008.02532.x. [DOI] [PubMed] [Google Scholar]
- 29.Sicard A, et al. Repeated evolutionary changes of leaf morphology caused by mutations to a homeobox gene. Curr Biol. 2014;24(16):1880–1886. doi: 10.1016/j.cub.2014.06.061. [DOI] [PubMed] [Google Scholar]
- 30.Ågren JA, et al. Mating system shifts and transposable element evolution in the plant genus Capsella. BMC Genomics. 2014;15(1):602. doi: 10.1186/1471-2164-15-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Becker D, et al. New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol. 1992;20(6):1195–1197. doi: 10.1007/BF00028908. [DOI] [PubMed] [Google Scholar]
- 32.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 33.Kuhl FP, Giardina CR. Elliptic Fourier features of a closed contour. Comput Graph Image Process. 1982;18:236–258. [Google Scholar]
- 34.Truernit E, et al. High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of Phloem development and structure in Arabidopsis. Plant Cell. 2008;20(6):1494–1503. doi: 10.1105/tpc.107.056069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horiguchi G, Fujikura U, Ferjani A, Ishikawa N, Tsukaya H. Large-scale histological analysis of leaf mutants using two simple leaf observation methods: Identification of novel genetic pathways governing the size and shape of leaves. Plant J. 2006;48(4):638–644. doi: 10.1111/j.1365-313X.2006.02896.x. [DOI] [PubMed] [Google Scholar]
- 36.Josephs EB, Lee YW, Stinchcombe JR, Wright SI. Association mapping reveals the role of purifying selection in the maintenance of genomic variation in gene expression. Proc Natl Acad Sci USA. 2015;112(50):15390–15395. doi: 10.1073/pnas.1503027112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.