Significance
Infection with human cytomegalovirus (HCMV) is a growing health problem, creating diagnostic and therapeutic challenges. Biomarkers for risk of infection are lacking, and the limited drugs that inhibit HCMV have major side effects. New strategies for virus control are needed. We report on the role of nucleotide-binding oligomerization domain 1 (NOD1), a cytoplasmic pattern recognition receptor, in HCMV suppression. NOD1 activation (through IKKα and IRF3) resulted in IFN response and HCMV inhibition. Specific mutations in NOD1 showed differential effects on HCMV replication in vitro. In a nested study of HCMV vaccine, specific polymorphisms in NOD1 were detected in HCMV-infected women compared with noninfected women. Our work provides direction for studies of innate immune response to HCMV and genetic susceptibility through NOD1.
Keywords: cytomegalovirus, NOD1, innate immune response, polymorphisms, RIPK2
Abstract
Induction of nucleotide-binding oligomerization domain 2 (NOD2) and downstream receptor-interacting serine/threonine-protein kinase 2 (RIPK2) by human cytomegalovirus (HCMV) is known to up-regulate antiviral responses and suppress virus replication. We investigated the role of nucleotide-binding oligomerization domain 1 (NOD1), which also signals through RIPK2, in HCMV control. NOD1 activation by Tri-DAP (NOD1 agonist) suppressed HCMV and induced IFN-β. Mouse CMV was also inhibited through NOD1 activation. NOD1 knockdown (KD) or inhibition of its activity with small molecule ML130 enhanced HCMV replication in vitro. NOD1 mutations displayed differential effects on HCMV replication and antiviral responses. In cells overexpressing the E56K mutation in the caspase activation and recruitment domain, virus replication was enhanced, but in cells overexpressing the E266K mutation in the nucleotide-binding domain or the wild-type NOD1, HCMV was inhibited, changes that correlated with IFN-β expression. The interaction of NOD1 and RIPK2 determined the outcome of virus replication, as evidenced by enhanced virus growth in NOD1 E56K mutant cells (which failed to interact with RIPK2). NOD1 activities were executed through IFN-β, given that IFN-β KD reduced the inhibitory effect of Tri-DAP on HCMV. Signaling through NOD1 resulting in HCMV suppression was IKKα-dependent and correlated with nuclear translocation and phosphorylation of IRF3. Finally, NOD1 polymorphisms were significantly associated with the risk of HCMV infection in women who were infected with HCMV during participation in a glycoprotein B vaccine trial. Collectively, our data indicate a role for NOD1 in HCMV control via RIPK2- IKKα-IRF3 and suggest that its polymorphisms predict the risk of infection.
Human cytomegalovirus (HCMV), a member of the herpesvirus family, induces complex innate immune responses (1, 2). Despite this effective and multifaceted induction, HCMV has developed strategies to counteract its recognition (3), allowing for its productive replication and the establishment of latency. Identification and characterization of HCMV-induced innate immune responses and resulting signaling pathways may provide novel strategies for its control.
Mounting evidence indicates that HCMV sensing is an intricate process involving activities of membrane, cytoplasmic, and nuclear receptors. Several HCMV-encoded proteins directly activate innate immune response molecules; the glycoprotein B (gB) binds to and activates TLR2 (4), and pp65 interacts with IFI16 (5). Other viral proteins, dsDNA, or dsRNA are likely to activate or inhibit host innate response molecules. Several previous reports have highlighted a complex role of the IFN pathway in response to HCMV. The activity of the promyeolcytic leukemia protein, a regulator of type I IFN response, is counteracted by HCMV-encoded immediate early 1 protein (IE1) (6). A cytoplasmic dsDNA sensor, ZBP1, activates IRF3 on infection, and its overexpression inhibits HCMV replication (7). IFN-inducible protein IFI16 modestly inhibits HCMV by blocking Sp1-mediated transcription of HCMV-encoded UL54 and UL44, which are involved in viral DNA synthesis (8).
The nucleotide-binding domain (NBD) and leucine-rich repeat-containing family (NLR) of receptors were originally reported to induce the NF-κB pathway in response to bacterial pathogens, but more recently induction of alternative signaling reminiscent of antiviral responses, including the IFN pathway and autophagy, have been reported (9–11). NLRC5 was found to be induced by HCMV within 24 h, and its knockdown (KD) impaired the up-regulation of IFN-α in response to HCMV (12). We reported on nucleotide-binding oligomerization domain 2 (NOD2) induction by HCMV, resulting in antiviral response and inhibition of virus replication (13). Induction of NOD2 by HCMV occurred starting at 24 h and resulted in activation of the receptor-interacting serine/threonine-protein kinase 2 (RIPK2), the major kinase downstream of NOD2. Overexpression of NOD2 or RIPK2 resulted in HCMV suppression. NOD2 activation by muramyl dipeptide (MDP), a peptidoglycan moiety of Gram-positive and Gram-negative bacteria, inhibited HCMV via an IFN-β pathway (14).
RIPK2 interacts with NOD1 and NOD2 through its caspase activation and recruitment domain (CARD), leading to its activation and downstream signaling. RIPK2’s role in NOD-dependent induction of innate and adaptive immunity has been reported previously (15). For some intracellular bacteria, collaboration between NOD1 and NOD2, rather than individual activation of each receptor, is important in host response (16). In addition, induction of tolerance to NOD2 activities resulted in increased activation of NF-κB in response to an NOD1 agonist, suggesting that cross-tolerization between NOD1 and NOD2 may result in improved recognition of bacteria (17–19).
To explore the interplay between NOD1 and NOD2 at the RIPK2 checkpoint during HCMV infection, we investigated the role of NOD1 in cellular defense against HCMV. Our data reveal that NOD1 plays an important role in HCMV suppression through the induction of antiviral responses. NOD1 KD or treatment with an NOD1 inhibitor, ML130, enhanced HCMV replication. Overexpression of NOD1 or pretreatment of human foreskin fibroblasts (HFFs) with l-Ala-γ-d-Glu-mDAP (Tri-DAP), a NOD1 activator present in the peptidoglycan of Gram-negative bacilli and certain Gram-positive bacteria, resulted in HCMV suppression. Mouse CMV (MCMV) was also inhibited after pretreatment with the NOD1 activator iE-DAP. HCMV inhibition through NOD1 required activation of the IFN pathway and was independent of the canonical NF-κB activation via IκB kinases (IKKs). Surprisingly, in IKKα KD cells, Tri-DAP lost its ability to inhibit HCMV, and neither infection nor Tri-DAP pretreatment resulted in nuclear translocation of IRF3. NOD1 and NOD2 collaborated in HCMV control, as evidenced by improved virus suppression when MDP and Tri-DAP were combined. Our data reveal different effects of specific NOD1 mutations on HCMV replication and antiviral signaling, pointing to the importance of the interaction between NOD1 and NOD2 with RIPK2 in HCMV control. Finally, single nucleotide polymorphisms (SNPs) in NOD1 were predictive of infection in a cohort of women with documented primary infection during their participation in a HCMV gB vaccine trial (20).
Results
NOD1 Activation Suppresses HCMV.
We previously reported that NOD2 expression was undetectable in noninfected HFFs but significantly induced after HCMV infection. NOD1 mRNA was abundant in noninfected HFFs and increased only modestly after infection (13). In the present study, the expression of NOD1 mRNA and protein was measured at 18 and 72 h postinfection (hpi) (Fig. 1 A and B). There was a twofold to fourfold increase in NOD1 mRNA at both time points. Tri-DAP pretreatment induced NOD1 mRNA to similar levels at 72 h. NOD1 protein was already expressed in noninfected cells, and no significant change in its expression was observed after infection; however, its activation by pretreatment with Tri-DAP (10 μg/mL) resulted in HCMV inhibition. Virus suppression was confirmed by decreased pp28-luciferase activity in second cycle infection (Fig. 1C), viral protein expression (Fig. 1D), and a plaque reduction assay using the Towne strain (Fig. 1E). The effect of Tri-DAP on HCMV replication was not secondary to cellular toxicity, as in treated HFFs during the same time frame. Tri-DAP did not affect cell viability (Fig. 1F). The effect of Tri-DAP was specific to HCMV, given that HSV-1 was not inhibited after Tri-DAP pretreatment (Fig. 1G).
Fig. 1.
NOD1 activation results in HCMV inhibition. HFFs were infected with HCMV Towne (MOI 1) or activated with Tri-DAP (10 μg/mL), and the expression level of NOD1 was measured by qRT-PCR (A) and Western blot analysis (B) at 18 and 72 hpi. Cells were pretreated (PT) with Tri-DAP for 72 h, followed by infection with pp28-luciferase HCMV or Towne HCMV. (D–E) Virus replication was measured by luciferase activity in the first cycle (96 hpi) and second cycle (72 hpi) (C), viral protein expression (D), and a plaque reduction assay (E). (F) Cell viability after 72 h of Tri-DAP treatment was determined by the MTT assay. (G) HFFs were pretreated with Tri-DAP for 72 h at the indicated concentrations followed by infection with a clinical isolate of HSV-1, and plaques were counted after 48 h. Data are mean ± SD from triplicate measurements. *P < 0.05, **P < 0.01, ***P < 0.001.
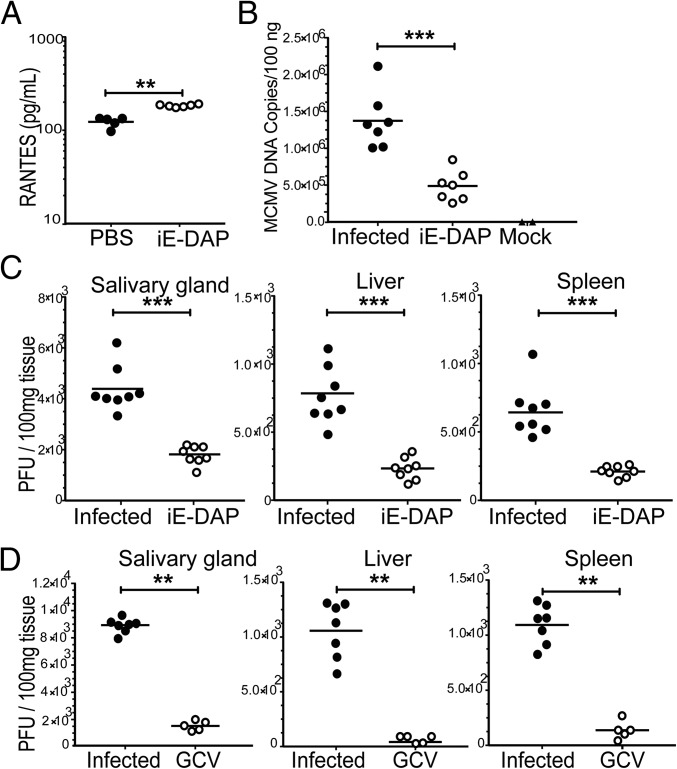
In Vivo NOD1-Dependent Anti-MCMV Activity.
BALB/c mice (3–4 wk) were pretreated with iE-DAP (Invivogen), 500 μg once daily for 2 d, followed by infection with MCMV at 106 PFU/mice. iE-DAP activity was confirmed by the induction of the chemokine RANTES in serum samples collected at 4 h after administration of the second dose (P < 0.01) (Fig. 2A). At 14 d postinfection, mice were killed, intracardiac blood samples were collected, and tissue homogenates were prepared for plaque assays. In iE-DAP–pretreated mice, real-time PCR for gB (P < 0.001) (Fig. 2B) and plaque numbers in salivary glands, liver, and spleen (P < 0.001) (Fig. 2C) were significantly reduced compared with values in infected-only mice. Ganciclovir (GCV), used as a direct antiviral agent, inhibited MCMV, as expected (Fig. 2D).
Fig. 2.
NOD1 activator, iE-DAP, inhibits MCMV replication. (A) BALB/c mice (age 3–4 wk) were pretreated with iE-DAP. Blood was collected at 4 h after the second dose of iE-DAP and RANTES levels were measured by ELISA in serum samples. (B and C) At 14 d postinfection, blood was collected for gB real-time PCR (B) and plaque assays were performed from salivary glands, liver, and spleen (C). (D) GCV was given after infection at 10 mg/kg twice daily for 5 d. Data are presented as mean ± SD of PFU/100 mg of tissue homogenate. P values were calculated using the two-tailed Mann–Whitney U test. **P < 0.01, ***P < 0.001.
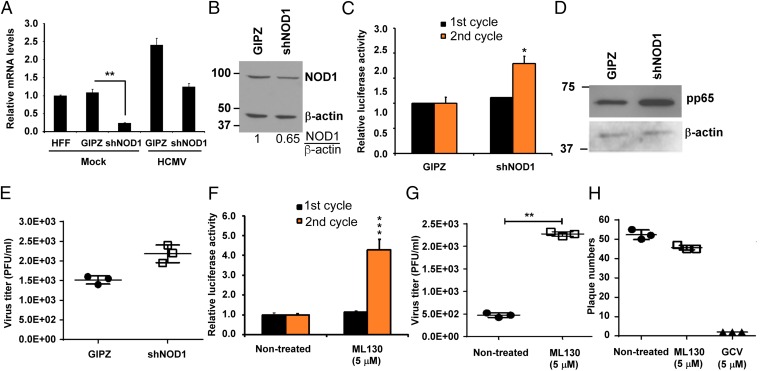
NOD1 KD or Inhibition of Its Activity Results in Enhanced HCMV Replication.
Given that NOD1 activation limited HCMV replication, we tested the effect of NOD1 KD or inhibition of its activity by the small molecule ML130. Using shRNA for NOD1, we found decreases in NOD1 mRNA of 80% in noninfected HFFs and 50% in HCMV-infected HFFs (Fig. 3A). On infection, a twofold to threefold reduction in NOD1 protein expression was observed in NOD1 KD cells compared with control cells (Fig. 3B). Luciferase activity from pp28 (Fig. 3C) and Western blot analysis for pp65 were measured in control (GIPZ) and NOD1 KD cells (Fig. 3D). During the first replication cycle, there was no difference in pp28-luciferase activity between control and NOD1 KD cells; however, after the second cycle, pp28-luciferase activity and pp65 expression were increased in NOD1 KD cells compared with control cells (Fig. 3 C and D). Virus titers measured using supernatants collected from the first cycle showed a mild (nonsignificant) increase in plaque numbers in NOD1 KD cells (Fig. 3E). Collectively, these data suggest that NOD1 might play a role in suppressing HCMV; however, because of its abundance in noninfected and infected cells, we suspected that the effects of its KD were moderate.
Fig. 3.
NOD1 KD or inhibition of NOD1 activity with small molecule ML130 results in enhanced HCMV replication. (A) HFFs stably expressing shRNA against NOD1 (shNOD1) were generated. Cells were infected with HCMV (MOI 1), and the expression level of NOD1 mRNA was measured by qRT-PCR at 24 hpi. (B) The expression level of NOD1 was determined by Western blot in HCMV-infected HFFs. (C) Cells were infected with pp28-luciferase Towne (MOI 1), and luciferase activity was measured at 96 hpi (first cycle) and the second cycle. (D) The expression of pp65 was determined by Western blot analysis after second cycle infection. (E) Virus titer was determined by plaque assay from supernatants collected after 72 h (first cycle). (F) HFFs were pretreated with ML130 (5 μM) for 72 h, and then infected with pp28-luciferase HCMV Towne for 96 h. (G) Cell-free supernatants were collected at 96 hpi from HCMV-infected cells and used to infect fresh HFFs for quantification of virus titer by plaque assay. (H) HSV-1 replication was determined by a plaque reduction assay in HFFs pretreated with ML130. Data are mean ± SD from triplicate measurements. *P < 0.05, **P < 0.01.
To achieve a more significant inhibition of NOD1 activity, we used ML130, a specific NOD1 inhibitor (21, 22). We found that ML130 did not affect cell viability even at a concentration of 100 µM, as determined by the 3-(4, 5-dimethyl-2-thiazolyl)-2, 5-diphenyl-2H-tetrazolim bromide (MTT) assay. HFFs were pretreated with ML130 for 72 h at a concentration sufficient to inhibit NOD1 activity (5 μM) but not NOD2 or TNF-α activity, followed by HCMV infection. A significant increase was observed in second cycle pp28-luciferase activity (Fig. 3F) and virus titer (Fig. 3G). The effect of ML130 was specific to HCMV, given that pretreatment of HFFs followed by HSV-1 infection did not change the number of plaques (Fig. 3H).
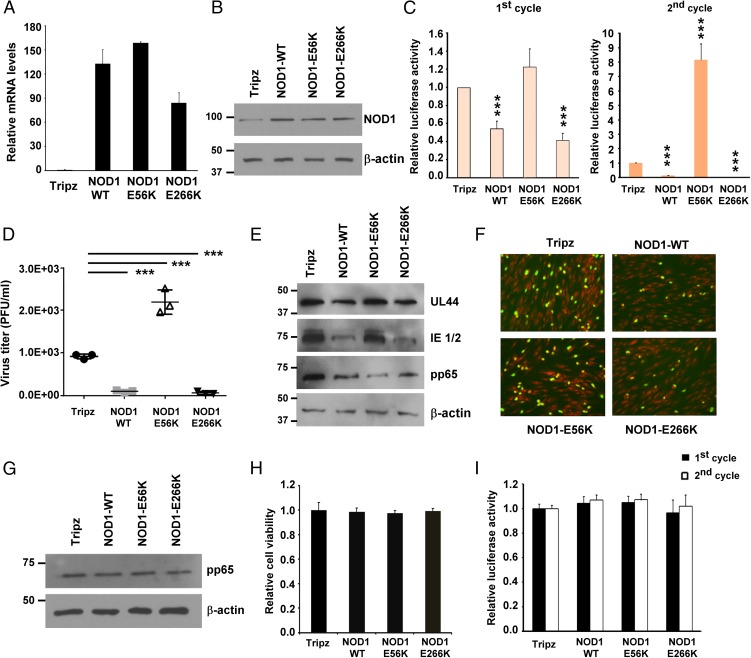
Differential Effects of NOD1 Mutations on HCMV Replication.
To determine whether mutations in specific regions of NOD1 affect HCMV replication, we generated stable cell lines overexpressing wild-type (WT) NOD1 and two NOD1 mutants, E56K (in the CARD) and E266K (in the NBD), using a doxycycline-inducible lentivirus system. The E56K mutation was reported to abrogate NOD1 signaling by abolishing its interaction with RIPK2 (23, 24), indicating a role for NOD1–RIPK2 interaction in executing downstream signaling. The E266K mutation in NOD1 has been suggested to increase the pathogenesis of Helicobacter pylori infection (25); however, its effect on NOD1 function remains undetermined.
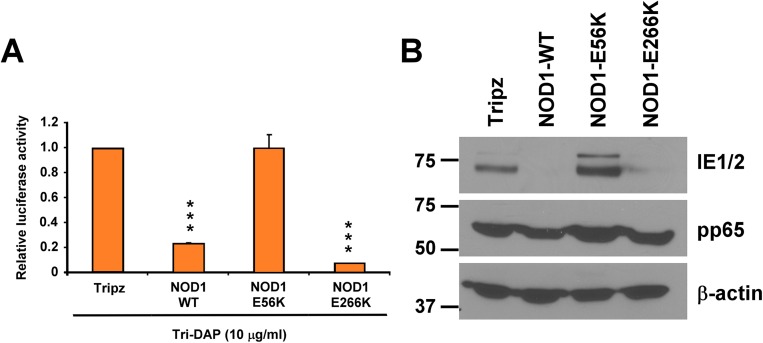
After doxycycline induction, NOD1 mRNA was induced by 75- to 160-fold (Fig. 4A), and the expression of NOD1 protein was increased in the overexpressing cell lines compared with Tripz control (Fig. 4B). We measured HCMV replication in the different NOD1-overexpressing cells and found that pp28-luciferase activity was reduced by 60% at 96 hpi in the WT and E266K-overexpressing cells, but was increased in the E56K- overexpressing cells (Fig. 4 C–F). Supernatants collected after the first cycle were used for second cycle infection and virus titration (Fig. 4 C and D). Significant virus inhibition was observed in the NOD1 WT and E266K cells, as opposed to increased HCMV replication in the E56K-overexpressing cells. The expression of HCMV proteins correlated with luciferase activity. Significant decreases in IE1/2, UL44, and pp65 were seen in cells overexpressing NOD1 WT or the E266K mutant; however, cells overexpressing the E56K NOD1 mutant consistently showed an inability to suppress HCMV or its protein expression (Fig. 4 C–E).
Fig. 4.
Differential effects of NOD1 polymorphisms on HCMV replication. (A and B) HFFs stably expressing empty vector (Tripz), NOD1-WT, E56K, or E266K mutants were induced with doxycycline (2 μg/mL) for 24 h, and the expression level of NOD1 was determined by qRT-PCR (A) and Western blot analysis (B). (C and D) Cells were induced with doxycycline (2 μg/mL) for 24 h, followed by infection with pp28-luciferase HCMV Towne. Luciferase activity was measured at 96 hpi as the first cycle. Cell-free supernatants were collected at 96 hpi from HCMV-infected cells and used to infect fresh HFFs as the second cycle (C) or to quantify virus titer by plaque assay (D). (E) The expression level of viral proteins was determined at 96 hpi by Western blot analysis. (F) The expression of viral IE1/2 was determined at 24 hpi by immunofluorescence assay. The primary antibody was IE1/2, the secondary antibody was goat anti-mouse (FITC, green) and nuclear stain (PI, red). Representative pictures from two independent experiments are shown. (G) HCMV entry into the different cell lines was determined by Western blot analysis for pp65 at 2 hpi. (H) Cell viability at 3 d after doxycycline induction was determined by the MTT assay. (I) HFFs stably overexpressing NOD1 WT and mutants were infected with HSV-1-luciferase for 24 h, and luciferase activity was measured in cell lysates. Data are mean ± SD from triplicate measurements. ***P < 0.001.
An immunofluorescence assay (IFA) for IE1/2 using a clinical isolate of HCMV showed reduced IE1/2 expression in NOD1 WT and E266K-overexpressing cells, but not in E56K-overexpressing cells (Fig. 4F). The changes in HCMV replication/protein expression were not secondary to lentivirus transduction or cellular toxicity; virus uptake was similar irrespective of the overexpressing cell line, based on pp65 level at 2 hpi (Fig. 4G), and the MTT assay revealed no effect on cell viability after 4 d of doxycycline induction (Fig. 4H). Furthermore, these effects were not secondary to altered cytokine expression induced by HCMV infection of the different cell lines; infection with purified HCMV Towne showed the same pattern of luciferase activity and viral protein expression depending on the cell line used (Fig. S1 A and B). Finally, HSV-1 replication was not altered in any of the overexpressing cell lines after 24 h (first cycle) or 48 h (second cycle; Fig. 4I), again indicating no role for NOD1 in controlling HSV-1 replication. Collectively, these data reveal that specific functional mutations in NOD1 may affect HCMV replication.
Fig. S1.
Infection of NOD1-overexpressing cell lines with purified HCMV. Purified HCMV Towne was used for infection of the different NOD1-overexpressing cells. Luciferase activity was measured after the first and second cycles (A), and pp65 expression was measured in cell lysates collected from second cycle infection (B). Data are mean ± SD from triplicate measurements. **P < 0.01, ***P < 0.001.
To further confirm that the observed antiviral activity in cells overexpressing the NOD1 WT or mutants was through NOD1, we performed Tri-DAP pretreatment. We found that in cells overexpressing the WT or E266K NOD1, luciferase activity was significantly inhibited (Fig. S2A) and the expression level of viral proteins was reduced (Fig. S2B); however, in the E56K-overexpressing cells, Tri-DAP pretreatment did not result in HCMV inhibition.
Fig. S2.
Tri-DAP pretreatment elicits enhanced HCMV suppression in specific NOD1-overexpressing cells. The different NOD1-overexpressing cells were pretreated with Tri-DAP for 72 h, followed by HCMV infection. Luciferase activity was measured at 72 hpi (A), and the expression level of viral proteins was determined by Western blot analysis (B). Data are mean ± SD from triplicate measurements. ***P < 0.001.
Signaling Downstream of NOD1 in HCMV-Infected Cells.
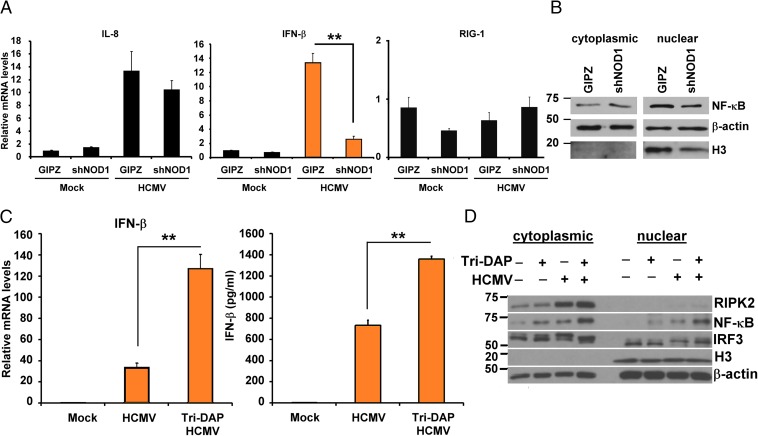
Tri-DAP activates a signaling pathway downstream of NOD1, through NF-κB (26), and the antiviral response to HCMV involves IRF3 (7). Because IFN-β is responsive to these transcription factors, we tested signaling in NOD1 KD and Tri-DAP–pretreated cells. We measured IL-8 and IFN-β transcripts in NOD1 KD and control cells at 24 hpi (Fig. 5A). Infection resulted in a 14-fold increase in both IL8 and IFN-β in GIPZ control cells, but only sixfold and twofold increases, respectively, in NOD1 KD cells. A significant reduction in IFN-β expression was observed in infected NOD1 KD cells compared with control cells (P < 0.01), but no change in RIG-I transcripts was observed, supporting the specificity of the NOD1 KD system (Fig. 5A, Right). We also measured NF-κB (p65) expression in cytoplasmic and nuclear extracts of infected cells. In control cells, HCMV infection resulted in NF-κB localization into the nucleus, but in the NOD1 KD cells, the changes in NF-κB localization were not as evident (Fig. 5B).
Fig. 5.
Downstream signaling in NOD1 KD and NOD1-activated HFFs. (A) NOD1 KD (shNOD1) and control (GIPZ) HFFs were infected with HCMV (MOI 1), and I-L8 and IFN-β mRNA were quantified by qRT-PCR at 24 hpi. RIG-I served as a control. (B) Expression of NF-κB was measured in cytoplasmic and nuclear fractions at 24 hpi. (C) HFFs were treated with Tri-DAP for 72 h, followed by HCMV infection. IFN-β mRNA (Left) and protein (Right) was measured by qRT-PCR and ELISA at 24 and 72 hpi, respectively. (D) HFFs were pretreated with Tri-DAP for 72 h, followed by HCMV infection, and expression levels of RIPK2, NF-κB, and IRF3 were measured in cytoplasmic and nuclear extracts at 24 hpi. The IRF3 antibody recognizes IRF3 and pIRF3. Data are mean ± SD from triplicate measurements. **P < 0.01.
NOD1 activation by Tri-DAP followed by HCMV infection induced IFN-β mRNA (Fig. 5C, Left), as well as secreted IFN-β (Fig. 5C, Right). We measured the expression of RIPK2, NF-κB, and IRF3 at 24 hpi in Tri-DAP–pretreated cells. Infection induced cytoplasmic expression of RIPK2, which was further induced in infected Tri-DAP–pretreated cells (Fig. 5D). Similar to the effect of infection, Tri-DAP induced NF-κB in both cytoplasmic and nuclear extracts. Tri-DAP treatment followed by infection further increased NF-κΒ in both fractions. The pattern of IRF3 activation differed from that of NF-κB, in that pretreatment with Tri-DAP without infection did not change nuclear IRF3 phosphorylation. The effect of Tri-DAP on IRF3 phosphorylation was enhanced only after infection (Fig. 5D). These data suggest that NOD1 activation results in enhanced downstream signaling, some independent of infection (NF-κB) and others triggered only by HCMV infection (IFN pathway).
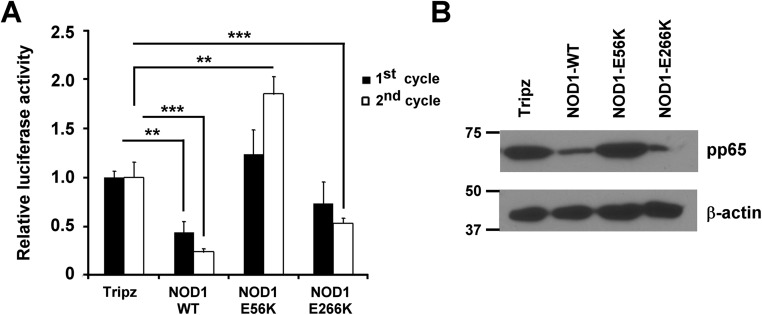
Differential Signaling Induced Downstream of NOD1 WT and NOD1 Mutant Cell Lines on HCMV Infection.
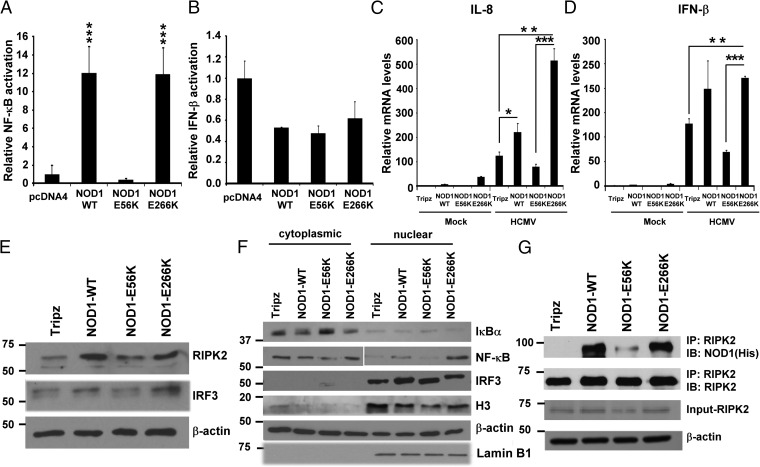
Given that HCMV replication was restricted in NOD1 WT and E266K mutant cell lines, but not in those overexpressing the E56K mutant, we tested the signaling induced by these constructs in transfected HEK293. Plasmids encoding for NOD1 WT, E56K, and E266K were cotransfected with NF-κB or IFN-β luciferase reporters. Transfection of NOD1 WT or E266K plasmid induced NF-κB activity in HEK293, but the E56K mutant failed to induce NF-κB (Fig. 6A). No induction of IFN-β was seen with any of the plasmids (Fig. 6B), in agreement with the data Fig. 5D, demonstrating that IRF3 activation through NOD1 occurred only on infection. IL-8 and IFN-β mRNA was measured in the stably transduced overexpressing cells at 24 h after infection. IL-8 mRNA was induced in control, NOD1 WT, and E266K-overexpressing cells, but enhanced induction was not observed in the E56K-overexpressing cells (Fig. 6C). Similarly, IFN-β was induced on infection of control cells, and enhanced induction was observed in WT and E266K-overexpressing cells, but not in E56K-overexpressing cells (Fig. 6D).
Fig. 6.
NOD1 downstream signaling in WT and mutant NOD1-overexpressing cells. (A and B) NF-κB (A) and IFN-β (B) luciferase reporter assays were performed in 293T cells. pcDNA-NOD1 WT and mutant plasmids were cotransfected with reporter plasmids. After 24 h, cells were lysed, and luciferase activity was determined. (C and D) NOD1-overexpressing cells were infected with HCMV (MOI 1) for 24 h, and IL-8 (C) and IFN-β (D) mRNA expression was measured by qRT-PCR. The depicted mRNA expression experiments represent mean ± SD from triplicate wells of two representative experiments. (E and F) The expression levels of NOD1-downstream signaling proteins were determined in total cell lysates (E) and cytoplasmic and nuclear fractions (F) at 3 hpi. β-actin served as a loading control; histone H3 and lamin B served as loading controls for nuclear proteins. (G) WT and NOD1 mutant-overexpressing cells were infected with HCMV Towne, and immunoprecipitation using anti-RIPK2 antibody, followed by immunoblotting for NOD1 using His antibody, were performed at 24 hpi. Data are mean ± SD from triplicate measurements. **P < 0.01, ***P < 0.001.
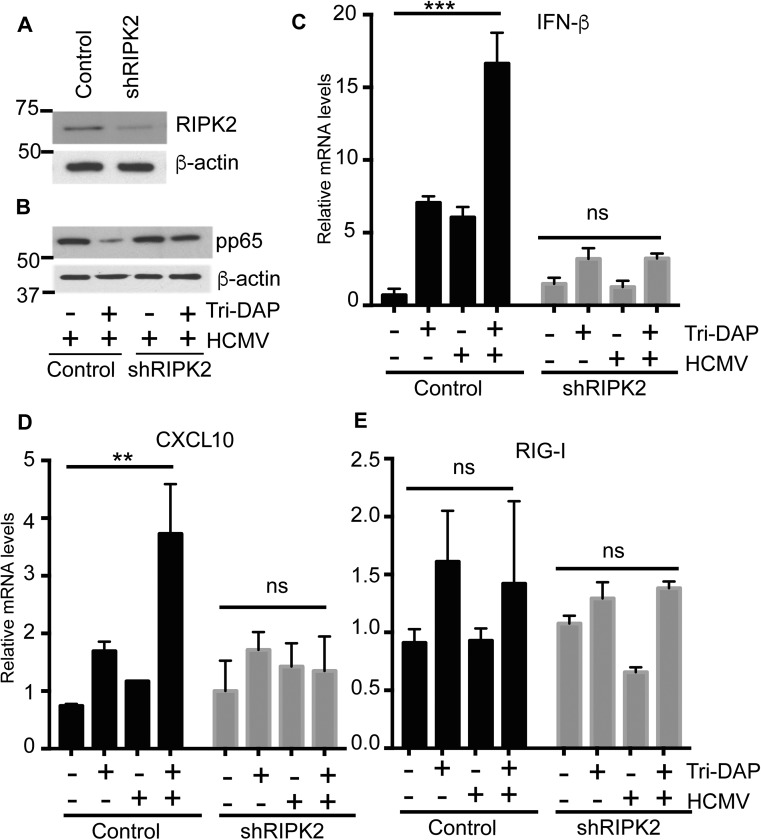
We measured the expression of proteins downstream of NOD1 in the HCMV-infected overexpressing cells. RIPK2 induction was observed in infected NOD1 WT and E266K-overexpressing cells, but not in E56K-overexpressing cells (Fig. 6E), and phospho-IRF3 was not induced in the latter (Fig. 6E). Nuclear translocation of NF-κB was observed on infection of WT and E266K-overexpressing cells, but not of E56K-overexpressing cells (Fig. 6F). Histone 3 levels also were reduced in the nuclear fraction of E56K (Fig. 6F), whereas lamin B levels were similar among all of the cell lines, possibly representing NF-κB–mediated changes in histone 3. NF-κB expression is regulated by inhibitory IκB proteins, which are regulated by upstream IKKs (27). Phosphorylation of IκB proteins results in their degradation and release of the NF-κB complex. Whereas IκBα was reduced in WT and E266K-overexpressing cells, its expression was increased in E56K-overexpressing cells, supporting the lack of nuclear translocation of NF-κB. Immunoprecipitation of RIPK2, followed by immunoblotting for NOD1-His, showed that an intact RIPK2–NOD1 interaction in all overexpressing cells except the E56K cells (Fig. 6G). Additional confirmation for the NOD1–RIPK2 interaction was obtained in RIPK2 KD cells (Fig. S3A). Tri-DAP pretreatment in these cells did not reduce CMV-pp65 expression (Fig. S3B). The expected induction of IFN-β and CXCL10 mRNA was observed in the control line, but not in the RIPK2 KD cells (Fig. S3 C and D). Taken together, these data indicate that NOD1 activation suppresses HCMV replication, and that mutations in NOD1 that potentially affect its interaction with RIPK2 and resulting downstream signaling will determine its capability to suppress HCMV.
Fig. S3.
Tri-DAP acts through NOD1-RIPK2 to inhibit HCMV. (A) KD of RIPK2 was performed in HFFs. (B) Control and shRIPK2 cells were treated with Tri-DAP for 72 h, followed by HCMV infection (MOI 2). The expression of HCMV pp65 in control and shRIPK2 cells was measured at 72 hpi. (C–E) IFN-β (C), CXCL10 (D), and RIG-I (E) mRNA were measured by qRT-PCR at 24 hpi. Data are mean ± SD from triplicate measurements. ns, nonsignificant. **P < 0.01, ***P < 0.001.
NOD1 and NOD2 Cooperate in HCMV Inhibition.
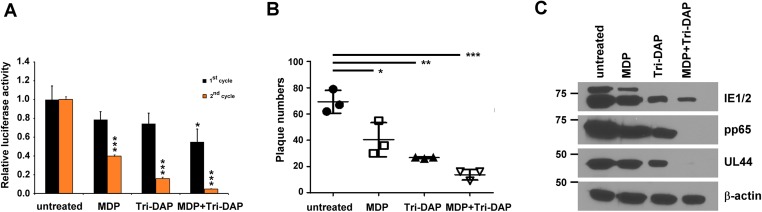
Our findings indicate that HCMV suppression is achieved through NOD1 activation and its interaction with RIPK2. Given our previous report of HCMV inhibition by the NOD2 activator, MDP (14), here we investigated the combined effect of NOD1 and NOD2 activation. Pretreatment of HFFs with MDP together with Tri-DAP augmented virus suppression to a greater degree than pretreatment with MDP or Tri-DAP alone, based on first and second replication cycle (Fig. S4A), plaque reduction (Fig. S4B), and viral protein expression (Fig. S4C) data.
Fig. S4.
Combined effect of MDP and Tri-DAP on HCMV replication. HFFs were pretreated with MDP, Tri-DAP, or Tri-DAP+MDP for 72 h, followed by infection (MOI 1). Virus replication was quantified by pp28-luciferase activity in the first and second cycles (A), plaque reduction for TB40 (B), and viral protein expression (C). Data are mean ± SD from triplicate measurements. *P < 0.05, **P < 0.01, ***P < 0.001.
HCMV Inhibition via NOD1 Requires IFN-β.
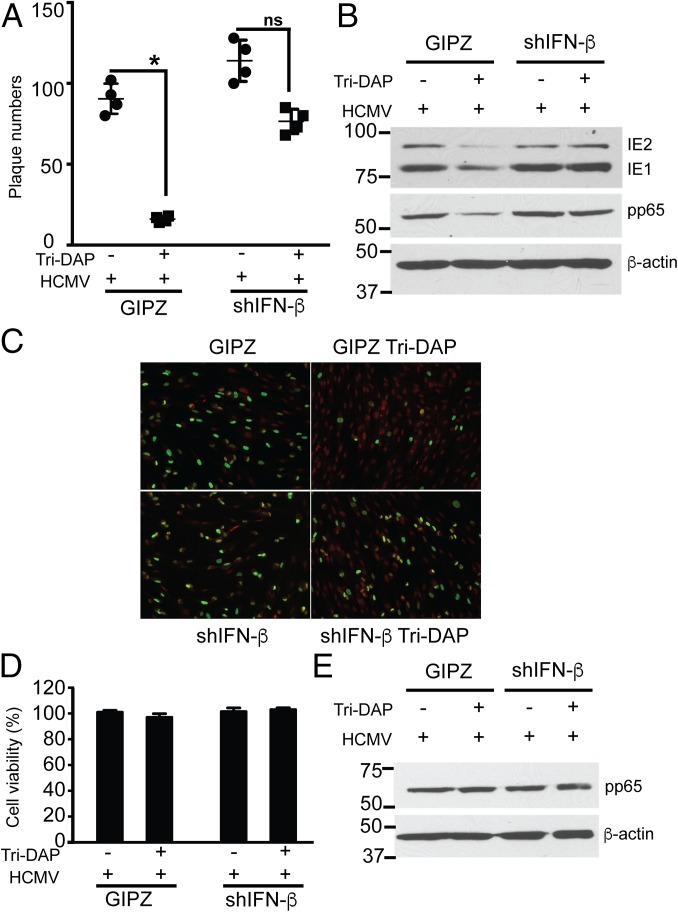
Because Tri-DAP pretreatment inhibited HCMV replication along with IFN-β induction, and because the NOD1-overexpressing cells exhibited differing effects on IFN-β mRNA, we tested whether the effects of Tri-DAP in HCMV-infected cells are IFN-β–dependent. For this, control and IFN-β KD cells were pretreated with Tri-DAP, followed by infection [at a multiplicity of infection (MOI) of 1]. Tri-DAP pretreatment reduced HCMV plaque formation and viral protein expression in control cells, but not in IFN-β KD cells (Fig. 7 A and B). IFA performed after infection with TB40 similarly showed reduced IE1/2 staining in control cells, but not in the IFN-β KD cells (Fig. 7C). None of the observed effects was secondary to cellular toxicity (Fig. 7D), and virus uptake was similar in the different cell lines (Fig. 7E).
Fig. 7.
IFN-β is required for HCMV inhibition by Tri-DAP. (A and B) HFFs were stably transduced with lentivirus expressing control (GIPZ) or shRNAs against IFN-β (shIFN-β), nontreated or pretreated with Tri-DAP, followed by HCMV infection (MOI 1) for 72 h. Plaque reduction (A), and expression level of viral proteins (B) were determined at 72 hpi. (C) IFA for IE1/2 was performed at 24 h in TB40-infected control or shIFN-β cells. The primary antibody was IE1/2, and the secondary antibody was goat anti-mouse (FITC, green) and nuclear stain (PI, red). (D) Cell viability with or without Tri-DAP pretreatment for 72 h was determined by an MTT assay. (E) For the virus entry assay, Tri-DAP–pretreated GIPZ control cells or shIFN-β cells were infected with HCMV Towne for 2 h at 37 °C and washed with citric acid buffer (pH 3) to strip off virus particles adhered to the cell surface, and pp65 was detected by Western blot analysis. Data are mean ± SD from triplicate measurements. ns, nonsignificant. *P < 0.05.
HCMV Inhibition via NOD1 Is Dependent on IKKα.
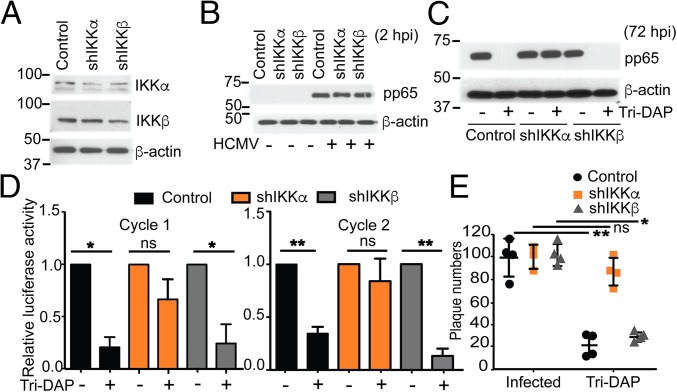
NOD1 activation by Tri-DAP results in signaling, leading to nuclear translocation of NF-κB accompanied by IKKα (27). Because Tri-DAP induced NF-κB in noninfected cells, we investigated whether NOD1 activity in HCMV suppression is dependent on the canonical pathway or on the alternative NF-κB pathway. For this, we performed Tri-DAP pretreatment, followed by infection, in IKKα KD, IKKβ KD, and control transduced cells. Once KD of IKKα and IKKβ (Fig. 8A) and similar virus entry into the three cell lines were confirmed (Fig. 8B), Tri-DAP pretreatment was performed, followed by infection. Virus replication was efficient in all three cell lines. In control and IKKβ KD cells, Tri-DAP pretreatment suppressed HCMV replication to a similar degree, as evidenced by viral protein expression (Fig. 8C), first and second cycle luciferase activity (Fig. 8D), and a plaque reduction assay using TB40 (Fig. 8E). However, in the IKKα KD cells, Tri-DAP could not suppress HCMV, suggesting that the anti-HCMV activity of Tri-DAP is independent of the IKKβ arm but requires the alternative IKKα pathway (Fig. 8 C–E). These results are in agreement with previous reports of the general mechanism of Tri-DAP showing the need for IKKα for translocation of NF-κB (28).
Fig. 8.
Effect of Tri-DAP on HCMV replication in IKKα and IKKβ KD cells. (A) KD of IKKα and IKKβ was determined by Western blot analysis in noninfected HFFs using anti-IKKα and IKKβ antibodies. (B) Virus entry into the IKKα, IKKβ KD (shIKKα, shIKKβ), and control cells was measured by Western blot analysis for pp65, as in Fig. 7E. (C–E) Cells were pretreated with Tri-DAP for 72 h, followed by infection with HCMV Towne (MOI 2). HCMV pp65 expression (C), pp28-luciferase activity after the first cycle (D) and after the second cycle and a plaque reduction assay (E) were measured in the respective cell lines. Data are mean ± SD from triplicate measurements. ns, nonsignificant. *P < 0.05, **P < 0.01.
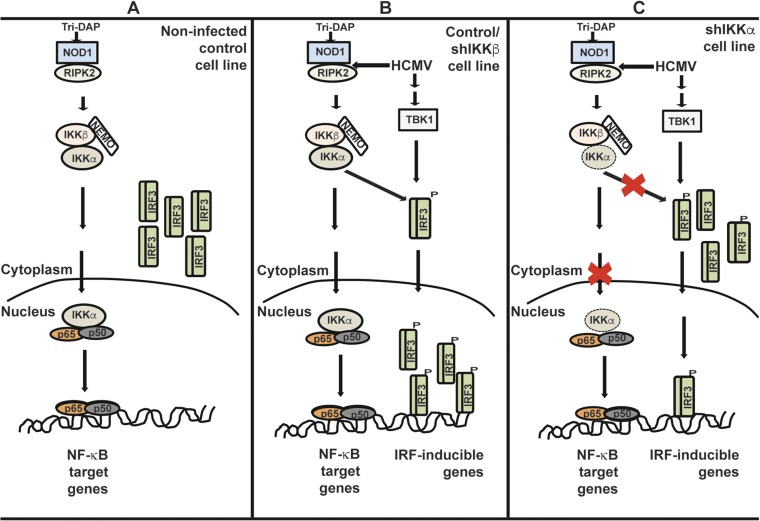
In control transduced cells, IKKα was detected in both cytoplasmic and nuclear fractions, whereas IKKβ was confined to the cytoplasm (Fig. 9A). Tri-DAP pretreatment increased the cytoplasmic expression of IKKα as well as pIKKα/β in the cytoplasmic and nuclear fractions. In control and IKKβ KD cells, Tri-DAP triggered NF-κB translocation into the nucleus, whereas in IKKα KD cells it did not. IRF3 phosphorylation in the different cell lines revealed an increase in the cytoplasm after Tri-DAP pretreatment, and a more significant increase in the nuclear fraction after Tri-DAP pretreatment and infection (Fig. 9A). Whereas Tri-DAP pretreatment followed by infection similarly induced nuclear translocation and phosphorylation of IRF3 in IKKβ KD cells, in the IKKα KD cells, IRF3 remained in the cytoplasm (Fig. 9 A–C). In agreement with these findings, mRNA expression of IFN-β and CXCL-10 was enhanced in control and IKKβ cells, but no such induction was observed in IKKα KD cells (Fig. 9 D–F). Thus, IKKα mediates an IRF3 effect in response to Tri-DAP that amplifies the antiviral cytokine response (Fig. S5, model).
Fig. 9.
Reduced IFN response in Tri-DAP–pretreated IKKα-infected cells. (A–C) Control transduced cells were pretreated with Tri-DAP, followed by infection with HCMV Towne (MOI 0.1). Expression levels of IKKα, IKKβ, pIKKα/β, p65, and IRF3 were measured by Western blot analysis in cytoplasmic and nuclear fractions at 24 hpi (A) and the expression pattern of IRF3 and p65 was determined in IKKα (B) and IKKβ (C) KD cells at 24 hpi. (D–F) mRNA levels of IFN- β, CXCL10, and RIG-I were measured after Tri-DAP pretreatment, followed by infection with HCMV Towne (MOI 0.1). Data are mean ± SD from triplicate measurements. ns, nonsignificant. *P < 0.05, ***P < 0.001.
Fig. S5.
Summary model of Tri-DAP activity and pathway utilization in HCMV-infected cells. (A) A model depicting the signaling events observed with Tri-DAP alone in control HFFs showing IKKα-mediated phosphorylation and nuclear translocation of NF-κB p65. (B) The cumulative effect of Tri-DAP stimulation and HCMV on NF-κB and IRF3 in control and IKKβ KD cells. (C) In IKKα KD cells, nuclear translocation of IKKα and NF-κB p65 is mostly inhibited, and there is only minimal infection-driven translocation of IRF3.
SNPs in NOD1 Are Significantly Associated with HCMV Infection.
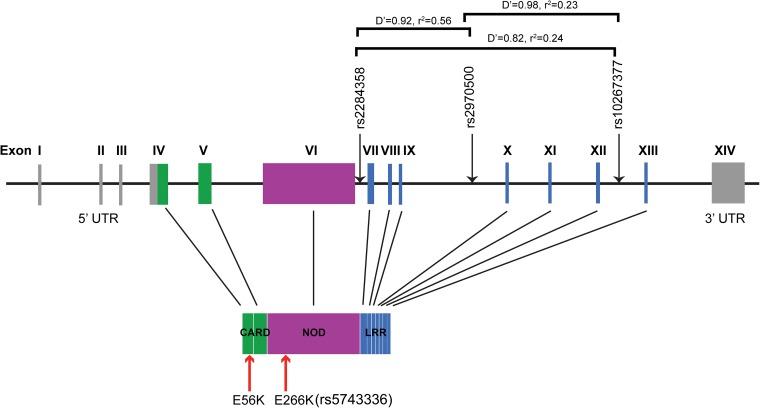
Finally, because mutations in NOD1 were seen to affect HCMV replication in vitro, we asked whether SNPs in NOD1 had clinical relevance for predicting the risk of HCMV infection. The HCMV gB vaccine trial provided a unique opportunity to address this question. Genomic data for 29 selected innate immune response genes and 768 SNPs were available from 383 women (152 who had received vaccine and 231 who had received placebo). Twenty women in the vaccine group and 32 women in the placebo group were infected with HCMV. A comparative analysis of SNPs in all infected and all noninfected women revealed that of six statistically significant SNPs, three were in introns 6, 9, and 12 of NOD1 (Fig. S6 and Table 1). SNPs in NOD1 were more significantly associated with HCMV infection compared with noninfected controls.
Fig. S6.
Schematic of NOD1 structure. The NOD1 gene has 14 exons. The 5′ UTR spans exons I–III and some part of exon IV, and the 3′ UTR is in exon XIV. The translated NOD1 protein has an N-terminal CARD, a central NBD, and seven C-terminal LRRs. Genetic polymorphisms identified in this study are indicated by black arrows pointing toward the line. The red arrows indicate the two mutations generated in the laboratory, of which E266K is available in the human database (rs5743336). Genetic positions are schematic rather than actual distances apart. Pairwise comparisons of linkage disequilibrium are indicated.
Table 1.
Significant SNPs in HCMV-infected and noninfected women
| Gene | rsID | Risk allele | Counts, all | Counts, infected | Counts, noninfected | OR | P value |
| NOD1 | rs2284358 | G | 10/117/256 | 5/19/28 | 5/96/225 | 2.70 | 0.00041 |
| LY96 | rs6472812 | A | 0/16/367 | 0/6/46 | 0/10/316 | 8.33 | 0.00063 |
| NOD1 | rs2970500 | C | 4/88/291 | 3/14/35 | 1/72/253 | 3.21 | 0.00069 |
| TLR1 | rs5743572 | A | 2/66/314 | 0/19/32 | 2/47/277 | 3.09 | 0.00069 |
| NOD1 | rs10267377 | C | 54/180/149 | 12/30/10 | 41/147/138 | 2.15 | 0.00103 |
| TLR10 | rs4513579 | G | 21/121/241 | 5/26/21 | 16/94/216 | 2.15 | 0.001380 |
Discussion
The innate immune response to HCMV involves an orchestrated system composed of multiple receptors residing in different cellular compartments (1, 4, 7, 29). Characterizing of these receptors and understanding their function and networking, as well as strategies used by HCMV to counteract their activities, are of paramount importance for HCMV control. In addition, pathways that are specific to HCMV and not shared by other herpesviruses may affect the targeting of unique host responses to HCMV. Toward this effort, here we report the role of NOD1 in HCMV suppression. NOD1 and NOD2 are the most well-studied NLRs in human disease. Both are expressed in monocytes, macrophages, and dendritic cells (30). NOD1 is also expressed in epithelial cells, and our results demonstrate its abundance in HFFs. NOD2 is induced by inflammatory signals, and we previously reported its significant induction in HCMV-infected HFFs starting at 24 hpi and thereafter (13). For NOD1, activation rather than induction appears to play a role in HCMV inhibition. Because of its abundance in HFFs, the response of NOD1 to HCMV was observed over a wider range of MOI in contrast to NOD2, which responded efficiently to a lower MOI (14). Pretreatment of mice with two doses of iE-DAP already initiated a sufficient signaling milieu that limited MCMV replication, although the exact balance of signaling activation and virus inhibition merits more detailed study.
The NOD1 protein contains an N-terminal CARD, an intermediary NBD that is required for nucleotide binding and self-oligomerization, and a C-terminal leucine-rich repeat domain (LRR) that detects conserved microbial patterns and modulates NLR activity (26, 31, 32). NOD2 recognizes MDP, which is present on most peptidoglycans (33). As bacterial sensors, NOD1 and NOD2 induce downstream signaling pathways. Although NF-κB is a major signaling pathway downstream of NOD1 and NOD2, type I IFNs were induced via NOD1 during infection with H. pylori, reminiscent of an antiviral response (34). NOD2-dependent IFN-β production during infection with Listeria resulted from synergy with other cytosolic microbial sensors (11). Evidence for IFN induction through NOD1 and NOD2 is also supported by reports of their ability to sense viruses. RNA viruses activated IRF3 in an NOD2- and mitochondrial antiviral signaling protein-dependent manner (35). NOD2-deficient mice had enhanced susceptibility to infection with respiratory syncytial virus (RSV), decreased IRF3 phosphorylation, and type I IFN production. Redundancy of innate immune response pathways to herpesviruses is well known, and some of the recently described pattern recognition receptors, such as IFI16 and cGMP-AMP synthase (cGAS), appear to be broad sensors of different herpesviruses (29, 36–41). In the case of NOD1, specific HCMV suppression through NOD1 activation (but not HSV-1 suppression) suggests the possible use of specialized pathways through HCMV which could be targeted for virus control.
On the basis of our previous finding that NOD2 induction by HCMV resulted in an antiviral response, in the present study we investigated the role of NOD1 in HCMV inhibition. NOD1 overexpression or activation by Tri-DAP inhibited HCMV, but not HSV-1. In addition, mutations in the CARD that interacts with RIPK2 abolished the inhibitory effect of NOD1 on HCMV. NOD1 activation resulted in induction of NF-κB and IFN-β signaling. The effects of Tri-DAP on NF-κB activation were observed in both noninfected and HCMV-infected cells, but changes in IFN-β were observed only in infected cells, supporting the model in which the NF-κB–dependent IFN-β pathway is required for NOD1 activities in infected cells. This hypothesis was confirmed by using IFN-β KD cells, in which HCMV suppression by Tri-DAP was abolished.
We tested the requirements of the IKKβ-dependent classical NF-κB pathway and the alternative IKKα-dependent pathway (42). In IKKβ KD cells, Tri-DAP inhibited HCMV, suggesting that the canonical NF-κB pathway is not required for Tri-DAP activity against HCMV. Although some remaining kinase activity could still induce NF-κB activation, it is unlikely that HCMV would be inhibited similarly in the respective cell lines. The activity of Tri-DAP against HCMV was significantly reduced in IKKα KD cells, however. There is only one published report of IRF3 activation by IKKα after its interaction with the NF-κB–inducing kinase (43). We found that nuclear translocation of IRF3 did not occur in IKKα KD cells in response to Tri-DAP treatment. Similarly, in another study, IKKβ was not required for NOD1 activation of IFN signaling in an H. pylori model. Although the role of IKKα was not studied in that model, the induction of IFN through NOD1 signaling was found to depend on TBK1 and IKKε (34). Nuclear translocation of NF-κB is a direct response to Tri-DAP–stimulated NOD1 (26), and is dependent on IKKα (28). Whereas IKKβ is predominantly cytoplasmic, IKKα shuttles between the nucleus and cytoplasm of cells (44). We observed an increase in both NF-κB and IKKα in response to Tri-DAP (Fig. 9). IKKα KD resulted in reduced IKKα-mediated nuclear translocation of NF-κB and IRF3 in response to Tri-DAP, indicating the requirement for IKKα in mediating an anti-HCMV response via NF-κB and IRF3. We propose a summary model of HCMV control by NOD1 through IKKα, leading to IRF3 activation and IFN-β induction (Fig. S5). In this model, IRF3 and NF-κB translocate to the nucleus in control and IKKβ KD cells, in response to HCMV infection and Tri-DAP, and a cumulative effect is observed when Tri-DAP precedes infection (Fig. S5 A and B). Activation of this pathway is IKKα-dependent; Tri-DAP stimulation results in increased NF-κB and IRF3 protein levels, but nuclear translocation does not occur in the absence of IKKα (Fig. S5C).
Mutations in NOD1 and NOD2 leading to loss or gain of function are associated with autoimmune and inflammatory diseases (19, 45–49). We previously reported that the NOD2 mutation associated with severe Crohn’s disease (3020C) results in enhanced HCMV replication in vitro (13). Here we provide in vitro evidence indicating that specific mutations in NOD1 result in either reduced or enhanced HCMV replication, as determined by NOD1 interaction with RIPK2. The laboratory-generated E56K mutation is an example that disrupts the interaction between NOD1 and RIPK2, but other mutations have been reported as well (24). Although a significant body of literature implicates associations between SNPs in NOD1 and several immune-related diseases, such as inflammatory bowel disease, atopic eczema, asthma, and rheumatoid arthritis (46–49), a link between these observed associations and specific NOD1 activity has not been established. Many of these genetic variants lie outside of protein-coding genes, and although they may or may not have a direct effect on protein structure, it is highly likely that cryptic splice sites are generated by these intronic polymorphisms, resulting in altered protein translation, stability, and expression of multiple isoforms. In fact, polymorphisms in the LRR domain of NOD1 that contribute to differences in expression levels of naturally occurring splice variants of NOD1 have been associated with differential inflammatory responses (48, 50).
Our genetic analysis of 29 selected innate immune response genes revealed that intronic SNPs in NOD1 were highly predictive of the risk of HCMV infection in humans. The majority of previous studies of host genetics and susceptibility to human herpesvirus infections have investigated SNPs in Toll-like receptors (TLRs) (46, 47); for example, an SNP in TLR2 was found to be associated with HCMV replication and disease in a small cohort of liver transplant recipients (51). Our data suggest a role for genetic variation in NOD1 as a predictor of the risk of HCMV acquisition, although its impact on virus replication and disease in a high-risk population remains to be studied. Thus, it is possible that a combination of NOD1 SNPs may determine protein folding/accessibility for interaction with RIPK2 and induction of antiviral responses. These human SNPs should be further investigated for their effect on LRR-mediated responses and the resulting NOD1-RIPK2 complex.
Collaboration between NOD1 and NOD2 has been identified in a Salmonella typhimurium colitis model. Mice deficient in either NOD1 or NOD2 were not susceptible to infection, but mice deficient in both NOD1 and NOD2 exhibited increased Salmonella colonization of the intestine (16). Similarly, it appears that for HCMV, collaboration between NOD1 and NOD2 may have an additive effect in virus suppression, with NOD1 activation inducing an early tier of innate immune response, followed by a second tier through NOD2. We previously reported that in IFN-β KD cells, pretreatment with MDP could not suppress HCMV or induce NOD2, suggesting that NOD2 activities require IFN-β (14). Similarly, IFN signaling was found to induce RIPK2 expression and downstream signaling in macrophages with a variety of stimuli (18). Our present data on the combined effect of MDP and Tri-DAP on HCMV replication, the lack of anti-HCMV activity of Tri-DAP in IFN-β KD cells, and the role of IKKα in inducing NF-κB and IRF3 downstream of NOD1 point to a model of initial activities through NOD1, resulting in IFN-β signaling leading to NOD2 induction and RIPK2 activation and further inhibiting HCMV replication.
In summary, here we provide information on a specific innate immune response pathway for HCMV control. Future studies will examine the role of NOD1 and NOD2 in vivo and with the aim of uncovering strategies used by HCMV to counteract activities through these receptors.
Materials and Methods
Chemicals and Proteins.
Tri-DAP, iE-DAP, and MDP were obtained from Invivogen. The NOD1 inhibitor ML130 was provided by Dr. G. Roth, Sanford Burnham Research Institute (21, 22). ML130’s high specificity against NOD1 has been confirmed by multiple downstream counterscreens that eliminated compounds impacting other NF-κB effectors, and its IC50 against NOD2 or TNF-α is >20 µM. iE-DAP was dissolved in PBS and used for experiments in mice. GCV was obtained from Sigma-Aldrich.
Cell Culture and Viruses.
HFFs were used for infection with HCMV and HSV-1 as described in SI Materials and Methods.
Generation of NOD1-Overexpressing Cells.
WT and mutant human NOD1 plasmids were constructed in pcDNA4/HisMax vector (Invitrogen), as described in SI Materials and Methods.
Additional information on procedures is provided in SI Materials and Methods.
Statistical Analysis.
All infection assays, qRT-PCR runs, and Western blot analyses were repeated three times unless stated otherwise. Statistical analyses were performed using two-tailed ANOVAs for comparisons between groups. For the animal studies, a two-tailed Mann–Whitney test was used with GraphPad Prism 7. A P value <0.05 was considered to indicate significance: *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Quantitative analysis of proteins detected by immunoblotting was performed by determining band intensities relative to β-actin using ImageJ version 1.48.
Study Population for SNP Studies.
Samples were collected from healthy women age 14–40 y (median age, 19.6 y) who enrolled in the gB vaccine trial (20). The majority of the study population (75%) was African American. The Institutional Review Boards of Johns Hopkins University School of Medicine and University of Alabama granted approval for this study.
SNP Selection.
Using a candidate gene approach, 29 genes shown to be associated with innate immune responses were selected: JUN, IKBKE, MYD88, TLR9, CD80, CD86, TLR10, TLR1, TLR6, NF-KB1, TLR2, TLR3, CD14, MXD3, MAPK14, MAP3K7, LY96, TLR4, MAPK8, CHU.K., TRAF6, IRAK4, TBK1, TICAM1, IRF3, CD40, TLR7, IRAK1. PDGFRA, integrins, and NOD1. Also included were 41 SNPs with previously reported associations in human diseases, 157 nonsynonymous SNPs, and 28 ancestry-informative markers (AIMs), for a total of 768 SNPs (52).
Genotyping Methods.
Genomic DNA (75-150 ng/μL) was obtained from frozen EDTA blood samples using Gentra Puregene extraction (Qiagen). Genotyping was performed using the Illumina GoldenGate chemistry as described previously (52). Genotypes were released for 714 SNPs (93% of those attempted), of which 694 were scored as high-quality SNPs.
Statistical Analysis of SNPs.
Statistical analysis of SNPs was done as reported previously (52). In brief, 28 AIMs were genotyped for evaluation of population stratification using principal components analysis in the statistical program Eigenstrat (53). Association analysis was done in PLINK version 1.062 (http://pngu.mgh.harvard.edu/purcell/plink) using linear regression and an additive model. A Hardy–Weinberg P value threshold of 10−3 and a minor allele frequency of >0.01 were used. A modified Bonferroni correction was used to correct for multiple comparisons based on the number of genes (owing to high LD), resulting in a threshold P value of 0.0017 for significance. SNP data were released for 383 women (99% of the attempted samples).
SI Materials and Methods
Cell Culture and Viruses.
HFFs, passage 12–16 [American Type Culture Collection (ATCC); CRL-2088], were grown in DMEM containing 10% (vol/vol) FBS (Gibco) in a 5% (vol/vol) CO2 incubator at 37 °C. At 1 d before infection or treatment, 8 × 104 cells were seeded into each well of 12-well tissue culture plates. Infection was carried out at an MOI of 1 PFU/cell or MOI 2 as specified for specific lentivirus-transduced cells. A pp28-luciferase Towne HCMV that expresses luciferase under the control of pp28 late promoter has been described previously (54). Luciferase activity was measured at 72 or 96 hpi using the GloMax-Multi+ Detection System (Promega). For virus replication assays, supernatants were collected at 96 hpi from cells infected with pp28-luciferase Towne (20% of total volume per well) and used for a second cycle infection of fresh cells. Luciferase activity was measured in cell lysates after 72 h. The HCMV TB40 strain (ATCC; VR-1578) was used for plaque reduction assays and IFA. Clinical isolates of HCMV and HSV-1 were used for IFA and plaque reduction assays, respectively. These isolates were obtained from the microbiology laboratory at Johns Hopkins Hospital with no identifiers that could be linked to a specific patient. The HSV-1 luciferase, KOS/Dlux/oriS, was kindly provided by David A. Leib, Dartmouth Medical School.
Plaque Assays.
HFFs were seeded into 12-well plates (8 × 104 cells/well) and treated with Tri-DAP, MDP, or combination of Tri-DAP and MDP (10 µg/mL) for 72 h, followed by infection with the TB40 performed in triplicates at ∼100 plaques per well. After 90 min, the medium was aspirated, and DMEM containing 0.5% carboxymethyl-cellulose and 4% (vol/vol) FBS were added. Following incubation at 37 °C for 10 d, the overlay was removed, and plaques were counted after crystal violet staining. Plaque assays using HCMV Towne were performed similarly in Tri-DAP–pretreated KD cells, including IFN-β, IKKα, and IKKβ (described below).
The effect of pretreatment with Tri-DAP or ML130 was tested in HSV-1–infected HFFs. A plaque assay was performed similar to that for HCMV, but with incubation for 72 h after infection.
Inhibition of MCMV Replication with iE-DAP.
All animal work was carried out in strict accordance with the recommendations in the National Institutes of Health’s Guide for the Care and Use of Laboratory Animals. The animal protocol (no. MO13M296) was approved by the Institutional Animal Care and Use Committee of Johns Hopkins University. For infection experiments, 3- to 4-wk-old BALB/c mice were purchased from Harlan Laboratories. After 2–3 d of adaptation to the housing environment, the mice were pretreated with iE-DAP (two daily doses of 500 μg each) (55) or saline intraperitoneally. Blood was collected at 4 h after the second dose of iE-DAP for measurement of the chemokine RANTES by ELISA (R&D Biosystems). The mice were then infected with 106 PFU of tissue culture-derived MCMV (Smith strain, VR-1399; ATCC) and killed after 14 d. Intracardiac blood was collected before sacrifice for a gB real-time PCR assay (56). Salivary glands, liver, and spleen were harvested and stored at −80 °C. Organs were homogenized in DMEM at a final concentration of 100 mg/mL. Two million mouse embryonic fibroblasts (MEFs) were seeded into 24-well plates. From each sample, 2.5% of the salivary gland or 10% of liver/spleen homogenate was used for infection of MEFs in triplicate. Plaques were counted after 3 d.
Cellular Toxicity.
WT, lentivirus-transduced GIPZ (control), and IFN-β KD cells were pretreated with Tri-DAP for 72 h, after which 20 µL of MTT (5 mg/mL in PBS) was added to each well. After being shaken at 150 rpm in a Gyrotory Shaker Model G2 (New Brunswick Scientific Company, Inc., Edison, NJ) for 5 min, the plates were incubated at 37 °C for 3 h. The conversion of yellow solution to dark-blue formazan by mitochondrial dehydrogenases of living cells was quantified by measuring absorbance at 560 nm.
Generation of Lentiviruses.
First, 21 µg of gag/pol, 7 µg of vesicular stomatitis virus glycoprotein, and 7 µg of shRNAmir plasmids were transfected into HEK293 cells using Lipofectamine 2000 (Invitrogen). After 48 h, the packaged lentivirus particles were concentrated from the medium. The supernatant was filtered and centrifuged at 1,750 × g for 30 min at 4 °C in an Amicon Ultra centrifugal filter unit with Ultracel 100 membrane (EMD Millipore). After centrifugation, 2 mL of cold PBS was added, and the tubes were centrifuged again for 20 min at 4 °C.
Generation of NOD1-Overexpressing Cells.
WT and mutant human NOD1 plasmids were constructed in pcDNA4/HisMax vector (Invitrogen). Primers for PCR amplification of the NOD1 gene were BamHI-NOD1*F, ACTAGGATCCATGGAAGAGCAGGGCCACAG and EcoRI-NOD1*R, CTAGGAATTCTCAGAAACAGATAATCCGCTTC (underscored nucleotides indicate the restriction enzyme sites of BamH I and EcoR I, respectively). PCR products were digested with BamH I and EcoR I and cloned into pcDNA4/HisMax vector. For DNA mutagenesis, overlapping primers containing the desired base change (in bold) were used for amplification of the upstream and downstream segment of NOD1, respectively. The full length of mutagenic NOD1 cDNA was amplified from the recombinant segments with the primer BamHI-NOD1*F and EcoRI-NOD1*R. The primers for generating NOD1 mutants were NOD1-E56K*F, CGAAGATGCGAAGATTGTGTGTGCCTGCCCC; NOD1-E56K*R, ACACACAATCTTCGCATCTTCGGCCGAGAAG and NOD1-E266K*F, GGACCCCAAGGAGGTGTTTGCCTTCC; NOD1-E266K*R, CAAACACCTCCTTGGGGTCCCGCTCTGGGTA.
The first and overlapping PCR program was performed at the following conditions: 95 °C for 5 min, 30 cycles of PCR (denaturation at 94 °C for 30 s, primer annealing at 55 °C for 30 s, extension at 72 °C for 2 min), and a final extension at 72 °C for 10 min. Constructs for lentivirus expression of WT and NOD1 mutants were amplified from the respective pcDNA4/HisMax plasmids with primers NOD1-TRIPZ*F, GGGATCCACCGGTCCATGGGGGGTTCTCATCATCATC and NOD1-TRIPZ*R, GGGATCCACGCGTTCATCAGAAACAGATAATCCGCTTC (underlined nucleotides indicate the restricted enzyme sites of Age I and Mlu I), and subcloned into Tripz lentiviral vector (Open Biosystems). The sequence of the new constructs was confirmed by DNA sequencing. Human NOD1-Tripz and Tripz control empty vector were packaged using lentivirus and transduced into HFFs. Equal numbers of NOD1-overexpressing and control cells were seeded into each well before infection.
RNA Isolation and qRT-PCR.
Total RNA was isolated from cells using TRIzol (Invitrogen). The RevertAid First-Strand cDNA Synthesis Kit (Thermo Fisher Scientific) was used to synthesize first-strand cDNA from total RNA using oligo-dT primers. Negative reverse-transcriptase reactions were included to ensure the specificity of qRT-PCR reactions. Synthesis of first-strand cDNA from mRNA template was carried out at 42 °C for 1 h. qRT-PCR was performed using specific primers and SYBR Green (Thermo Fisher Scientific) with a two-step cycling protocol (95 °C for 15 s, 60 °C for 1 min). All reactions were performed in triplicate, with GAPDH as an internal control. The expression levels of the tested genes were normalized to the expression of GAPDH. Primer sequences were as follows: NOD1 forward, 5′-CCTGAGCAACAACAATCTCAACGACTA-3′, NOD1 reverse, 5′-TTTACCCCACCGTCAGTGATC- 3′; IFN-β forward, 5′-GATTCATCTAGCACTGGCTGG-3′, IFN-β reverse, 5′-CTTCAGGTAATGCAGAATCC-3′; IL-8 forward, 5′-TGCAGCTCTGTGTGAAGGTGCAGT-3′, IL-8 reverse, 5′-CAGTGTGGTCCACTCTCAATCACTC-3′; RIG-I forward, 5′-ATCCCAACCGATATCATTTCTGATC-3′, RIG-I reverse, 5′-TTCCACCAATTTCTCTGCACCTGC-3′; GAPDH forward, 5′-TTGGTATCGTGGAAGGACTC-3′, GAPDH reverse, 5′- ACAGTCTTCTGGGTGGCAGT-3′.
Lentivirus-Mediated KD of NOD1, IFN-β, IKKα, IKKβ, and NOD2.
Human GIPZ lentiviral shRNAmir constructs (Open Biosystems) were used for NOD1 KD in HFFs. Two clones (shNOD1#2: V2LHS_69528 and shNOD1#5: V3LHS_309375) targeting different regions of NOD1 mRNA were combined to generate stable cell lines. A control nontargeting GIPZ plasmid was used to rule out nonspecific effects of shRNAmir. Individual shRNAmir constructs were packaged using lentivirus as described previously (57). Lentivirus particles containing shRNAmir were transduced into HFFs. Then 0.5 ×106 cells were plated onto T-25 flasks, and 40 µL of concentrated virus and polybrene (final concentration, 8 µg/mL) were added to the cells, followed by incubation for 4 h. After transduction, puromycin (2 µg/mL) was added to select for stably transduced cells. Control and NOD1 KD cells were counted, and an equal number of cells was plated into each well before infection. The KD of IFN-β and NOD2 in HFFs was recently reported from our laboratory (14). Validated TRC shRNA clones for RIPK2 (N0000218097, N0000218326, and N0000230255) were obtained from Sigma-Aldrich. The TRC control (pLKO.1) was obtained from Addgene. Plasmids for IKKα and IKKβ KD and control lentivirus (pYNC352) were kindly provided by Dr. Young Choi, Johns Hopkins University School of Medicine, and have been described previously (58).
SDS/PAGE and Immunoblot Analysis.
Cell lysates containing equivalent amount of proteins were mixed with an equal volume of sample buffer [125 mM Tris⋅HCl pH 6.8, 4% (vol/vol) SDS, 20% (vol/vol) glycerol, and 5% (vol/vol) β-mercaptoethanol], followed by boiling at 100 °C for 10 min. Denatured proteins were resolved in SDS polyacrylamide gels (8–15%) and transferred to PVDF membranes (Bio-Rad) by electroblotting. Membranes were incubated in blocking solution [5% wt/vol nonfat dry milk and 0.1% Tween-20 in PBS (PBST)] for 1 h, washed with PBST, and then incubated with antibody at 4 °C overnight. Membranes were washed with PBST and then incubated with HRP-conjugated secondary antibodies in blocking solution for 1 h at room temperature. After washing with PBST, protein bands were visualized by chemiluminescence using SuperSignal West Dura and Pico reagents (Thermo Fisher Scientific).
Antibodies for HCMV proteins were mouse anti-IE1&2 (MAB810, 1:2,000; EMD Millipore), mouse anti-HCMV UL83 (pp65, 1:2,000; Vector Laboratories), and mouse anti-UL44 (1:2,000; Santa Cruz Biotechnology). Mouse anti-human β-actin (1:5,000) was obtained from Sigma-Aldrich. Rabbit anti-RIPK2 (1:5,000), mouse anti–NF-κB (p65, 1:1,000), and rabbit anti-IRF3 (1:1,000) were obtained from Santa Cruz Biotechnology. Rabbit anti human-NOD1 antibody (1:2,000), rabbit monoclonal anti-pIκBα (1:2,000), rabbit anti-IKKα and anti-IKKβ, rabbit mAb 2697 anti-pIKKα/β (Ser176/180; 1:1,000), and rabbit anti-histone H3 (1:2,000) were purchased from Cell Signaling Technology. Rabbit anti-lamin B (1:1,000; Abcam) served as a nuclear loading control. HRP-conjugated anti-rabbit and anti-mouse IgG were obtained from Cell Signaling Technology and GE Healthcare, respectively.
Preparation of Cytoplasmic and Nuclear Extracts.
Cytoplasmic and nuclear fractions were isolated from HCMV-infected or mock-infected HFFs at 24 hpi as reported previously (13). Cells were washed twice with ice-cold PBS and resuspended on ice for 15 min in buffer A containing 10 mM Hepes (pH 7.9), 10 mM KCl, 0.1 mM EDTA, 1 mM DTT, protease, and phosphatase inhibitors. Cells were then lysed by adding 0.1% Nonidet P-40, and cytosolic supernatants were obtained by centrifugation at 9,600 × g for 30 s. Crude nuclei were washed twice with buffer A to prevent cytosolic contamination, and the nuclear proteins were extracted by resuspending cell pellets with buffer C, containing 20 mM Hepes (pH 7.9), 400 mM NaCl, 1 mM EDTA, 1 mM DTT, protease, and phosphatase inhibitors. The mixture was incubated for 15 min with vigorous shaking on a rocker at 4 °C and then centrifuged at 13,000 × g at 4 °C for 10 min to obtain the nuclear proteins.
IFA for HCMV IE1/2.
HFFs were seeded into four-well chamber slides (5 × 104 cells/well) and pretreated with Tri-DAP (10 µg/mL) for 72 h. Infection was performed with a clinical isolate of HCMV or TB40 (MOI 1). At 24 hpi, cells were fixed with methanol/acetone (1:1) at −20 °C for 10 min. Cells were then blocked with 7.5% (wt/vol) BSA and incubated for 1 h with anti-human CMV IE1/2 antibody (MAB810; 1:500 in 0.5% BSA), followed by incubation with FITC-anti mouse antibody (F8521, 1:500 in 0.5% BSA; Sigma-Aldrich). Images were obtained with a Nikon Eclipse TS100 microscope.
Transfection of NOD1 Plasmids.
HEK293 cells were used for cotransfection. The NOD1 plasmids (WT, E56K, and E266K) were cotransfected with NF-κB (Stratagene) and IFN-β luciferase reporter plasmids (59) using Lipofectamine 2000 (Thermo Fisher Scientific). The reporter plasmids were used at 100 ng, and all NOD1 plasmids were used at 500 ng. After 24 h, cells were lysed, and luciferase activity was determined.
SNP Selection for the Genetic Study.
A total of 29 genes known to be associated with innate immune responses were selected, including TLRs and associated intracellular signaling molecules. We identified tag SNPs within the gene and in the 10 kb flanking each side of the genes, using the LDselect algorithm in individuals from the publicly available HapMap Yoruba, Nigeria population. For tagging SNPs, we used an R2 value >0.8. SNPs with a high R2 value are correlated and can give information about one another, and thus they are grouped together and one SNP—termed a tag SNP—is selected to represent them all. Selection was restricted to those SNPs with a minor allele frequency >1% in the African population. In addition, all SNPs were evaluated using a proprietary algorithm from Illumina that assesses the likelihood that an assay will succeed on the Illumina genotyping platform. Only those with a design score >0.8 were selected. For nonsynonymous SNPs, a more liberal cutoff score >0.4 was used.
Forty-one additional SNPs with previously reported associations in a variety of human diseases, 157 nonsynonymous SNPs, and 28 AIMs were also typed, for a total of 768 SNPs. African Americans are typically of a mixed ethnic background. Allele frequencies of SNPs can vary by race and ethnicity; therefore, it is important to assess the degree of mixture in an admixed individual when analyzing SNP data. If the cases and controls have differing amounts of admixture, then variation in allele frequency by race can cause the appearance of an association with the phenotype of interest, when actually the association is with race. Including AIMs in the SNP panel allows an assessment of admixture and allows differing amounts of admixture to be controlled for in the data analysis.
Acknowledgments
We thank Dr. David A. Leib (Dartmouth Medical School) for providing the HSV-1 luciferase (KOS/Dlux/oriS) and Dr. Young Choi (Johns Hopkins University School of Medicine) for providing the IKKα and IKKβ KD plasmids. Dr. Greg Roth (now deceased), Sanford Burnham Research Institute, Orlando, FL, provided the ML130 compound. This work was supported by the Johns Hopkins Institute of Clinical and Translational Research. Genotyping services were provided by Johns Hopkins University under Contract NO1-HV-48195 from the National Heart, Lung, and Blood Institute.
Footnotes
Conflict of interest statement: Two US applications are currently pending in connection with this paper: US application 15/026,863, Compositions and Methods for Prediction and Treatment of Human Cytomegalovirus Infections, and US Application 15/215,711, Vaccine Adjuvants for Cytomegalovirus Prevention and Treatment.
This article is a PNAS Direct Submission. M.N. is a Guest Editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1611711113/-/DCSupplemental.
References
- 1.La Rosa C, Diamond DJ. The immune response to human CMV. Future Virol. 2012;7(3):279–293. doi: 10.2217/fvl.12.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rossini G, et al. Interplay between human cytomegalovirus and intrinsic/innate host responses: A complex bidirectional relationship. Mediators Inflamm. 2012;2012:607276. doi: 10.1155/2012/607276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marshall EE, Geballe AP. Multifaceted evasion of the interferon response by cytomegalovirus. J Interferon Cytokine Res. 2009;29(9):609–619. doi: 10.1089/jir.2009.0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boehme KW, Guerrero M, Compton T. Human cytomegalovirus envelope glycoproteins B and H are necessary for TLR2 activation in permissive cells. J Immunol. 2006;177(10):7094–7102. doi: 10.4049/jimmunol.177.10.7094. [DOI] [PubMed] [Google Scholar]
- 5.Cristea IM, et al. Human cytomegalovirus pUL83 stimulates activity of the viral immediate-early promoter through its interaction with the cellular IFI16 protein. J Virol. 2010;84(15):7803–7814. doi: 10.1128/JVI.00139-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim YE, Ahn JH. Positive role of promyelocytic leukemia protein in type I interferon response and its regulation by human cytomegalovirus. PLoS Pathog. 2015;11(3):e1004785. doi: 10.1371/journal.ppat.1004785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeFilippis VR, Alvarado D, Sali T, Rothenburg S, Früh K. Human cytomegalovirus induces the interferon response via the DNA sensor ZBP1. J Virol. 2010;84(1):585–598. doi: 10.1128/JVI.01748-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gariano GR, et al. The intracellular DNA sensor IFI16 gene acts as restriction factor for human cytomegalovirus replication. PLoS Pathog. 2012;8(1):e1002498. doi: 10.1371/journal.ppat.1002498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanneganti TD. Central roles of NLRs and inflammasomes in viral infection. Nat Rev Immunol. 2010;10(10):688–698. doi: 10.1038/nri2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watanabe T, et al. Activation of type I IFN signaling by NOD1 mediates mucosal host defense against Helicobacter pylori infection. Gut Microbes. 2011;2(1):61–65. doi: 10.4161/gmic.2.1.15162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herskovits AA, Auerbuch V, Portnoy DA. Bacterial ligands generated in a phagosome are targets of the cytosolic innate immune system. PLoS Pathog. 2007;3(3):e51. doi: 10.1371/journal.ppat.0030051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuenzel S, et al. The nucleotide-binding oligomerization domain-like receptor NLRC5 is involved in IFN-dependent antiviral immune responses. J Immunol. 2010;184(4):1990–2000. doi: 10.4049/jimmunol.0900557. [DOI] [PubMed] [Google Scholar]
- 13.Kapoor A, Forman M, Arav-Boger R. Activation of nucleotide oligomerization domain 2 (NOD2) by human cytomegalovirus initiates innate immune responses and restricts virus replication. PLoS One. 2014;9(3):e92704. doi: 10.1371/journal.pone.0092704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kapoor A, Fan YH, Arav-Boger R. Bacterial muramyl dipeptide (MDP) restricts human cytomegalovirus replication via an IFN-β–dependent pathway. Sci Rep. 2016;6:20295. doi: 10.1038/srep20295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Magalhaes JG, et al. Essential role of Rip2 in the modulation of innate and adaptive immunity triggered by Nod1 and Nod2 ligands. Eur J Immunol. 2011;41(5):1445–1455. doi: 10.1002/eji.201040827. [DOI] [PubMed] [Google Scholar]
- 16.Geddes K, et al. Nod1 and Nod2 regulation of inflammation in the Salmonella colitis model. Infect Immun. 2010;78(12):5107–5115. doi: 10.1128/IAI.00759-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim YG, Park JH, Daignault S, Fukase K, Núñez G. Cross-tolerization between Nod1 and Nod2 signaling results in reduced refractoriness to bacterial infection in Nod2-deficient macrophages. J Immunol. 2008;181(6):4340–4346. doi: 10.4049/jimmunol.181.6.4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim YG, et al. Viral infection augments Nod1/2 signaling to potentiate lethality associated with secondary bacterial infections. Cell Host Microbe. 2011;9(6):496–507. doi: 10.1016/j.chom.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaw MH, Reimer T, Kim YG, Nuñez G. NOD-like receptors (NLRs): Bona fide intracellular microbial sensors. Curr Opin Immunol. 2008;20(4):377–382. doi: 10.1016/j.coi.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pass RF, et al. Vaccine prevention of maternal cytomegalovirus infection. N Engl J Med. 2009;360(12):1191–1199. doi: 10.1056/NEJMoa0804749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khan PM, et al. Identification of inhibitors of NOD1-induced nuclear factor-κB activation. ACS Med Chem Lett. 2011;2(10):780–785. doi: 10.1021/ml200158b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Correa RG, et al. Discovery and characterization of 2-aminobenzimidazole derivatives as selective NOD1 inhibitors. Chem Biol. 2011;18(7):825–832. doi: 10.1016/j.chembiol.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manon F, Favier A, Núñez G, Simorre JP, Cusack S. Solution structure of NOD1 CARD and mutational analysis of its interaction with the CARD of downstream kinase RICK. J Mol Biol. 2007;365(1):160–174. doi: 10.1016/j.jmb.2006.09.067. [DOI] [PubMed] [Google Scholar]
- 24.Mayle S, et al. Engagement of nucleotide-binding oligomerization domain-containing protein 1 (NOD1) by receptor-interacting protein 2 (RIP2) is insufficient for signal transduction. J Biol Chem. 2014;289(33):22900–22914. doi: 10.1074/jbc.M114.557900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kara B, et al. The significance of E266K polymorphism in the NOD1 gene on Helicobacter pylori infection: An effective force on pathogenesis? Clin Exp Med. 2010;10(2):107–112. doi: 10.1007/s10238-009-0077-6. [DOI] [PubMed] [Google Scholar]
- 26.Girardin SE, et al. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science. 2003;300(5625):1584–1587. doi: 10.1126/science.1084677. [DOI] [PubMed] [Google Scholar]
- 27.Karin M. The beginning of the end: IkappaB kinase (IKK) and NF-kappaB activation. J Biol Chem. 1999;274(39):27339–27342. doi: 10.1074/jbc.274.39.27339. [DOI] [PubMed] [Google Scholar]
- 28.Kim ML, Jeong HG, Kasper CA, Arrieumerlou C. IKKα contributes to canonical NF-κB activation downstream of Nod1-mediated peptidoglycan recognition. PLoS One. 2010;5(10):e15371. doi: 10.1371/journal.pone.0015371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li T, Chen J, Cristea IM. Human cytomegalovirus tegument protein pUL83 inhibits IFI16-mediated DNA sensing for immune evasion. Cell Host Microbe. 2013;14(5):591–599. doi: 10.1016/j.chom.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanneganti TD, Lamkanfi M, Núñez G. Intracellular NOD-like receptors in host defense and disease. Immunity. 2007;27(4):549–559. doi: 10.1016/j.immuni.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 31.Inohara N, et al. Nod1, an Apaf-1-like activator of caspase-9 and nuclear factor-kappaB. J Biol Chem. 1999;274(21):14560–14567. doi: 10.1074/jbc.274.21.14560. [DOI] [PubMed] [Google Scholar]
- 32.Chamaillard M, et al. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol. 2003;4(7):702–707. doi: 10.1038/ni945. [DOI] [PubMed] [Google Scholar]
- 33.Girardin SE, et al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem. 2003;278(11):8869–8872. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- 34.Watanabe T, et al. NOD1 contributes to mouse host defense against Helicobacter pylori via induction of type I IFN and activation of the ISGF3 signaling pathway. J Clin Invest. 2010;120(5):1645–1662. doi: 10.1172/JCI39481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sabbah A, et al. Activation of innate immune antiviral responses by Nod2. Nat Immunol. 2009;10(10):1073–1080. doi: 10.1038/ni.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lio CW, et al. cGAS-STING signaling regulates initial innate control of cytomegalovirus infection. J Virol. 2016;90(17):7789–7797. doi: 10.1128/JVI.01040-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang G, et al. Cytoplasmic isoforms of Kaposi sarcoma herpesvirus LANA recruit and antagonize the innate immune DNA sensor cGAS. Proc Natl Acad Sci USA. 2016;113(8):E1034–E1043. doi: 10.1073/pnas.1516812113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu JJ, et al. Inhibition of cGAS DNA sensing by a herpesvirus virion protein. Cell Host Microbe. 2015;18(3):333–344. doi: 10.1016/j.chom.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma Z, et al. Modulation of the cGAS-STING DNA sensing pathway by gammaherpesviruses. Proc Natl Acad Sci USA. 2015;112(31):E4306–E4315. doi: 10.1073/pnas.1503831112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orzalli MH, et al. cGAS-mediated stabilization of IFI16 promotes innate signaling during herpes simplex virus infection. Proc Natl Acad Sci USA. 2015;112(14):E1773–E1781. doi: 10.1073/pnas.1424637112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ansari MA, et al. Herpesvirus genome recognition-induced acetylation of nuclear IFI16 is essential for its cytoplasmic translocation, inflammasome and IFN-β responses. PLoS Pathog. 2015;11(7):e1005019. doi: 10.1371/journal.ppat.1005019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Häcker H, Karin M. Regulation and function of IKK and IKK-related kinases. Sci STKE. 2006;2006(357):re13. doi: 10.1126/stke.3572006re13. [DOI] [PubMed] [Google Scholar]
- 43.Wang RP, et al. Differential regulation of IKK alpha-mediated activation of IRF3/7 by NIK. Mol Immunol. 2008;45(7):1926–1934. doi: 10.1016/j.molimm.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 44.Albanese C, et al. IKKalpha regulates mitogenic signaling through transcriptional induction of cyclin D1 via Tcf. Mol Biol Cell. 2003;14(2):585–599. doi: 10.1091/mbc.02-06-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen G, Shaw MH, Kim YG, Nuñez G. NOD-like receptors: Role in innate immunity and inflammatory disease. Annu Rev Pathol. 2009;4:365–398. doi: 10.1146/annurev.pathol.4.110807.092239. [DOI] [PubMed] [Google Scholar]
- 46.McGovern DP, et al. Association between a complex insertion/deletion polymorphism in NOD1 (CARD4) and susceptibility to inflammatory bowel disease. Hum Mol Genet. 2005;14(10):1245–1250. doi: 10.1093/hmg/ddi135. [DOI] [PubMed] [Google Scholar]
- 47.Weidinger S, et al. Association of NOD1 polymorphisms with atopic eczema and related phenotypes. J Allergy Clin Immunol. 2005;116(1):177–184. doi: 10.1016/j.jaci.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 48.Hysi P, et al. NOD1 variation, immunoglobulin E and asthma. Hum Mol Genet. 2005;14(7):935–941. doi: 10.1093/hmg/ddi087. [DOI] [PubMed] [Google Scholar]
- 49.Plantinga TS, et al. Role of NOD1 polymorphism in susceptibility and clinical progression of rheumatoid arthritis. Rheumatology (Oxford) 2013;52(5):806–814. doi: 10.1093/rheumatology/kes404. [DOI] [PubMed] [Google Scholar]
- 50.Girardin SE, et al. Identification of the critical residues involved in peptidoglycan detection by Nod1. J Biol Chem. 2005;280(46):38648–38656. doi: 10.1074/jbc.M509537200. [DOI] [PubMed] [Google Scholar]
- 51.Kijpittayarit S, Eid AJ, Brown RA, Paya CV, Razonable RR. Relationship between Toll-like receptor 2 polymorphism and cytomegalovirus disease after liver transplantation. Clin Infect Dis. 2007;44(10):1315–1320. doi: 10.1086/514339. [DOI] [PubMed] [Google Scholar]
- 52.Arav-Boger R, et al. Polymorphisms in Toll-like receptor genes influence antibody responses to cytomegalovirus glycoprotein B vaccine. BMC Res Notes. 2012;5(1):140. doi: 10.1186/1756-0500-5-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 54.He R, et al. Recombinant luciferase-expressing human cytomegalovirus (CMV) for evaluation of CMV inhibitors. Virol J. 2011;8:40. doi: 10.1186/1743-422X-8-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cardenas I, et al. Nod1 activation by bacterial iE-DAP induces maternal-fetal inflammation and preterm labor. J Immunol. 2011;187(2):980–986. doi: 10.4049/jimmunol.1100578. [DOI] [PubMed] [Google Scholar]
- 56.Vliegen I, Herngreen S, Grauls G, Bruggeman C, Stassen F. Improved detection and quantification of mouse cytomegalovirus by real-time PCR. Virus Res. 2003;98(1):17–25. doi: 10.1016/j.virusres.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 57.Tiscornia G, Singer O, Ikawa M, Verma IM. A general method for gene knockdown in mice by using lentiviral vectors expressing small interfering RNA. Proc Natl Acad Sci USA. 2003;100(4):1844–1848. doi: 10.1073/pnas.0437912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Choi YB, Harhaj EW. HTLV-1 tax stabilizes MCL-1 via TRAF6-dependent K63-linked polyubiquitination to promote cell survival and transformation. PLoS Pathog. 2014;10(10):e1004458. doi: 10.1371/journal.ppat.1004458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin R, Génin P, Mamane Y, Hiscott J. Selective DNA binding and association with the CREB binding protein coactivator contribute to differential activation of alpha/beta interferon genes by interferon regulatory factors 3 and 7. Mol Cell Biol. 2000;20(17):6342–6353. doi: 10.1128/mcb.20.17.6342-6353.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]