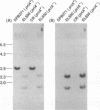
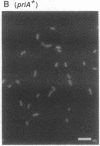
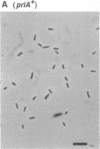
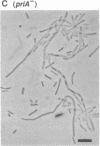
Abstract
The priA gene of Escherichia coli encodes the protein that initiates assembly of the promosome, the entity essential for the replication of phage phi X174 and ColE1-like plasmids in vitro. We have prepared a null priA mutant to assess its role in vivo in replication of phages, plasmids, and the host chromosome. Extracts of this mutant are inert in the initial conversion of the phi X174 viral strand to the duplex form, confirming the absence of the PriA activity. In vivo, the priA mutant fails to produce phi X174 phage and, remarkably, is unable to maintain plasmids that depend on the E. coli chromosome origin as well as those of ColE1. Deficiencies in cell growth and cell division are also manifest.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abarzúa P., Soeller W., Marians K. J. Mutational analysis of primosome assembly sites. I. Distinct classes of mutants in the pBR322 Escherichia coli factor Y DNA effector sequences. J Biol Chem. 1984 Nov 25;259(22):14286–14292. [PubMed] [Google Scholar]
- Arai K., McMacken R., Yasuda S., Kornberg A. Purification and properties of Escherichia coli protein i, a prepriming protein in phi X174 DNA replication. J Biol Chem. 1981 May 25;256(10):5281–5286. [PubMed] [Google Scholar]
- Baker T. A., Kornberg A. Transcriptional activation of initiation of replication from the E. coli chromosomal origin: an RNA-DNA hybrid near oriC. Cell. 1988 Oct 7;55(1):113–123. doi: 10.1016/0092-8674(88)90014-1. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Hirota Y., Mordoh J., Jacob F. On the process of cellular division in Escherichia coli. 3. Thermosensitive mutants of Escherichia coli altered in the process of DNA initiation. J Mol Biol. 1970 Nov 14;53(3):369–387. doi: 10.1016/0022-2836(70)90072-0. [DOI] [PubMed] [Google Scholar]
- Imber R., Low R. L., Ray D. S. Identification of a primosome assembly site in the region of the ori 2 replication origin of the Escherichia coli mini-F plasmid. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7132–7136. doi: 10.1073/pnas.80.23.7132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lark C. A., Riazi J., Lark K. G. dnaT, dominant conditional-lethal mutation affecting DNA replication in Escherichia coli. J Bacteriol. 1978 Dec;136(3):1008–1017. doi: 10.1128/jb.136.3.1008-1017.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasken R. S., Kornberg A. The primosomal protein n' of Escherichia coli is a DNA helicase. J Biol Chem. 1988 Apr 25;263(12):5512–5518. [PubMed] [Google Scholar]
- Lee E. H., Masai H., Allen G. C., Jr, Kornberg A. The priA gene encoding the primosomal replicative n' protein of Escherichia coli. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4620–4624. doi: 10.1073/pnas.87.12.4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low R. L., Shlomai J., Kornberg A. Protein n, a primosomal DNA replication protein of Escherichia coli. Purification and characterization. J Biol Chem. 1982 Jun 10;257(11):6242–6250. [PubMed] [Google Scholar]
- Masai H., Arai K. Escherichia coli dnaT gene function is required for pBR322 plasmid replication but not for R1 plasmid replication. J Bacteriol. 1989 Jun;171(6):2975–2980. doi: 10.1128/jb.171.6.2975-2980.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H., Arai K. Leading strand synthesis of R1 plasmid replication in vitro is primed by primase alone at a specific site downstream of oriR. J Biol Chem. 1989 May 15;264(14):8082–8090. [PubMed] [Google Scholar]
- Masai H., Nomura N., Kubota Y., Arai K. Roles of phi X174 type primosome- and G4 type primase-dependent primings in initiation of lagging and leading strand syntheses of DNA replication. J Biol Chem. 1990 Sep 5;265(25):15124–15133. [PubMed] [Google Scholar]
- Matson S. W., Kaiser-Rogers K. A. DNA helicases. Annu Rev Biochem. 1990;59:289–329. doi: 10.1146/annurev.bi.59.070190.001445. [DOI] [PubMed] [Google Scholar]
- Meijer M., Beck E., Hansen F. G., Bergmans H. E., Messer W., von Meyenburg K., Schaller H. Nucleotide sequence of the origin of replication of the Escherichia coli K-12 chromosome. Proc Natl Acad Sci U S A. 1979 Feb;76(2):580–584. doi: 10.1073/pnas.76.2.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minden J. S., Marians K. J. Replication of pBR322 DNA in vitro with purified proteins. Requirement for topoisomerase I in the maintenance of template specificity. J Biol Chem. 1985 Aug 5;260(16):9316–9325. [PubMed] [Google Scholar]
- Nomura N., Low R. L., Ray D. S. Identification of ColE1 DNA sequences that direct single strand-to-double strand conversion by a phi X174 type mechanism. Proc Natl Acad Sci U S A. 1982 May;79(10):3153–3157. doi: 10.1073/pnas.79.10.3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P., DiGate R. J., Zavitz K. H., Marians K. J. Molecular cloning and DNA sequence analysis of Escherichia coli priA, the gene encoding the primosomal protein replication factor Y. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4615–4619. doi: 10.1073/pnas.87.12.4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell C. B., Thaler D. S., Dahlquist F. W. Chromosomal transformation of Escherichia coli recD strains with linearized plasmids. J Bacteriol. 1989 May;171(5):2609–2613. doi: 10.1128/jb.171.5.2609-2613.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassanfar M., Roberts J. W. Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J Mol Biol. 1990 Mar 5;212(1):79–96. doi: 10.1016/0022-2836(90)90306-7. [DOI] [PubMed] [Google Scholar]
- Shlomai J., Kornberg A. A prepriming DNA replication enzyme of Escherichia coli. II. Actions of protein n': a sequence-specific, DNA-dependent ATPase. J Biol Chem. 1980 Jul 25;255(14):6794–6798. [PubMed] [Google Scholar]
- Shlomai J., Kornberg A. An Escherichia coli replication protein that recognizes a unique sequence within a hairpin region in phi X174 DNA. Proc Natl Acad Sci U S A. 1980 Feb;77(2):799–803. doi: 10.1073/pnas.77.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomai J., Polder L., Arai K., Kornberg A. Replication of phi X174 dna with purified enzymes. I. Conversion of viral DNA to a supercoiled, biologically active duplex. J Biol Chem. 1981 May 25;256(10):5233–5238. [PubMed] [Google Scholar]
- Van Houten B. Nucleotide excision repair in Escherichia coli. Microbiol Rev. 1990 Mar;54(1):18–51. doi: 10.1128/mr.54.1.18-51.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G. C. Inducible DNA repair systems. Annu Rev Biochem. 1985;54:425–457. doi: 10.1146/annurev.bi.54.070185.002233. [DOI] [PubMed] [Google Scholar]
- van der Ende A., Baker T. A., Ogawa T., Kornberg A. Initiation of enzymatic replication at the origin of the Escherichia coli chromosome: primase as the sole priming enzyme. Proc Natl Acad Sci U S A. 1985 Jun;82(12):3954–3958. doi: 10.1073/pnas.82.12.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]