Significance
Elevated levels of N-Myc protein (the product of the MYCN oncogene) drive cancers such as neuroblastoma. Accumulation of N-Myc in these cancer cells depends upon the formation of a complex with the protein kinase Aurora-A in which the N-Myc is not properly degraded. We mapped the region of N-Myc that interacts with Aurora-A and determined the molecular structure of the complex. Because this region also interacts with cellular machinery that targets N-Myc for degradation, we sought to understand the mechanism by which N-Myc stabilizes Aurora-A. The structure explains how compounds that induce distorted conformations of Aurora-A are able to disrupt the interaction with N-Myc. This understanding may provide a basis for designing better compounds that work in this way for the treatment of neuroblastoma.
Keywords: structural biology, Aurora-A kinase, protein–protein interaction, Myc, neuroblastoma
Abstract
Myc family proteins promote cancer by inducing widespread changes in gene expression. Their rapid turnover by the ubiquitin–proteasome pathway is regulated through phosphorylation of Myc Box I and ubiquitination by the E3 ubiquitin ligase SCFFbxW7. However, N-Myc protein (the product of the MYCN oncogene) is stabilized in neuroblastoma by the protein kinase Aurora-A in a manner that is sensitive to certain Aurora-A–selective inhibitors. Here we identify a direct interaction between the catalytic domain of Aurora-A and a site flanking Myc Box I that also binds SCFFbxW7. We determined the crystal structure of the complex between Aurora-A and this region of N-Myc to 1.72-Å resolution. The structure indicates that the conformation of Aurora-A induced by compounds such as alisertib and CD532 is not compatible with the binding of N-Myc, explaining the activity of these compounds in neuroblastoma cells and providing a rational basis for the design of cancer therapeutics optimized for destabilization of the complex. We also propose a model for the stabilization mechanism in which binding to Aurora-A alters how N-Myc interacts with SCFFbxW7 to disfavor the generation of Lys48-linked polyubiquitin chains.
Myc proteins are transcription factors that markedly alter gene expression through both activation and repression of transcription (1, 2). Three Myc protein family members can be aberrantly expressed in human cancers. Cellular Myc (c-Myc) was first discovered as the cellular homolog of the viral Myc (v-Myc) oncoprotein and is frequently deregulated in a wide range of human cancers (3–5). N-Myc and L-Myc were subsequently identified as the products of amplified genes MYCN in neuroblastoma and MYCL in small cell lung cancer, respectively (6–8). Inhibition of Myc is a validated therapeutic strategy, but efforts to develop clinical compounds that target Myc proteins directly have failed (9).
c-Myc, N-Myc, and L-Myc have regions of sequence homology that mediate interactions with critical partner proteins such as Max, WDR5, TRRAP, PAF1C, and the protein kinase Aurora-A (10). The most C-terminal of these regions forms an essential DNA-binding domain through formation of a basic helix–loop–helix leucine zipper domain in complex with Max (9, 11). Other conserved sequence motifs called “Myc boxes” (MB0–IV) serve as docking sites for protein–protein interactions (10, 12, 13). The Myc transactivation domain (TAD) spans the N-terminal conserved motifs MB0, MBI, and MBII. The TAD of c-Myc is intrinsically disordered, as reported by circular dichroism and NMR spectroscopy, but there are transient secondary structure elements that in some cases become stable in complex with binding partners (12, 14, 15).
The stability of Myc proteins is regulated by phosphorylation within MBI, which targets the protein for ubiquitinylation and proteolysis. For example, N-Myc is first phosphorylated on Ser62 by Cdk1/cyclin B and is then phosphorylated on Thr58 by Gsk3β (16). Dephosphorylation of Ser62 by PP2A directs the activity of the E3 ubiquitin ligase SCFFbxW7 to modify N-Myc with K48-linked ubiquitin chains (17, 18). In neuroblastoma cells, the Ser/Thr protein kinase Aurora-A blocks this process, resulting in an excess of N-Myc protein (19). Aurora-A binds to the N-Myc/SCFFbxW7 complex and reduces the proportion of K48 linkages in the polyubiquitin chains. Catalytic activity of Aurora-A is not required for N-Myc stabilization, and the underlying mechanism is unclear. Some Aurora-A inhibitors such as MLN8237/alisertib and CD532 can destabilize N-Myc by disrupting the complex, whereas other Aurora-A inhibitors have no effect (19, 20). The current hypothesis is that the destabilizing inhibitors alter the conformation of Aurora-A in ways that disrupt the complex, but inhibitors that compete with ATP without causing a conformational change leave the complex intact (20–22).
We set out to investigate the structural basis of Aurora-A stabilization of N-Myc and the effect of Aurora-A inhibitors on the complex. Here we show that the catalytic domain of Aurora-A interacts directly with N-Myc through binding sites that flank either side of MBI. We present a crystal structure of the complex between Aurora-A and a fragment of N-Myc corresponding to the region immediately C-terminal to MBI which reveals Aurora-A in a fully active conformation that is incompatible with inhibitors of Aurora-A that disrupt the complex. Biochemical studies show an interaction between SCFFbxW7 and the same region of N-Myc, and we propose that the way in which Aurora-A interferes with this interaction changes N-Myc ubiquitination to promote stability.
Results and Discussion
Structural Basis of the Interaction Between N-Myc and Aurora-A.
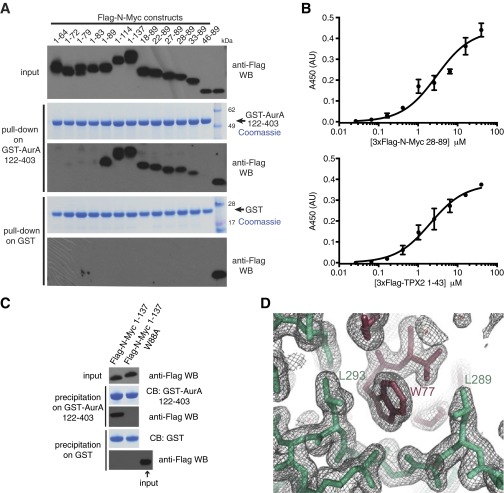
To show that Aurora-A and N-Myc interact directly, we carried out a coprecipitation experiment using a GST–Aurora-A kinase domain fusion protein incubated with a range of purified fragments from the N-Myc TAD (Fig. S1A). This method identified residues 28–89 of N-Myc as the minimal Aurora-A–interaction region (AIR), which spans MB0 through MBI but does not include MBII or beyond (Fig. 1A). The binding affinity of Aurora-A for the AIR was quantified as 2.9 ± 0.5 μM by ELISA (Fig. S1B) and as 1.0 ± 0.3 μM by competition AlphaScreen assay (Fig. 1B). The presence of a Trp residue toward the C terminus of this N-Myc fragment was striking, and mutation of Trp88 to Ala abrogated the interaction in the context of the entire N-Myc TAD fragment (residues 1–137) in a precipitation experiment with GST-tagged Aurora-A catalytic domain (residues 122–403); the TAD of N-Myc did not interact with GST alone. Taken together, these results indicate that the interaction is specific (Fig. S1C).
Fig. S1.
Interaction between N-Myc and Aurora-A. (A) Coprecipitation of fragments of the N-Myc transactivation domain with the GST–Aurora-A kinase domain. Flag-tagged N-Myc fragments were incubated with GST–Aurora-A 122–403 D274N or GST alone immobilized on glutathione Sepharose beads. Coprecipitated N-Myc fragments were visualized by anti-Flag Western blot (WB). (B) Binding isotherms measured by ELISA quantifying the interaction of 3xFlag-N-Myc 28–89 (Upper) and 3xFlag-N-Myc TPX2 1–43 (Lower) with immobilized biotinyl Avi-tagged–Aurora-A 122–403. Data points represent the mean of three experiments; error bars indicate SD. (Affinities of 2.9 ± 0.5 μM and 2.0 ± 0.3 μM were measured for 3xFlag-N-Myc 28–89 and 3xFlag-N-Myc TPX2 1–43, respectively.) (C) The specific interaction between the GST–Aurora-A catalytic domain and Flag-tagged N-Myc TAD is abrogated by a single point mutation (Trp88). Glutathione resin precipitates were separated by SDS/PAGE followed by Western blot analysis and Coomassie Blue (CB) staining. (D) A representative region of the electron density map of the 1.72-Å crystal structure of the Aurora-A/N-Myc complex showing the interaction of Trp77 of N-Myc (dark red) with a hydrophobic pocket on Aurora-A (pale green). The mesh represents a 2mFo-DFc map contoured at 1.0 σ.
Fig. 1.
Crystal structure of the Aurora-A/N-Myc complex. (A) Domain structure of N-Myc with conserved regions indicated as boxes and the AIR marked with a red line. (B) Quantification of the binding affinity between the Aurora-A kinase domain and the N-Myc AIR by competition AlphaScreen assay. Data represent the mean of three experiments ± SD. (C) Crystal structure of the Aurora-AC290A/C393A catalytic domain (AurA, green cartoon) in complex with N-Myc residues 61–89 (N-Myc, red cartoon). (D) Magnified view of the structure with key residues shown as sticks.
To elucidate the structural basis of the Aurora-A/N-Myc interaction, we crystallized a complex between the Aurora-A kinase domain with surface Cys mutated to enhance stability (Aurora-AC290A/C393A) and a synthetic N-Myc peptide corresponding to residues 28–89. The crystals yielded diffraction data to 1.72-Å resolution, and we solved the structure by molecular replacement using an existing structure of Aurora-AC290A/C393A (Table S1 and Fig. S1D). Residues 28–60 of N-Myc are not observed in the structure, whereas residues 61–89 are associated with the cleft between the N- and C-lobes of the Aurora-A kinase domain formed by the αB/αC helices, the activation loop, and the αG helix (Fig. 1C). N-Myc residues 76–89 form an α-helix, which packs onto the C-lobe of Aurora-A at a surface formed by Tyr334, Gln335, and Tyr338 with its N terminus pointing toward the substrate-binding region. Both ends of the α-helix are capped by Trp residues (Trp77 and Trp88), whose side chains pack against the surface of Aurora-A. Key intermolecular interactions include a salt bridge between Glu73 of N-Myc and Lys143 from the Gly-rich loop on the N-lobe of Aurora-A, an additional interaction between Gln335 of Aurora-A and Trp88 of N-Myc, and the insertion of the Trp77 side chain of N-Myc into the hydrophobic P+1 pocket in the activation loop region of Aurora-A (Fig. 1D). A pair of prolines (Pro74 and Pro75) breaks the α-helix at its N terminus and directs the chain toward the Aurora-A N-lobe. Residues 69–71 are hydrogen-bonded into a turn, and residues 61–67 are bound into a groove between the N-lobe and the surface formed by the activation loop.
Table S1.
Crystallographic data collection, refinement, and structure validation statistics
| AurA/N-Myc complex | |
| Lattice parameters | |
| Space group | P32 2 1 |
| Cell dimensions | |
| a, b, c, Å | 86.52, 86.52, 92.21 |
| α, β, γ, degrees | 90.00, 90.00, 120.00 |
| Data collection | |
| Resolution range, Å | 74.93–1.72 (1.76–1.72)* |
| Rmerge, % | 6.3 (74.3)* |
| I/σI | 19.8 (3.4)* |
| Completeness, % | 100 (100)* |
| Redundancy | 9.7 (9.7)* |
| Refinement | |
| Resolution, Å | 74.3–1.72 |
| No. reflections | 42,820 |
| Rwork/Rfree | 17.62/19.51 |
| No. atoms | |
| Protein | 2,362 |
| ADP/Mg2+ | 29 |
| Water | 197 |
| Mean B-factors | |
| Protein | 28.09 |
| ADP/Mg2+ | 19.16 |
| Water | 37.17 |
| Wilson B-factor | 25.63 |
| Rmsd | |
| Bond lengths, Å | 0.005 |
| Bond angles, degrees | 1.043 |
| MolProbity analysis | |
| All-atom clash-score | 2.34 |
| Rotamers outliers, % | 0 |
| Ramachandran outliers, % | 0 |
| Ramachandran favored, % | 97.25 |
| MolProbity score | 1.15 |
Values in parentheses refer to the highest-resolution shell.
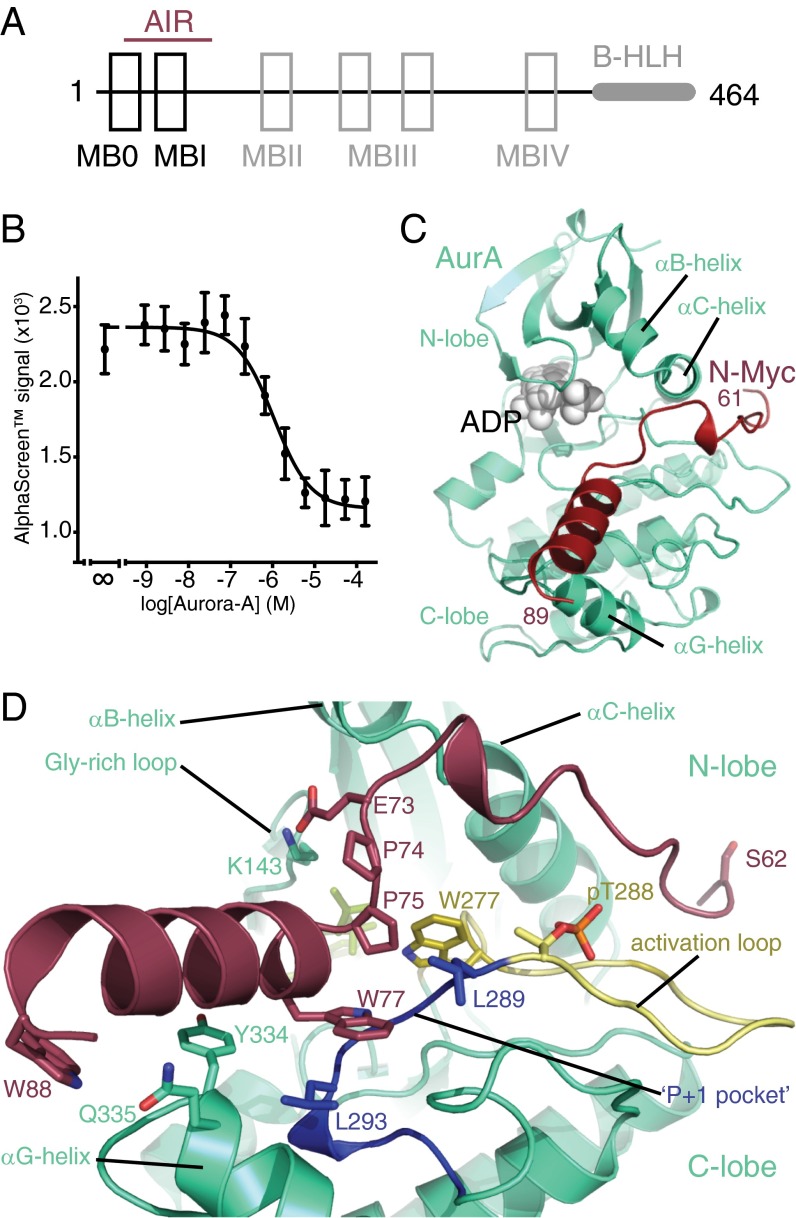
The region of N-Myc observed in the crystal structure starts at the C terminus of MBI and is not conserved in c-Myc (Fig. S2A). Our initial mapping suggested that this region was insufficient for binding, so we used a more sensitive assay based on changes in fluorescence polarization (FP) using synthetic peptides of N-Myc to confirm that residues 61–89 of N-Myc bind Aurora-A independently, with a measured Kd of 12 μM (Fig. S2B). N-Myc peptides with E73K or W77A mutations reduced binding to Aurora-A, consistent with their observed contributions to the interface in the crystal structure (Fig. S2B). Similarly, the contributions of Aurora-A Y334 and Q335 to the interaction were confirmed.
Fig. S2.
Verification of two binding sites for Aurora-A on N-Myc. (A) Sequence alignment of N-Myc and c-Myc in the Aurora-A interaction region. Asterisks, colons, and periods mark identical, conserved, and conservatively substituted residues, respectively. Conserved regions MB0 and MBI (incorporating the phospho-degron) are boxed, and the two binding sites for Aurora-A within N-Myc, corresponding to the peptides used in B, are marked with dashed (19–47) and solid (61–89) red lines. (B) Binding isotherms measured by FP quantifying the interaction between the Aurora-A catalytic domain and N-Myc residues 61–89. Data points represent the mean of three experiments; error bars indicate SD. (Affinities of 12.1 ± 0.6 μM and 12.8 ± 0.7 μM were measured for binding of the phosphorylated and dephosphorylated Aurora-A, respectively, to wild-type peptide. N-Myc 61–89 peptides containing mutations E73K and W77A interacted with wild-type Aurora-A with affinities of 81.1 ± 3.4 μM and 106 ± 4.5 μM, respectively. Measured affinities of Aurora-A mutants Y334A and Q335A for wild-type N-Myc 61–89 were 22.6 ±1.1 μM and 16.1 ± 0.8 μM, respectively.) (C) Coprecipitation experiments showing the interaction of Aurora-A kinase domain with biotinyl–N-Myc peptides. (Note: Some peptides do not stain with Coomassie Blue.) (D) Binding isotherms measured by FP quantifying the interaction between the Aurora-A catalytic domain and N-Myc residues 19–47. Data points represent the mean of three experiments; error bars indicate SD. (Affinities of 11.1 ± 0.4 μM and 9.2 ±0.3 μM were measured for the binding of the phosphorylated and dephosphorylated Aurora-A, respectively, to wild-type N-Myc 19–47 peptide. Affinities of 18.6 ± 0.6 μM and 23.7 ± 0.8 μM were measured for the binding of 19–47 peptides containing the mutations F28A and F35A, respectively, whereas a peptide containing both of these mutations bound with a Kd of 114.0 ± 4.0 μM. A Y29A, Y36A double-mutant 19–47 peptide bound Aurora-A with a Kd of 68.8 ± 2.3 μM.) (E) Coprecipitation experiments showing the interaction of the Aurora-A kinase domain with biotinyl–N-Myc 28–89 peptides carrying different phosphorylation marks within the region corresponding to the phospho-degron. Also included in the experiment is 52–56_polyA, a nonphosphorylated 28–89 peptide with residues 52–56 mutated to Ala. The sequence of N-Myc in this region, 51-KKFELLP-57, bears some resemblance to an Aurora-A consensus phosphorylation site (K/R/N-R-XS/T-B), with E54 in place of the P1 Ser/Thr. The 28–89 52–56_polyA peptide was used to investigate the possibility that a pseudosubstrate interaction between this region of N-Myc and Aurora-A might contribute to the binding.
The section of the AIR that was not resolved in the crystal structure, residues 28–60 of N-Myc, includes the MB0 and MBI regions and is conserved in c-Myc (Fig. S2A). Having shown that the C-terminal part of the AIR was able to bind Aurora-A independently, we looked for other subfragments that might contribute to binding with a sensitive pull-down assay using peptides spanning the region, under less stringent conditions than the first set of pull-downs used for mapping the interaction (Fig. S2C). In addition to residues 61–89, a region corresponding to MB0 was also capable of independent interaction with Aurora-A. Using the FP assay, we confirmed that a peptide spanning residues 19–47 of N-Myc, including MB0, binds independently to Aurora-A (Fig. S2D). The presence of conserved aromatic residues in this region is striking (F28, Y29, F35, Y36), and these residues were shown to contribute to the interaction with Aurora-A (Fig. S2D). In contrast to the established roles of the regions flanking it, MBI itself does not appear to contribute to the interaction with Aurora-A, because a peptide corresponding to residues 44–64 showed no binding in the peptide coprecipitation assay (Fig. S2C). Consistent with this result, neither the phosphorylation status of the phosphodegron residues Thr58 and Ser62 nor mutation of residues 52–56 to Ala affected Aurora-A binding of the N-Myc 28–89 peptide (Fig. S2E). We postulate that the flanking regions on either side of MBI form specific interactions with Aurora-A, linked by the MBI region itself, which interacts in a phosphorylation-dependent manner with FbxW7 but makes no critical interactions with Aurora-A.
N-Myc Activates Aurora-A and Competes with TPX2, the Targeting Protein for Xklp2.
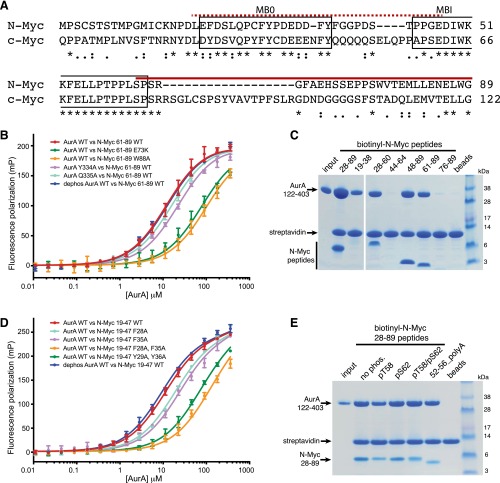
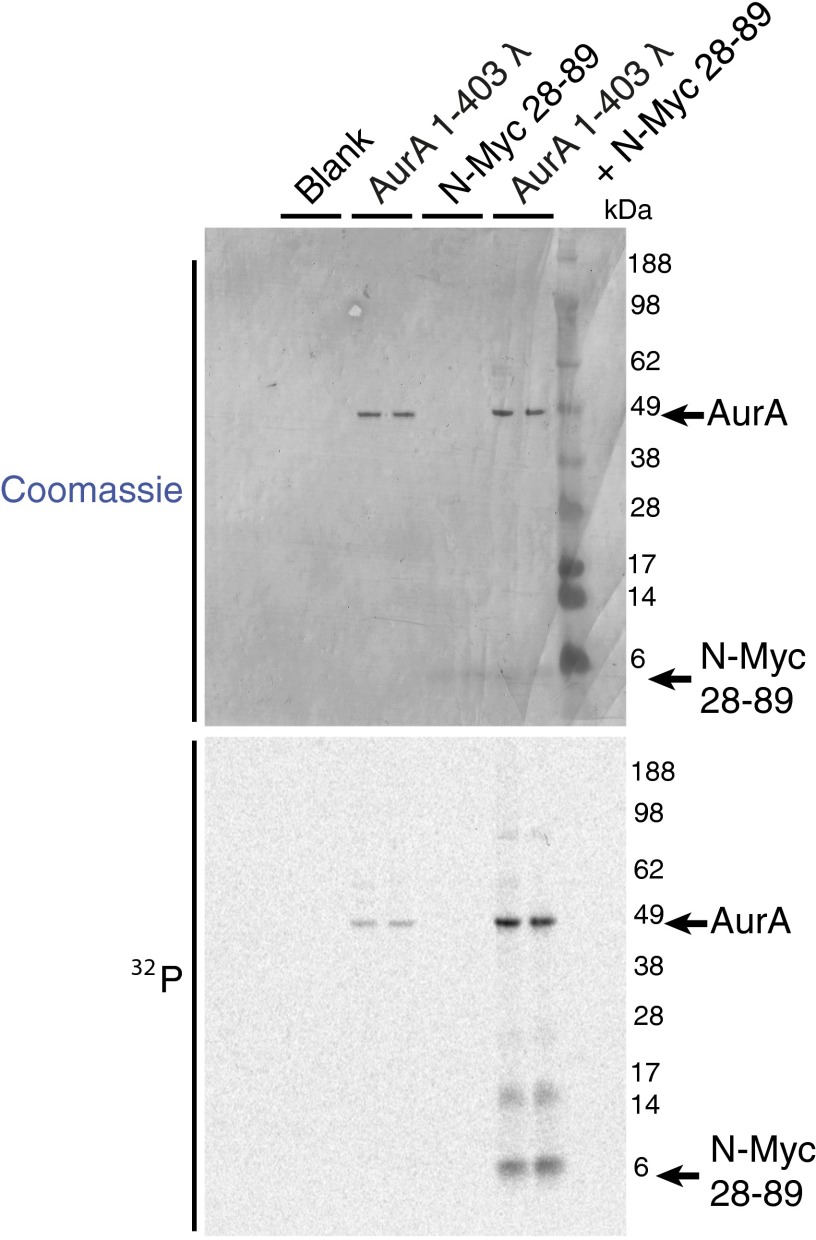
When bound to N-Myc, Aurora-A is in a fully active conformation similar to that observed for the Aurora-A/TPX2 complex (Fig. 2A) (23). The microtubule-associated protein-targeting protein for Xklp2 (TPX2) binds to Aurora-A through its first 43 residues: amino acids 7–21 at a site on the N-lobe and amino acids 30–43 at a site between the two lobes. The conformation adopted by Aurora-A when bound to TPX2 is incompatible with the interaction of N-Myc residues 61–67 (Fig. 2B). There is a clear steric clash, because Leu61 of N-Myc binds to the same pocket on the Aurora-A surface as Trp34/Phe35 of TPX2. To accommodate the marked increase in size in going from the single Leu side chain of N-Myc to the two bulky side chains of TPX2, the side chain of Aurora-A residue His187 rotates out of the pocket in the TPX2-bound conformation. Side chains on the αC-helix of Aurora-A such as His176 and Arg179 are also observed in different positions, suggesting that the N-terminal stretch of TPX2 (Tyr8/Tyr10) that binds on one side of the αC-helix is incompatible with binding of N-Myc to the other side of the αC-helix. Consistent with this structural analysis, the first 43 amino acids of TPX2 and the AIR of N-Myc compete for binding to Aurora-A (Fig. 2C), and they also have comparable affinities, because Kd values of 2–3 μM were measured for both proteins in ELISAs using immobilized Aurora-A (Fig. S1B). Competition with TPX2 was observed for the individual regions of the AIR both N- and C-terminal to MB1 (Fig. 2C), suggesting that the binding site for MB0 on Aurora-A also overlaps with that of TPX2. The AIR of N-Myc, like TPX2, initially activates unphosphorylated Aurora-A, so there is also functional overlap between these two Aurora-A–binding proteins (Fig. 2D and Fig. S3). The crystal structure reported here does not reveal the mechanism by which N-Myc activates Aurora-A, because residues 61–89 of N-Myc were not sufficient to activate Aurora-A, and we used Aurora-A prephosphorylated on Thr288 to form the complex. However, it is clear that N-Myc, like TPX2, is able to trigger Aurora-A activation through protein–protein interactions, and the result is a stabilized conformation of Aurora-A in which the activation loop forms a platform for the binding of N-Myc residues 61–89 (Fig. 2E). Furthermore, as shown in Fig. S3, Aurora-A is able to phosphorylate residues within the 28–89 region of N-Myc efficiently in vitro.
Fig. 2.
Structural and functional comparison of Aurora-A interactions with N-Myc and TPX2. (A) Superposition of crystal structures of Aurora-A bound to N-Myc (colored red) and TPX2 (colored pink). Aurora-A bound to TPX2 is colored a lighter shade of green. (B) Magnified view of the superposed structures. The side-chain motions accompanying the transition from the N-Myc to the TPX2 complex are indicated by black arrows. (C) Coprecipitation experiments to investigate competition between TPX2 (residues 1–43) and N-Myc fragments for binding to Aurora-A (residues 122–403), analyzed by SDS/PAGE. The gel shows proteins coprecipitated with biotinyl–N-Myc peptides (as indicated) immobilized on streptavidin beads following incubation with 12-μM Aurora-A catalytic domain in the presence or absence of a fivefold molar excess of TPX2 1–43 protein. (D) Kinase assays to measure the activity of the 0.625-μM initially unphosphorylated Aurora-A catalytic domain in the presence of [32P]ATP and varying concentrations N-Myc peptides (residues 28–89, 19–47, or 61–89) or TPX2 (residues 1–43), reported by scintillation counting. Data are the mean of two experiments ± SE. (E) Schematic illustration of the activating interactions of N-Myc and TPX2 with Aurora-A. The unphosphorylated activation loop of Aurora-A is flexible (dashed black line) but becomes ordered upon kinase activation through the binding of protein partners and autophosphorylation.
Fig. S3.
Autoradiograph showing activation of Aurora-A by N-Myc 28–89 in the presence of [32P]ATP. Initially unphosphorylated full-length Aurora-A shows enhanced autophosphorylation in the presence of N-Myc 28–89 peptide. The N-Myc peptide itself also becomes phosphorylated. Reactions contained 0.625-μM dephosphorylated full-length Aurora-A and 10-μM N-Myc 28–89. Samples were separated by SDS/PAGE and 32P-incorporation into individual proteins visualized by autoradiography.
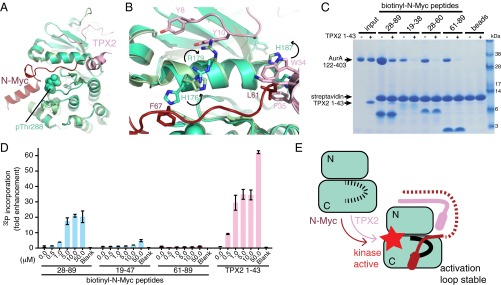
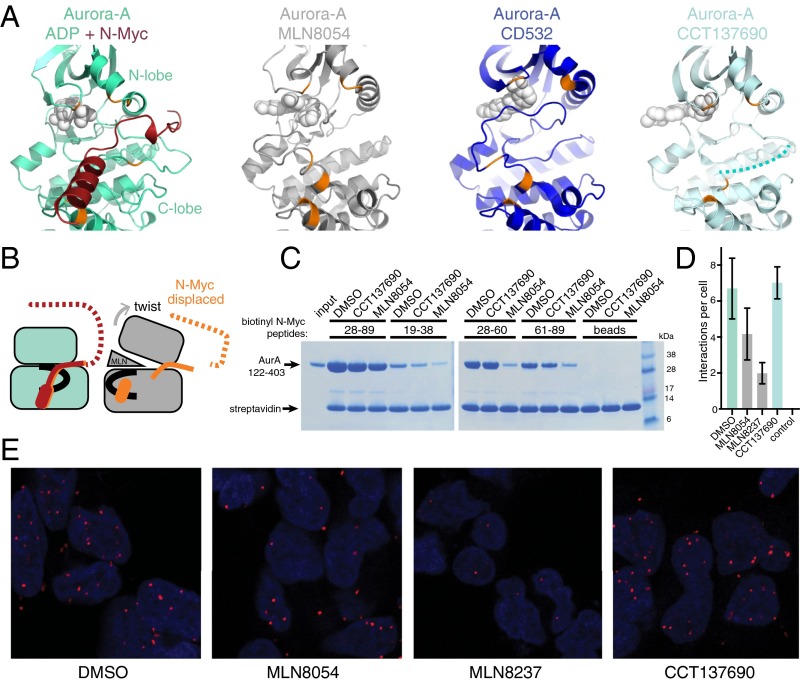
The Conformation of Aurora-A Bound to N-Myc Is Incompatible with Inhibitors That Destabilize the Interaction.
Previous studies showed that ATP-competitive inhibitors of Aurora-A such as MLN8054, MLN8237, and CD532 disrupt the formation of the Aurora-A/N-Myc complex, resulting in destabilization of N-Myc in cell models. Crystal structures of Aurora-A in complex with MLN8054 or CD532 show distorted conformations of the kinase, notably in the positions of the activation loop and the Gly-rich loop (20, 21). In contrast, most Aurora-A inhibitors, such as CCT137690, do not substantially affect the structure of the protein (24). There is no available crystal structure of MLN8237 bound to Aurora-A, but the chemical structures of MLN8054 and MLN8237 are almost identical, and it is likely that MLN8237 induces a similar conformation in Aurora-A. We compared the structures of Aurora-A in complex with N-Myc, MLN8054, CD532, and CCT137690 (Fig. 3A). Aurora-A grips N-Myc through interactions involving both the N- and C-lobes of the kinase (colored orange in Fig. 3A). Binding of CCT137690 does not affect the relative orientation of the two kinase lobes. The activation loop of Aurora-A is disordered in the Aurora-A/CCT137690 structure because it is neither phosphorylated nor stabilized by a protein such as TPX2 or N-Myc, and there is no obvious mechanism by which CCT137690 could affect the activation loop. However, when Aurora-A is bound to MLN8054 or CD532, the surfaces that form the binding site for residues 61–89 of N-Myc are moved apart through motions that twist the two kinase lobes relative to one another and displace the activation loop (Fig. 3B). The rearrangement of this binding site provides a possible mechanism by which compounds such as MLN8054 disrupt the Aurora-A/N-Myc complex by acting as a wedge that forces the kinase into an inactive conformation. The key differences between CCT137690 and MLN8054 are that only the latter makes a specific contact with the displaced activation loop; although both compounds contact both the hinge and the Gly-rich loop, the wedge effect is a function of the 3D shape of MLN8054 in contrast to the flatter scaffold of CCT137690.
Fig. 3.
The conformation of Aurora-A required for N-Myc binding is incompatible with the conformation induced by kinase inhibitors that disrupt the interaction. (A) Views of Aurora-A bound to N-Myc (PDB ID code 5G1X), MLN8054 (PDB ID code 2WTV), CD532 (PDB ID code 4J8M), and CCT137690 (PDB ID code 2X6E). Key residues of Aurora-A (orange) that grasp N-Myc (dark red) remain in place in the presence of CCT137690 but are moved apart in the presence of MLN8054/8237 and are moved even further apart in the presence of CD532. The dotted line in the rightmost panel indicates the disordered region of the activation loop. (B) Schematic model to explain the disruption of the N-Myc binding site by ATP-competitive inhibitors. The activation loop is shown in black. (C) Competition coprecipitation experiments using the untagged Aurora-A catalytic domain and biotinylated N-Myc peptides immobilized on streptavidin beads in the presence of 20-μM MLN8054 or CCT137690; 0.2% DMSO was used as control. (D) Incidence of complex formation between N-Myc and Aurora-A observed in situ by proximity ligation assay in Kelly neuroblastoma cells treated with compounds as indicated. Data represent the mean ± SD. No antibodies were added to the control cells. (E) Representative images of Kelly cells used in D. Nuclei are indicated by DAPI staining (blue). Red foci indicate the position of Aurora-A/N-Myc complexes.
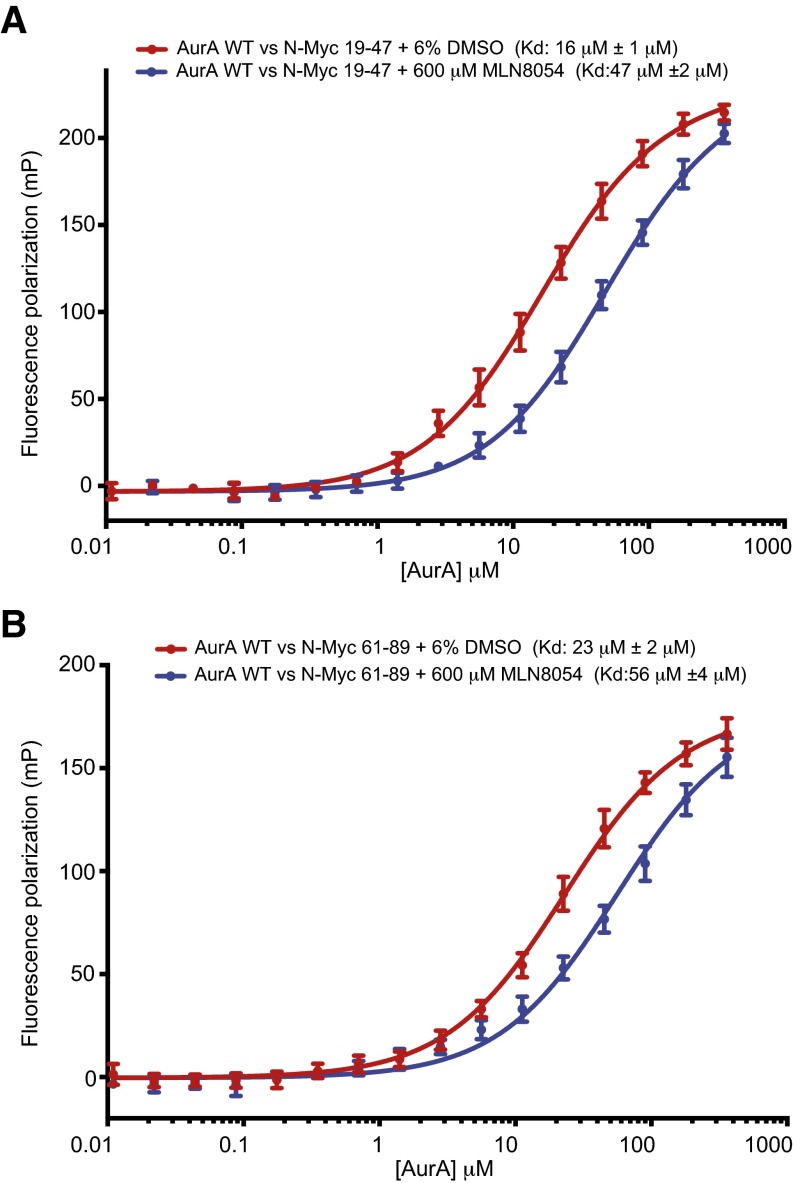
Consistent with the structural analysis, MLN8054 but not CCT137690 disrupted the direct interaction of the catalytic domain of Aurora-A with its binding sites within the AIR (Fig. 3C). Indeed, Aurora-A bound to MLN8054 displayed two- to threefold reduced affinity for both of the binding sites within the AIR of N-Myc compared with Aurora-A alone (Fig. S4). Next, we used proximity ligation assays to quantify complex formation between endogenous N-Myc and Aurora-A proteins in a neuroblastoma cell line (Fig. 3 D and E) and observed significantly fewer interactions in cells treated with MLN8054 or MLN8237 than in untreated cells, whereas the number of interactions was unaffected by treatment with CCT137690.
Fig. S4.
Aurora-A ATP-competitive inhibitors and N-Myc interaction. Binding isotherms measured by FP quantifying the interaction of the Aurora-A catalytic domain (red) and the Aurora-A catalytic domain/MLN8054 complex (blue) with N-Myc residues 19–47 (A) and with N-Myc residues 61–89 (B). Data points represent the mean of three experiments; error bars indicate SD; binding affinities between Aurora-A and N-Myc peptides derived from curve-fitting are stated as Kd ± SE.
A Model for the Regulation of N-Myc Ubiquitination by Aurora-A.
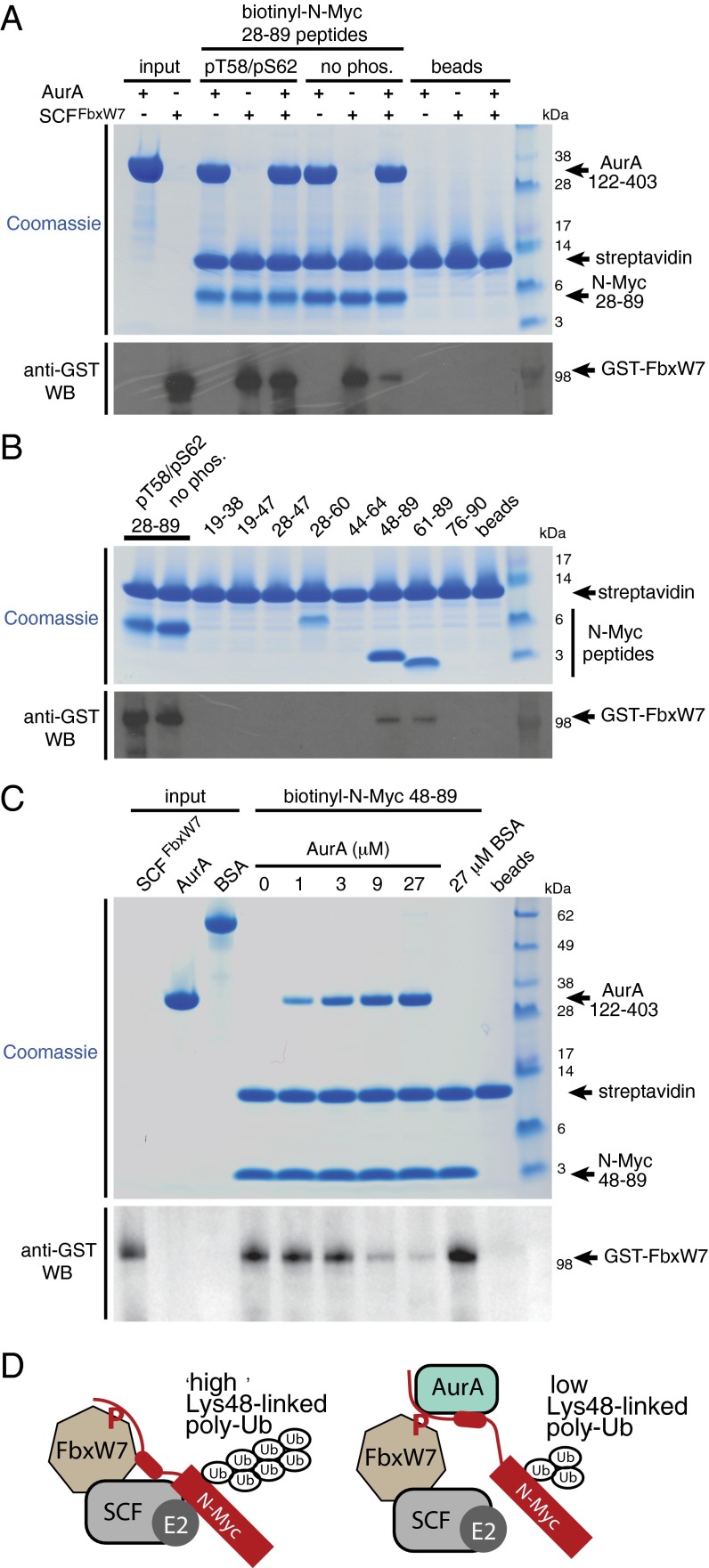
In this study, we showed that Aurora-A binds N-Myc irrespective of its phosphorylation state and interacts with regions of N-Myc that flank MBI. The binding interaction between the Aurora-A catalytic domain and N-Myc residues 61–89 that we have resolved is immediately adjacent to the phospho-degron motif centered on Thr58. Phosphorylation of this motif is required for recognition of N-Myc by SCFFbxW7. Thus, binding of Aurora-A might affect the interaction of N-Myc with SCFFbxW7. Using purified proteins, we discovered that SCFFbxW7, like Aurora-A, interacts with N-Myc irrespective of the phosphorylation state of Thr58/Ser62 (Fig. 4A). Binding of SCFFbxW7 to N-Myc that is phosphorylated on Thr58/Ser62 was not affected by Aurora-A, as is consistent with nonoverlapping binding sites on N-Myc. In contrast, Aurora-A competed with SCFFbxW7 for binding to unphosphorylated N-Myc. This observation suggested that the interaction of SCFFbxW7 with unphosphorylated N-Myc might depend on a binding site that overlaps with one or both of the Aurora-A binding sites that flank MBI. Binding of SCFFbxW7 was mapped to residues 61–89 of N-Myc; binding to residues 48–89 of N-Myc was also observed, but an unphosphorylated MBI peptide (residues 44–64) did not bind (Fig. 4B). Aurora-A efficiently competed with SCFFbxW7 for binding to residues 48–89 of N-Myc in a dose-dependent manner, whereas a control protein bovine serum albumin (BSA) did not compete (Fig. 4C). Taken together, these results show that there is competition between Aurora-A and SCFFbxW7 for binding to residues 61–89 of N-Myc, the region bound to Aurora-A in the crystal structure.
Fig. 4.
Aurora-A alters the interaction of N-Myc with SCFFbxW7. (A, Upper) Competition coprecipitation experiments using biotinylated N-Myc AIR peptides immobilized on streptavidin beads and incubated with 2 μg of the SCF complex incorporating GST-tagged FbxW7 in the presence or absence of 12-μM untagged Aurora-A catalytic domain. Equal loading of resin was assessed by Coomassie Blue staining. (Lower) Binding of SCFFbxW7 to N-Myc was visualized by Western blot (WB) using anti-GST antibody. (B, Upper) Coprecipitation experiments using biotinylated N-Myc AIR fragment peptides immobilized on streptavidin beads and incubated with 2 μg SCF complex incorporating GST-tagged FbxW7. Equal loading of resin was assessed by Coomassie Blue staining. (Lower) Binding of SCFFbxW7 to N-Myc peptides was visualized by Western blot using an anti-GST antibody. Note that in A and B, despite the uneven appearance of the peptides in the upper panels because of differences in migration and sensitivity to Coomassie Blue staining, equal amounts were loaded into the experiments. (C) Coprecipitation experiments showing competition between SCFFbxW7 and Aurora-A for binding to N-Myc 48–89. Biotinylated N-Myc 48–89 peptide was immobilized on streptavidin beads and incubated with 2 μg of the SCFFbxW7 complex in the presence of Aurora-A at a range of concentrations or with BSA. FbxW7 coprecipitating with the peptide was visualized by anti-GST Western blot. (D) A working model to explain how the interaction between Aurora-A and N-Myc residues 61–89 may reduce ubiquitination to stabilize N-Myc. In this model, a low-affinity interaction between the 61–89 region and SCFFbxW7 is required for N-Myc to be modified effectively with K48-linked polyubiquitin chains, and competitive binding of Aurora-A to this region interferes with K48-linked polyubiquitination of N-Myc, leading to reduced proteasomal degradation.
These observations suggest a working model by which Aurora-A could stabilize N-Myc by altering its interaction with SCFFbxW7; we put forward this model as a basis for further investigation (Fig. 4D). Recruitment of N-Myc to SCFFbxW7 is driven by the well-characterized interaction between the phospho-degron and FbxW7 that is regulated by phosphorylation of Thr58. However, a second interaction is also formed between N-Myc residues 61–89 and an unknown site on SCFFbxW7. Aurora-A binds to N-Myc at sites that flank the phospho-degron, disrupting the interaction between residues 61–89 of N-Myc and SCFFbxW7 but not the phospho-degron region; thus a complex of Aurora-A, N-Myc, and SCFFbxW7 is formed. We postulate that the interaction between N-Myc residues 61–89 and SCFFbxW7 is important for efficient polyubiquitination of N-Myc and that disruption of this interaction by Aurora-A reduces the formation of Lys48-linked chains on N-Myc, as previously reported (19).
The dysregulation of Myc proteins is a feature of several cancers that are refractory to treatment, and targeting Myc is a validated therapeutic strategy (9, 25–27). Unfortunately, there are currently no drug-like molecules that target Myc directly. Myc family proteins are intrinsically disordered proteins that dynamically adopt many conformations and therefore lack “druggable” pockets. This property has frustrated attempts to develop a drug-like molecule that directly targets the interaction of Myc with its binding partners. Here we have characterized the complex with Aurora-A in which N-Myc is trapped in an ordered conformation. The interaction can be disrupted by targeting the ATP-binding site of Aurora-A to induce a conformation that is incompatible with the binding of N-Myc, and early clinical trial data suggest these compounds may provide patient benefit when combined with chemotherapy (20, 22, 28). However, this approach also would target the main function of Aurora-A in the proper assembly and the function of the mitotic spindle, and it is too early to know whether the potential benefit of this in targeting dividing cancer cells offsets the accompanying damage to normal tissue. Alternatively, it may be possible to inhibit the protein–protein interaction directly by targeting the interface. The structure of the Aurora-A/N-Myc complex provides a rational basis for the development of inhibitors that destabilize N-Myc and molecular coordinates of the interface that will facilitate structure-guided approaches to target the binding sites for ATP or N-Myc on Aurora-A.
Materials and Methods
Crystallography.
Aurora-A 122–403 containing the mutations C290A and C393A was expressed and purified as described previously (29) and was gel-filtrated into 20 mM Tris (pH 7.5), 200 mM NaCl, 5 mM MgCl2, 5 mM β-mercaptoethanol, and 10% (vol/vol) glycerol. An N-biotinyl peptide corresponding to residues 28–89 of N-Myc and carrying a phosphothreonine at position 58 (Peptide Synthetics) was dissolved to 10 mM in DMSO. A mixture containing 300-μM Aurora-A, 5 mM adenosine diphosphate, and 500-μM N-Myc peptide [with a final DMSO concentration of 5% (vol/vol)] was incubated on ice for 1 h and then mixed 1:1 with reservoir solution [100 mM Bicine (pH 9.0), 20% (wt/vol) PEG 6000]. Crystals were obtained by vapor diffusion at 18 °C within a few days and were frozen in reservoir solution supplemented with 25% (vol/vol) ethylene glycol. Diffraction data were collected from a single crystal at Diamond I03 and processed with xia2 (30) (Table S1). The structure of the Aurora-A/N-Myc complex was solved by molecular replacement phasing using Phaser (31) with an existing Aurora-A structure [Protein Data Bank (PDB) ID code 4CEG] (29) as the search model. Iterative modeling and refinement were carried out using Coot (32) and phenix.refine (33). Structure validation was carried out using MolProbity (34). Crystallographic data, refinement statistics, and details of structure validation analysis are summarized in Table S1. Structure figures were prepared using PyMOL (www.pymol.org).
Coprecipitation Assay.
For in vitro peptide coprecipitation assays, 40 μL of Strep-Tactin Sepharose beads (IBA) were incubated with 900 μL of 10-μM N-biotinyl N-Myc peptides for 1 h and then were washed three times with pull-down buffer [50 mM Tris (pH 7.5), 150 mM NaCl, 5 mM MgCl2, 5 mM β-mercaptoethanol, 0.02% Tween-20]. The beads were then incubated with 900 μL of a solution containing analyte proteins, competing proteins, or compounds as required for 2 h at 4 °C and were washed three times with 900 μL pull-down buffer. Finally, the beads were eluted by boiling with 80 μL SDS-loading buffer, and the eluted proteins were analyzed by SDS/PAGE. Aurora-A 122–403 was used at a final concentration of 12 μM. SCFFbxW7 incorporating GST-tagged FbxW7 (Millipore) was used at a final concentration of 5.5 ng/μL and was visualized in precipitates by Western blotting using an anti-GST antibody (GE Healthcare).
Competition AlphaScreen Assay.
Biotinyl Avi-tagged Aurora-A 122–403 was produced as described previously (35). Appropriate concentrations of biotinyl Aurora-A 122–403 and 3xFlag-N-Myc 28–89 for use in competition experiments were determined in preliminary cross-titration experiments over a range of 0–300 nM. The assay buffer was 50 mM Tris (pH 7.5), 150 mM NaCl, 5 mM MgCl2, 5 mM β-mercaptoethanol, 0.02% Tween-20, and 0.1% BSA. For competition assays, 10 nM biotinyl Aurora-A 122–403, 0.3 nM 3xFlag-N-Myc 28–89, and 0–160-μM Aurora-A 122–403 were mixed together and incubated at RT for 1 h in white, opaque 96-well half-area plates. Streptavidin donor beads (20 μg/mL) and anti-Flag acceptor beads (PerkinElmer) were added simultaneously to each reaction under subdued lighting conditions, and the plate was incubated for a further 90 min. The signal from each reaction was read using an EnVision multilabel plate-reader (PerkinElmer). Data points represent the mean of three experiments; error bars indicate SD. Data were analyzed using GraphPad Prism 6, and the dependence of the AlphaScreen signal on the concentration of competing Aurora-A 122–403 was fitted by nonlinear regression to a one-site logIC50 equation in which, under the selected reaction conditions, the IC50 value approximates the Kd. Results are reported as Kd ± SE.
In Situ Proximity Ligation Assay.
Kelly cells were cultured in RPMI 1640 supplemented with heat-inactivated FBS. Compounds (as indicated) at 500 nM were added 4 h before fixation. Cells were fixed with 4% (vol/vol) paraformaldehyde for 30 min and permeabilized with PBS containing 0.2% Triton X-100 for a further 30 min. Cells were incubated with Duolink In situ blocking solution (Olink Bioscience) for 1 h and then with antibodies against Aurora-A (GeneTex) and N-Myc (Calbiochem) diluted into the same buffer for an additional hour. In situ proximity ligation assays were carried out using the Duolink kit (Olink Bioscience) according to the manufacturer’s instructions. Images were collected and analyzed using a Humphrey 730 field analyzer system (Zeiss). The average number of interactions detected per cell was calculated, and results are reported as the mean of three independent experiments ± SD.
Kinase Assay.
[32P]ATP kinase assays were performed as described (36) with modifications as follows. Measurement of Aurora-A activity was carried out using reactions containing 0.625-μM unphosphorylated Aurora-A catalytic domain, 40-μM myelin basic protein (Sigma), and 0–50-μM N-Myc 28–89, N-Myc 61–89, or TPX2 1–43 peptides. Reactions were incubated at 21 °C for 45 min, and total incorporation of 32P radioisotope into Aurora-A, N-Myc 28–89, and myelin basic protein was then measured by scintillation counting. Results are reported as the mean of two independent experiments ± SE.
SI Materials and Methods
Protein Production and Coprecipitation Assay.
TPX2 1–43 and GST-Aurora-A 122–403 D274N were expressed and purified as described previously (36). Constructs encoding 3xFlag-tagged fragments of the N-Myc transactivation domain and TPX2 1–43 were expressed as His-NusA fusions in Escherichia coli BL21 (DE3)-RIL cells and were purified on 5 mL chelating Sepharose columns (GE Healthcare) charged with nickel; the His-NusA tags were cleaved away using tobacco etch virus protease and were removed by nickel affinity chromatography. The remaining 3xFlag-tagged polypeptides were purified further by anion exchange on a Resource Q column (GE Healthcare) over a 50–500 mM NaCl gradient in 20 mM Tris (pH 7.5), 1 mM EDTA and then by gel filtration into Tris-buffered saline using a Superdex 75 column (GE Healthcare). GST-Aurora-A 122–403 D274N (250 μg) or GST (100 μg) was immobilized on 40 μL of glutathione Sepharose beads (GE Healthcare) and incubated with 900 μL of 35-μM 3xFlag-N-Myc fragments (including compounds as required) for 2 h in pull-down buffer. The beads were washed four times with 900 μL pull-down buffer. Bound proteins were analyzed by SDS/PAGE and visualized by Coomassie Blue staining and by anti-Flag immunoblotting (M2; Sigma).
ELISA.
Preblocked HBC Streptavidin plates (Pierce) were incubated with 10 μg/mL biotinyl Avi-tagged-Aurora-A 122–403 for 30 min at RT. Wells were washed three times with 300 μL PBS supplemented with 0.1% Tween-20 (PBST). 3xFlag-N-Myc 28–89 (0–40 μM) was added to the wells and incubated for 1 h at RT. Negative control reactions containing no immobilized Aurora-A were also performed. Plates were washed with PBST followed by the addition of 1:5,000 HRP-conjugated anti-Flag antibody (ab49763; Abcam). Reactions were incubated at RT for a further 30 min and washed again with PBST. Binding of 3xFlag-N-Myc 28–89 was resolved by the addition of 3,3′,5,5′-Tetramethylbenzidine, and the reaction was quenched by the addition of 0.5 M H2SO4. Absorbance at 450 nm was read for each reaction. Data points represent the mean of three experiments; error bars indicate SD. The data were fitted to a one-site–specific binding equation by nonlinear regression using GraphPad Prism 6. Results are reported as Kd ± SE.
FP Assay.
Fluorescein-labeled N-Myc (amino acids 61–89) (50 nM) or peptides (amino acids 19–47) (Pepceuticals) were incubated at 21 °C with Aurora-A kinase domain protein (amino acids 122–403) at a range of concentrations in a buffer containing 25 mM Hepes (pH 7.5), 150 mM NaCl, 5 mM MgCl2, 5% (vol/vol) glycerol, and 0.02% Tween-20 in a flat-bottomed, black 96-well plate at a total volume of 50 μL per sample. The dephosphorylated Aurora-A catalytic domain was made by coexpression with lambda phosphatase and purified in the same way as non-dephosphorylated protein (36). To investigate the influence of MLN8054 on the interaction, the compound was dissolved in DMSO to a concentration of 10 mM and was added to all samples at a final concentration of 600 μM; an equivalent quantity of DMSO [6% (vol/vol)] was added to corresponding compound-free samples. FP was measured using a Victor X5 instrument (PerkinElmer) at excitation and emission wavelengths of 480 nm and 535 nm, respectively. Data points represent the mean of three experiments; error bars indicate SD. The dependence of FP upon Aurora-A concentration was fitted to a one-site total binding model by nonlinear regression using GraphPad Prism 6. Results are reported as Kd ± SE.
Acknowledgments
We thank the beamline scientists of Diamond I03 for assistance with data collection. This work was funded by European Research Council Grants “AuroMYC” (to M.E. and R.B.), a grant from Deutsche Krebshilfe (to M.E.), and Cancer Research UK Grant C24461/A12772 (to R.B.). L.C. and E.P. acknowledge funding from Cancer Research UK and Children with Cancer UK.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Atomic coordinates and structure factors have been deposited in the Protein Data Bank (ID code 5G1X).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1610626113/-/DCSupplemental.
References
- 1.Kress TR, Sabò A, Amati B. MYC: Connecting selective transcriptional control to global RNA production. Nat Rev Cancer. 2015;15(10):593–607. doi: 10.1038/nrc3984. [DOI] [PubMed] [Google Scholar]
- 2.Wolf E, Lin CY, Eilers M, Levens DL. Taming of the beast: Shaping Myc-dependent amplification. Trends Cell Biol. 2015;25(4):241–248. doi: 10.1016/j.tcb.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vennstrom B, Sheiness D, Zabielski J, Bishop JM. Isolation and characterization of c-myc, a cellular homolog of the oncogene (v-myc) of avian myelocytomatosis virus strain 29. J Virol. 1982;42(3):773–779. doi: 10.1128/jvi.42.3.773-779.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ciriello G, et al. Emerging landscape of oncogenic signatures across human cancers. Nat Genet. 2013;45(10):1127–1133. doi: 10.1038/ng.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohl NE, et al. Transposition and amplification of oncogene-related sequences in human neuroblastomas. Cell. 1983;35(2 Pt 1):359–367. doi: 10.1016/0092-8674(83)90169-1. [DOI] [PubMed] [Google Scholar]
- 7.Schwab M, et al. Amplified DNA with limited homology to myc cellular oncogene is shared by human neuroblastoma cell lines and a neuroblastoma tumour. Nature. 1983;305(5931):245–248. doi: 10.1038/305245a0. [DOI] [PubMed] [Google Scholar]
- 8.Nau MM, et al. L-myc, a new myc-related gene amplified and expressed in human small cell lung cancer. Nature. 1985;318(6041):69–73. doi: 10.1038/318069a0. [DOI] [PubMed] [Google Scholar]
- 9.Soucek L, et al. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455(7213):679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tu WB, et al. Myc and its interactors take shape. Biochim Biophys Acta. 2015;1849(5):469–483. doi: 10.1016/j.bbagrm.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 11.Nair SK, Burley SK. X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell. 2003;112(2):193–205. doi: 10.1016/s0092-8674(02)01284-9. [DOI] [PubMed] [Google Scholar]
- 12.Helander S, et al. Pre-anchoring of Pin1 to unphosphorylated c-Myc in a fuzzy complex regulates c-Myc activity. Structure. 2015;23(12):2267–2279. doi: 10.1016/j.str.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaenicke LA, et al. Ubiquitin-dependent turnover of MYC antagonizes MYC/PAF1C complex accumulation to drive transcriptional elongation. Mol Cell. 2016;61(1):54–67. doi: 10.1016/j.molcel.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 14.Fladvad M, et al. N and C-terminal sub-regions in the c-Myc transactivation region and their joint role in creating versatility in folding and binding. J Mol Biol. 2005;346(1):175–189. doi: 10.1016/j.jmb.2004.11.029. [DOI] [PubMed] [Google Scholar]
- 15.Pineda-Lucena A, et al. A structure-based model of the c-Myc/Bin1 protein interaction shows alternative splicing of Bin1 and c-Myc phosphorylation are key binding determinants. J Mol Biol. 2005;351(1):182–194. doi: 10.1016/j.jmb.2005.05.046. [DOI] [PubMed] [Google Scholar]
- 16.Sjostrom SK, Finn G, Hahn WC, Rowitch DH, Kenney AM. The Cdk1 complex plays a prime role in regulating N-myc phosphorylation and turnover in neural precursors. Dev Cell. 2005;9(3):327–338. doi: 10.1016/j.devcel.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 17.Welcker M, et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci USA. 2004;101(24):9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6(8):635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- 19.Otto T, et al. Stabilization of N-Myc is a critical function of Aurora A in human neuroblastoma. Cancer Cell. 2009;15(1):67–78. doi: 10.1016/j.ccr.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 20.Gustafson WC, et al. Drugging MYCN through an allosteric transition in Aurora kinase A. Cancer Cell. 2014;26(3):414–427. doi: 10.1016/j.ccr.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dodson CA, et al. Crystal structure of an Aurora-A mutant that mimics Aurora-B bound to MLN8054: Insights into selectivity and drug design. Biochem J. 2010;427(1):19–28. doi: 10.1042/BJ20091530. [DOI] [PubMed] [Google Scholar]
- 22.Brockmann M, et al. Small molecule inhibitors of aurora-a induce proteasomal degradation of N-myc in childhood neuroblastoma. Cancer Cell. 2013;24(1):75–89. doi: 10.1016/j.ccr.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bayliss R, Sardon T, Vernos I, Conti E. Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol Cell. 2003;12(4):851–862. doi: 10.1016/s1097-2765(03)00392-7. [DOI] [PubMed] [Google Scholar]
- 24.Bavetsias V, et al. Imidazo[4,5-b]pyridine derivatives as inhibitors of Aurora kinases: Lead optimization studies toward the identification of an orally bioavailable preclinical development candidate. J Med Chem. 2010;53(14):5213–5228. doi: 10.1021/jm100262j. [DOI] [PubMed] [Google Scholar]
- 25.Gustafson WC, Weiss WA. Myc proteins as therapeutic targets. Oncogene. 2010;29(9):1249–1259. doi: 10.1038/onc.2009.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barone G, Anderson J, Pearson AD, Petrie K, Chesler L. New strategies in neuroblastoma: Therapeutic targeting of MYCN and ALK. Clin Cancer Res. 2013;19(21):5814–5821. doi: 10.1158/1078-0432.CCR-13-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hill RM, et al. Combined MYC and P53 defects emerge at medulloblastoma relapse and define rapidly progressive, therapeutically targetable disease. Cancer Cell. 2015;27(1):72–84. doi: 10.1016/j.ccell.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DuBois SG, et al. Phase I Study of the Aurora A Kinase Inhibitor Alisertib in Combination With Irinotecan and Temozolomide for Patients With Relapsed or Refractory Neuroblastoma: A NANT (New Approaches to Neuroblastoma Therapy) Trial. J Clin Oncol. 2016;34(12):1368–1375. doi: 10.1200/JCO.2015.65.4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burgess SG, Bayliss R. The structure of C290A:C393A Aurora A provides structural insights into kinase regulation. Acta Crystallogr F Struct Biol Commun. 2015;71(Pt 3):315–319. doi: 10.1107/S2053230X15002290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winter G, Lobley CM, Prince SM. Decision making in xia2. Acta Crystallogr D Biol Crystallogr. 2013;69(Pt 7):1260–1273. doi: 10.1107/S0907444913015308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 11):1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 34.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burgess SG, et al. Allosteric inhibition of Aurora-A kinase by a synthetic vNAR domain. Open Biol. 2016;6(7):160089. doi: 10.1098/rsob.160089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burgess SG, et al. Aurora-A-dependent control of TACC3 influences the rate of mitotic spindle assembly. PLoS Genet. 2015;11(7):e1005345. doi: 10.1371/journal.pgen.1005345. [DOI] [PMC free article] [PubMed] [Google Scholar]