Significance
Many bacteria are adapted to live in specific locations in animal hosts, even while these same locations exclude colonization by most microorganisms. However, the genetic underpinnings enabling successful colonization are largely unclear. We developed a system to genetically manipulate Snodgrassella alvi, a bacterium restricted to bees, and explored the factors permitting S. alvi to establish within its natural habitat in the bee digestive tract. Using high-throughput methods that screen the entire genome, we find that host colonization is dependent on genes mediating cell surface interactions (e.g., adhesion), metabolism under nutrient limitation, and responses to various stresses. This study demonstrates the genetic tractability of the bee gut microbiota, an emerging system with parallels to the human microbiome.
Keywords: type IV pilus, transposon mutagenesis, microbiota, symbiosis, Neisseriaceae
Abstract
Animal guts are often colonized by host-specialized bacterial species to the exclusion of other transient microorganisms, but the genetic basis of colonization ability is largely unknown. The bacterium Snodgrassella alvi is a dominant gut symbiont in honey bees, specialized in colonizing the hindgut epithelium. We developed methods for transposon-based mutagenesis in S. alvi and, using high-throughput DNA sequencing, screened genome-wide transposon insertion (Tn-seq) and transcriptome (RNA-seq) libraries to characterize both the essential genome and the genes facilitating host colonization. Comparison of Tn-seq results from laboratory cultures and from monoinoculated worker bees reveal that 519 of 2,226 protein-coding genes in S. alvi are essential in culture, whereas 399 are not essential but are beneficial for gut colonization. Genes facilitating colonization fall into three broad functional categories: extracellular interactions, metabolism, and stress responses. Extracellular components with strong fitness benefits in vivo include trimeric autotransporter adhesins, O antigens, and type IV pili (T4P). Experiments with T4P mutants establish that T4P in S. alvi likely function in attachment and biofilm formation, with knockouts experiencing a competitive disadvantage in vivo. Metabolic processes promoting colonization include essential amino acid biosynthesis and iron acquisition pathways, implying nutrient scarcity within the hindgut environment. Mechanisms to deal with various stressors, such as for the repair of double-stranded DNA breaks and protein quality control, are also critical in vivo. This genome-wide study identifies numerous genetic networks underlying colonization by a gut commensal in its native host environment, including some known from more targeted studies in other host–microbe symbioses.
Many animal species, including humans, have highly specific associations with a core gut microbiota consisting of host-adapted bacteria (1). Although many other microbial species are routinely ingested with food, few are able to stably colonize the gut. Considerable progress has been made in identifying the genes and mechanisms enabling bacterial colonization and persistence in hosts (e.g., refs. 2–7), but the extent to which common processes play a role for gut symbionts in different host taxa is unclear.
Honey bees (Apis mellifera) resemble mammals in possessing characteristic gut bacteria acquired through social contact; however, the bee gut is dominated by only approximately eight species, in contrast to the hundreds of species typically found in mammalian guts (8). The distribution of these bacteria across related bee species (8) indicates a long evolutionary association, another feature shared with some mammalian gut associates (9). An abundant constituent of the honey bee and bumble bee gut microbiota is Snodgrassella alvi (10), a member of Betaproteobacteria that is related to human commensals in the genus Neisseria (11). S. alvi colonizes the hindgut ileum, a region analogous to the proximal large intestine in humans and is normally found in contact with the intima lining, of the ileum (12, 13).
S. alvi is cultivable in the laboratory, and cultured strains can be used to experimentally colonize germ-free adult worker bees with high efficiency (14) (Fig. 1). Experimental transfers of S. alvi strains between honey bee and bumble bee hosts indicate that strains are specialized to colonize their native host species (14). Although diverse bacteria are initially present in guts of young bees, only core species such as S. alvi can stably colonize (8). Neither the basis for host specificity nor the basis for S. alvi’s ability to persist in bee guts is known. To determine the genes that enable S. alvi to colonize the bee gut, we used high-throughput sequencing of a saturated transposon mutant library (Tn-seq) to screen tens of thousands of random insertion mutants across the S. alvi genome during colonization in vivo and in laboratory culture. This technique allowed us to assay the contribution of each gene toward the establishment of this symbiosis. We find that extensive genetic networks involved in extracellular interaction, metabolism, and stress response have strong fitness benefits in vivo and likely play key roles in facilitating gut colonization.
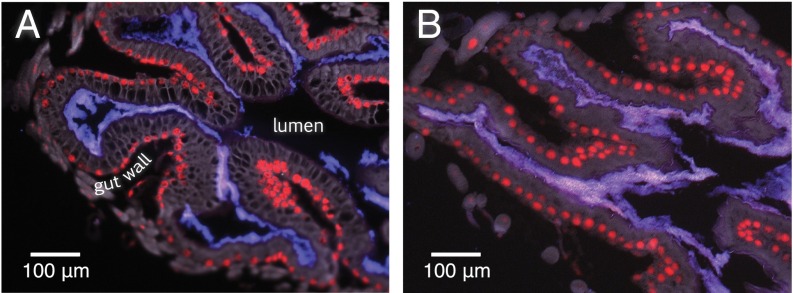
Fig. 1.
Cross-sections of the honey bee ileum, showing colonization by S. alvi in 4-d-old workers inoculated (A) experimentally with laboratory-cultured S. alvi wkB2 and (B) naturally within the hive. S. alvi localization (purple) is derived from an overlay of a DNA stain (red) and an rRNA probe specific to S. alvi (blue). Methods and B are adapted from ref. 14.
Results and Discussion
To identify genes needed for growth and genes contributing to gut colonization, we generated a transposon insertion library in S. alvi strain wkB2 (SI Appendix). The library comprised ∼74,000 individual mutants, representing on average one insertion per 34 bp across the S. alvi genome and thus ensuring that every gene was inactivated by multiple mutants (Fig. 2A and SI Appendix, Table S1).
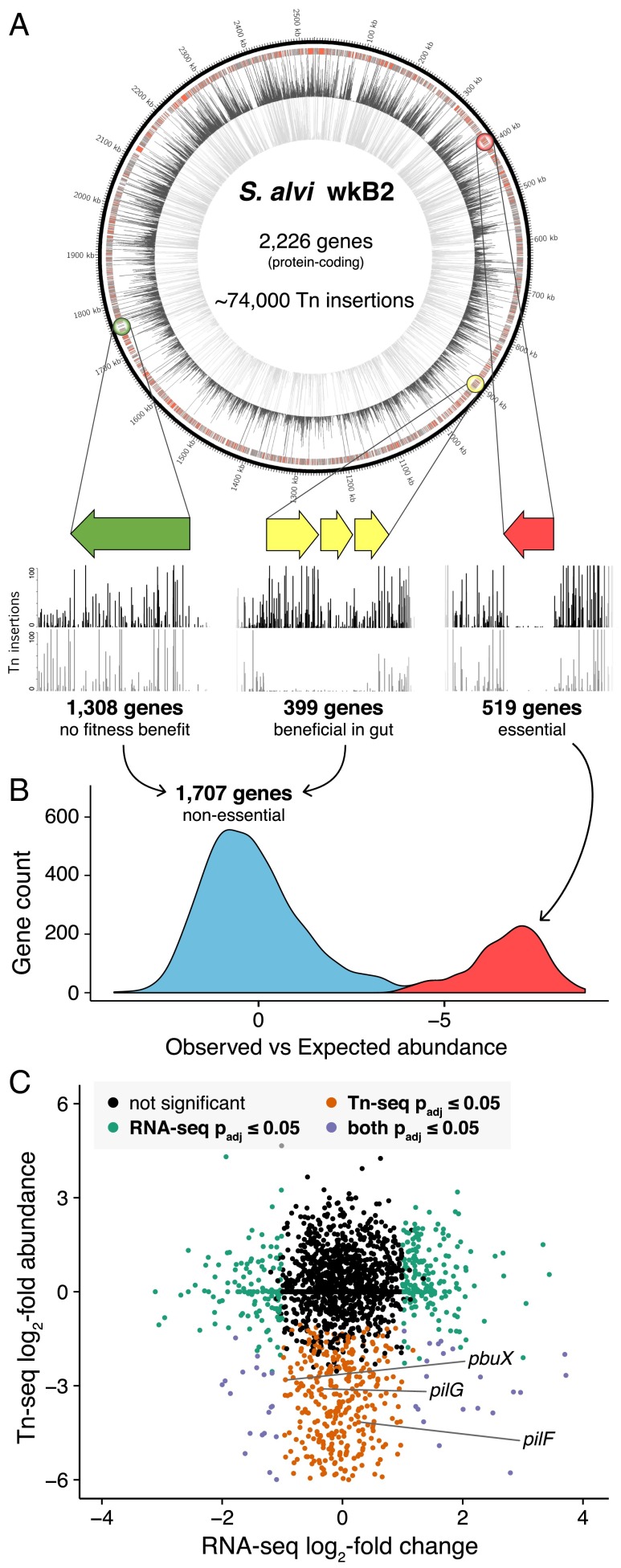
Fig. 2.
Determining the S. alvi gene set essential for growth and beneficial for gut colonization. (A) Genome of S. alvi strain wkB2 with locations of transposon insertions in bee gut samples (inside track) and on culture media (outside track). Callouts illustrate three categories of genes identified from this approach. (B) The essential genome of S. alvi wkB2 on laboratory culture plates. (C) Tn-seq and RNA-seq results for S. alvi genes in the bee gut compared with culture plate growth. For Tn-seq, only genes with significant fitness benefit are highlighted.
Genes Contributing to Growth in Laboratory Culture.
We identified genes tolerating far fewer transposon insertions than expected at random during growth in laboratory culture (SI Appendix). Of 2,226 protein-coding genes, 519 are essential or highly beneficial under our culture conditions (Padj ≤ 0.05) (Fig. 2B and Dataset S1C). Most of these genes participate in highly conserved cellular functions such as central informational processes (replication, transcription, and translation), respiration (electron transport chain), and biosynthesis of cell constituents (including peptidoglycan and lipopolysaccharide) (SI Appendix, Fig. S1). Genes of the gluconeogenesis pathway are essential, reflecting the fact that S. alvi lacks the ability to take up exogenous sugars (14). Genes required or highly beneficial for growth in culture included 37 encoding proteins of unknown function.
Genes Promoting Gut Colonization.
We compared genome-wide transposon insertion frequencies in our mutant library fed to germ-free adult worker bees to the frequencies obtained from laboratory cultures (SI Appendix). This approach allowed us to query the 1,707 protein-coding genes considered “nonessential” in culture, for roles in gut colonization. Of these, 399 genes were underrepresented in the library recovered from guts (cutoff criteria of log2-fold decrease ≥ 1 and Padj ≤ 0.05) (Fig. 2C and Dataset S1B). Of 399 genes conferring a benefit in vivo, 109 could not be confidently ascribed a function. The remaining 290 genes were inspected and placed into pathways based on similarity to characterized homologs in other bacteria. This analysis reveals the presence of extensive, interconnected gene networks that underlie host colonization (Fig. 3) and gives insight into the demands of the gut environment. Most genes conferring fitness benefits during colonization fall into three broad functional categories: extracellular interaction, metabolism, and stress response.
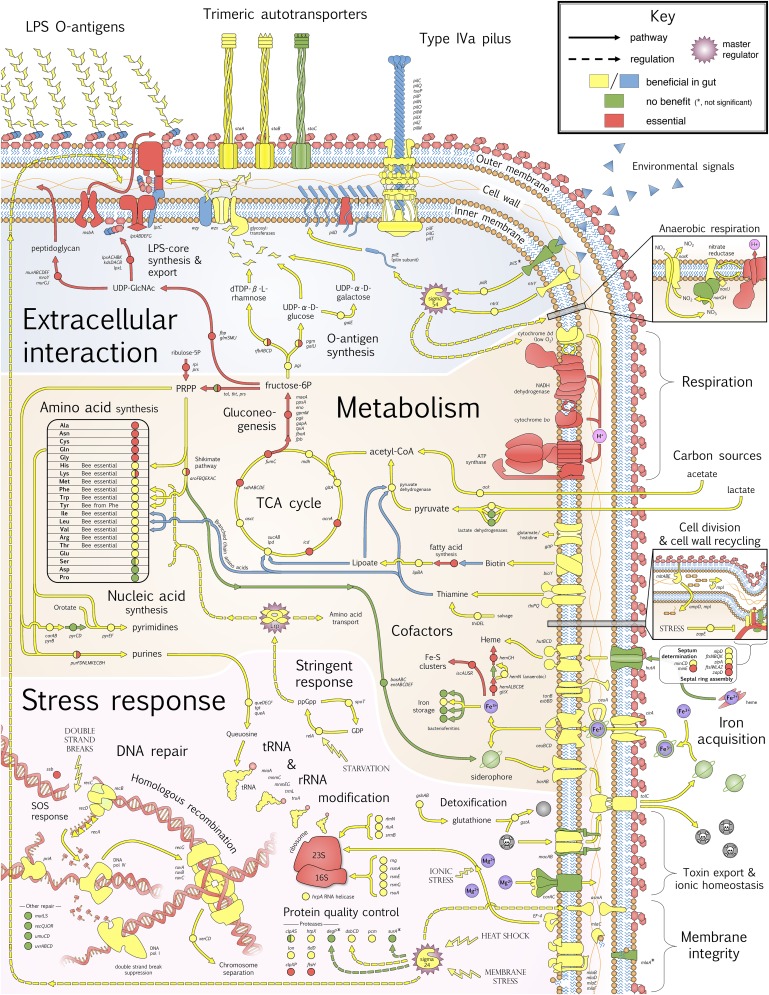
Fig. 3.
Genes required or beneficial for S. alvi colonization of the honey bee ileum. Pathways and regulation inferred from characterized homologs in other bacterial species (e.g., E. coli and Neisseria spp.). Gene products and pathways that confer fitness benefits in the gut are colored yellow or blue (two colors are used to facilitate visualization of different pathways and gene products within complexes). Genes that may be beneficial, but do not reach significance, are denoted by an asterisk. Dataset S2 and SI Appendix, Figs. S2 and S3 provide details.
Extracellular interaction.
Extracellular components mediate direct, physical contact between a bacterium and its environment, which potentially includes both host cells and other bacteria (15). In S. alvi, as for most Gram-negative bacteria, lipopolysaccharide (LPS) is the major constituent of the outer leaf of the outer membrane, and its synthesis from the carbohydrate products of gluconeogenesis, as well as its export (via the lpt gene products) were essential for cell viability under all conditions (Fig. 3). However, in vivo, the LPS core alone was insufficient for colonization. O antigens (polysaccharide additions to LPS) appear crucial; mutants in any of the pathways leading to O-antigen precursor synthesis (dTDP-β-l-rhamnose, UDP-α-d-glucose, and UDP-α-d-galactose) and assembly (wzx and wzy) were noncompetitive in the bee gut. O antigens can confer benefits by enabling evasion of host immunity, binding to host receptors, and promoting cell envelope resilience against external insults (16).
Adhesion and biofilm formation are often important for host colonization (15). S. alvi grows in a distinct layer adjacent to the gut epithelium, suggesting adherence (12, 13) (Fig. 1). Two of the three trimeric autotransporter adhesin (TAA) genes in S. alvi, staA and staB, appear crucial for gut colonization. TAAs are a family of diverse, but structurally similar, adhesion factors that are widespread in bacteria, including human-associated Yersinia, Neisseria, and Bartonella (17). Consisting of a sticky head for adhering to host cells or extracellular matrices, and connected to the outer membrane by a lengthy stalk, these proteins are encoded by some of the largest S. alvi genes (∼10 kb). Both staA and staB are also present in S. alvi strains from bumble bees (14), suggesting a conserved role in colonization.
S. alvi encodes a complete type IV pilus (T4P) (18), plus a number of accessory or duplicated components. The T4P is a versatile bacterial cellular machine that can facilitate adhesion, biofilm formation, motility, secretion, and DNA uptake (19). In S. alvi, only pilD (prepilin peptidase) was essential in vitro. However, in vivo, mutations to all core T4P structural components (pilFGMNOPQW and tsaP) were highly detrimental. Among T4P genes with multiple variant copies, such as the pilus retraction/twitch motility motor pilT, and the pilus tip adhesin pilC, our screen discerned the variant most important for colonization (Dataset S2).
The pilus itself is chiefly composed of major pilin subunits, encoded by pilE. Pilin expression is triggered by environmental cues; in S. alvi, this is likely controlled by the two-component pilR/S system, as documented in the related species Kingella kingae (20). Transposon insertions in both pilR and pilS were underrepresented in vivo; although, as in Kingella, nonfunctionalization of the sensor pilS was more tolerated than that of pilR (20). The pilE gene in many bacteria, including commensal Neisseria species (21), is directly regulated by the products of pilR and rpoN (encoding the master transcriptional regulator σ-54), another gene required for gut colonization in our screen. A role for rpoN in pilus formation is likely in S. alvi, which has the canonical σ-54 promoter upstream of the pilE start codon.
Metabolism.
S. alvi is an obligate aerobe, but potentially experiences periods of suboptimal O2 concentrations in the gut, as some insect guts have anaerobic regions (22). Correspondingly, the ntrX/ntrY two-component oxygen sensor (23), was crucial for gut colonization. Regulatory responses to ntrX/ntrY are mediated by σ-54 and potentially lead to the expression of microaerobic/anaerobic respiratory components such as cytochrome bd, the nitrite/nitrate antiporter narK, and nitrate reductase. These products may accord S. alvi flexibility in meeting energy requirements in light of fluctuating gut O2 levels.
In terms of carbon utilization, genes responsible for acetate and lactate catabolism had strong fitness benefits in the gut (Fig. 3). These compounds likely represent major carbon/energy sources in vivo, being converted to acetyl-CoA and fed into the tricarboxylic acid (TCA) cycle for energy production and generation of biosynthetic precursors (e.g., via gluconeogenesis). All enzymes of the TCA cycle were needed for gut colonization, but not for growth in vitro, possibly due to compensation by TCA intermediates (e.g., succinate, citrate, malate) present in our culture media. The essentiality of TCA-mediated acetate and lactate utilization in vivo was also supported by the fact that mutants in the uptake and synthesis of cofactors for key enzymes in this pathway (e.g., lipolate and thiamine) were similarly strongly selected against in the gut (Fig. 3).
De novo biosynthetic pathways for amino acids and nucleic acids were required for host colonization, further implying low nutrient conditions within the bee hindgut. Most genes for purine and pyrimidine synthesis were nonessential in culture but needed in vivo. Similarly, most amino acid synthesis pathways were not required for growth in vitro, suggesting that S. alvi preferentially uptakes exogenous nutrients when available. In the gut, however, insertions in all genes involved in synthesizing 12 of the 20 amino acids were detrimental (Fig. 3). Intriguingly, this set of 12 contains the “essential amino acids” that cannot be synthesized by animals (including bees) (24); their scarcity may be due to efficient host absorption in the midgut. The distal hindgut is nutritionally impoverished in many animals due to upstream absorption. Nonetheless, the largest gut bacterial communities are typically found in hindguts, as exemplified by the human colon (25), the termite paunch (22), and the cecum of certain herbivores (26). The honey bee gut microbiota is also mainly in the hindgut (12), and our results show that a variety of pathways are needed for tolerating the limited nutrient availability there.
Another vital micronutrient is iron, which is scarce in host-associated environments (27). That S. alvi possesses multiple systems for iron uptake (Fig. 3 and SI Appendix, Fig. S2) is indicative of its importance, particularly for iron bound in soluble complexes, the dominant form in aerobic environments. Three iron importers, for heme-bound Fe2+ and siderophore-bound Fe3+, and associated genes (tonB, exeBD, and fepA), were necessary in vivo, as were the exporters barAB and tolC to avoid toxic buildup of siderophores (28). Siderophore synthesis, however, was not essential, presumably because mutants can “cheat” and take up siderophores secreted by nonmutants in the community. Indeed, mutants for siderophore synthesis were overrepresented in guts (Dataset S1H), suggesting that such cheaters prosper when they are at low frequencies by avoiding the costs of production.
Stress response.
Many genes involved in stress responses confer strong fitness benefits in vivo, suggesting that the bee gut is a challenging environment in which S. alvi is exposed to toxins, low nutrient availability, and fluctuating O2, pH, and temperature. Among genes required in vivo were those underlying the ppGpp stringent response and Lrp, a master regulator that is induced by ppGpp upon amino acid starvation in other bacteria (29). In turn, Lrp can modulate expression of various amino acid synthesis enzymes and transporters to ensure cell survival (30).
Twenty-three S. alvi genes beneficial in the gut function to modify rRNA (16S and 23S rRNA subunits) and tRNA (e.g., via queuosine incorporation), with the likely effect of improving translational efficiency and fidelity. In other bacteria, these genes have been shown to enhance fitness under nutrient and temperature stress (31) (Dataset S2). Protein quality control, involving genes for protein recycling (clpS, htpX, lon, and tldD) and stabilization (dsbC and pcm), also appeared important in vivo, as were genes for the synthesis (gshAB) and activity (gstA) of glutathione, an antioxidant that contributes to general stress tolerance and detoxification (32).
A gene set with strong fitness benefits in the gut was that for the repair of double-stranded DNA breaks. This pathway includes genes for SOS response (recA and recX), recombinational repair (recBCD), D-loop extension (priA, DNA pol IV), resolution of Holliday junctions (ruvABC and recG), and postrepair chromosomal separation (xerCD) (Fig. 3 and SI Appendix, Fig. S3). DNA breaks may be induced by reactive oxygen species released by the dual oxidase component of the host immune system, which regulates gut microbiota in Drosophila (33) and necessitates DNA repair in the human gut bacterium Helicobacter pylori (34).
Response to membrane stress (e.g., via temperature, osmotic, ionic, pH shock, or LPS perturbation) was another critical feature for S. alvi gut colonization. A central component of this response appears to be the σ-24 factor (rpoE), which regulates, among other targets, protein quality control and LPS synthesis and export (35). The mla system, for maintaining outer membrane LPS integrity, and the poorly characterized membrane organizational protein, AsmA, were also important in vivo. Finally, a number of genes involved in cell division were intolerant of mutation in vivo; these may play a role in determining appropriate conditions for cellular replication, the regulation of which is likely crucial under stressful conditions (e.g., for zapE) (36).
We note that some mutants, such as those affecting genes underlying siderophore production, were overrepresented in the gut, suggesting that these genes hinder gut colonization under our experimental conditions (upper points of Fig. 2C and Dataset S1H).
Changes in Gene Expression upon Gut Colonization.
In addition to the Tn-seq analysis, we used transcriptome sequencing to identify genes differentially expressed in the bee gut relative to our culture conditions. A total of 369 protein-coding genes were significantly up- or down-regulated twofold or higher (Dataset S1D). Only 22 of these were among the 399 genes found to promote gut colonization by Tn-seq (Fig. 2C). These include genes underlying branched-chain amino acid synthesis, iron acquisition, and short-chain fatty acid utilization (SI Appendix, Fig. S1). Other up-regulated genes include genes coding for transporters involved in nutrient acquisition, and the type VI secretion system, implicated in interbacterial competition.
Testing Candidates from Tn-Seq for Roles in Gut Colonization.
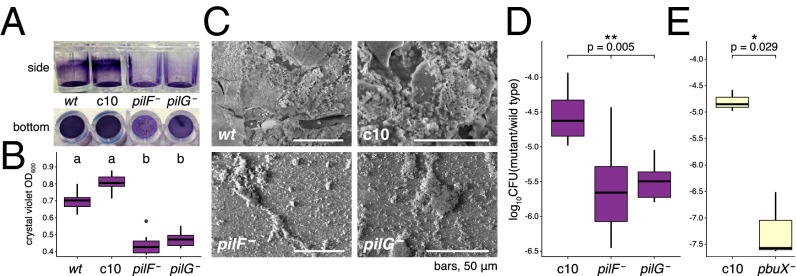
We tested colonization abilities of three insertion mutants with strong fitness detriment in the gut, by comparing their colonization success to that of a neutral transposon mutant, c10, in one-on-one competitions with wild-type strain wkB2. Two T4P structural mutants (pilF− and pilG−) were examined in detail. In vitro evaluations showed that both pilF− and pilG− had reduced abilities to produce surface biofilms compared with wild-type S. alvi or to c10 (Fig. 4 A and B and SI Appendix for explanation of screen). Scanning electron microscopy showed biofilms of multilayered composition along with prolific extracellular matrix in wild-type and c10, whereas the T4P mutants had small and isolated aggregations of cells (Fig. 4C). Lack of T4P-mediated adhesion was detrimental in vivo, as pilF− and pilG− mutants introduced to the bee gut had lower proportional recovery compared with the c10 mutant (Fig. 4D).
Fig. 4.
In vivo competitive ability of mutants implicated in Tn-seq screen as having roles in gut colonization. (A–D) Roles of pilF and pilG in biofilm formation and host colonization. (A) Crystal violet-stained surface biofilms in polypropylene wells. (B) Mean solubilized crystal violet-stained biofilm, as absorbance at OD600 (n = 16 wells per strain, P = 1.91 × 10−13, Kruskal–Wallis multiple comparisons). (C) Scanning electron micrographs of surface biofilms at the air/liquid interface of strains grown in vitro. (D and E) Ratio of viable transposon mutants to wild-type following coinoculation of bee guts. *P < 0.029, **P < 0.005. (D) n = 7 bees per condition (Kruskal–Wallis test). (E) n = 4 bees per condition (Mann–Whitney test).
A mutant of a third gene, pbuX, was tested. This poorly characterized gene encodes a permease that is likely involved in xanthine/purine metabolism. Results verified that disruption of pbuX reduced colonization ability (Fig. 4E).
Comparison of S. alvi wkB2 Gene Sets to Those of Related Bacteria with Different Hosts.
To better understand the basis for host specificity in S. alvi strains, we compared wkB2 to S. alvi wkB12, a strain restricted to Bombus hosts that is unable to colonize A. mellifera (14) (Dataset S1G). Of 2,226 wkB2 genes, 80% (1,762) are found in wkB12; of 519 essential wkB2 genes, 95% (497) are in wkB12; and of the 399 wkB2 genes beneficial for host colonization, all but 40 are present in wkB12. These 40 genes are potential determinants of host specificity. Most (26 of 40) are uncharacterized, but one of these is the major pilin subunit, pilE (SALWKB2_RS07815), which is highly expressed by wkB2 and significantly up-regulated in the gut.
We compared the genes of S. alvi wkB2 to those of Neisseria gonorrheae MS11, a human pathogen that is a close relative, for which a similar transposon mutant dataset is available (37) (Dataset S1F). Only 49% (1,085) of S. alvi’s 2,226 genes have orthologs in N. gonorrheae, but 90% (465) of the 519 S. alvi wkB2 essential genes are present, with 337 scored as essential. Among S. alvi genes important in gut colonization, 126 genes lack orthologs in N. gonorrheae, and these are candidates for mechanisms enabling colonization of insect guts.
Conclusions
Our results provide a genome-scale view of pathways that enable host colonization by a specialized gut symbiont. We find that colonization depends on distinct processes, including cell surface modifications and biofilm formation, metabolic capabilities enabling growth in low nutrient conditions, and responses to diverse stressors. Some of these capabilities reflect the particular conditions of the bee gut ileum; for example, many amino acids appear to be scarce, possibly due to upstream host absorption, requiring de novo biosynthesis on the part of S. alvi. The retention of genes for operating under low O2 conditions (e.g., the cytochrome bd complex, nitrate reductase, and the anaerobic hemN heme synthesis pathway) may likewise be explained by fluctuating O2 availability at the gut wall, where S. alvi lives.
The ability of S. alvi to form resident populations in a persistent physical layer along the ileum is likely facilitated by T4P. T4Ps are known to play central roles in host colonization for other proteobacterial taxa, including close relatives of Snodgrassella such as Neisseria meningitidis and N. gonorrheae, via surface adhesion and motility, and are thus fundamental to biofilm organization and attachment to the host epithelium (19). Two other major cell surface components, trimeric autotransporters and LPS O antigens, are also likely candidates for participation in adhesion, as well as in specific interactions with host receptors or evasion of host immune mechanisms (16, 17). These latter functions may give rise to the observed host specificity of particular S. alvi strains to particular host species (14).
The incongruence between the RNA-seq and Tn-seq results was not surprising, as transcriptional responses correlate poorly with gene essentiality (38), potentially reflecting either constitutive expression or very transitory expression of many genes. For instance, genes for adhesion might only be highly expressed during an initial critical stage of laying down biofilm. Nonetheless, our transcriptomic results reinforce the importance of nutrient uptake and amino acid synthesis by S. alvi in the gut. Some genes, such as RTX toxins and type VI secretion systems, are not essential for gut colonization, but were up-regulated in vivo. These features may affect colonization in the presence of other members of the bee gut microbiome by mediating interbacterial interactions. Studying the responses of S. alvi as part of the wider gut community is a logical next step, as other bacterial species may considerably influence the gut environment by liberating nutrients from pollen (39), eliciting host responses (40), altering pH and O2 concentration, or engaging in direct competition.
To date, there have been few genome-wide studies of the factors underlying colonization by gut commensals in their native host environment. Direct experimental approaches are not feasible for human gut symbionts, but rodent models have been used with considerable success (2–7). Several pathogens have been examined using Tn-seq in vivo, within different tissue types (38, 41–43). Some critical mechanisms for S. alvi colonization of the bee gut, including LPS modifications, membrane integrity components, the stringent response, proteases, and DNA break repair, are also important for infection by human pathogens (41–43). Similarly, colonization of light-producing organs by the squid symbiont Vibrio fisheri depends on genes dictating biofilm formation and cellular stress responses (44). In aquatic Vibrio colonizing zebra fish intestines, genes underlying cell envelope, chemosensory and motility functions seem crucial (45). Hence, many animal-associated bacteria, from pathogens to mutualists, likely face common challenges and rely on a similar set of genes to establish in the host. Whereas many of these genes are part of well-characterized pathways, others remain poorly described despite their importance across a range of host–microbe systems. Examples of enigmatic genes that strongly contribute to S. alvi fitness in the bee gut, and also in other animal symbionts, include the ntrXY two-component system, the T4P-associated fimV, the RNA helicase hrpA, the DNA repair regulator yebC, the protein stabilizer dsbC, and asmA (Dataset S2).
High-throughput screening of mutant libraries derived from bacterial colonizers in vivo opens a window into the gut milieu as well as the adaptations required to live under such conditions. We uncovered gene networks involved in extracellular interaction, metabolism, and stress responses that appear critical for gut colonization; these genes may be of general importance to host-associated microbiota, and are ideal candidates for future analyses. Finally, our study demonstrates the genetic tractability of S. alvi, a species only recently discovered (10), and paves the way for new applications and engineering of the bee microbiome.
Materials and Methods
Detailed protocols are available in SI Appendix. Strains are listed in SI Appendix, Table S2. S. alvi wkB2 was mutagenized with a Mariner-derived transposon bearing a gentamycin resistance marker, delivered by a counterselectable Escherichia coli strain via conjugation with plasmid pBT20 (SI Appendix, Figs. S4 and S5). Experiments verified that this transposon made single random insertions in the wkB2 genome and that strains carrying this marker can colonize the bee gut to the same abundance as the wild type (SI Appendix, Fig. S6).
A saturated transconjugant library was constructed and harvested from replicates from rich agar-based media (control group) or from the ileums of pooled cohorts of worker bees 5 d after inoculation (experimental group). Recovered mutant libraries were prepared for Illumina MiSeq sequencing and analyzed using methods adapted from Turner et al. (46). To score essential genes, we tallied mutant abundances from the control group. We then examined the fitness effects of insertions by comparing the experimental and control groups. A gene was considered to contribute to gut colonization if mutant frequency showed a decrease of ≥2 with P ≤ 0.05 (adjusted for false discovery rate). Tn-seq results were matched to RNA-seq data that compared transcripts from plated bacteria to those from inoculated worker ileums (Fig. 2C and Dataset S1D).
Transconjugant clones were arrayed from the mutant library and their phenotypic characteristics were observed in vitro. Transposon–genome junctions were sequenced for select clones, and three mutants, pilF–, pilG– , and pbuX–, were chosen for further experiments based on the importance of these genes as indicated by the Tn-seq fitness analysis.
Supplementary Material
Acknowledgments
We thank Keith Turner and Marvin Whiteley for protocols, scripts, and advice; Andrew Goodman for advice, strains, and vectors; Margaret Steele for OD600 enumeration curves; and Kim Hammond for assistance with experiments and manuscript preparation. This work was supported by NIH Award 1R01-GM108477-01 (to N.A.M.), a Canadian Natural Sciences and Engineering Research Council postgraduate scholarship (to W.K.K.), and the Swiss National Science Foundation and the European Molecular Biology Organization (P.E.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited under NCBI BioProject, www.ncbi.nlm.nih.gov (accession no. PRJNA319191).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1610856113/-/DCSupplemental.
References
- 1.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489(7415):220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee SM, et al. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501(7467):426–429. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goodman AL, et al. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe. 2009;6(3):279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu Y, Waldor MK, Mekalanos JJ. Tn-Seq analysis of Vibrio cholerae intestinal colonization reveals a role for T6SS-mediated antibacterial activity in the host. Cell Host Microbe. 2013;14(6):652–663. doi: 10.1016/j.chom.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu M, et al. Genetic determinants of in vivo fitness and diet responsiveness in multiple human gut Bacteroides. Science. 2015;350(6256):aac5992. doi: 10.1126/science.aac5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coyne MJ, Chatzidaki-Livanis M, Paoletti LC, Comstock LE. Role of glycan synthesis in colonization of the mammalian gut by the bacterial symbiont Bacteroides fragilis. Proc Natl Acad Sci USA. 2008;105(35):13099–13104. doi: 10.1073/pnas.0804220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rakoff-Nahoum S, Foster KR, Comstock LE. The evolution of cooperation within the gut microbiota. Nature. 2016;533(7602):255–259. doi: 10.1038/nature17626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwong WK, Moran NA. Gut microbial communities of social bees. Nat Rev Microbiol. 2016;14(6):374–384. doi: 10.1038/nrmicro.2016.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moeller AH, et al. Cospeciation of gut microbiota with hominids. Science. 2016;353(6297):380–382. doi: 10.1126/science.aaf3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwong WK, Moran NA. Cultivation and characterization of the gut symbionts of honey bees and bumble bees: Description of Snodgrassella alvi gen. nov., sp. nov., a member of the family Neisseriaceae of the Betaproteobacteria, and Gilliamella apicola gen. nov., sp. nov., a member of Orbaceae fam. nov., Orbales ord. nov., a sister taxon to the order ‘Enterobacteriales’ of the Gammaproteobacteria. Int J Syst Evol Microbiol. 2013;63(Pt 6):2008–2018. doi: 10.1099/ijs.0.044875-0. [DOI] [PubMed] [Google Scholar]
- 11.Liu G, Tang CM, Exley RM. Non-pathogenic Neisseria: Members of an abundant, multi-habitat, diverse genus. Microbiology. 2015;161(7):1297–1312. doi: 10.1099/mic.0.000086. [DOI] [PubMed] [Google Scholar]
- 12.Martinson VG, Moy J, Moran NA. Establishment of characteristic gut bacteria during development of the honeybee worker. Appl Environ Microbiol. 2012;78(8):2830–2840. doi: 10.1128/AEM.07810-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engel P, et al. Standard methods for research on Apis mellifera gut symbionts. J Apic Res. 2013;52(4):1–24. [Google Scholar]
- 14.Kwong WK, Engel P, Koch H, Moran NA. Genomics and host specialization of honey bee and bumble bee gut symbionts. Proc Natl Acad Sci USA. 2014;111(31):11509–11514. doi: 10.1073/pnas.1405838111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ribet D, Cossart P. How bacterial pathogens colonize their hosts and invade deeper tissues. Microbes Infect. 2015;17(3):173–183. doi: 10.1016/j.micinf.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 16.Lerouge I, Vanderleyden J. O-antigen structural variation: Mechanisms and possible roles in animal/plant-microbe interactions. FEMS Microbiol Rev. 2002;26(1):17–47. doi: 10.1111/j.1574-6976.2002.tb00597.x. [DOI] [PubMed] [Google Scholar]
- 17.Linke D, Riess T, Autenrieth IB, Lupas A, Kempf VA. Trimeric autotransporter adhesins: Variable structure, common function. Trends Microbiol. 2006;14(6):264–270. doi: 10.1016/j.tim.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 18.Chang YW, et al. Architecture of the type IVa pilus machine. Science. 2016;351(6278):aad2001. doi: 10.1126/science.aad2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Craig L, Pique ME, Tainer JA. Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol. 2004;2(5):363–378. doi: 10.1038/nrmicro885. [DOI] [PubMed] [Google Scholar]
- 20.Kehl-Fie TE, Porsch EA, Miller SE, St Geme JW., 3rd Expression of Kingella kingae type IV pili is regulated by sigma54, PilS, and PilR. J Bacteriol. 2009;191(15):4976–4986. doi: 10.1128/JB.00123-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rendón MA, Hockenberry AM, McManus SA, So M. Sigma factor RpoN (σ54) regulates pilE transcription in commensal Neisseria elongata. Mol Microbiol. 2013;90(1):103–113. doi: 10.1111/mmi.12350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brune A. Symbiotic digestion of lignocellulose in termite guts. Nat Rev Microbiol. 2014;12(3):168–180. doi: 10.1038/nrmicro3182. [DOI] [PubMed] [Google Scholar]
- 23.Carrica MdelC, Fernandez I, Martí MA, Paris G, Goldbaum FA. The NtrY/X two-component system of Brucella spp. acts as a redox sensor and regulates the expression of nitrogen respiration enzymes. Mol Microbiol. 2012;85(1):39–50. doi: 10.1111/j.1365-2958.2012.08095.x. [DOI] [PubMed] [Google Scholar]
- 24.de Groot AP. Protein and amino acid requirements of the honey bee (Apis mellifera L.) Phys Comp Oec. 1953;3:197–285. [Google Scholar]
- 25.Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol. 2016;14(1):20–32. doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dougal K, et al. Identification of a core bacterial community within the large intestine of the horse. PLoS One. 2013;8(10):e77660. doi: 10.1371/journal.pone.0077660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andrews SC, Robinson AK, Rodríguez-Quiñones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27(2–3):215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 28.Jones CM, et al. Self-poisoning of Mycobacterium tuberculosis by interrupting siderophore recycling. Proc Natl Acad Sci USA. 2014;111(5):1945–1950. doi: 10.1073/pnas.1311402111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Traxler MF, et al. Discretely calibrated regulatory loops controlled by ppGpp partition gene induction across the ‘feast to famine’ gradient in Escherichia coli. Mol Microbiol. 2011;79(4):830–845. doi: 10.1111/j.1365-2958.2010.07498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho BK, Federowicz S, Park YS, Zengler K, Palsson BØ. Deciphering the transcriptional regulatory logic of amino acid metabolism. Nat Chem Biol. 2011;8(1):65–71. doi: 10.1038/nchembio.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cantara WA, et al. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res. 2011;39(Database issue):D195–D201. doi: 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smirnova GV, Oktyabrsky ON. Glutathione in bacteria. Biochemistry (Mosc) 2005;70(11):1199–1211. doi: 10.1007/s10541-005-0248-3. [DOI] [PubMed] [Google Scholar]
- 33.Ha EM, et al. Coordination of multiple dual oxidase-regulatory pathways in responses to commensal and infectious microbes in drosophila gut. Nat Immunol. 2009;10(9):949–957. doi: 10.1038/ni.1765. [DOI] [PubMed] [Google Scholar]
- 34.Wang G, Maier RJ. 2011. A recombination puzzle solved: Role for new DNA repair systems in Helicobacter pylori diversity/persistence. DNA Repair, ed Kruman I (InTech, Rijeka, Croatia), Chapter 1.
- 35.Rowley G, Spector M, Kormanec J, Roberts M. Pushing the envelope: Extracytoplasmic stress responses in bacterial pathogens. Nat Rev Microbiol. 2006;4(5):383–394. doi: 10.1038/nrmicro1394. [DOI] [PubMed] [Google Scholar]
- 36.Marteyn BS, et al. ZapE is a novel cell division protein interacting with FtsZ and modulating the Z-ring dynamics. MBio. 2014;5(2):e00022–14. doi: 10.1128/mBio.00022-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Remmele CW, et al. Transcriptional landscape and essential genes of Neisseria gonorrhoeae. Nucleic Acids Res. 2014;42(16):10579–10595. doi: 10.1093/nar/gku762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turner KH, Everett J, Trivedi U, Rumbaugh KP, Whiteley M. Requirements for Pseudomonas aeruginosa acute burn and chronic surgical wound infection. PLoS Genet. 2014;10(7):e1004518. doi: 10.1371/journal.pgen.1004518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee FJ, Rusch DB, Stewart FJ, Mattila HR, Newton IL. Saccharide breakdown and fermentation by the honey bee gut microbiome. Environ Microbiol. 2015;17(3):796–815. doi: 10.1111/1462-2920.12526. [DOI] [PubMed] [Google Scholar]
- 40.Engel P, Bartlett KD, Moran NA. The bacterium Frischella perrara causes scab formation in the gut of its honeybee host. MBio. 2015;6(3):e00193–15. doi: 10.1128/mBio.00193-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gawronski JD, Wong SM, Giannoukos G, Ward DV, Akerley BJ. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc Natl Acad Sci USA. 2009;106(38):16422–16427. doi: 10.1073/pnas.0906627106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skurnik D, et al. A comprehensive analysis of in vitro and in vivo genetic fitness of Pseudomonas aeruginosa using high-throughput sequencing of transposon libraries. PLoS Pathog. 2013;9(9):e1003582. doi: 10.1371/journal.ppat.1003582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang N, Ozer EA, Mandel MJ, Hauser AR. Genome-wide identification of Acinetobacter baumannii genes necessary for persistence in the lung. MBio. 2014;5(3):e01163–14. doi: 10.1128/mBio.01163-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brooks JF, 2nd, et al. Global discovery of colonization determinants in the squid symbiont Vibrio fischeri. Proc Natl Acad Sci USA. 2014;111(48):17284–17289. doi: 10.1073/pnas.1415957111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stephens WZ, et al. Identification of population bottlenecks and colonization factors during assembly of bacterial communities within the zebrafish intestine. MBio. 2015;6(6):e01163–e15. doi: 10.1128/mBio.01163-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turner KH, Wessel AK, Palmer GC, Murray JL, Whiteley M. Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc Natl Acad Sci USA. 2015;112(13):4110–4115. doi: 10.1073/pnas.1419677112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.