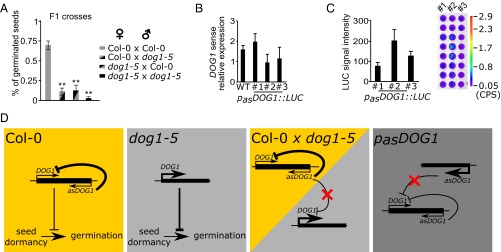
Significance
Sequential developmental transitions in plant life cycle are tightly controlled by dynamic regulation of key genes. Seed dormancy release is probably the first developmental transition in a plant’s life cycle, and it is regulated by the Delay of Germination 1 (DOG1) gene. Here we demonstrate that a nonprotein-coding antisense transcript originating from a conserved at DNA—but not protein level—DOG1 region is a negative regulator of DOG1 expression and seed dormancy establishment. We show that this antisense transcript negatively regulates DOG1 expression in cis. This mechanism is presumably conserved across the Brassicaceae, given the evolutionary conservation of the antisense DOG1 promoter.
Keywords: seed dormancy, DOG1 gene, cis-acting ncRNA, antisense transcript
Abstract
Seed dormancy is one of the most crucial process transitions in a plant’s life cycle. Its timing is tightly controlled by the expression level of the Delay of Germination 1 gene (DOG1). DOG1 is the major quantitative trait locus for seed dormancy in Arabidopsis and has been shown to control dormancy in many other plant species. This is reflected by the evolutionary conservation of the functional short alternatively polyadenylated form of the DOG1 mRNA. Notably, the 3′ region of DOG1, including the last exon that is not included in this transcript isoform, shows a high level of conservation at the DNA level, but the encoded polypeptide is poorly conserved. Here, we demonstrate that this region of DOG1 contains a promoter for the transcription of a noncoding antisense RNA, asDOG1, that is 5′ capped, polyadenylated, and relatively stable. This promoter is autonomous and asDOG1 has an expression profile that is different from known DOG1 transcripts. Using several approaches we show that asDOG1 strongly suppresses DOG1 expression during seed maturation in cis, but is unable to do so in trans. Therefore, the negative regulation of seed dormancy by asDOG1 in cis results in allele-specific suppression of DOG1 expression and promotes germination. Given the evolutionary conservation of the asDOG1 promoter, we propose that this cis-constrained noncoding RNA-mediated mechanism limiting the duration of seed dormancy functions across the Brassicaceae.
Plants have evolved elaborate adaptation mechanisms to cope with unexpected and rapid changes in their natural environment (1). The division of the plant life cycle into consecutive developmental phases can be viewed as one such mechanism. This compartmentalization allows plants to focus their resources on particular tasks. The most pronounced developmental phases in plant development are seed dormancy, the juvenile phase, vegetative growth, flowering, and senescence (2). The transition between each successive phase has to be tightly controlled and aligned with the plant’s internal metabolic state and external conditions.
Seeds are characterized by their remarkable ability to withstand harsh environmental conditions (3). This is in part because of a seed dormancy mechanism that imposes a block on the ability of seeds to sense permissive conditions and initiate germination (4, 5). This mechanism allows seeds to temporarily bypass favorable conditions to germinate in an environment that will support the entire plant life cycle. Seed dormancy is under strong evolutionary selection because the improper timing of germination often results in immediate death (6). In addition, from an agronomical point of view, seed dormancy has been a subject of intensive selection, because on the one hand strong dormancy leads to uneven germination, but on the other hand weak dormancy may result in preharvest sprouting because of germination on the mother plant (7).
An analysis of seed dormancy variability among Arabidopsis thaliana accessions identified the Delay of Germination 1 (DOG1) gene as the major quantitative trait locus (QTL) controlling this phenotype (8). DOG1 is a member of a small gene family (9). The molecular function of the DOG1 protein in seed dormancy control is currently unclear. However, the DOG1 protein has been shown to self-dimerize and to be a highly stable protein that is posttranslationaly modified during after-ripening and germination (10, 11). Genetically, DOG1 has been shown to control a gibberellin-dependent endosperm-weakening mechanism in Arabidopsis and other Brassicaceae family members (12). DOG1 is also required for the miRNA156-mediated delay of flowering and germination (13).
The level of DOG1 mRNA is tightly correlated with the strength of seed dormancy. Its expression is seed-specific and shows a strong peak during seed maturation when dormancy is established (9). The DOG1 gene is alternatively spliced, leading to the production of four mRNA isoforms. DOG1 alternative splicing is strongly coupled to the speed of polymerase II (PolII) elongation. Transcription factor (TF)IIS knockout and TFIIS dominant-negative mutation lead to the slowdown of PolII elongation, which enhances proximal splice site selection on DOG1. In contrast, mutation of the spliceosome cofactor AtNTR1, which increases the rate of PolII elongation, results in the selection of distal splice sites (14). In addition to changes in DOG1 splice site selection, tfIIs and atntr1 mutants display low DOG1 expression and consequently a weak seed dormancy phenotype (14–16). Other known factors required for high DOG1 expression include the histone H2B ubiquitin transferases HUB1 and HUB2 (17), and histone methyltransferase SDG8 (18). The only known example of a negative regulator of DOG1 expression is histone H3 lysine 9 methyltransferase KYP/SUVH4 (19).
DOG1 is also subject to alternative polyadenylation. Use of the distal polyadenylation site results in the production of a three-exonic long mRNA lgDOG1 that is poorly expressed and presumably not translated in vivo (11, 20). Selection of the proximal polyA site leads to the production of a two-exon short mRNA shDOG1 that is translated and can complement the weak dormancy phenotype of the dog1 mutant (20). The function of the lgDOG1-specific exon 3 region, which encodes a nonconserved polypeptide sequence and is probably not translated, is unclear. We and others have shown that a transfer (T)-DNA insertion in exon 3, rather than diminishing seed dormancy, produces a very strong dormancy phenotype, indicating that this region may negatively regulate seed dormancy strength (13, 20).
Developmental transitions are often controlled by elaborate mechanisms that center on a small set of key regulators allowing the plant to integrate diverse positive and negative cues (18, 21). In Arabidopsis, this is probably best exemplified by the regulation of Flowering Locus C (FLC), a QTL for the transition from vegetative to generative development (22). The majority of known positive and negative regulators that converge on FLC control its expression. These regulators include different noncoding RNAs (ncRNAs), including a small RNA targeting the FLC 3′ end, an intron-derived sense ncRNA participating in the cold-sensing mechanism, and a long-noncoding antisense transcript. This antisense transcript, named COOLAIR, originates from the 3′ end of FLC and has a dual function in both cold perception and autonomous pathway-mediated regulation of FLC (23–26).
ncRNAs have emerged as important players in the regulation of gene expression (27, 28). Many of these are antisense transcripts, such as Hidden Treasure 1 (HID1) (29), cis-NATPHO1;2 (30), and asHSFB2a (31). In these examples, expression of the antisense transcript from an independently integrated transgene led to phenotypic changes, showing that the ncRNA can act in trans. In contrast, FLC silencing during vernalization has been shown to be cis-controlled because two FLC alleles can be silenced independently. Thus, it has been hypothesized that the antisense transcript COOLAIR acts in cis. Indeed, recent single-molecule RNA FISH showed that COOLAIR and FLC sense transcription is mutually exclusive and confirmed that COOLAIR acts in cis to silence FLC during cold (32). Comparison of the A. thaliana and Arabidopsis alpina FLC antisense promoter regions revealed DNA sequence conservation, and an analogous antisense transcript was also detected in the latter species (33).
In the present study, we show that a conserved element in the region of the proximal polyA site of DOG1 serves as an autonomous ncRNA promoter. Its activity leads to the production of a long antisense transcript that is both 5′ capped and polyadenylated, and displays a developmentally regulated expression pattern that is different from the DOG1 sense transcript. This antisense transcript (asDOG1) acts as a negative regulator of seed dormancy that strongly suppresses DOG1 expression during dormancy establishment, thereby controlling this important developmental transition. Interestingly, asDOG1-mediated control of DOG1 expression and seed dormancy appears to be cis-restricted, suggesting a mechanism that may involve asDOG1 transcription rather than the resulting RNA.
Results
The DOG1 Exon 3 Region Shows Conservation at the DNA Level but the Encoded Polypeptide Sequence Is Not Conserved.
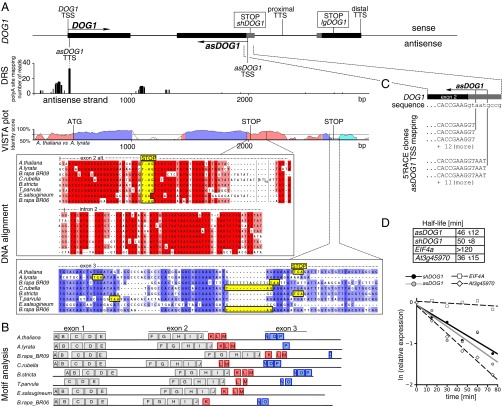
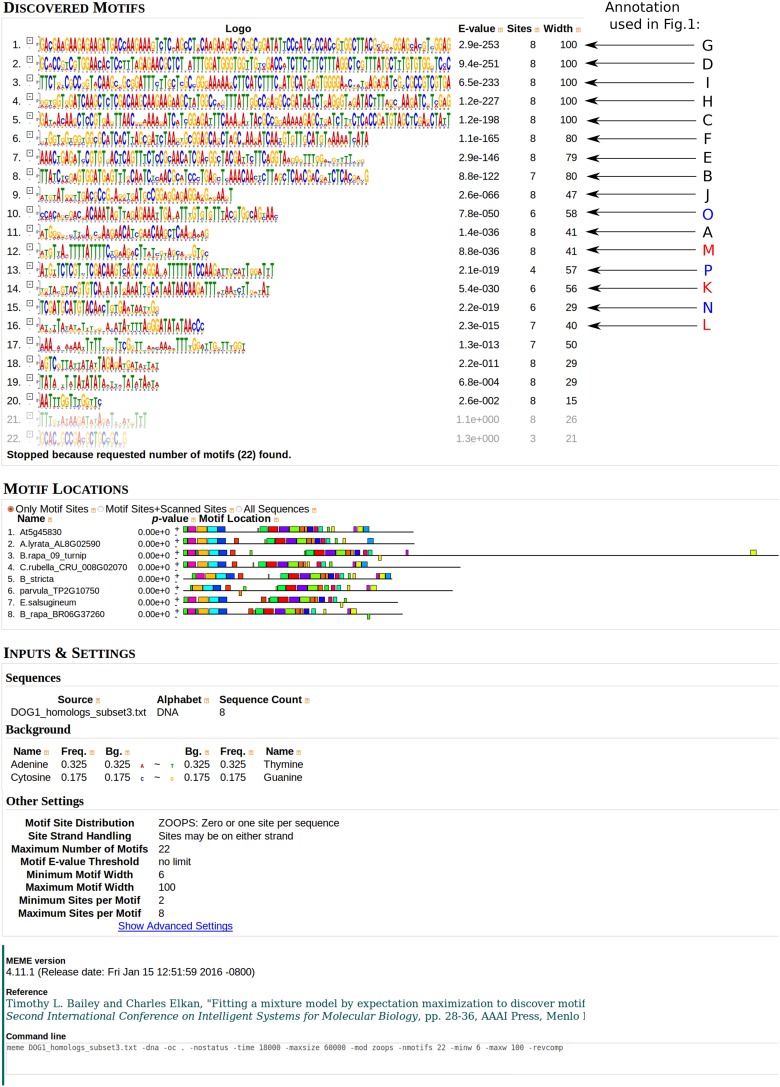
We recently reported that the DOG1 transcript displays alternative polyadenylation and that the short two-exonic proximally polyadenylated DOG1 mRNA is functional in the establishment of seed dormancy (20). In contrast, the long three-exonic DOG1 transcript is unable to complement the dog1 mutant and does not appear to be translated in vivo (20). DNA sequence alignment of the exon 3 region from selected members of the Brassicaceae revealed mutations, including single-nucleotide insertions and deletions that introduce stop codons (e.g., in DOG1 of Arabidopsis lyrata) (Fig. 1A). Despite the apparent lack of evolutionary pressure for ORF integrity, the DNA sequence of exon 3 showed substantial sequence conservation. In addition, a Vista plot of an alignment of the DOG1 genes from A. thaliana and A. lyrata suggested the presence of a conserved DNA sequence element within intron 2 (Fig. 1A). This element extends beyond the recently described short DOG1 protein ORF generated by use of the proximal polyA site located in intron 2. A multiple alignment of intron 2 revealed extensive DNA sequence conservation of this region in other plants (Fig. 1A). Furthermore, whole DOG1 locus DNA motif analysis, which does not require sequence colinearity, confirmed the presence of conserved DNA motifs corresponding to the A. thaliana intron 2–exon 3 element (Fig. 1B).
Fig. 1.
The DOG1 3′ region is conserved at the DNA level (but the encoded polypeptide is not conserved) and colocalizes with an asDOG1 TSS. (A) DOG1 panel: schematic diagram of DOG1 gene organization. lgDOG1, long three exonic DOG1 transcript; shDOG1, short two exonic DOG1 transcript; STOP, end of ORF; black boxes, exon sequences; gray boxes, alternative exonic regions, sense and antisense transcripts are marked with arrows. DRS panel: reanalysis of polyA site mapping by Direct RNA sequencing. Reads mapped to the antisense strand represent sites where polyadenylation occurs (TTS antisense). VISTA panel: plot of a pairwise alignment between A. thaliana and A. lyrata DOG1 orthologs. The curve is calculated using default VISTA thresholds based on percentage identity shown on y axis and base pair position on the x axis (Materials and Methods). Regions longer than 100 bp with average conservation score above threshold of 70% were colored (exons in blue, introns and promoter in pink, and UTR in light blue). Alternative splicing is not marked. DNA alignment panel: multiple alignment of DOG1 orthologs from seven Brassicaceae members, colored according to percentage identity. Exon/intron annotation refers to the A. thaliana DOG1 gene. Stop codons are marked for each ortholog based on the in silico ORF prediction. (B) Motif analysis: DNA motifs in DOG1 orthologs, identified with MEME software. Only motifs with an e-value < 1 × 10−14 and P < 1 × 10−14 are shown. Exon annotation refers to the A. thaliana DOG1 gene. Alternative exon 2 is marked with a dotted line. Red boxes, motifs found in intron 2; blue boxes, motifs found in exon 3. (C) 5′ RACE sequencing results revealed two TSS of a 5′ capped antisense transcript originating from the end of exon 2. Individual DNA amplicons were cloned and sequenced. (D) RNA stability assay performed on Col-0 WT seedlings. Half-life (see table) was calculated based on degradation curves after cordycepin treatment (plot) for asDOG1, the sense shDOG1 transcript, the stable transcript of housekeeping gene EIF4A and a short-lived mRNA transcribed from gene At3g45970. Presented values are averages with SD from three independent experiments.
In contrast, the sequence of the polypeptide encoded by A. thaliana DOG1 exon 3 is not conserved, as evident by the presence of multiple, independently acquired stop codons in relative species (Fig. 1A) (exon 3, alignment). The lack of protein conservation of the DOG1 exon 3 region is also supported by protein sequence alignment, as published previously (20). However, we now show that, together with the intron 2 region, this exon comprises a larger element that is evolutionarily conserved at the DNA sequence level. This is reminiscent of a conserved nonprotein coding sequence element, and may indicate the presence of a potential regulatory element or ncRNA in this region (34).
The DOG1 Gene Is Transcribed in the Antisense Orientation.
We hypothesized that this conserved 3′ region of the DOG1 gene may encode a nonprotein-coding RNA or contain the promoter mediating the transcription of such an RNA. Reanalysis of strand-specific direct RNA sequencing (DRS) -based mapping of polyadenylation sites in the Arabidopsis genome (35) revealed several polyadenylation sites on the DOG1 antisense strand (Fig. 1A). The most prominent of these is in the DOG1 promoter, but a signal could also be detected in intron 1 (Fig. 1A). Using 5′ RACE, we demonstrated the presence of a 5′ capped antisense transcript originating from the second intron of the DOG1 gene (Fig. 1C and Fig. S1). This antisense transcription start site (TSS) coincides with the proximal polyadenylation site of the short sense DOG1 transcript and the aforementioned conserved intron 2-exon 3 DNA sequence element (Fig. S1) (20).
Fig. S1.
The DOG1 antisense TSS. Aligned sequences of 5′ RACE clones with DOG1 genomic sequence shown above. Identified TTS marked as “^”.
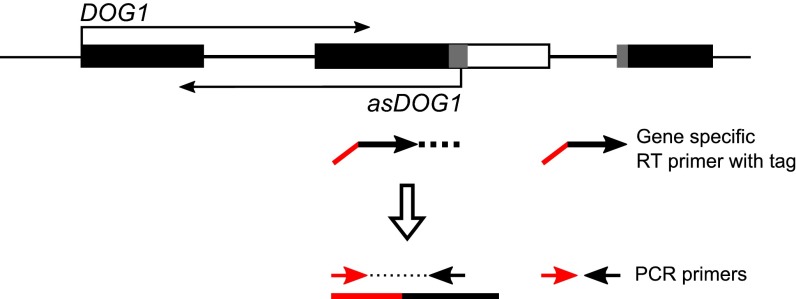
Our RACE experiments did not reveal any splicing of asDOG1 transcripts. Therefore, we developed an adapter-mediated RT-PCR assay that allowed us to specifically detect asDOG1 in the presence of the complementary DOG1 sense mRNA (36) (Fig. S2). Altogether, DRS, 5′ RACE, and the adapter-mediated RT-PCR assay showed that DOG1 is transcribed in the antisense orientation, leading to the production of a 5′ capped and polyadenylated transcript. The observation that this antisense transcript’s 3′ end extends to the TSS of the DOG1 sense mRNA (compare sense and antisense in Fig. 1A, Top) suggests that it may potentially be able to regulate DOG1 gene expression by a cis-acting mechanism like that described for H3K4me2-depositing transcripts in yeast (37).
Fig. S2.
Schematic diagram of the DOG1 gene showing the tag-based strand-specific RT-PCR assay used to specifically detect asDOG1.
To further characterize the DOG1 antisense transcript, we analyzed its stability using a cordycepin-dependent assay (38). This approach demonstrated that asDOG1 has a half-life of about 46 min (Fig. 1D), compared with 50 min for the shDOG1 sense transcript and 36 min for the short-lived At3g45970 mRNA. The relatively long (for ncRNA) half-life of asDOG1, which is close to that of short-lived protein-coding transcripts, indicated that it may function at a posttranscriptional level, for example by recruiting specific regulators (28). This comparatively high stability of DOG1 antisense is therefore more consistent with a trans-acting mode of action, which often requires an RNA molecule to diffuse over a considerable distance in the nucleus (28). In contrast, ncRNAs that act at the level of transcription are often highly unstable (39, 40).
asDOG1 Transcription Is Controlled by an Independent Promoter.
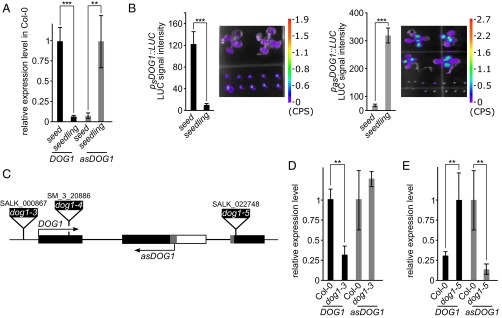
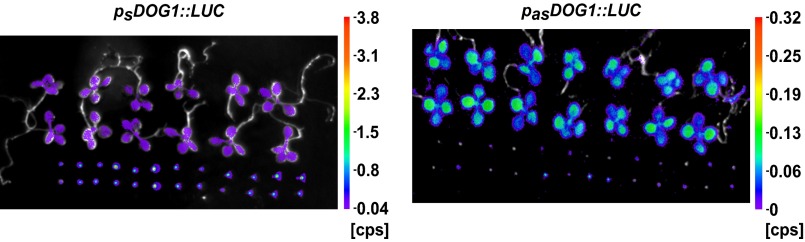
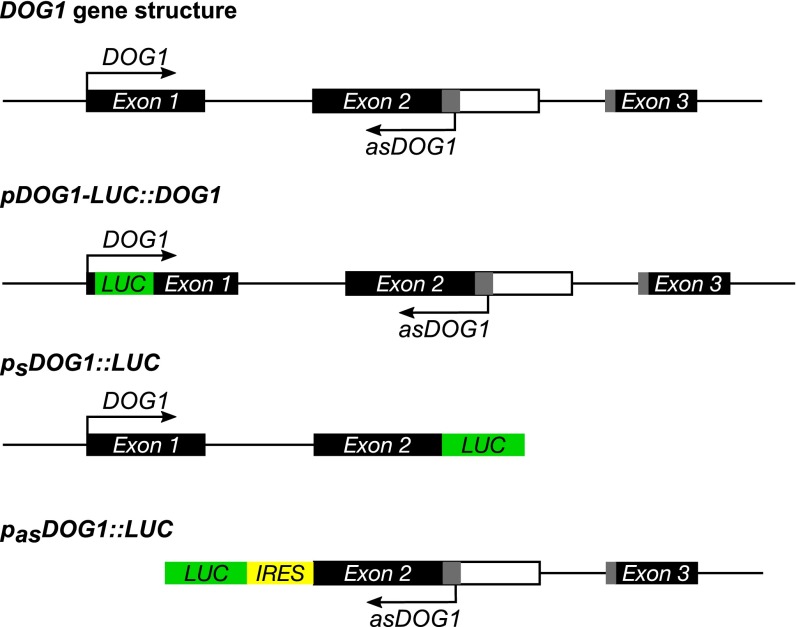
The DOG1 gene is the major QTL for seed dormancy in Arabidopsis and, as might be expected, the DOG1 sense transcript is highly expressed in seeds in comparison with seedlings (9) (Fig. 2A). Using the tag-based strand-specific quantitative RT-PCR (RT-qPCR) method, we found that the antisense and sense transcripts of DOG1 show reciprocal expression profiles (Fig. 2A). Therefore, we questioned whether production of the antisense transcript might occur independently of the DOG1 promoter and sense transcription. Assuming that the conserved noncoding region detected by our bioinformatic analysis contains a promoter for antisense transcription, we cloned the DOG1 antisense transcript, including its putative promoter region (exon 2–intron 2–exon 3) and fused it to an internal ribosomal entry site–luciferase gene (IRES-LUC) reporter cassette in the antisense orientation (pasDOG1::LUC). Separately, we prepared a construct with LUC linked to the short DOG1 sense gene (psDOG1::LUC), including the sense promoter (Figs. S3 and S4). Stable transgenic lines in the Col-0 (WT) background were produced using both constructs. The native DOG1 promoter gave an expression pattern similar to that of the endogenous DOG1 transcript (compare Fig. 2 A and B and Fig. S3). Interestingly, a LUC signal was also detected in “antisense” lines, indicating that there is indeed a transcriptionally active antisense promoter within the DOG1 gene. Moreover, the expression pattern produced using the antisense promoter was similar to the endogenous transcript profile determined by adapter-mediated RT-qPCR (compare Fig. 2 A and B and Fig. S3). These data confirmed that the DOG1 downstream region is sufficient to activate antisense transcription and may serve as a stand-alone promoter.
Fig. 2.
Tissue-specific expression pattern of asDOG1. DOG1 and asDOG1 show opposite patterns of expression in freshly harvested seeds and seedlings, when (A) assayed in Col-0 (WT) using RT-qPCR or (B) by measuring luciferase activity in transgenic plants expressing DOG1 (Left) or asDOG1 (Right), fused with LUC. Luciferase activity was quantified and normalized to the signal area. Images of seedlings (Upper) and seeds (Lower) expressing luciferase were taken with a NightSHADE (Berthold) camera. Colored bar next to image represents light intensity scale in counts per second (CPS). Error bars for B represent 95% confidence interval (CI). (C) Schematic diagram of DOG1 gene structure; black boxes, exon sequences; gray boxes, alternative exonic regions; white box region included in alternatively polyadenylated DOG1 short transcript; arrows show the sense and antisense transcripts. The positions of T-DNA insertions are indicated by black rectangles: dog1-3 (SALK_000867), dog1-4 (SM_3_20886) and dog1-5 (SALK_022748). (D) Down-regulation of the DOG1 sense transcript in dog1-3 does not affect the expression of asDOG1. Relative expression levels in freshly harvested seeds measured by RT-qPCR. (E) The dog1-5 mutant shows up-regulation of short DOG1 and down-regulation of asDOG1 expression. Relative expression levels in freshly harvested seeds measured by RT-qPCR. **P < 0.01 and ***P < 0.001. RT-qPCR results were normalized against the UBC mRNA level and are the means of three biological replicates with error bars representing SD.
Fig. S3.
Freshly harvested seeds and 10-day-old seedlings of plants expressing sense or antisense DOG1 transcripts fused with luciferase (LUC). Full-sized image from Fig. 2B of plants examined using a NightSHADE (Berthold) camera.
Fig. S4.
Schematic diagrams of constructs of DOG1 (pDOG1-LUC::DOG1 and pSDOG1::LUC) and asDOG1 (pasDOG1::LUC) fused with luciferase (LUC). The asDOG1 construct contains an additional IRES sequence.
To independently corroborate our conclusion, we took advantage of a previously reported loss-of-function mutant dog1-3, which has a T-DNA insertion in the DOG1 sense promoter region (9) (Fig. 2C). RT-qPCR analysis confirmed that the DOG1 sense transcript was clearly reduced in the dog1-3 mutant, but the level of the asDOG1 transcript remained unaffected compared with Col-0 WT plants (Fig. 2D). This result indicated that sense promoter activity is not required for antisense promoter-driven transcription.
Taken together, our results show that the DOG1 antisense transcript originates from an independent promoter. The conservation of the DNA sequence in this region (but not the encoded polypeptide sequence) could therefore reflect evolutionary conservation of this promoter.
Disruption of the asDOG1 Promoter Causes Down-Regulation of Antisense Transcription and Increases the DOG1 Sense mRNA Level.
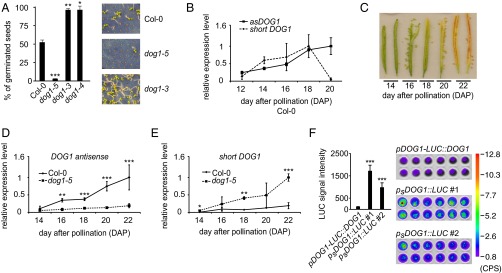
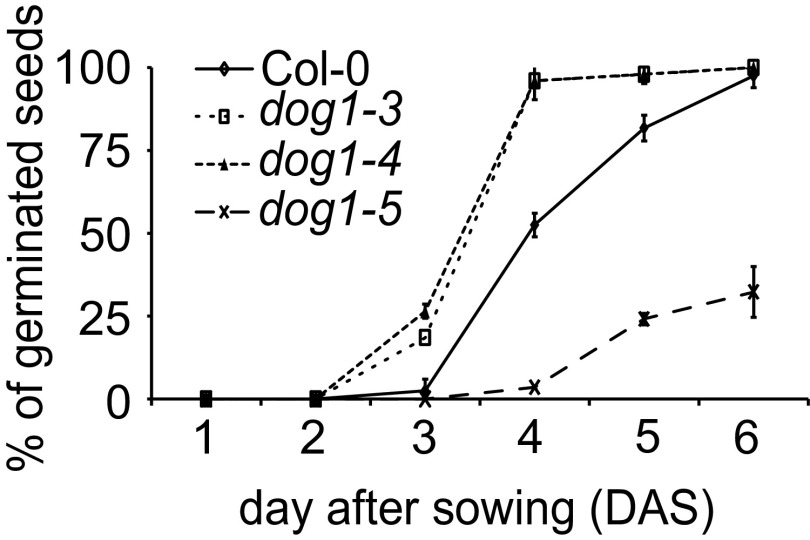
Although the level of antisense transcript in Col-0 WT plants is low in seeds compared with seedlings, it was further reduced in seeds of the dog1-5 mutant (Fig. 2 A and E). The dog1-5 mutant allele was originally described by our group (20) and, as recently confirmed by others (13), it acts as a gain-of-function mutant showing enhanced seed dormancy (Fig. 3A and Fig. S5). In agreement with this phenotype, dog1-5 showed significant up-regulation of the short proximally polyadenylated DOG1 transcript (Fig. 2E) and raised levels of the DOG1 protein (20). The dog1-5 mutant carries a T-DNA insertion within DOG1 exon 3. This exon is included in the long- but absent from the short-DOG1 transcript. Considering the location of the asDOG1 TSS, the T-DNA insertion in dog1-5 was predicted to affect the asDOG1 promoter (Fig. 2C). Indeed, we found a reduction in the asDOG1 transcript level in freshly harvested dog1-5 seeds (Fig. 2E).
Fig. 3.
Seed dormancy is negatively regulated by asDOG1. (A) The dog1-5 mutant shows strong seed dormancy. Freshly harvested seeds of Col-0 (WT) and different DOG1 mutants were scored for germination ability: germination efficiency (%) of tested seeds scored as radical protrusion. P value was calculated in comparison with Col-0 (WT) (Left); representative seed dormancy test result (Right). (Magnification: 0.9×.) (B) The expression profiles of the shDOG1 and asDOG1 transcripts during silique development in Col-0 (WT) plants show an increase in the antisense transcription in the late stages of seed maturation. (C) Siliques from manually pollinated flowers collected at different days after pollination. (D) The level of asDOG1 is decreased in the dog1-5 mutant during all tested seed developmental stages. (E) The dog1-5 mutant shows strongly increased shDOG1 expression during silique development. (F) Removal of the asDOG1 promoter results in increased DOG1 expression. LUC intensities produced by the psDOG1::LUC construct, in which asDOG1 was deleted, and by the pDOG1-LUC::DOG1 construct, containing the whole DOG1 gene, were quantified (Left) or visualized (Right) in freshly harvested seeds from the indicated transgenic lines, error bars for this panel represent 95% CI. P value of both psDOG1::LUC lines was calculated in comparison with pDOG1-LUC::DOG1. RT-qPCR data show relative expression levels normalized against UBC mRNA and are the means of three biological replicates with error bars representing SD. *P < 0.05, **P < 0.01, and ***P < 0.001.
Fig. S5.
Germination efficiency (%) of freshly harvested seeds of DOG1 T-DNA insertion mutants and Col-0 WT plants. The graph shows mean values from three biological replicates of each line with error bars representing SD.
This finding led us to speculate that the strong seed dormancy phenotype and underlying up-regulation of the DOG1 sense mRNA level in the dog1-5 mutant is actually a secondary effect of the loss of DOG1 antisense function.
DOG1 Antisense Is a Suppressor of Seed Dormancy in Arabidopsis.
The DOG1 gene is mainly expressed during seed maturation. So far, all tested DOG1 mRNA isoforms have displayed parallel expression patterns, consisting of a slow increase phase with a peak that coincides with the seed maturation stage, and a phase of rapid decline at the seed desiccation stage (9–11, 20). Strand-specific RT-qPCR showed that DOG1 antisense has an expression pattern similar to the DOG1 sense transcript in the early stages of seed development. However, whereas DOG1 sense expression reached a peak and began to decline, the level of asDOG1 continued to increase up to the end of silique maturation (Fig. 3 B and C).
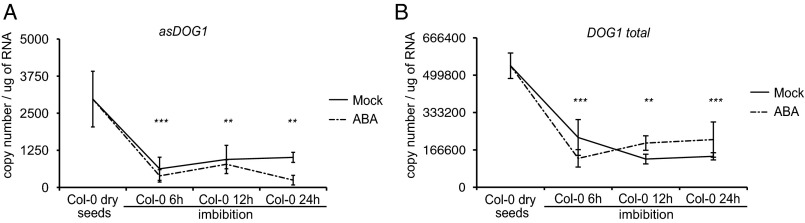
DOG1 expression has been shown to strongly decrease in response to imbibition (9). In contrast addition of ABA during imbibition diminished the DOG1 mRNA reduction in Lepidium sativum (41). Using RT-qPCR we have confirmed that DOG1 mRNA is strongly reduced in Col-0 (WT) during imbibition. Our data show that DOG1 antisense expression exhibits similar strong reduction (Fig. S6). The use of an RNA standard curve, as described in Materials and Methods, allowed us to conclude that the sense transcript is ∼400 times more abundant than the antisense transcript in mature freshly harvested seeds (Fig. S6). Our data show that addition of exogenous ABA did not change the observed reduction of DOG1 sense and DOG1 antisense during imbibition at the concentration used by us (Fig. S6). Given that DOG1 antisense and sense transcripts have differential behavior in seed development and similar expression patterns during imbibition, we focused on the seed-dormancy establishment stage.
Fig. S6.
(A) asDOG1 and (B) DOG1 sense levels are strongly reduced after imbibition without or with addition of ABA at a final concentration of 10 μM. asDOG1 and total DOG1 sense levels measured by RT-qPCR and normalized against a standard curve prepared with sense and antisense RNA as described in methods with primers shown in Dataset S1. *P < 0.05, **P < 0.01, and ***P < 0.001. P value was calculated in comparison with the level of expression in dry seeds. RT-qPCR results were normalized against the UBC mRNA level and are the means of five biological replicates with error bars representing SD.
The asDOG1 transcript level was strongly reduced in seeds of the dog1-5 mutant compared with those of Col-0 (WT), showing a fivefold decrease by the late developmental stages (Fig. 3D). This finding is in agreement with the low levels of antisense transcript observed in dry seeds of this mutant (Fig. 2E). Concurrently, the short DOG1 transcript was strongly up-regulated (fivefold) in dog1-5 in comparison with Col-0 (WT) (Fig. 3E). Moreover, the average fold-change in asDOG1 between the WT and the dog1-5 mutant was similar to that for the short DOG1 transcript.
DOG1 protein has been reported to be highly stable (10, 12). Therefore, the strong up-regulation of the DOG1 mRNA in the dog1-5 mutant during the final stages of seed maturation may account for the substantial overabundance of DOG1 protein in this mutant’s mature seeds, providing an explanation for the extremely strong seed-dormancy phenotype of the dog1-5 mutant (Fig. 3A) (20).
To independently confirm the negative influence of asDOG1 on DOG1 expression, we compared the expression of LUC driven by full-length genomic DOG1 (pDOG1-LUC::DOG1) and by a truncated version of this gene that lacks the DOG1 antisense promoter region (pSDOG1::LUC). In agreement with the dog1-5 mutant analysis, the pSDOG1::LUC construct showed much higher expression than the pDOG1-LUC::DOG1 control construct (Fig. 3F).
In summary, the elimination of DOG1 antisense expression by T-DNA insertion (dog1-5 mutant) or deletion of the asDOG1 promoter (pSDOG1::LUC construct) resulted in significant up-regulation of DOG1 sense transcription. Therefore, asDOG1 acts as a negative regulator of DOG1 expression. Furthermore, based on the enhanced seed-dormancy phenotype of dog1-5, we conclude that by controlling DOG1 expression, asDOG1 suppresses seed dormancy.
DOG1 Antisense Acts in cis.
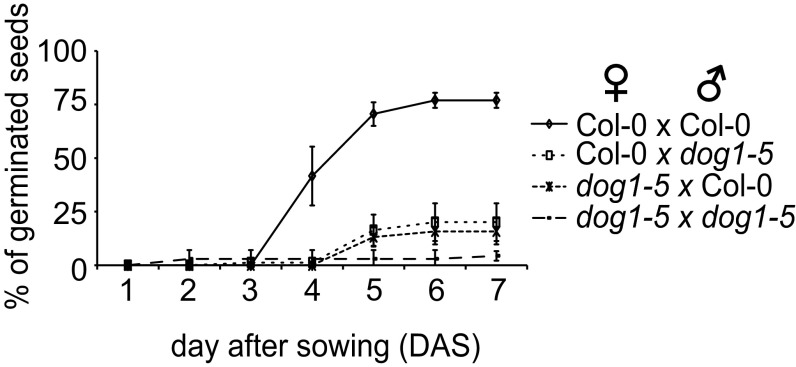
Although our data showed that the DOG1 antisense transcript acts as a suppressor of DOG1 expression, the mechanism of this regulation remained uncharacterized (42). To determine whether asDOG1 acts in cis or in trans, we crossed the dog1-5 mutant with Col-0 (WT) plants and obtained heterozygous F1 seeds with two copies of the DOG1 gene: one from Col-0 (WT) transcribed in both sense and antisense orientations, and the other from the dog1-5 mutant, generating mostly sense transcripts. The resulting dog1-5 × Col-0 seeds displayed seed dormancy that was only slightly weaker than that of dog1-5 plants, indicating that the mutant allele can still confer strong seed dormancy in the presence of an antisense transcript originating from the WT DOG1 gene. This effect was independent of whether the antisense transcript was provided by the maternal or paternal copy of DOG1 (Fig. 4A and Fig. S7).
Fig. 4.
DOG1 antisense acts in cis. (A) Germination efficiency (%) of freshly harvested F1 seeds from a dog1-5 × Col-0 cross. P values were calculated in comparison with Col-0 × Col-0. Mean values from at least three biological replicates with error bars representing SD. **P < 0.01. (B) An additional copy of asDOG1 introduced into Col-0 (WT) by transformation does not affect DOG1 expression. RT-qPCR was normalized against UBC mRNA; data are the means of three biological replicates with error bars representing SD. t tests show no statistical difference between WT and pasDOG1::LUC lines. (C) asDOG1 expressed from construct pasDOG1::LUC introduced into Col-0 (WT). The luciferase signal was quantified (Left) and visualized (Right) in freshly harvested seeds from the indicated transgenic lines. (D) Summary of evidence supporting the proposed model of cis-restricted asDOG1 regulation of seed dormancy. Yellow box: DOG1 antisense activity in Col-0 (WT) plants results in limited DOG1 expression leading to moderate seed dormancy strength. Gray box: the absence of asDOG1 in mutant dog1-5 results in high level expression of shDOG1, which strongly inhibits the transition to germination. Yellow/gray box: in Col-0 × dog1-5 F1 seeds the asDOG1 originating from Col-0 (WT) DOG1 is unable to silence the dog1-5 antisense-less copy of DOG1 in trans. Dark gray box: the addition of a construct expressing additional asDOG1 does not result in reduced DOG1 sense expression in trans.
Fig. S7.
Germination efficiency (%) of freshly harvested seeds from F1 generation prepared by crossing of dog1-5 mutant with Col-0 (WT) in both directions. The graph shows mean values from three biological replicates of each line with error bars representing SD.
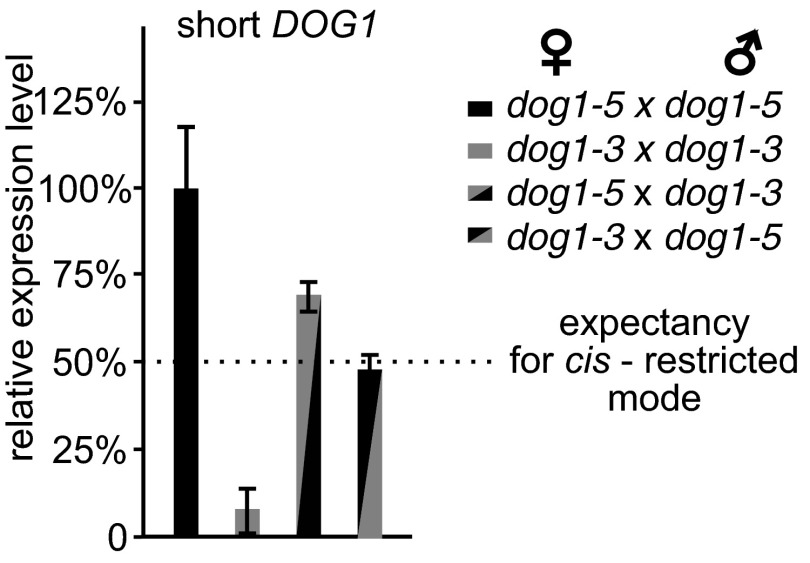
To define each allele contribution to the DOG1 mRNA pool in Col-0 × dog1-5 F1 seeds, we sought to do an allele-specific RT-qPCR in this genetic background. However, it was not possible because of lack of sequence difference between a short DOG1 mRNA transcript derived from the WT and dog1-5 copy. We therefore have combined the dog1-5 antisense-deficient allele with the dog1-3 allele that produces little sense transcript but a nearly WT level of antisense (Fig. 2D). The resulting F1 seeds allowed us to assay the effect of antisense derived from the dog1-3 on the short DOG1 sense mRNA level from the dog1-5 (Fig. S8). We found that in dog1-3 × dog1-5 F1 plants, shDOG1 expression was reduced no more than 50% compared to dog1-5 × dog1-5 F1 plants, which is a level predicted by a loss of one sense DOG1-producing copy (Fig. S8). One-sample Student’s t test showed that the null hypothesis about the 50% reduction cannot be rejected (P > 0.05). This result is in agreement with previous Col-0 × dog1-5 F1 analysis and indicates that asDOG1 is not able to reduce sense DOG1 mRNA level if expressed in trans from a different allele.
Fig. S8.
DOG1 expression is reduced no more than 50% in seeds of dog1-3 × dog1-5 and dog1-5 × dog1-3 F1 seeds when compared to dog1-5 parent, indicating allele-specific cis-restricted control of DOG1 expression by antisense. DOG1 expression was measured using RT-qPCR in seeds of indicated backgrounds. One-sample Student’s t test showed that the 50% reduction null hypothesis (expected by the loss of one DOG1-producing allele in dog1-5 heterozygote plants) cannot be rejected (P > 0.05). One-sample t test was calculated using the R-project t.test function.
However, our observation of the inability of the antisense originating from a Col-0 (WT) or dog1-3 allele to silence an antisense defective dog1-5 allele could be also interpreted as a dosage/dilution effect, rather than a lack of trans silencing. Therefore, to further confirm the inability of asDOG1 to act in trans, we examined DOG1 expression in seeds of transgenic plants carrying an additional copy of DOG1 antisense (Fig. 4 B and C; see Fig. S4 for construct description). RT-qPCR analysis of three independent lines showed that endogenous sense DOG1 expression was unaffected by the presence of a cassette expressing asDOG1 in trans.
Discussion
Seed Dormancy Is Controlled by Antisense ncRNA.
The Arabidopsis life cycle, like that of many other plants, is composed of a series of developmental transitions (2), starting with the transition from a dormant to a nondormant seed state that allows the embryo to germinate in permissive external conditions. To ensure a plant’s survival and reproduction in a changing environment, these transitions are tightly controlled, including regulation by ncRNA. For example, small micro and tasiRNAs control juvenile phase length, and the long antisense transcript COOLAIR controls flowering time (24, 43, 44).
Here, we describe ncRNA-mediated control of seed dormancy. The DOG1 gene is a central positive regulator of seed dormancy strength in Arabidopsis (9). We show that seed dormancy and DOG1 expression are negatively regulated by an antisense transcript. This long ncRNA (lncRNA) is initiated in the region of the proximal transcription termination site (TTS) of the DOG1 gene and terminates around the DOG1 TSS (Fig. 1 A and C). Therefore, we named this transcript asDOG1 or 1GOD. The transcript is both 5′ capped and polyadenylated (Fig. 1 A and C), which suggests that it is transcribed by DNA-dependent RNA polII. This antisense transcript has a tissue-specific expression pattern, indicating that it is not generated by spurious transcriptional noise (45) (Fig. 2A). In agreement with this notion, we cloned the antisense promoter that drives its transcription (Fig. 2B). Most importantly, the elimination of DOG1 antisense by T-DNA insertion or deletion resulted in strong DOG1 sense expression. This finding confirmed that the asDOG1 transcript acts as a negative regulator of DOG1 expression and seed dormancy. DOG1 antisense is strongly induced at the end of seed maturation when levels of the DOG1 mRNA are reduced (Fig. 3B). Thus, the negative effect of asDOG1 on DOG1 expression is most prominent at the end of seed maturation. At this time we observed a fivefold increase in DOG1 mRNA level in the Arabidopsis dog1-5 mutant, in which antisense production is severely compromised in seeds (Fig. 3E).
This finding is corroborated by strong induction of DOG1 protein level in freshly harvested seeds of dog1-5, as shown by us previously (20). In agreement, we and others have shown that the dog1-5 mutant has very strong seed dormancy (13, 20). Given the absence of a long-3 exonic transcript of DOG1 in dog1-5 seeds, the presence of a single overabundant DOG1 antibody reactive band in Western blot (20), we previously suggested that the short DOG1 transcript is the source of the majority of DOG1 protein in vivo, and that it is the short DOG1 protein that controls the seed dormancy (20). Others have shown that DOG1 proteins dimerize and that the multiple splicing/polyadenylation isoforms presented could be required for DOG1 activity in controlling seed dormancy (11). Our published results are partially incompatible with this notion; therefore, more experimental work is needed to resolve those differences (11, 20).
Nevertheless, the DOG1 antisense has the potential to regulate the DOG1 alternative processing, including alternative splicing and alternative polyadenylation (9, 11, 20). Unfortunately, the antisense TSS is located in a very close proximity to the alternative splice sites and alternative polyA sites of the DOG1 gene (Fig. 1A), making the study of antisense effect on DOG1 alternative processing difficult.
Our data clearly show that DOG1 antisense acts as a negative regulator of the short, two exonic (shDOG1) transcript during seed dormancy establishment and, in agreement with this dog1-5 mutant, show strong seed dormancy (13, 20). Given that the DOG1 expression in seeds is controlled by seed maturation temperature (46–49), it is possible that DOG1 antisense may act as a sensor for external and internal stimuli, as shown for other antisense transcripts (26, 30).
DOG1 expression is strongly down-regulated during seed imbibition (9). Our data show a parallel reduction of DOG1 sense and antisense transcripts during imbibition (Fig. S6), suggesting that DOG1 antisense may not be involved at this stage but may have functions restricted to seed maturation.
Seed dormancy control by asDOG1 resembles the regulation of flowering time by COOLAIR. Both antisense transcripts act as negative regulators, are 5′ capped, polyadenylated, and originate from the TTS region of the gene they regulate (25). In the case of FLC, the COOLAIR TSS is located downstream of the FLC protein-coding region. In DOG1, the antisense transcript is initiated from the proximal TTS of the sense transcript of DOG1, and the asDOG1 promoter overlaps the DOG1 exon 3 region. Recently, we showed that the short form of the DOG1 transcript, comprising only exons 1 and 2, is able to complement the dog1 mutant phenotype and that the amino acid sequence of the encoded protein is evolutionarily conserved. Because we were unable to identify a function for the long form of the DOG1 protein, we focused on DOG1 exon 3, which encodes its C-terminal region. Previously we reported that the amino acid sequence encoded by exon 3 shows very weak conservation (20). However, despite numerous insertions and deletions resulting in premature stop codons, the exon 3 DNA sequence is relatively well conserved. Using motif analysis we showed that exon 3, together with the intron 2 region, form part of a larger DNA element that could be considered a conserved nonprotein coding-sequence element. The high level of DNA sequence conservation and lack of homology of the encoded polypeptides may be explained by the function of this element as a promoter for DOG1 antisense transcription (Fig. 1A). This finding is reminiscent of the highly conserved FLC antisense transcript COOLAIR promoter region found in the Brassicaceae family (33).
DOG1 Antisense Regulates Seed Dormancy in cis.
Antisense transcripts represent a substantial proportion of reported lncRNAs. To distinguish them from artificially introduced antisense transcripts used for genetic modification, they have been named natural antisense transcripts (50, 51). In plants, most of the well-characterized natural antisense transcripts act in trans, so they are able to regulate their targets when transcribed from a different location in the genome. Examples include: HID1, involved in photomorphogenesis (29), cis-NATPHO1;2 regulating phosphate homeostasis in rice (30), and asHSFB2a, controlling gametophyte development in Arabidopsis (31). In all of the above cases, expression of the antisense transcript in trans led to phenotypic changes. Similar trans-acting lncRNAs have been reported in other kingdoms: for example, human HOTAIR (52) and TY1 CUT in Saccharomyces cerevisiae (53).
Trans-acting antisense transcripts predominantly act as relatively stable RNA molecules that sequester, recruit, or scaffold trans-acting factors to regulate their targets (28). Our data show that asDOG1 is relatively stable compared with the DOG1 sense transcript and other short-lived protein-coding mRNAs (Fig. 1D). This finding indicated that part of the activity of asDOG1 could be dependent on the RNA molecule itself. Alternatively, the observed stability of asDOG1 might simply be a consequence of the presence of a 5′ cap and a polyA tail stabilizing this transcript, and does not necessarily indicate that this RNA is functional.
When we expressed the DOG1 antisense transcript in trans (Fig. 4B), no changes in DOG1 sense expression were observed, suggesting that its main mode of activity is in cis. However, it is possible that some of the antisense elements missing in our construct are required for its action. To exclude this possibility, we show that asDOG1 transcribed from its native WT location was also unable to silence the dog1-5 allele that is deficient in DOG1 antisense production (Fig. 4A). Moreover, a DOG1 transgene lacking the asDOG1 promoter was highly overexpressed, despite the presence of an antisense transcript originating from a WT DOG1 copy. Thus, the endogenous antisense RNA was unable to silence the transgenic DOG1 copy. Taken together, these data strongly suggest that asDOG1 acts in cis in seed dormancy control. Examples of cis-acting antisense mechanisms include Xist-mediated allele-specific X-chromosome inactivation in humans and PHO gene regulation in yeast (54, 55). In addition, it has been suggested that in plants the antisense transcript COOLAIR acts via the process of its transcription or by formation of an RNA cloud, as seen in single-molecule FISH suppressing FLC transcription in cis (22, 32).
In contrast to published examples of trans-acting noncoding antisense transcripts in plants (29–31), the function of asDOG1 is cis-restricted: it suggests that in seed dormancy, the act of asDOG1 transcription, rather than the asDOG1 RNA molecule, may be important. This suggestion is consistent with the strong requirement for transcription elongation factors in DOG1 expression (14, 16, 17). However, this does not exclude the possibility that asDOG1 may regulate other targets in trans, or act in trans to regulate DOG1 in other tissues or situations. In agreement with this finding, in fission yeast the same ncRNA trigger has been shown to act in cis or trans based on its expression level (56) and the local chromatin context of the target (57).
Seed dormancy release represents a key developmental transition in plants and one that is subject to very strong selection. To ensure optimal timing of seed germination, there is a strong counter-selection against inadequate germination (6, 58). For this reason, the expression of DOG1, the main QTL for seed dormancy in Arabidopsis, is tightly regulated. In the present study we have demonstrated that DOG1 expression and seed dormancy are controlled by a cis-acting antisense transcript. Given the conserved function of DOG1 in seed dormancy and the evolutionary conservation of the DOG1 antisense promoter described here, we expect this mechanism to be active in other plants.
Materials and Methods
Plant Materials and Growth Conditions.
A. thaliana plants were grown in soil in a greenhouse with a long-day (LD) photoperiod (16-h light/8-h dark) at 22 °C/18 °C. For all experiments Col-0 was used as the WT background. The DOG1 T-DNA insertion mutants, dog1-3 (SALK_000867), dog1-4 (SM_3_20886), and dog1-5 (SALK_022748) were purchased from The European Arabidopsis Stock Centre (NASC). The sequences of the examined DOG1 gene can be found in The Arabidopsis Information Resource (TAIR) data library under the accession number At5g45830. Material for transcript expression level analysis during seed development was performed as described previously (20). Briefly, flowers were manually emasculated and pollinated, then siliques were collected at the indicated number of days after pollination (DAP).
Seed Dormancy Tests.
We used in our study Col-0 and mutants derived from it. Col-0 show a relatively weak seed dormancy phenotype, making the use of standard after-ripening treatment difficult (10). We have therefore taken advantage of a seed dormancy test based on the speed of germination of freshly harvested seeds because it shows a good correlation with DOG1 transcript and protein levels. Briefly, freshly harvested seeds were sown on wet blue germination paper (Anchor) and germinated in a LD photoperiod (16-h light/8-h dark) at 22 °C/18 °C. Pictures were taken each day using a high-resolution camera and seed radical perfusion was calculated as described previously (59).
Imbibition Experiments.
Approximately 150 seeds were imbibed in 300 μL of water with or without 10 μM final ABA concentration. Dry seeds were used as a control. The experiment was performed in a LD photoperiod (16-h light/8-h dark) in 22 °C on a rotating wheel. Samples were collected during the daytime, after 6, 12, and 24 h of imbibition.
RNA Extraction, cDNA Synthesis, and PCR Analysis.
RNA extraction and cDNA synthesis were performed as previously described (20). Briefly, RNA was extracted using a phenol-chloroform protocol and treated with DNase (TURBO DNA-free kit, Life Technologies). Reverse transcription of 2.5 μg of RNA was performed using a RevertAid First Strand cDNA Synthesis kit (Fermentas). For cDNA synthesis of sense DOG1, oligo(dT) primers were used. The sequences of all primers are given in Dataset S1.
Adapter-Mediated RT-qPCR Assay.
RNA extracted as described above was used in cDNA synthesis with a DOG1 antisense specific primer with tag as shown in Fig. S3. Subsequently, the qPCR was performed as described above with tag-specific primer (AS_SS_RT) and DOG1 primers (AS_F, AS_R), as shown in Dataset S1. RT-qPCR was performed using a LightCycler 480 real-time system (Roche) with SYBR Green mix (Roche). RT-qPCR results were normalized against the expression level of the Arabidopsis UBC gene (60). P values presented on graphs indicate *P < 0.05, **P < 0.01, and ***P < 0.001.
Absolute Quantification of DOG1 Sense and Antisense Transcripts.
The absolute level of sense and antisense DOG1 transcripts was calculated based on standard curve method, as described previously (61). First, the PCR fragment produced from a cDNA sample with primers DOG1_total_F and DMD3 was cloned into pJET 1.2 blunt-end cloning vector (all primer sequences are listed in Dataset S1). Resulting plasmids with insert in direct (for sense DOG1 RNA) and reverse (for antisense DOG1 RNA) orientation were linearized with a NcoI restriction enzyme and used as templates for in vitro transcription with T7 RNA polymerase. After synthesis, RNA products were recovered using LiCl precipitation and digested with recombinant DNaseI (Roche). Samples were checked for DNA-template contamination by PCR. Subsequently, RNA products were confirmed as nondegraded single-bands on a half-denaturing agarose gel and RNA concentration was measured using Qubit 2.0 fluorimetric assay. Next, dilution series ranging from 10 to 109 ng/μL were prepared for both sense and antisense RNAs, each dilution spiked with total yeast RNA to 750 ng/μL. These were used for cDNA synthesis and subsequent qPCR reactions, in triplicate for each dilution. Based on qPCR results, the curve parameters were calculated and used for standardization of data obtained from biological samples. RT-qPCR assays on imbibed seeds were performed using the same primers and procedures as those for standard curve preparation. The obtained Cp values were used to calculate absolute quantities in number of molecules per qPCR reaction. These values were than standarized according to normalized relative levels of the reference transcript UBC.
Evolutionary Conservation.
DOG1 ortholog sequences were retrieved from the PLAZA3.0 dicots database (62) and from the National Center for Biotechnology Information GenBank. A pairwise alignment of the A. thaliana and A. lyrata DOG1 orthologs was obtained from the VISTA database of prealigned genomes and visualized using VISTApoint with default settings (63). In brief, for each base pair position, the 100-bp window-averaged identity score was calculated. Regions where the score was higher than 70% were considered as conserved and colored in blue (exons), pink (introns), or light-blue (UTRs). Multiple alignments were prepared using the procoffee algorithm available at the T-coffee web server (tcoffee.crg.cat). This algorithm is specifically designed for the alignment of promoter sequences (64). DNA motifs were identified using the MEME Suite (65). The original MEME output is presented in Fig. S9.
Fig. S9.
Full MEME results used to construct Fig. 1B.
RNA Stability Assay.
A cordycepin-dependent RNA stability assay was performed as described previously (20). Col-0 (WT) seedlings were grown on 1/2 Murashige and Skoog Basal Medium with 1% sucrose for 2 weeks in a growth chamber at a LD photoperiod (16-h light/8-h dark) at 22 °C/18 °C. All seeds were stratified at 4 °C for 2 days before sowing. Whole plants were collected and transferred to a flask containing incubation buffer (1 mM Pipes, pH 6.25, 1 mM trisodium citrate, 1 mM KCl, 15 mM sucrose). After 30 min of incubation, cordycepin was added to a final concentration of 150 mg/mL and vacuum-infiltrated (2 × 5 min). At each time-point (0, 15, 30, 45, 60, 75, and 120 min), seedlings representing ∼0.05 g were collected and frozen in liquid nitrogen. Samples were analyzed in triplicate. RNA extraction was performed as described above, and RT-qPCR analysis with primers specific for EIF4A and At3g45970 were used in control reactions for mRNAs showing high and low stability, respectively (38).
5′ RACE.
The 5′ RACE was performed using the Invitrogen 5′ RACE System, including dephosphorylation and subsequent TAP treatment. Individual clones were sequenced. Sequencing results are presented in Fig. S1.
Vectors and Plant Transformation.
To prepare pSDOG1::LUC and pasDOG1::LUC constructs, cloning was performed using the Gateway system (Life Technologies) according to the standard protocol. Plasmid pSDOG1::LUC was prepared using donor (pDONR201) and destination (pGWB635_LUC) vectors. Plasmid pasDOG1::LUC was made using donor (pENTR/D-TOPO) and destination (pGWB635_LUC) vectors. All constructs based on vector pGWB635_LUC were introduced into Agrobacterium tumefaciens strain GV3101 by electroporation. Col-0 (WT) plants were transformed by the floral-dip method (66).
Luciferase Measurement.
Seedlings were sprayed with 0.5 mM luciferin, held in darkness for 1 h, and then emitted light was measured using a NightSHADE LB985 camera, with an exposure time of 10 min. For seeds, about 100 were placed in a well of a white 96-well PCR plate and covered with 10 μL of 1 mM luciferin. After incubating overnight in darkness, emitted light was measured using the NightSHADE camera with an exposure time of 10 min.
Supplementary Material
Acknowledgments
We thank Paweł Mikulski for inventing the DOG1 antisense name 1GOD. H.F., M.P., K.K., L.B., R.Y., Z.P., S.K., and S.S. were supported by National Science Centre, Poland (Grant 2011/01/D/NZ8/03690) and Ministry of Science and Higher Education, Poland (Grant IdP2011 000461). L.B. was supported by Ministry of Science and Higher Education, Poland (Grant IP2011 004671).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1608827113/-/DCSupplemental.
References
- 1.Fournier-Level A, et al. Predicting the evolutionary dynamics of seasonal adaptation to novel climates in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2016;113(20):E2812–E2821. doi: 10.1073/pnas.1517456113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bäurle I, Dean C. The timing of developmental transitions in plants. Cell. 2006;125(4):655–664. doi: 10.1016/j.cell.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 3.van Zanten M, et al. Seed maturation in Arabidopsis thaliana is characterized by nuclear size reduction and increased chromatin condensation. Proc Natl Acad Sci USA. 2011;108(50):20219–20224. doi: 10.1073/pnas.1117726108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holdsworth MJ, Finch-Savage WE, Grappin P, Job D. Post-genomics dissection of seed dormancy and germination. Trends Plant Sci. 2008;13(1):7–13. doi: 10.1016/j.tplants.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 5.Koornneef M, Bentsink L, Hilhorst H. Seed dormancy and germination. Curr Opin Plant Biol. 2002;5(1):33–36. doi: 10.1016/s1369-5266(01)00219-9. [DOI] [PubMed] [Google Scholar]
- 6.Huang X, et al. The earliest stages of adaptation in an experimental plant population: Strong selection on QTLS for seed dormancy. Mol Ecol. 2010;19(7):1335–1351. doi: 10.1111/j.1365-294X.2010.04557.x. [DOI] [PubMed] [Google Scholar]
- 7.Gubler F, Millar AA, Jacobsen JV. Dormancy release, ABA and pre-harvest sprouting. Curr Opin Plant Biol. 2005;8(2):183–187. doi: 10.1016/j.pbi.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 8.Alonso-Blanco C, Bentsink L, Hanhart CJ, Blankestijn-de Vries H, Koornneef M. Analysis of natural allelic variation at seed dormancy loci of Arabidopsis thaliana. Genetics. 2003;164(2):711–729. doi: 10.1093/genetics/164.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bentsink L, Jowett J, Hanhart CJ, Koornneef M. Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc Natl Acad Sci USA. 2006;103(45):17042–17047. doi: 10.1073/pnas.0607877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakabayashi K, et al. The time required for dormancy release in Arabidopsis is determined by DELAY OF GERMINATION1 protein levels in freshly harvested seeds. Plant Cell. 2012;24(7):2826–2838. doi: 10.1105/tpc.112.100214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakabayashi K, Bartsch M, Ding J, Soppe WJJ. Seed dormancy in Arabidopsis requires self-binding ability of DOG1 protein and the presence of multiple isoforms generated by alternative splicing. PLoS Genet. 2015;11(12):e1005737. doi: 10.1371/journal.pgen.1005737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graeber K, et al. Spatiotemporal seed development analysis provides insight into primary dormancy induction and evolution of the Lepidium delay of germination1 genes. Plant Physiol. 2013;161(4):1903–1917. doi: 10.1104/pp.112.213298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huo H, Wei S, Bradford KJ. DELAY OF GERMINATION1 (DOG1) regulates both seed dormancy and flowering time through microRNA pathways. Proc Natl Acad Sci USA. 2016;113(15):E2199–E2206. doi: 10.1073/pnas.1600558113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dolata J, et al. NTR1 is required for transcription elongation checkpoints at alternative exons in Arabidopsis. EMBO J. 2015;34(4):544–558. doi: 10.15252/embj.201489478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grasser M, et al. Transcript elongation factor TFIIS is involved in Arabidopsis seed dormancy. J Mol Biol. 2009;386(3):598–611. doi: 10.1016/j.jmb.2008.12.066. [DOI] [PubMed] [Google Scholar]
- 16.Mortensen SA, Grasser KD. The seed dormancy defect of Arabidopsis mutants lacking the transcript elongation factor TFIIS is caused by reduced expression of the DOG1 gene. FEBS Lett. 2014;588(1):47–51. doi: 10.1016/j.febslet.2013.10.047. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y, Koornneef M, Soppe WJJ. The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. Plant Cell. 2007;19(2):433–444. doi: 10.1105/tpc.106.049221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bassel GW, et al. Genome-wide network model capturing seed germination reveals coordinated regulation of plant cellular phase transitions. Proc Natl Acad Sci USA. 2011;108(23):9709–9714. doi: 10.1073/pnas.1100958108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng J, et al. A novel role for histone methyltransferase KYP/SUVH4 in the control of Arabidopsis primary seed dormancy. New Phytol. 2012;193(3):605–616. doi: 10.1111/j.1469-8137.2011.03969.x. [DOI] [PubMed] [Google Scholar]
- 20.Cyrek M, et al. Seed dormancy in Arabidopsis is controlled by alternative polyadenylation of DOG1. Plant Physiol. 2016;170(2):947–955. doi: 10.1104/pp.15.01483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wagner GP, Zhang J. The pleiotropic structure of the genotype-phenotype map: The evolvability of complex organisms. Nat Rev Genet. 2011;12(3):204–213. doi: 10.1038/nrg2949. [DOI] [PubMed] [Google Scholar]
- 22.Hepworth J, Dean C. Flowering Locus C’s lessons: Conserved chromatin switches underpinning developmental timing and adaptation. Plant Physiol. 2015;168(4):1237–1245. doi: 10.1104/pp.15.00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heo JB, Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 2011;331(6013):76–79. doi: 10.1126/science.1197349. [DOI] [PubMed] [Google Scholar]
- 24.Liu F, Marquardt S, Lister C, Swiezewski S, Dean C. Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science. 2010;327(5961):94–97. doi: 10.1126/science.1180278. [DOI] [PubMed] [Google Scholar]
- 25.Swiezewski S, et al. Small RNA-mediated chromatin silencing directed to the 3′ region of the Arabidopsis gene encoding the developmental regulator, FLC. Proc Natl Acad Sci USA. 2007;104(9):3633–3638. doi: 10.1073/pnas.0611459104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swiezewski S, Liu F, Magusin A, Dean C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature. 2009;462(7274):799–802. doi: 10.1038/nature08618. [DOI] [PubMed] [Google Scholar]
- 27.Liu X, Hao L, Li D, Zhu L, Hu S. Long non-coding RNAs and their biological roles in plants. Genomics Proteomics Bioinformatics. 2015;13(3):137–147. doi: 10.1016/j.gpb.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17(1):47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, et al. Arabidopsis noncoding RNA mediates control of photomorphogenesis by red light. Proc Natl Acad Sci USA. 2014;111(28):10359–10364. doi: 10.1073/pnas.1409457111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jabnoune M, et al. A rice cis-natural antisense RNA acts as a translational enhancer for its cognate mRNA and contributes to phosphate homeostasis and plant fitness. Plant Cell. 2013;25(10):4166–4182. doi: 10.1105/tpc.113.116251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wunderlich M, Gross-Hardt R, Schöffl F. Heat shock factor HSFB2a involved in gametophyte development of Arabidopsis thaliana and its expression is controlled by a heat-inducible long non-coding antisense RNA. Plant Mol Biol. 2014;85(6):541–550. doi: 10.1007/s11103-014-0202-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosa S, Duncan S, Dean C. Mutually exclusive sense-antisense transcription at FLC facilitates environmentally induced gene repression. Nat Commun. 2016;7:13031. doi: 10.1038/ncomms13031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Castaings L, et al. Evolutionary conservation of cold-induced antisense RNAs of FLOWERING LOCUS C in Arabidopsis thaliana perennial relatives. Nat Commun. 2014;5:4457. doi: 10.1038/ncomms5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Freeling M, Subramaniam S. Conserved noncoding sequences (CNSs) in higher plants. Curr Opin Plant Biol. 2009;12(2):126–132. doi: 10.1016/j.pbi.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 35.Sherstnev A, et al. Direct sequencing of Arabidopsis thaliana RNA reveals patterns of cleavage and polyadenylation. Nat Struct Mol Biol. 2012;19(8):845–852. doi: 10.1038/nsmb.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lanford RE, Sureau C, Jacob JR, White R, Fuerst TR. Demonstration of in vitro infection of chimpanzee hepatocytes with hepatitis C virus using strand-specific RT/PCR. Virology. 1994;202(2):606–614. doi: 10.1006/viro.1994.1381. [DOI] [PubMed] [Google Scholar]
- 37.Kim T, Buratowski S. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell. 2009;137(2):259–272. doi: 10.1016/j.cell.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Golisz A, Sikorski PJ, Kruszka K, Kufel J. Arabidopsis thaliana LSM proteins function in mRNA splicing and degradation. Nucleic Acids Res. 2013;41(12):6232–6249. doi: 10.1093/nar/gkt296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szczepińska T, et al. DIS3 shapes the RNA polymerase II transcriptome in humans by degrading a variety of unwanted transcripts. Genome Res. 2015;25(11):1622–1633. doi: 10.1101/gr.189597.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tani H, et al. Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res. 2012;22(5):947–956. doi: 10.1101/gr.130559.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Graeber K, et al. Cross-species approaches to seed dormancy and germination: Conservation and biodiversity of ABA-regulated mechanisms and the Brassicaceae DOG1 genes. Plant Mol Biol. 2010;73(1-2):67–87. doi: 10.1007/s11103-009-9583-x. [DOI] [PubMed] [Google Scholar]
- 42.Koerner MV, Pauler FM, Huang R, Barlow DP. The function of non-coding RNAs in genomic imprinting. Development. 2009;136(11):1771–1783. doi: 10.1242/dev.030403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004;18(19):2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu G, et al. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell. 2009;138(4):750–759. doi: 10.1016/j.cell.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Z, Gerstein M, Snyder M. RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chiang GCK, et al. DOG1 expression is predicted by the seed-maturation environment and contributes to geographical variation in germination in Arabidopsis thaliana. Mol Ecol. 2011;20(16):3336–3349. doi: 10.1111/j.1365-294X.2011.05181.x. [DOI] [PubMed] [Google Scholar]
- 47.Footitt S, Müller K, Kermode AR, Finch-Savage WE. Seed dormancy cycling in Arabidopsis: Chromatin remodelling and regulation of DOG1 in response to seasonal environmental signals. Plant J. 2015;81(3):413–425. doi: 10.1111/tpj.12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Graeber K, Nakabayashi K, Miatton E, Leubner-Metzger G, Soppe WJJ. Molecular mechanisms of seed dormancy. Plant Cell Environ. 2012;35(10):1769–1786. doi: 10.1111/j.1365-3040.2012.02542.x. [DOI] [PubMed] [Google Scholar]
- 49.Kendall SL, et al. Induction of dormancy in Arabidopsis summer annuals requires parallel regulation of DOG1 and hormone metabolism by low temperature and CBF transcription factors. Plant Cell. 2011;23(7):2568–2580. doi: 10.1105/tpc.111.087643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;10(9):637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lapidot M, Pilpel Y. Genome-wide natural antisense transcription: coupling its regulation to its different regulatory mechanisms. EMBO Rep. 2006;7(12):1216–1222. doi: 10.1038/sj.embor.7400857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berretta J, Pinskaya M, Morillon A. A cryptic unstable transcript mediates transcriptional trans-silencing of the Ty1 retrotransposon in S. cerevisiae. Genes Dev. 2008;22(5):615–626. doi: 10.1101/gad.458008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell. 2007;131(4):706–717. doi: 10.1016/j.cell.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 55.Galupa R, Heard E. X-chromosome inactivation: New insights into cis and trans regulation. Curr Opin Genet Dev. 2015;31:57–66. doi: 10.1016/j.gde.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 56.Iida T, Kawaguchi R, Nakayama J. Conserved ribonuclease, Eri1, negatively regulates heterochromatin assembly in fission yeast. Curr Biol. 2006;16(14):1459–1464. doi: 10.1016/j.cub.2006.05.061. [DOI] [PubMed] [Google Scholar]
- 57.Simmer F, et al. Hairpin RNA induces secondary small interfering RNA synthesis and silencing in trans in fission yeast. EMBO Rep. 2010;11(2):112–118. doi: 10.1038/embor.2009.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Donohue K, et al. The evolutionary ecology of seed germination of Arabidopsis thaliana: Variable natural selection on germination timing. Evolution. 2005;59(4):758–770. [PubMed] [Google Scholar]
- 59.Müller K, Tintelnot S, Leubner-Metzger G. Endosperm-limited Brassicaceae seed germination: Abscisic acid inhibits embryo-induced endosperm weakening of Lepidium sativum (cress) and endosperm rupture of cress and Arabidopsis thaliana. Plant Cell Physiol. 2006;47(7):864–877. doi: 10.1093/pcp/pcj059. [DOI] [PubMed] [Google Scholar]
- 60.Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible W-R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139(1):5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol. 2000;25(2):169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- 62.Proost S, et al. PLAZA 3.0: An access point for plant comparative genomics. Nucleic Acids Res. 2015;43(Database issue) D1:D974–D981. doi: 10.1093/nar/gku986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004;32(Web Server issue):W273–279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Erb I, et al. Use of ChIP-Seq data for the design of a multiple promoter-alignment method. Nucleic Acids Res. 2012;40(7):e52. doi: 10.1093/nar/gkr1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bailey TL, et al. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server issue):W202–208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.