Significance
DNA-mediated nanoparticle assembly is an emerging concept to design drug delivery vehicles that can modify their structure or function in response to the in vivo environment. However, better understanding of their interactions with different tissues and organs is needed to establish specific design criteria. Here, we perform a systematic investigation of the molecular properties and mechanisms responsible for serum degradation of DNA-assembled structures. We show that the degradation process is determined by the combined contributions of the surface chemistry of nanoparticles and their supramolecular arrangement. The results also present a strategy for using physiological fluid degradation as the mechanism of controlled drug release. Our findings provide a general framework to study biological interactions of DNA nanostructures.
Keywords: nanoparticle assembly, DNA nanostructures, serum stability, serum resistance, controlled cargo release
Abstract
Understanding the interaction of molecularly assembled nanoparticles with physiological fluids is critical to their use for in vivo delivery of drugs and contrast agents. Here, we systematically investigated the factors and mechanisms that govern the degradation of DNA on the nanoparticle surface in serum. We discovered that a higher DNA density, shorter oligonucleotides, and thicker PEG layer increased protection of DNA against serum degradation. Oligonucleotides on the surface of nanoparticles were highly resistant to DNase I endonucleases, and degradation was carried out exclusively by protein-mediated exonuclease cleavage and full-strand desorption. These results enabled the programming of the degradation rates of the DNA-assembled nanoparticle system from 0.1 to 0.7 h−1 and the engineering of superstructures that can release two different preloaded dye molecules with distinct kinetics and half-lives ranging from 3.3 to 9.8 h. This study provides a general framework for investigating the serum stability of DNA-containing nanostructures. The results advance our understanding of engineering principles for designing nanoparticle assemblies with controlled in vivo behavior and present a strategy for storage and multistage release of drugs and contrast agents that can facilitate the diagnosis and treatment of cancer and other diseases.
Increased understanding of nanoparticle interactions with biological systems enables the construction of safe and efficacious nanomedicine design. One emergent concept is the need for dynamic nanoparticle systems that are capable of changing their physicochemical properties within the physiological environment (1–3). This idea builds on the culmination of experimental evidence showing that cells and tissues interact with engineered nanoparticles as a function of their size, shape, and surface chemistry (4) but that these parameters must adapt to the evolution of in vivo barriers that nanoparticles encounter en route to their intended destination (2, 5).
DNA-functionalized nanoparticles are potentially powerful building blocks for engineering dynamic delivery systems. DNA can be tethered onto a variety of nanoparticle formulations (e.g., inorganic, polymer, lipid, etc.) to program their assembly into precise shapes and sizes based on Watson–Crick hybridization (6, 7). DNA-assembled nanoparticles retain their large surface area to volume ratios for functionalization with molecular payload (8, 9) and can be designed to generate novel properties that derive from particle–particle coupling (10, 11). DNA-assembled nanoparticle systems are highly modular, allowing combinations of varying nanoparticle compositions, surface coatings, and assembly architectures to be rapidly prototyped and evaluated for their biological properties and applications (5, 12, 13). Finally, biodegradation of DNA results in nanoparticle disassembly and release of tethered components, which provides a natural mechanism for bioelimination and/or drug release (2).
As we advance the concept of DNA-assembled nanoparticles into real world clinical applications, there is a need to better understand and control the degradation properties of such molecularly assembled constructs to engineer their biological delivery. Although there have been some in vitro studies on the degradation of DNA-functionalized nanoparticles in simplified biological medium [e.g., phosphate-buffered saline (PBS) spiked with nucleases or cell culture medium containing 10% (vol/vol) serum] (14–17), there is little knowledge linking the physicochemical properties of DNA-coated and nucleotide-assembled nanoparticles to their biodegradation behavior in complex physiological fluids, such as whole blood or serum. Here, we provide systematic mapping of the serum breakdown rates and degradation processes as they relate to nanoparticle design parameters. This information allows us to control the stability and degradability of the assembled system in vivo. The morphology, size, shape, and surface chemistry of nanoparticles may determine their in vivo behavior (e.g., pharmacokinetics, biodistribution, and toxicity). Although premature degradation would alter these parameters and make it difficult to predict biological function, temporally programmed breakdown can serve as a mechanism for controlled cargo release, presentation of targeting ligand, or postdelivery bioelimination of nanoparticle components. This study, thus, provides design parameters and mechanisms to engineer DNA-assembled nanoparticle systems for biological delivery.
Results and Discussion
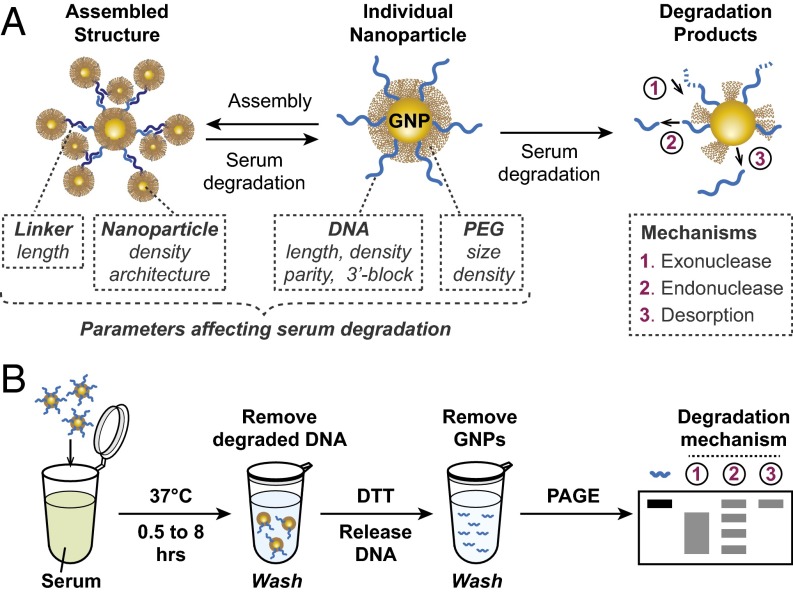
We hypothesized that serum degradation of DNA-assembled nanoparticles is a multistep process governed by two sets of design parameters: (i) the physicochemical properties of DNA-functionalized gold nanoparticles (GNP-DNA), including the composition, size, and density of surface ligands; and (ii) the overall supramolecular architecture of how these nanoparticles are assembled together (Fig. 1A). To assess the relative significance of these parameters, we examined the serum stability of DNA-functionalized nanoparticles in their nonassembled state. GNP-DNAs were synthesized using a published method (18) and stabilized by polyethylene glycol (PEG) to prevent serum-induced aggregation (SI Appendix, SI Discussion S1 and Fig. S1). All oligos used in this study are listed in SI Appendix, Table S1.
Fig. 1.
(A) Schematics of serum degradation. DNA linkers mediate assembly of GNP-DNA into supramolecular structures. Incubation with serum causes assembly breakdown into individual component nanoparticles followed by the degradation of surface-immobilized DNA. Various parameters and mechanisms determine the rate of this process. (B) Experimental setup. PAGE band patterns corresponding to different degradation mechanisms are indicated (A).
Serum Degradation of DNA-Functionalized Nanoparticles.
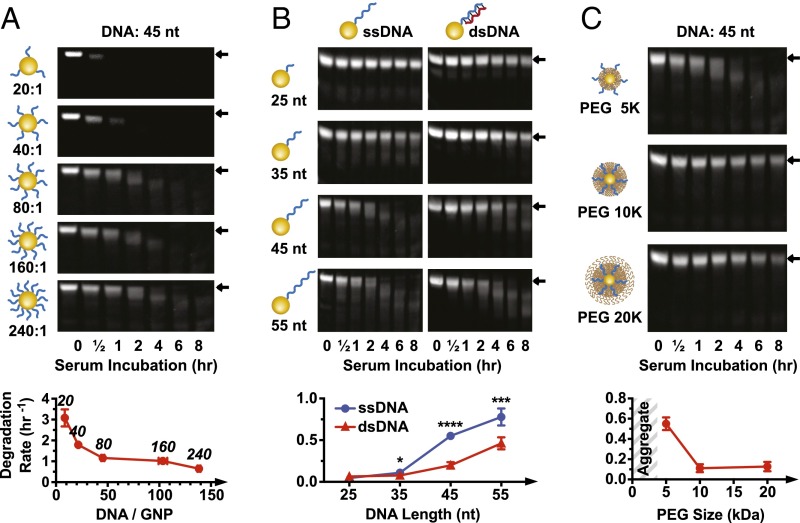
We examined the effects of DNA density, length, and parity [single stranded (ss) or double stranded (ds)] on serum stability of GNP-DNAs; 15 nm GNPs were grafted with a 45-nt sequence (GNP-DNA45) at 20, 40, 80, 160, or 240 strands per particle. Backfilling with 5 kDa PEG resulted in mild DNA displacement (∼17%,), with the final number of 8–138 strands per particle (SI Appendix, Fig. S2 A–C). Serum stability was evaluated by incubating GNP-DNAs in undiluted mouse serum for 0.5, 1, 2, 4, 6, or 8 h (experimental scheme is in Fig. 1B). Particles were then treated with proteinase K and sodium dodecyl sulfate (SDS) to degrade proteins and washed by centrifugation, and DNA remaining on nanoparticles was released by 1,4-dithiothreitol (DTT) treatment and visualized on native polyacrylamide gel electrophoresis (PAGE). DNA degradation was quantified by densitometry of the gel band associated with the full-length DNA strand, and the rate of degradation was obtained from a one-phase decay curve fitting. Degradation rate varied from 0.6 to 3.1 h−1 and was inversely correlated with DNA grafting density (Fig. 2A and SI Appendix, Fig. S2D). The relationship was more pronounced at lower densities, increasing from 1.2 h−1 (45 strands per particle) to 3.1 h−1 (8 strands per particle). Additional increases in DNA grafting ratios had more modest impact on degradation rate, rising only by 0.6 h−1 from 45 to 138 strands per particle. In contrast to nanoparticle-bound DNA, free strands degraded rapidly in serum (SI Appendix, Fig. S3A). These findings suggest that high grafting density of DNA on a nanoparticle surface increases its resistance to serum degradation, consistent with what was observed for GNP-DNA degradation by DNase I (14, 15) and GNP-RNA serum degradation (19–22).
Fig. 2.
Effect of DNA density, length, and PEG-layer thickness on serum degradation. (A) Degradation of nanoparticles with different DNA grafting ratios. Degradation rates (densitometry curves are in SI Appendix, Fig. S2D) plotted against DNA loading (quantified in SI Appendix, Fig. S2A). The number above each data point indicates the grafting ratio. All nanoparticles are backfilled with 5 kDa PEG. (B) Serum degradation of nanoparticles with ssDNA or dsDNA of variable length. Degradation rates based on densitometry curves are in SI Appendix, Fig. S4A. All nanoparticles are backfilled with 5 kDa PEG. Statistical significance between ssDNA and dsDNA for each oligonucleotide length was determined by unpaired t test. *P ≤ 0.05; ***P ≤ 0.001; ****P ≤ 0.0001. (C) Serum degradation of nanoparticles with ssDNA and 5, 10, or 20 kDa PEG backfill (grafted at 16,000 PEG GNP). Densitometry curves are in SI Appendix, Fig. S4B. Error bars are 95% confidence intervals of curve fitting based on three experimental replicates; representative PAGE gel data are shown.
We next examined the effect of DNA length and parity on the rate of degradation. Four GNP-DNAs were synthesized with oligos of 25- (DNA25), 35- (DNA35), 45- (DNA45), and 55-nt long (DNA55). We altered DNA parity by designing complementary strands (cDNA) that bind the variable length regions of the GNP-DNA to form DNA duplexes. Degradation rates were found to increase nonlinearly as a function of DNA length (Fig. 2B and SI Appendix, Fig. S4A). Oligos of 25- and 35-nt long showed slow degradation and little or no difference between single-stranded DNA (ssDNA) and double-stranded DNA (dsDNA). Much faster rates and significantly higher degradation of ssDNA were observed as DNA strand length extended to 45 or 55 nt. These results are consistent with our findings that degradation rate correlates inversely with DNA density. Because of the high curvature of nanoparticles, DNA density drops rapidly away from the nanoparticle surface (9, 23). Therefore, longer DNA strands experience lower density-dependent protective effect than their shorter counterpart. In contrast, addition of the cDNA strand increases the DNA density, resulting in improved protection. Another important factor is the effect of PEG backfill layer, which blocks enzymatic access to DNA (17, 24–27). DNA strand that is shorter than the thickness of the PEG layer is protected from protein binding by being fully submersed within the PEG molecules. In such configuration, additional increase in DNA density through addition of the complementary strand has little effect. This result is seen for GNP-DNA45 with 10 and 20 kDa PEG (Fig. 2C and SI Appendix, Fig. S4B) and GNP-DNA25/35 with 5 kDa PEG in Fig. 2B. In contrast, when the DNA extends beyond the PEG brush layer, it is exposed to rapid enzymatic cleavage, which can be mitigated by the cDNA addition. This result is the case for GNP-DNA45 with 5 kDa PEG in Fig. 2C. Full discussion regarding the determination of PEG-layer thickness can be found in SI Appendix, SI Discussion S2 and Fig. S5D.
The role of PEG in mediating GNP-DNA stability prompted us to investigate the effect of PEG density. GNP-DNA45 were backfilled with 5 kDa PEG at grafting densities ranging from 8,000 to 32,000 PEG per nanoparticle. We did not observe correlation between PEG grafting density and the rate of degradation (SI Appendix, Fig. S6 A and B). Quantification of PEG loading showed fairly slow increase in surface grafting density from 2.7 to 3.8 PEG per 1 nm2 (SI Appendix, Fig. S6C), indicating that PEG density on the surface was close to saturation. Our results are in agreement with the observation that a relatively low PEG density of 0.64/nm2 was sufficient to block adsorption of the majority of serum proteins (25).
Mechanism of DNA Degradation.
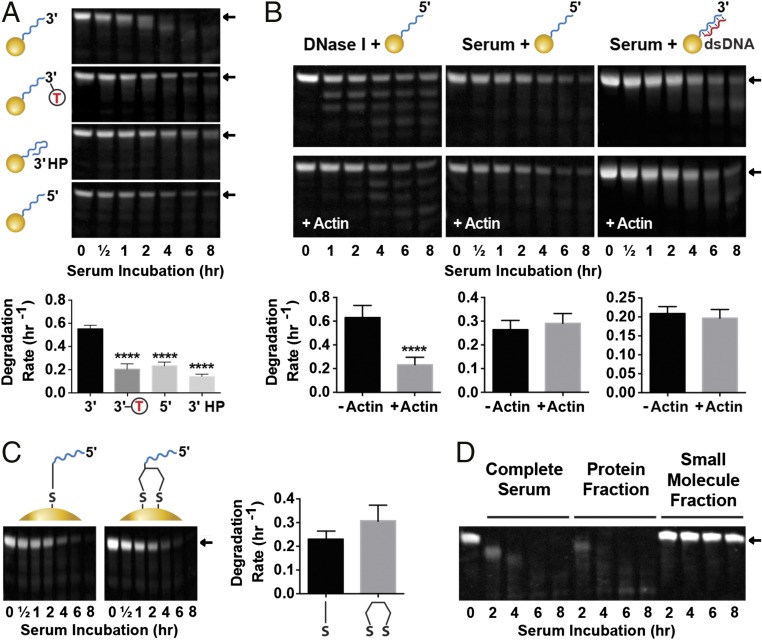
Three mechanisms can contribute to DNA degradation on the nanoparticle surface: (i) degradation from the ends of the strand by exonucleases, (ii) internal cleavage by endonucleases, and (iii) desorption of the whole strand from the surface through thiol–gold bond disruption (Fig. 1A). There is no clear consensus in the literature on the exact serum degradation pathway of free DNA, with some studies reporting exclusive 3′ exonuclease activity (28), and others showing combined exo- and endonuclease degradation (29–31). Our own data showed that free DNA protected from 3′ exonuclease degradation by introduction of inverted deoxythymidine base (invT) at its 3′ end (32) slowed down but did not inhibit serum degradation, indicating the presence of endonuclease or 5′ exonuclease activity (SI Appendix, Fig. S3B). Strand desorption from the surface is a nanoparticle-specific process that could not be predicted from free DNA studies.
We investigated the mechanism of degradation by examining the gel band pattern of the DNA remaining on the nanoparticle surface after serum incubation. Exonucleases are expected to produce band smear and a shift to lower molecular mass (28), endonucleases should result in the appearance of discrete bands corresponding to preferential cleavage sites (30, 33), and full-strand desorption should not generate degradation bands but lead to gradual disappearance of the full-size band (Fig. 1B). Band patterns associated with serum incubation of DNA45 and DNA55 (both with exposed 3′ termini) indicated degradation by 3′ exonucleases (Fig. 2B). Also, in agreement with 3′ exonuclease activity, which is significantly reduced in the case of dsDNA (28), hybridization of the complementary strand slowed the rate of degradation. We probed this further by modifying the DNA45 strand to protect its 3′ end with inverted deoxythymidine base (DNA45-invT) or a hairpin (DNA45-HP) (34) or inverting strand polarity to hide the 3′ end, exposing the 5′ terminus (DNA45-5′). Full-strand desorption patterns were observed when these oligos were incubated in serum (Fig. 3A and SI Appendix, Fig. S7), confirming that the degradation of DNA45 was mainly carried out by 3′ exonucleases. Much slower degradation of DNA45-5′ and its corresponding desorption band pattern indicated that 5′ exonucleases do not play a significant role in DNA breakdown. In contrast to longer DNA strands, DNA25 and DNA35 with exposed 3′ ends showed slow rate of degradation for both their ssDNA and dsDNA forms with characteristic strand desorption rather than exonuclease cleavage patterns (Fig. 2B). For these strands, DNA degradation rates based on lane densitometry (which includes the original size band and all degradation products) matched those of the band densitometry (SI Appendix, Fig. S8), confirming desorption mechanism (DNA cleavage would lead to slower lane degradation compared with band degradation). Protecting DNA25 from 3′ exonucleases by inverting the strand to expose the 5′ end (DNA25-5′) had no effect on the degradation (SI Appendix, Fig. S9). This result suggested that, when DNA strands are shorter than the thickness of the PEG layer, they are protected from exonucleases and can only be removed by desorption, and therefore, 3′ end protection is not useful. Interestingly, desorption rate of the shorter DNA25-5′ was noticeably slower compared with DNA45-5′, which extends above the PEG brush layer. This result indicates that access to DNA is a key parameter affecting the rate of desorption.
Fig. 3.
Mechanisms of serum DNA degradation on nanoparticle surface. (A) 3′ Exonucleases play a major role in degradation. Degradation of DNA45 compared with end-protected DNA45-invT, DNA45-HP, and inverted DNA45-5′. Densitometry curves are in SI Appendix, Fig. S7. (B) DNase I contribution to serum degradation of GNP-DNA is not significant. Nanoparticles with ssDNA45-5′ or dsDNA45 are subjected to degradation by DNase I (at 500× serum concentration) or serum with and without DNase I inhibitor actin (densitometry curves are in SI Appendix, Fig. S10). Actin inhibited activity of concentrated DNase I but had no effect on serum degradation. ****P ≤ 0.0001. (C) Dithiol linker does not prevent serum-induced DNA desorption. Comparison of serum degradation of DNA45-5′ strands adsorbed onto nanoparticle with a monothiol or dithiol linker (densitometry curves are in SI Appendix, Fig. S13). (D) Proteins and not small molecules carry out serum degradation. Nanoparticles incubated with complete serum or its protein and small molecule fractions (SI Appendix, Fig. S16). Error bars are 95% confidence intervals of curve fitting based on three experimental replicates; representative PAGE gel data are shown. Statistical significance was determined by (A) ordinary one-way ANOVA or (B and C) unpaired t test. All nanoparticles had 5 kDa PEG backfill.
Next, we confirmed that the pattern observed for exonuclease protected strands is indeed caused by strand desorption. If endonuclease cleaving resulted in short DNA fragments, their staining on the gel could be weak, and results seem to mimic desorption. DNase I was used as the model endonuclease, because it is the major serum and tissue endonuclease (30, 35), and resistance to DNase I degradation is often used to predict biological stability of DNA-containing nanoparticles (14, 15, 17, 36–38). We subjected exonuclease-resistant nanoparticles with DNA45-5′ to DNase I degradation and observed the expected multiband pattern (Fig. 3B and SI Appendix, Fig. S10). We found that 50 U/mL DNase I was needed for clearly observable degradation (SI Appendix, Fig. S11), which is 500 times higher than its physiological mouse serum concentration of 37 ng/mL (∼0.1 U/mL using the conversion 1 mg = ∼2,500 U) (31). These data agree with past studies that DNA immobilized on the nanoparticle surface is highly resistant to DNase I degradation (14, 15). Actin is a known inhibitor of DNase I activity (39). The addition of 0.25 mg/mL actin to samples treated with 50 U/mL DNase I showed clear inhibition of the enzyme. However, addition of the same amount of actin, which is orders of magnitude higher than that required for complete DNase I inhibition in mouse serum (31), had no effect on serum degradation of nanoparticles with ssDNA45-5′ or dsDNA45 (Fig. 3B). These results confirm that nanoparticle-immobilized DNA is strongly protected from DNase I cleavage, and therefore, this enzyme does not play a significant role in serum degradation.
The degradation rates and patterns associated with the different end modifications indicated that DNA desorption is the dominant mechanism of exonuclease-resistant strand removal from the nanoparticle surface. It has been previously shown that the rate of desorption of DNA from gold nanoparticles by small thiol-containing molecules, such as DTT, can be reduced by replacing a monothiol with a dithiol linker (40–42). We observed that, although introducing a dithiol linker at the 3′ end of DNA45-5′ strongly protected nanoparticles from salt-induced aggregation in 10 mM DTT (SI Appendix, Fig. S12), no resistance to serum degradation was observed (Fig. 3C and SI Appendix, Fig. S13). DNA25 modified with dithiol yielded similar results (SI Appendix, Fig. S14). This discrepancy could be explained by observations that the majority of thiol-containing small molecules in serum, such as glutathione (GSH) or cysteine, are present in the disulphide protein-bound state, leaving few free thiols to interact with nanoparticle (43, 44). To test this idea, we incubated DNA45-5′ nanoparticles in PBS supplemented with normal serum levels of 20 μM cysteine and 1 μM GSH (43). No DNA desorption was observed (SI Appendix, Fig. S15). We also incubated DNA45 or DNA45-5′ nanoparticles with protein and small molecule serum components (Fig. 3D and SI Appendix, Fig. S16). Incubation with protein fraction showed a degradation pattern similar to that of complete serum, but no degradation was observed for small molecule components or PBS control. These results indicate that DNA desorption in serum is mediated exclusively by proteins without the involvement of small molecules and cannot be inhibited by the introduction of dithiol linkers. Interestingly, this process contrasts with our observation of serum aggregation of nanoparticles lacking protective PEG backfill, which involves the combined interaction of proteins and small molecules (SI Appendix, SI Discussion S1 and Fig. S1).
Finally, we investigated the effect of serum type on degradation. Fetal bovine serum (FBS) showed a exonuclease-dominated degradation profile of DNA45 similar to that of the mouse serum but a slower removal of DNA45-5′ strand, suggesting that desorption processes are weaker in FBS compared with mouse serum (SI Appendix, Fig. S17). Surprisingly, stability of nanoparticle-immobilized DNA was found to be ∼10 times higher in human serum. Little difference was observed between the 3′- and 5′-terminated strand degradation profiles, and only a weak exonuclease degradation pattern was observed for DNA45. This result suggests that, in contrast to mouse serum, GNP-DNA degradation in human serum is a much slower process that is dominated by desorption rather than exonuclease degradation. These results suggest that serum type has a significant effect on the degradation of DNA-containing nanostructures. This finding is expected to have significant implications when GNP-DNA–containing nanostructures are developed for human applications.
Controlling Degradation of DNA-Assembled Nanoparticles.
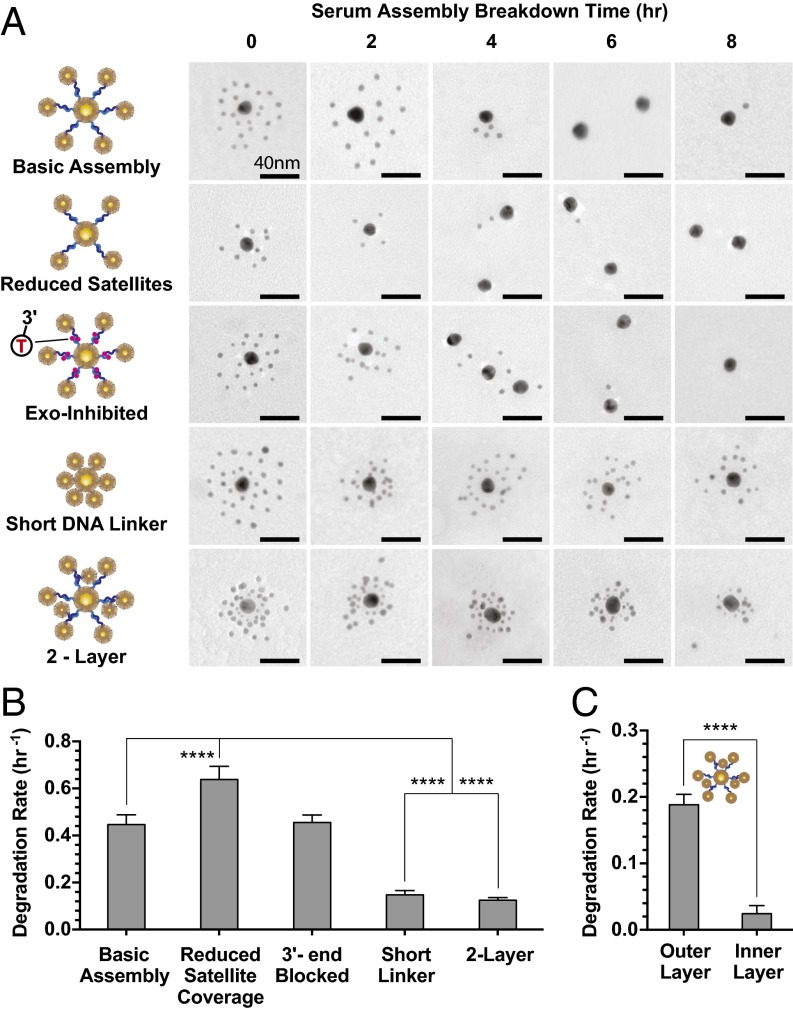
The results obtained from a single-particle system indicated that increased serum protection was associated with (i) higher DNA density, (ii) proximity between the DNA strands and nanoparticle surface, and (iii) increased thickness of PEG backfill layer. To show how these criteria can be applied to control serum degradation of more complex nanoparticle assemblies, we designed a library of core satellite structures with different architectures and surface chemistries (molecular details are in SI Appendix, Fig. S18 and Table S1). “Basic Assembly” structures contained 18.9 ± 2.1 satellite particles of 5-nm diameter linked to a 15-nm core by a 67-nt-long linker DNA, 30 bases of which were present in a ds form, and backfilled with 10 kDa PEG. The structures were incubated with whole serum, and degradation was quantified using transmission electron microscopy (TEM) by counting the number of satellites remaining attached to the core as a function of time (Fig. 4A). The rate of degradation was found to be 0.45 h−1 (Fig. 4B), which is in the range of the rates calculated for a single-particle system densely packed with DNA (Fig. 2B).
Fig. 4.
Controlling serum degradation rates of GNP-DNA assemblies. (A) Representative TEM (zoomed out TEM is shown in SI Appendix, Figs. S22–S26) of five assembled structures (details are in SI Appendix, Fig. S18) incubated in serum. Two-layer structure: 5 kDa PEG; all other structures: 10 kDa PEG. (B) Associated serum degradation rates. Number of satellites per core nanoparticle quantified by direct TEM counting. Degradation rates are based on single-phase decay fits of these data (SI Appendix, Fig. S19A). Increase in satellite density correlates with improved protections against serum breakdown. (C) Degradation rates of the two-layer structures; 5-nm satellites in the outer layer were quantified separately from the inner-layer 3-nm satellites. Degradation of the outer layer occurs much more rapidly that of the inner layer. Degradation rates are based on single-phase decay fits of these data (SI Appendix, Fig. S19B); error bars are 95% fitting confidence intervals. Statistical significance was determined by (B) ordinary one-way ANOVA or (C) unpaired t test. ****P ≤ 0.0001.
Next, we introduced a number of modifications predicted to affect the rate of degradation. First, the number of satellites per core was reduced to 11.0 ± 2.9 to increase the rate of degradation caused by reduced DNA density and longer average distance between DNA and nanoparticle surface. Second, another modification was to block exposed 3′ ends of all oligonucleotides with invT to prevent degradation by exonucleases. The last two modifications aimed to increase resistance to serum degradation by maximizing the density of DNA and PEG-layer coverage and reducing the distances between nanoparticles and DNA strands. This design was achieved by either shortening the linker DNA by 17 nt to bring the satellites closer to the core or introducing an extra layer of 3-nm satellites between the outer layer of 5-nm particles and the core. Fig. 4 and SI Appendix, Fig. S19 show results of incubating these assembled structures in serum. Reduced satellite coverage increased the rate of degradation by 1.5-fold (0.64 h−1) relative to Basic Assembly (Fig. 4B). In contrast, structures with shorter linker or two layers showed significant protection from serum degradation (0.15 and 0.12 h−1, respectively), leading to the observation of partially intact structures even after 8 h of incubation (Fig. 4A). Interestingly, the 3′ end blocking did not have an observable effect. The likely explanation for this is that the core satellite architecture results in the 3′ ends of the DNA strands being located within a distance of 25 nt of 15-nm core nanoparticles’ surface. At this distance, they are protected by the PEG layer, which was shown in Fig. 2B. The ends are present in more resistant dsDNA forms. The addition of actin as a DNase I inhibitor did not have any effect on serum degradation of the structures, indicating that, similar to a single-particle system, DNase I contributes little to the degradation of nanoparticle assemblies (SI Appendix, Fig. S20). Therefore, strand desorption seems to be the main mechanism of breakdown of core satellite structures. Finally, the inner and outer satellite layers of the two-layer assembly degraded with distinctly different rates of 0.02 and 0.19 h−1, respectively (Fig. 4C). These results show that control of the density of DNA-functionalized particles in assembled structures is an efficient method to modulate their serum degradation rates.
Serum Degradation as a Mechanism for Controlled Cargo Release.
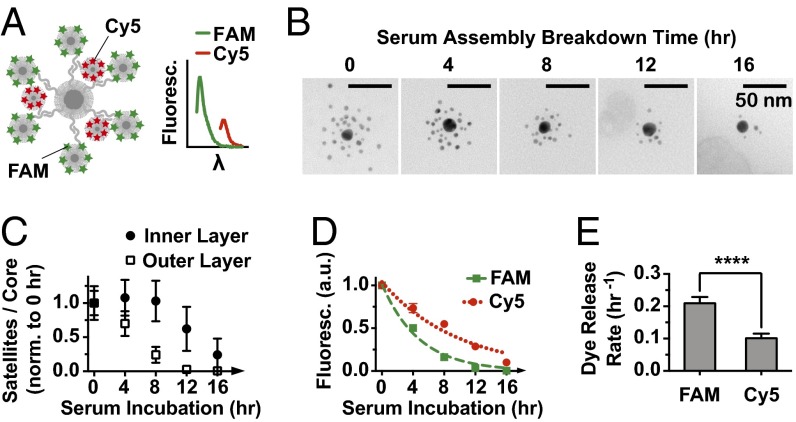
We used the principles discovered in this study to design a multistage cargo release system with programmable degradation rates. Controlled release rates may mediate the therapeutic response of nanoparticles as well as determine the toxicity of a medical delivery vehicle. We designed a two-layer superstructure construct and loaded two types of fluorescent dyes onto the two sets of satellites by conjugating them to thiolated DNA. FAM (6-fluorescein amidate) was loaded onto the outer layer, whereas cyanine 5 (Cy5) was loaded onto the inner layer (Fig. 5A and SI Appendix, Fig. S18). To ensure that the degradation of the highly stable inner layer could be observed, serum incubation was extended to 16 h. According to TEM data, the outer layer began degrading immediately, whereas the inner layer remained mostly intact for the first 8 h (Fig. 5 B and C). Only after almost complete removal of the outer layer, the inner 3-nm nanoparticles started to get ejected from the assemblies. Interestingly, although the breakdown profile was offset for the two layers by around 8 h, their degradation slopes looked similar. This result suggests that the outer-layer structures prevented the serum factors responsible for assembly degradation from reaching the inner layer. Otherwise, the two layers represented similar environments in terms of oligonucleotide density, average DNA to nanoparticle distance, and PEG-layer protection. For the assembly structures backfilled with 5 kDa PEG, the release of the FAM dye was two times faster than that of Cy5, corresponding to half-lives of 3.3 and 6.9 h, respectively (Fig. 5 D and E). The release profiles were modestly steeper than the ejection of satellite nanoparticles, suggesting that the carrier DNA or the dyes were released from the satellites before the nanoparticle itself was disassembled from the structure. The use of longer 10 kDa PEG further increased FAM and Cy5 half-lives to 5.0 and 9.8 h, respectively (SI Appendix, Fig. S21). Therefore, by applying the criteria governing serum degradation rates, we engineered assembly structures that could provide serum-mediated two-stage cargo release with tunable half-lives ranging from 3 to 10 h.
Fig. 5.
Controlling the rate of serum breakdown of two-layer assemblies backfilled with 5 kDa PEG as the mechanism of multistage cargo release. (A) Fluorescent dyes FAM and Cy5 loaded onto the outer and inner layers of the structure, respectively (details are in SI Appendix, Fig. S18). Fluorescence plotted against wavelength (λ) shows peaks corresponding to emissions of both dyes. (B) Representative TEM images (zoomed out TEM is shown in SI Appendix, Fig. S27) of structure breakdown in serum. (C) Quantification of breakdown by separate TEM counting of outer-layer 5-nm satellites and inner-layer 3-nm satellites. Data are normalized to 0-h counts. The outer layer breaks down much faster than the inner layer. (D) Decrease in the fluorescence of assemblies over time as a result of dye release from the surface. Data are normalized to 0 h and fitted with single-phase decay curves. (E) Dye release rates are based on decay curves in D. FAM is released significantly faster than Cy5. Error bars are (C and D) SDs or (E) 95% fitting confidence intervals. Statistical significance was determined by unpaired t test based on three experimental replicates. ****P ≤ 0.0001.
Conclusions
Self-assembling nanoparticle systems are emerging as a design concept for biomedical applications (6, 7, 12), but our understanding of their in vivo design criteria remains at a nascent stage. These assemblies are complex composite structures with multiple components, which makes their interactions with biological systems challenging to evaluate. Nevertheless, using serum interactions of DNA-assembled nanoparticles as an example, we showed that different aspects of these interactions can be investigated separately to identify the relative importance of each parameter. The optimized structural and surface chemistry design criteria can then be combined to generate complex structures with well-defined and programmable behavior. We found that higher DNA density, shorter DNA strands, and longer thickness of PEG increased resistance of DNA to serum degradation. Backfilling DNA-functionalized nanoparticles with 5 kDa or larger PEG protected them from aggregation mediated by serum proteins and small molecules. GNP-DNAs were found to be highly resistant to DNase I cleavage, and the dominant mechanisms of serum degradation were attributable to 3′ exonucleases and protein-mediated strand desorption. Serum type was found to have a significant effect on GNP-DNA degradation, with as much as 10 times slower degradation in human compared with mouse serum. Applying these design criteria to engineer DNA-assembled nanoparticles, we showed assembled structures with tunable serum stability ranging from 0.1 to 0.7 h−1 in addition to structures capable of releasing two model drugs at distinct kinetics. In future work, we plan to explore these engineering principles for multidrug therapeutic targeting of cells in vitro and tumors in vivo. The results of this study offer useful information for designing DNA-assembled nanoparticles for in vivo applications (2, 5) and show an experimental framework that could be applied to investigate serum stability of other types of DNA nanostructures (45, 46).
Materials and Methods
The list of chemicals, materials, and buffer recipes is provided in SI Appendix. All DNA sequences are listed in SI Appendix, Table S1. Methods to synthesize DNA-functional nanoparticles and -assembled structures are described in SI Appendix, Fig. S18.
Serum Incubation of DNA-Functionalized GNPs.
Four microliters 350 nM GNP-DNA was incubated for 0.5, 1, 2, 4, 6, or 8 h at 37 °C with one of the following: (i) 96 μL serum; (ii) 96 μL PBS; (iii) 91 μL serum and 5 μL 1 mg/mL actin (or 5 μL PBS as control); (iv) 5 μL 1U/μL DNase I, 10 μL 10× DNase I buffer, 71 μL PBS, and 10 μL 1 mg/mL actin (or 10 μL ultrapure water); (v) 96 μL protein or small molecule serum fraction (purification details are in SI Appendix); (vi) 10 μL 10× PBS, 10 μL 100 mM DTT, and 76 μL water; or (vii) 10 μL 10× PBS, 10 μL 200 μM l-cysteine, 10 μL 210 μM GSH, and 66 μL water. Also, 4 μL 100 μM free DNA was incubated with 96 μL serum for 0.5, 1, 2, 4, 6, or 8 h at 37 °C. At the end of incubation, 3.9 μL 20 mg/mL Proteinase K, 47 μL 5% SDS, and 310 μL PBS supplemented with 0.01% Tween-20 were added; the solution was incubated for 60 min at 37 °C, and nanoparticles were washed by centrifugation (two times at 16,000 × g for 35 min each) and resuspended in 100 μL Tris-borate-EDTA buffer (TBE) supplemented with 0.01% Tween-20. Remaining DNA strands were released by DTT (10 μL 1 M DTT incubated at 60 °C for 35 min), nanoparticles were removed by centrifugation (21,000 × g for 35 min), and DNA in supernatant was visualized by 20% TBE PAGE gel. Additional details are in SI Appendix.
Serum Incubation of Assembled Structures.
Four microliters ∼80 nM assembled structures were incubated with 96 μL serum for 2, 4, 6, 8, 12, or 16 h at 37 °C, washed by centrifugation in PBS (two times at 12,000 × g for 35 min each), resuspended in 10 μL PBS, and characterized by TEM. Fluorescently labeled two-layer structures were instead resuspended in 100 μL PBS, 10 μL of which were used for TEM characterization. The remaining 90 μL were mixed with 10 μL 1 M DTT and incubated for 30 min at 60 °C to release all remaining DNA-conjugated dyes into solution. Nanoparticles were removed by centrifugation (21,000 × g for 30 min), and the amount of the dye in the supernatant was measured by spectrophotometry (emission and absorbance were set to 488 and 512 nm for FAM and 640 and 670 nm for Cy5, respectively). Additional details are in SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by Natural Sciences and Engineering Research Council of Canada (NSERC, RGPIN288231) and Canadian Institute of Health Research (MOP 130143 and GCS105653-1). K.Z. and L.Y.T.C. acknowledge a fellowship from NSERC. L.Y.T.C. acknowledges a Banting Fellowship and a Canadian Breast Cancer Foundation Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.E.D. is a Guest Editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1610028113/-/DCSupplemental.
References
- 1.Ohta S, Glancy D, Chan WC. DNA-controlled dynamic colloidal nanoparticle systems for mediating cellular interaction. Science. 2016;351(6275):841–845. doi: 10.1126/science.aad4925. [DOI] [PubMed] [Google Scholar]
- 2.Chou LY, Zagorovsky K, Chan WC. DNA assembly of nanoparticle superstructures for controlled biological delivery and elimination. Nat Nanotechnol. 2014;9(2):148–155. doi: 10.1038/nnano.2013.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim Y, Macfarlane RJ, Jones MR, Mirkin CA. Transmutable nanoparticles with reconfigurable surface ligands. Science. 2016;351(6273):579–582. doi: 10.1126/science.aad2212. [DOI] [PubMed] [Google Scholar]
- 4.Albanese A, Tang PS, Chan WCW. The effect of nanoparticle size, shape, and surface chemistry on biological systems. Annu Rev Biomed Eng. 2012;14(1):1–16. doi: 10.1146/annurev-bioeng-071811-150124. [DOI] [PubMed] [Google Scholar]
- 5.Wang C, et al. Stimuli-responsive plasmonic core-satellite assemblies: i-Motif DNA linker enabled intracellular pH sensing. Chem Commun (Camb) 2013;49(51):5739–5741. doi: 10.1039/c3cc80005a. [DOI] [PubMed] [Google Scholar]
- 6.Tan SJ, Campolongo MJ, Luo D, Cheng W. Building plasmonic nanostructures with DNA. Nat Nanotechnol. 2011;6(5):268–276. doi: 10.1038/nnano.2011.49. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Lu F, Yager KG, van der Lelie D, Gang O. A general strategy for the DNA-mediated self-assembly of functional nanoparticles into heterogeneous systems. Nat Nanotechnol. 2013;8(11):865–872. doi: 10.1038/nnano.2013.209. [DOI] [PubMed] [Google Scholar]
- 8.Hurst SJ, Lytton-Jean AK, Mirkin CA. Maximizing DNA loading on a range of gold nanoparticle sizes. Anal Chem. 2006;78(24):8313–8318. doi: 10.1021/ac0613582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hill HD, Millstone JE, Banholzer MJ, Mirkin CA. The role radius of curvature plays in thiolated oligonucleotide loading on gold nanoparticles. ACS Nano. 2009;3(2):418–424. doi: 10.1021/nn800726e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature. 1996;382(6592):607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 11.Chou LYT, Song F, Chan WCW. Engineering the structure and properties of DNA-nanoparticle superstructures using polyvalent counterions. J Am Chem Soc. 2016;138(13):4565–4572. doi: 10.1021/jacs.6b00751. [DOI] [PubMed] [Google Scholar]
- 12.Mu CJ, Lavan DA, Langer RS, Zetter BR. Self-assembled gold nanoparticle molecular probes for detecting proteolytic activity in vivo. ACS Nano. 2010;4(3):1511–1520. doi: 10.1021/nn9017334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu X, Rosi NL, Wang Y, Huo F, Mirkin CA. Asymmetric functionalization of gold nanoparticles with oligonucleotides. J Am Chem Soc. 2006;128(29):9286–9287. doi: 10.1021/ja061980b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seferos DS, Prigodich AE, Giljohann DA, Patel PC, Mirkin CA. Polyvalent DNA nanoparticle conjugates stabilize nucleic acids. Nano Lett. 2009;9(1):308–311. doi: 10.1021/nl802958f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cutler JI, et al. Polyvalent nucleic acid nanostructures. J Am Chem Soc. 2011;133(24):9254–9257. doi: 10.1021/ja203375n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alexander CM, Dabrowiak JC, Maye MM. Investigation of the drug binding properties and cytotoxicity of DNA-capped nanoparticles designed as delivery vehicles for the anticancer agents doxorubicin and actinomycin D. Bioconjug Chem. 2012;23(10):2061–2070. doi: 10.1021/bc3002634. [DOI] [PubMed] [Google Scholar]
- 17.Song L, et al. Terminal PEGylated DNA-gold nanoparticle conjugates offering high resistance to nuclease degradation and efficient intracellular delivery of DNA binding agents. ACS Appl Mater Interfaces. 2015;7(33):18707–18716. doi: 10.1021/acsami.5b05228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X, Servos MR, Liu J. Instantaneous and quantitative functionalization of gold nanoparticles with thiolated DNA using a pH-assisted and surfactant-free route. J Am Chem Soc. 2012;134(17):7266–7269. doi: 10.1021/ja3014055. [DOI] [PubMed] [Google Scholar]
- 19.Patel PC, Hao L, Yeung WS, Mirkin CA. Duplex end breathing determines serum stability and intracellular potency of siRNA-Au NPs. Mol Pharm. 2011;8(4):1285–1291. doi: 10.1021/mp200084y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giljohann DA, Seferos DS, Prigodich AE, Patel PC, Mirkin CA. Gene regulation with polyvalent siRNA-nanoparticle conjugates. J Am Chem Soc. 2009;131(6):2072–2073. doi: 10.1021/ja808719p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barnaby SN, Lee A, Mirkin CA. Probing the inherent stability of siRNA immobilized on nanoparticle constructs. Proc Natl Acad Sci USA. 2014;111(27):9739–9744. doi: 10.1073/pnas.1409431111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prigodich AE, Alhasan AH, Mirkin CA. Selective enhancement of nucleases by polyvalent DNA-functionalized gold nanoparticles. J Am Chem Soc. 2011;133(7):2120–2123. doi: 10.1021/ja110833r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tambasco M, Kumar SK, Szleifer I. Quantitatively modeling the equilibrium properties of thiol-decorated gold nanoparticles. Langmuir. 2008;24(16):8448–8451. doi: 10.1021/la8008046. [DOI] [PubMed] [Google Scholar]
- 24.Perrault SD, Walkey C, Jennings T, Fischer HC, Chan WCW. Mediating tumor targeting efficiency of nanoparticles through design. Nano Lett. 2009;9(5):1909–1915. doi: 10.1021/nl900031y. [DOI] [PubMed] [Google Scholar]
- 25.Walkey CD, Olsen JB, Guo H, Emili A, Chan WCW. Nanoparticle size and surface chemistry determine serum protein adsorption and macrophage uptake. J Am Chem Soc. 2012;134(4):2139–2147. doi: 10.1021/ja2084338. [DOI] [PubMed] [Google Scholar]
- 26.Dai Q, Walkey C, Chan WCW. Polyethylene glycol backfilling mitigates the negative impact of the protein corona on nanoparticle cell targeting. Angew Chem Int Ed Engl. 2014;53(20):5093–5096. doi: 10.1002/anie.201309464. [DOI] [PubMed] [Google Scholar]
- 27.Larson TA, Joshi PP, Sokolov K. Preventing protein adsorption and macrophage uptake of gold nanoparticles via a hydrophobic shield. ACS Nano. 2012;6(10):9182–9190. doi: 10.1021/nn3035155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eder PS, DeVine RJ, Dagle JM, Walder JA. Substrate specificity and kinetics of degradation of antisense oligonucleotides by a 3′ exonuclease in plasma. Antisense Res Dev. 1991;1(2):141–151. doi: 10.1089/ard.1991.1.141. [DOI] [PubMed] [Google Scholar]
- 29.Chu BC, Orgel LE. The stability of different forms of double-stranded decoy DNA in serum and nuclear extracts. Nucleic Acids Res. 1992;20(21):5857–5858. doi: 10.1093/nar/20.21.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Napirei M, Wulf S, Mannherz HG. Chromatin breakdown during necrosis by serum Dnase1 and the plasminogen system. Arthritis Rheum. 2004;50(6):1873–1883. doi: 10.1002/art.20267. [DOI] [PubMed] [Google Scholar]
- 31.Macanovic M, Lachmann PJ. Measurement of deoxyribonuclease I (DNase) in the serum and urine of systemic lupus erythematosus (SLE)-prone NZB/NZW mice by a new radial enzyme diffusion assay. Clin Exp Immunol. 1997;108(2):220–226. doi: 10.1046/j.1365-2249.1997.3571249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ortigão JFR, et al. Antisense effect of oligodeoxynucleotides with inverted terminal internucleotidic linkages: A minimal modification protecting against nucleolytic degradation. Antisense Res Dev. 1992;2(2):129–146. doi: 10.1089/ard.1992.2.129. [DOI] [PubMed] [Google Scholar]
- 33.Napirei M, Ludwig S, Mezrhab J, Klöckl T, Mannherz HG. Murine serum nucleases--contrasting effects of plasmin and heparin on the activities of DNase1 and DNase1-like 3 (DNase1l3) FEBS J. 2009;276(4):1059–1073. doi: 10.1111/j.1742-4658.2008.06849.x. [DOI] [PubMed] [Google Scholar]
- 34.Abdelgany A, Wood M, Beeson D. Hairpin DNAzymes: A new tool for efficient cellular gene silencing. J Gene Med. 2007;9(8):727–738. doi: 10.1002/jgm.1061. [DOI] [PubMed] [Google Scholar]
- 35.Samejima K, Earnshaw WC. Trashing the genome: The role of nucleases during apoptosis. Nat Rev Mol Cell Biol. 2005;6(9):677–688. doi: 10.1038/nrm1715. [DOI] [PubMed] [Google Scholar]
- 36.Yuan Y, Tan J, Wang Y, Qian C, Zhang M. Chitosan nanoparticles as non-viral gene delivery vehicles based on atomic force microscopy study. Acta Biochim Biophys Sin (Shanghai) 2009;41(6):515–526. doi: 10.1093/abbs/gmp038. [DOI] [PubMed] [Google Scholar]
- 37.Han G, Martin CT, Rotello VM. Stability of gold nanoparticle-bound DNA toward biological, physical, and chemical agents. Chem Biol Drug Des. 2006;67(1):78–82. doi: 10.1111/j.1747-0285.2005.00324.x. [DOI] [PubMed] [Google Scholar]
- 38.Kabbaj M, Phillips NC. Anticancer activity of mycobacterial DNA: Effect of formulation as chitosan nanoparticles. J Drug Target. 2001;9(5):317–328. doi: 10.3109/10611860108998768. [DOI] [PubMed] [Google Scholar]
- 39.Su X, et al. Simultaneous fluorescence imaging of the activities of DNases and 3′ exonucleases in living cells with chimeric oligonucleotide probes. Anal Chem. 2013;85(20):9939–9946. doi: 10.1021/ac402615c. [DOI] [PubMed] [Google Scholar]
- 40.Li F, Zhang H, Dever B, Li X-F, Le XC. Thermal stability of DNA functionalized gold nanoparticles. Bioconjug Chem. 2013;24(11):1790–1797. doi: 10.1021/bc300687z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dougan JA, Karlsson C, Smith WE, Graham D. Enhanced oligonucleotide-nanoparticle conjugate stability using thioctic acid modified oligonucleotides. Nucleic Acids Res. 2007;35(11):3668–3675. doi: 10.1093/nar/gkm237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Z, Jin R, Mirkin CA, Letsinger RL. Multiple thiol-anchor capped DNA-gold nanoparticle conjugates. Nucleic Acids Res. 2002;30(7):1558–1562. doi: 10.1093/nar/30.7.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iciek M, Chwatko G, Lorenc-Koci E, Bald E, Włodek L. Plasma levels of total, free and protein bound thiols as well as sulfane sulfur in different age groups of rats. Acta Biochim Pol. 2004;51(3):815–824. [PubMed] [Google Scholar]
- 44.Toohey JI. Sulphane sulphur in biological systems: A possible regulatory role. Biochem J. 1989;264(3):625–632. doi: 10.1042/bj2640625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chao J, Lin Y, Liu H, Wang L, Fan C. DNA-based plasmonic nanostructures. Mater Today. 2015;18(6):326–335. [Google Scholar]
- 46.Chen Y-J, Groves B, Muscat RA, Seelig G. DNA nanotechnology from the test tube to the cell. Nat Nanotechnol. 2015;10(9):748–760. doi: 10.1038/nnano.2015.195. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.