Significance
Plants require precise control over growth regulators during development and in their responses to biotic and abiotic stresses. One strategy for modulating levels of bioactive phytohormones is to conjugate these molecules to amino acids using acyl acid amido synthetases of the Gretchen Hagen 3 (GH3) protein family. Typically, GH3 proteins modify one type of phytohormone. Structural studies, along with in vitro and in planta biochemical analyses, reveal that the GH3.5 protein from the model plant Arabidopsis thaliana conjugates multiple molecules from various phytohormone pathways. This activity mediates crosstalk between auxin developmental and pathogen response pathways.
Keywords: protein structure, plant hormone, Arabidopsis, plant biochemistry, auxin
Abstract
In Arabidopsis thaliana, the acyl acid amido synthetase Gretchen Hagen 3.5 (AtGH3.5) conjugates both indole-3-acetic acid (IAA) and salicylic acid (SA) to modulate auxin and pathogen response pathways. To understand the molecular basis for the activity of AtGH3.5, we determined the X-ray crystal structure of the enzyme in complex with IAA and AMP. Biochemical analysis demonstrates that the substrate preference of AtGH3.5 is wider than originally described and includes the natural auxin phenylacetic acid (PAA) and the potential SA precursor benzoic acid (BA). Residues that determine IAA versus BA substrate preference were identified. The dual functionality of AtGH3.5 is unique to this enzyme although multiple IAA-conjugating GH3 proteins share nearly identical acyl acid binding sites. In planta analysis of IAA, PAA, SA, and BA and their respective aspartyl conjugates were determined in wild-type and overexpressing lines of A. thaliana. This study suggests that AtGH3.5 conjugates auxins (i.e., IAA and PAA) and benzoates (i.e., SA and BA) to mediate crosstalk between different metabolic pathways, broadening the potential roles for GH3 acyl acid amido synthetases in plants.
Plants adjust to environmental and developmental cues through control of phytohormone networks. Metabolism of growth regulators during plant development is critical for establishing active hormone responses and biological effects. The tight control of phytohormone levels occurs not only through their synthesis and degradation but also via conjugation to a variety of different molecules (1). One important form of conjugation is the attachment of amino acids to carboxylic acid-containing hormones via formation of an amide bond (2). The roles of the amino acid-conjugated phytohormones vary depending on the hormone and the amino acid. For example, conjugation of isoleucine to jasmonic acid (JA) leads to the formation of the bioactive form of the hormone (i.e., JA-Ile) (3–5), whereas conjugation of amino acids to indole-3-acetic acid (IAA) leads to the inactivation of the major auxin plant growth regulator (6). In addition, the amino acid attached to IAA changes the fate of the conjugate. Attachment of either aspartate or glutamate leads to degradation of IAA (6). Unlike IAA-Asp and IAA-Glu, the conjugates IAA-Ala and IAA-Val can be hydrolyzed back to the active free IAA form and provide inactive storage forms of the hormone (6).
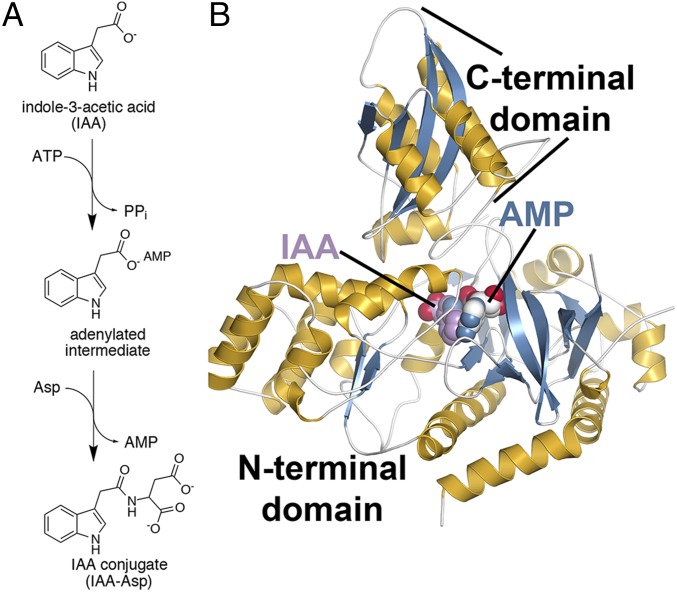
Gretchen Hagen 3 (GH3) acyl acid amido synthetases contribute to regulating levels of phytohormones, including JA and IAA. These proteins modulate the phytohormone signaling pathways responsible for plant growth, seed development, light signaling, drought response, and pathogen resistance (3, 7–9). The GH3 enzymes belong to the acyl-CoA synthetase, nonribosomal peptide synthetase, and luciferase (ANL) superfamily (10). ANL enzymes work through a two-step reaction mechanism with an adenylated intermediate (Fig. 1A). For the GH3 proteins, the first step involves adenylation of an acyl acid hormone and release of pyrophosphate (11, 12). The second transferase step has the amine group of an amino acid nucleophilically displace AMP, yielding the conjugated acyl acid. X-ray crystal structures of two GH3 proteins from Arabidopsis thaliana, i.e., the JA-Ile biosynthesis enzyme AtGH3.11/JAR1 (9) and the benzoate-using AtGH3.12/PBS3 (9), and an auxin-specific grape GH3 protein VvGH3.1 (13) provided the first molecular insights into how these proteins conjugate amino acids to diverse acyl acid substrates. These structures and small-angle X-ray scattering analysis show that conformational changes in the C-terminal domain toggles the active site between open (ATP-bound) and closed (AMP-bound) forms during the adenylation and transferase half-reactions, respectively (9, 14). Subsequent biochemical studies delineated the roles of active site residues in each half-reaction (9).
Fig. 1.
Reaction and 3D structure of AtGH3.5. (A) Overall reaction catalyzed by AtGH3.5 and other IAA-conjugating acyl acid amido synthetases. (B) 3D structure of AtGH3.5. The ribbon diagram shows the N- and C-terminal domains with α-helices (gold) and β-strands (blue). Ligands are shown as space-filling models.
In Arabidopsis, six GH3 proteins, AtGH3.1 (7), AtGH3.2/YDK1 (15), AtGH3.5/WES1 (16, 17), AtGH3.6/DFL1 (18), AtGH3.9 (19), and AtGH3.17/VAS2 (20), are linked to IAA activity in plant growth and development. Of these proteins, AtGH3.5 is also implicated in salicylic acid (SA)-linked pathogen responses. Studies with the loss-of-function mutant wes1 and two gain-of-function mutants wes1-D and gh3.5-1D indicate that AtGH3.5 contributes to both IAA and SA responses (16, 17, 21–23). The gain-of-function lines displayed both low auxin phenotypes along with increased disease resistance (16, 17). Additionally, either IAA or SA treatment induced gh3.5 gene expression (16, 17). Initial biochemical studies suggested that AtGH3.5, along with the other GH3 proteins that accepted IAA as a substrate, also could adenylate other auxins, including phenylacetic acid (PAA) and indole-3-butyric acid, but, unlike other Arabidopsis GH3 proteins, AtGH3.5 also adenylated SA (7).
We report the 3D structure of AtGH3.5, which suggests the molecular basis for its dual IAA and SA activity and its ability to impact both auxin and SA homeostasis. Kinetic analysis demonstrates that the substrate preference of AtGH3.5 is wider than originally described. The dual functionality of AtGH3.5 is unique to this enzyme, even though multiple IAA-conjugating GH3 proteins share nearly identical acyl acid binding sites. In vitro and in planta analyses suggest that AtGH3.5 conjugates multiple auxins and modulates levels of SA and the SA precursor benzoic acid (BA) in Arabidopsis. These data broaden the roles that GH3 acyl acid amido synthetases play in plant metabolism and suggest that AtGH3.5 mediates metabolic crosstalk between the auxin and SA response pathways.
Results
3D Structure of AtGH3.5.
Previous work described AtGH3.5 as a dual-function protein that accepts IAA and SA as substrates and is involved in auxin- and SA-mediated plant responses (7, 16, 17, 21–23). To understand the molecular basis of the substrate specificity of AtGH3.5, we determined the X-ray crystal structure of this protein with IAA and AMP bound (Fig. 1B and Table S1). Comparison of AtGH3.5 with other GH3 protein structures using DALI reveals the highest similarity with VvGH3.1 (7) (PDB ID code 4B2G; Z = 59.8; 0.9 Å2 rmsd for 552 Cα atoms; 68% identity), AtGH3.12 (9, 14) (PDB: 4EG4; 1.8 Å rmsd for 545 Cα atoms; 50% identity), and AtGH3.11 (9) (PDB: 4EPL; 3.7 Å rmsd for 546 Cα atoms; 40% identity). The GH3 protein structure, like that of other adenylating enzymes, is defined by a large (∼450 aa) N-terminal α/β fold domain that provides a platform for ligand binding and a smaller (∼160 aa) C-terminal domain that is conformationally flexible (9, 14). The C-terminal domain centers on a four-stranded β-sheet flanked by two pairs of α-helices and can adopt two conformations that differ by a 180° rotation. This rotation allows different sets of residues to interact with substrates during the adenylation and transferase half-reactions (9). The C-terminal domain of the AtGH3.5 structure in complex with AMP and IAA reported here adopts the closed active site conformation associated with the second half-reaction.
Table S1.
Summary of crystallographic statistics: AtGH3.5•AMP•IAA
| Data collection | |
| Space group | P21 |
| Cell dimensions | a = 91.61 Å, b = 143.5 Å, c = 102.3 Å; β = 114.7° |
| Wavelength, Å | 0.979 |
| Resolution, Å (highest shell) | 42.2–2.20 (2.25–2.20) |
| Reflections, total/unique | 254,788/108,566 |
| Completeness, % (highest shell) | 89.5 (77.3) |
| <I/σ> (highest shell) | 11.2 (2.0) |
| Rsym, % (highest shell) | 9.3 (51.8) |
| Refinement | |
| Rcryst/Rfree, % | 20.5/24.5 |
| No. of protein atoms | 18,186 |
| No. of waters | 1,002 |
| No. of ligand atoms | 154 |
| Rmsd, bond lengths | 0.008 |
| Rmsd, bond angles, degrees | 1.167 |
| Average B-factor, Å2: protein, water, ligand | 30.7, 28.3, 26.4 |
| Stereochemistry: most favored, allowed, disallowed, % | 97.3, 2.5, 0.2 |
AtGH3.5 Nucleotide and Acyl Acid Binding Sites.
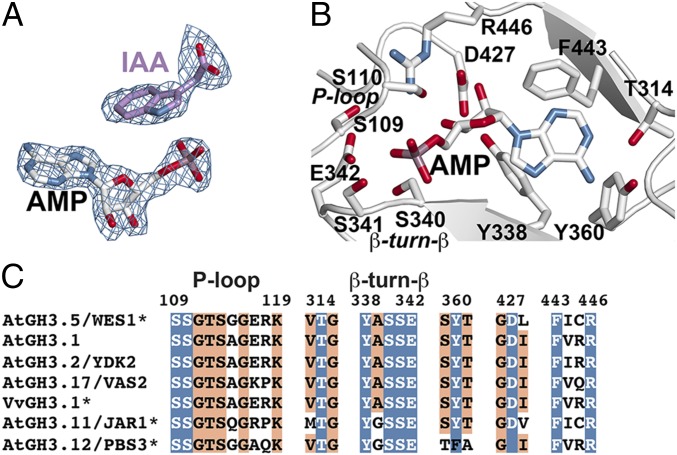
Clear electron density for AMP and IAA in the AtGH3.5 structure (Fig. 2A) defines the location of the active site along the interface of the N- and C-terminal domains (Fig. 1B). Within the active site, a canonical P-loop (residues 109–119), the β8-turn-β9 loop (residues 338–342), and multiple residues that surround the adenine ring contribute to nucleotide binding (Fig. 2B). The architecture of the AMP binding site is nearly identical to those described for previously reported GH3 protein structures (9, 13, 14). Sequence comparison of AtGH3.5 with the IAA-conjugating AtGH3.1, AtGH3.2, AtGH3.17, and VvGH3.1, the JA-Ile biosynthesis enzyme AtGH3.11, and the benzoate-conjugating AtGH3.12 emphasizes the similarity of this site across GH3 proteins that recognize diverse acyl acid substrates (Fig. 2C).
Fig. 2.
AtGH3.5 nucleotide binding site. (A) Electron density for IAA and AMP is shown as a 2Fo-Fc omit map (1.5 σ). (B) AMP and nucleotide binding site residues are shown as stick models. Regions corresponding to the P-loop and the β-turn-β motif are indicated. (C) Targeted sequence comparison of functionally characterized GH3 proteins from Arabidopsis and grape (VvGH3.1). Residues of the P-loop and the β-turn-β motif are noted. Numbering corresponds to AtGH3.5. Residues with side chains shown in B are colored in blue with white text. Other conserved positions are highlighted in orange. Asterisks indicate proteins with 3D structures (9, 13).
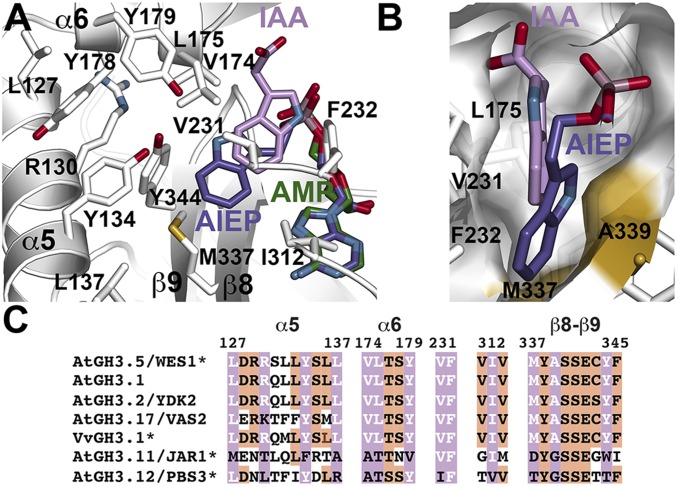
The position of IAA defines the acyl acid binding site of AtGH3.5 as a pocket formed by multiple residues from α5, α6, β8, and β9 (Fig. 3A). Leu127, Arg130, Tyr134, and Leu137 from α5 form the back wall of the site. Val174, Leu175, and Tyr179 from α6, Met337 from β8, and Tyr344 from β9 sandwich the indole ring of IAA with Val231, Phe232, and Ile312 providing additional van der Waals contacts. The carboxylate group of IAA is oriented toward α6 and away from the phosphate group of AMP. This orientation would be a catalytically nonproductive binding orientation and likely results from charge repulsion between the negatively charged carboxylate of IAA and the phosphate of AMP. Peat et al. (13) reported the structure of VvGH3.1 in complex with an inhibitor (adenosine-5′-[2-(1H-indol-3-yl)ethyl]phosphate; AIEP) that mimics the IAA reaction intermediate. An overlay of VvGH3.1 and AtGH3.5 highlights the similarity in IAA binding (Fig. 3 A and B). For clarity, the protein structure of VvGH3.1 is not shown, but, as discussed below, the acyl acid binding site of VvGH3.1 is nearly identical to that of AtGH3.5. The positions of AMP and the AMP-derived portion of AIEP in the two structures are nearly superimposable, but the indole ring of AIEP in VvGH3 binds deeper in the active site toward Met337 and Ala339 than in IAA.
Fig. 3.
AtGH3.5 acyl acid binding site. (A) Residues in the acyl acid binding site of AtGH3.5 are shown as stick drawings. IAA (pink) and AMP (green) in the AtGH3.5 structure are indicated. For comparison, the position of the adenylated intermediate mimic AIEP (purple) from the VvGH3.1 crystal structure (13) is shown. (B) Comparison of IAA and AIEP from the AtGH3.5 and VvGH3.1 structures, respectively. The surface corresponds to AtGH3.5 with the positions of Met337 and Ala339, residues that alter IAA versus BA preference, colored in gold. (C) Targeted sequence comparison of functionally characterized GH3 proteins from Arabidopsis and grape. Residues of α5, α6, and β8-turn-β9 are noted. Numbering corresponds to AtGH3.5. Residues with side chains shown in A are colored in pink with white text. Other conserved positions are highlighted in orange. Asterisks indicate proteins with 3D structures (9, 13).
The acyl acid binding sites of the IAA-conjugating GH3 proteins from Arabidopsis that have been biochemically characterized to date (i.e., AtGH3.5, AtGH3.1, AtGH3.2, and AtGH3.17) and VvGH3.1 are nearly invariant (Fig. 3C). Variations occur in residues along α5, with AtGH3.17 having the largest number of differences. Comparison of AtGH3.5 with other GH3 proteins highlights multiple differences throughout the acyl acid binding site (Fig. S1). The varied acyl acid binding sites accommodate different substrates for a common chemical reaction. For example, the AtGH3.5 acyl acid site (Fig. S1A) is more constrained than the same region of AtGH3.11 (Fig. S1B). Sequence differences in AtGH3.11 deepen the active site to provide space for the oxylipin tail of JA. Although a physiological substrate for AtGH3.12 remains to be identified (9), binding of the inhibitor SA in the acyl acid site suggests that a larger, but as yet unidentified, molecule may be the substrate of this protein (Fig. S1C).
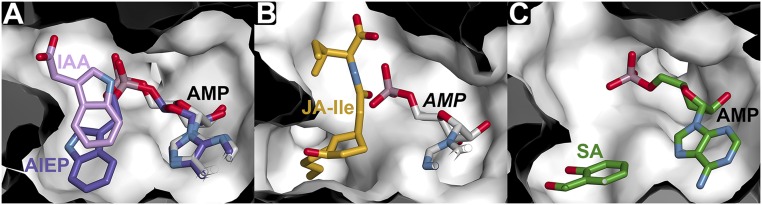
Fig. S1.
Comparison of GH3 protein acyl acid binding sites. Surface views of the acyl acid binding sites of AtGH3.5 (A), AtGH3.11 (B), and AtGH3.12 (C) highlight the structural difference in each enzyme. Each view is oriented to show the binding of acyl acids relative to the nucleotide site. (A) AtGH3.5 active site. The positions of IAA (pink) and AMP (white) were determined crystallographically (Figs. 2 and 3) with the position of AIEP (purple) from the VvGH3.1 structure (13) overlaid on the AtGH3.5•AMP•IAA complex structure. (B) AtGH3.11 active site. Binding of JA-Ile in the crystal structure reveals a larger active site to accommodate the oxylipin moiety of JA (9). The location of AMP is modeled based on the AtGH3.5 structure and is shown only for reference. (C) The AtGH3.12 active site. The crystallographically determined positions of SA, an inhibitor of AtGH3.12, and AMP (both ligands in green) are shown (9).
AtGH3.5 Amino Acid and Acyl Acid Specificity.
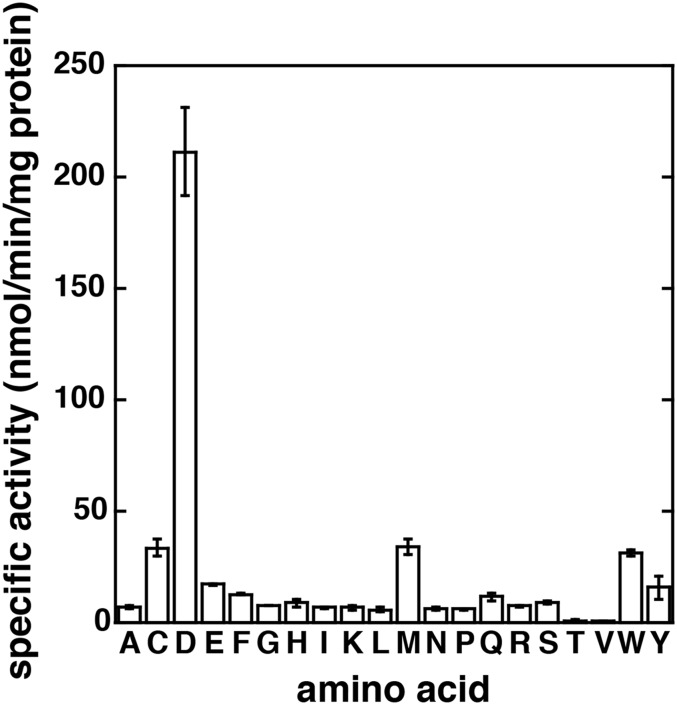
A spectrophotometric assay was used to quantify the substrate specificity of AtGH3.5. AtGH3.5 displayed a clear preference for aspartate (211 ± 20 nmol⋅min−1⋅mg protein−1) as the amino acid substrate but also had appreciable rates for cysteine (33 ± 4 nmol⋅min−1⋅mg protein−1), glutamate (17 ± 1 nmol⋅min−1⋅mg protein−1), methionine (34 ± 4 nmol⋅min−1 mg⋅protein−1), and tryptophan (31 ± 1 nmol⋅min−1⋅mg protein−1) (Fig. S2). AtGH3.5 was active with a range of auxin and benzoate substrates (Table S2). With the natural auxins IAA and PAA, AtGH3.5 had comparable catalytic efficiencies (kcat/Km). The synthetic auxin naphthaleneacetic acid (NAA) was a substrate but had a twofold lower catalytic efficiency than IAA. Surprisingly, SA was a poor substrate with a 17-fold lower kcat/Km than IAA. In contrast, conjugation of BA occurred with a catalytic efficiency 18-fold higher than that of SA but similar to that of IAA. The nonphysiological substrate 4-hydroxybenzoic acid (4-HBA), previously used to monitor the activity of AtGH3.12 (9), showed a kcat/Km similar to that of NAA. No activity was detected with JA.
Fig. S2.
Amino acid screen of AtGH3.5. Activity assays of AtGH3.5 with 1 mM IAA, 1 mM ATP, and 5 mM amino acid. Each assay was performed in triplicate; data are shown as mean ± SD.
Table S2.
AtGH3.5 acyl acid specificity
| Substrate | kcat, min−1 | Km, μM | kcat/Km, M−1⋅s−1 |
| IAA | 14.5 ± 1.4 | 770 ± 110 | 314 |
| PAA | 28.0 ± 2.0 | 1,840 ± 233 | 253 |
| NAA | 2.3 ± 0.3 | 220 ± 70 | 174 |
| SA | 0.8 ± 0.1 | 700 ± 130 | 19 |
| BA | 16.0 ± 0.8 | 790 ± 60 | 338 |
| 4-HBA | 2.5 ± 0.3 | 300 ± 120 | 139 |
Average values ± SD (n = 3–5) are shown. Assays were performed as described with fixed concentrations of ATP (1 mM) and Asp (5 mM) and with varied concentrations of acyl acid substrates (up to 15 mM depending on the Km value).
AtGH3.5: IAA Versus BA Specificity.
To probe IAA versus BA specificity, a series of point mutants (Y134W, V174L, L175F, V231L, F232W, M337L, A339V, and Y344W) were generated by PCR to introduce changes that reduced the size of the acyl acid binding site pocket. All the mutants except L175F and F232W were expressed and purified as soluble monomeric proteins. Kinetic parameters of the Y134W, V174L, V231L, M337L, A339V, and Y344W mutants were determined using IAA and BA as substrates (Table S3). All the mutants displayed decreased catalytic efficiency with IAA compared with the wild-type enzyme. The Y134W, V174L, V213L, and Y344W substitutions also reduced kcat/Km with BA by 9- to 43-fold. Although the M337L and A339V mutants were less efficient with IAA, both retained comparable kcat/Km using BA as a substrate versus wild-type. Thus, both mutants showed a three- to fourfold preference for BA over IAA. Given the positions of Met337 and Ala339 in the acyl acid site (Fig. 3B), the longer side chains of M337L and A339V likely sterically occlude the indole ring of IAA compared with the phenyl ring of BA.
Table S3.
Steady-state kinetics of AtGH3.5 acyl binding site mutants
| Protein | kcat IAA, min−1 | KmIAA, μM | kcat/KmIAA, M−1⋅s−1 | kcat BA, min−1 | KmBA, μM | kcat/KmBA, M−1⋅s−1 |
| AtGH3.5 | 14.5 ± 1.4 | 770 ± 110 | 314 | 16.0 ± 0.8 | 790 ± 60 | 338 |
| Y134W | 3.7 ± 0.3 | 3,060 ± 310 | 20 | 7.7 ± 0.4 | 6,680 ± 700 | 19 |
| V174L | 7.2 ± 2.8 | 6,720 ± 330 | 18 | 3.1 ± 0.3 | 7,910 ± 1,250 | 7 |
| V231L | 22.0 ± 5.4 | 4,380 ± 126 | 84 | 9.4 ± 0.3 | 4,120 ± 270 | 38 |
| M337L | 13.5 ± 1.5 | 2,930 ± 470 | 77 | 100 ± 47 | 5,650 ± 310 | 295 |
| A339V | 18.0 ± 5.1 | 3,770 ± 130 | 80 | 21.0 ± 0.7 | 1,320 ± 90 | 265 |
| Y344W | 4.8 ± 1.3 | 3,600 ± 100 | 21 | 6.1 ± 0.6 | 4,550 ± 620 | 22 |
AtGH3.5 values are from Table S2 and are shown for comparison with the mutant enzymes. Average values ± SD (n = 3–5) are shown for steady-state kinetic parameters. Assays were performed as described with fixed concentrations of ATP (1 mM) and Asp (5 mM) and with varied concentrations of IAA and BA.
IAA and BA Specificity in Other Arabidopsis GH3 Proteins.
Sequence comparison of AtGH3.5 with other IAA-conjugating GH3 proteins (Fig. 3C) suggests that the acyl acid binding site is highly conserved. To test if the substrate profile of AtGH3.5 is similar to other IAA-conjugating GH3 proteins from Arabidopsis, AtGH3.1, AtGH3.2, and AtGH3.17 were expressed in Escherichia coli and were purified for kinetic analysis. AtGH3.6 and AtGH3.9 were also cloned but were not assayed because of protein stability issues. AtGH3.1, AtGH3.2, and AtGH3.17 favored IAA over BA as a substrate by 14-, 70-, and 50-fold, respectively (Table S4). These three enzymes were not active with SA using the spectrophotometric assay. Each enzyme also conjugated PAA with varying efficiency (Table S4). Although the acyl acid sites of the IAA-conjugating GH3 proteins from Arabidopsis are highly conserved, there are distinct preferences for the acyl acid substrate.
Table S4.
IAA and BA kinetics of IAA-conjugating GH3 proteins
| Kinetic parameter | AtGH3.5 | AtGH3.1 | AtGH3.2 | AtGH3.17 |
| kcat IAA, min−1 | 14.5 ± 1.4 | 5.7 ± 0.6 | 17.0 ± 1.6 | 2.7 ± 0.1 |
| KmIAA, μM | 770 ± 110 | 530 ± 150 | 510 ± 105 | 68.0 ± 6.0 |
| kcat/KmIAA, M−1⋅s−1 | 314 | 179 | 556 | 662 |
| kcat PAA, min−1 | 28.0 ± 2.0 | 4.8 ± 0.8 | 19.5 ± 0.5 | 3.0 ± 0.1 |
| KmPAA, μM | 1,840 ± 233 | 3,075 ± 760 | 2,525 ± 174 | 268 ± 7.8 |
| kcat/KmPAA, M−1⋅s−1 | 253 | 26 | 125 | 187 |
| kcat BA, min−1 | 16.0 ± 0.8 | 2.7 ± 0.7 | 2.2 ± 0.6 | 0.87 ± 0.08 |
| KmBA, μM | 790 ± 60 | 3,350 ± 1,340 | 4,760 ± 285 | 1,090 ± 222 |
| kcat/KmBA, M−1⋅s−1 | 338 | 13 | 77 | 13 |
AtGH3.5 values are from Table S2 and are shown for comparison with other A. thaliana GH3 proteins. Average values ± SD (n = 3–5) are shown for steady-state kinetic parameters. Assays were performed as described with fixed concentrations of ATP (1 mM) and Asp (10 mM for AtGH3.5 and AtGH3.2), Asn (10 mM for AtGH3.1), or Glu (10 mM for AtGH3.17) and with varied concentrations of IAA, PAA, or BA (up to 10 x Km value).
AtGH3.5: In Planta Conjugation of IAA, PAA, SA, and BA.
Earlier studies of AtGH3.5 in A. thaliana used an activation tagging line and focused on IAA homeostasis and pathogen-related responses (16, 17). Given the substrate preference of AtGH3.5 (Table S2), its in vivo function was reexamined. AtGH3.5 was expressed as an N-terminal FLAG-tagged protein under control of the 35S promoter in Arabidopsis. Independent T2 lines (OE1–4) were used for analysis of free and conjugated IAA, PAA, SA, and BA. The overexpression lines displayed the same severely dwarfed phenotype (Fig. S3A) described for the activation-tagged wes1-D line (16, 17). Immunoblot analysis confirmed the expression of FLAG-tagged AtGH3.5 protein in each line (Fig. S3B).
Fig. S3.
AtGH3.5-overexpressing plants. (A) Comparison of 1-mo-old A. thaliana wild-type Col-0 rosettes (Left) and 1-mo-old 35S:Atgh3.5 rosettes for overexpressing lines OE1–4 (Right). (B) Immunoblot analysis of AtGH3.5::FLAG (transgenic lines OE1–4) expression in transgenic A. thaliana plants. Wild-type A. thaliana Col-0 is included as a negative control. The molecular weight (MW) ladder is shown. Equal amounts (20 μg) of total soluble protein extracted from 1-mo-old leaf tissues were separated by SDS/PAGE, electrotransferred, and probed with anti-FLAG M2 antibody to detect FLAG-tagged proteins.
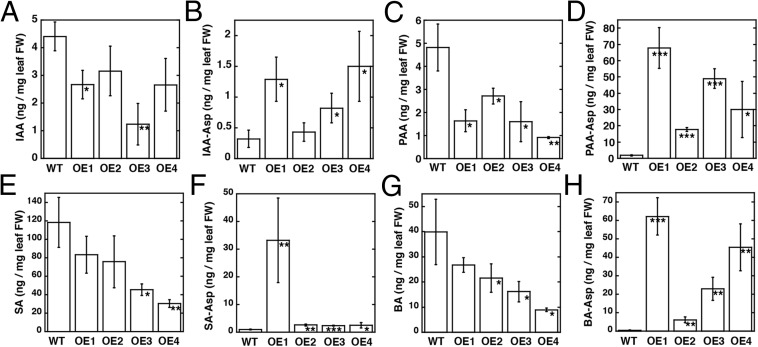
Mass spectrometry determined free and aspartyl-conjugated IAA, PAA, SA, and BA levels in leaves from A. thaliana AtGH3.5-overexpressing lines (Fig. 4 and Table S5). Purified AtGH3.5 was used to generate PAA-Asp, BA-Asp, and SA-Asp for quantification by mass spectrometry using multiple reaction monitoring; other metabolites were commercially available (Fig. S4 and Table S6). AtGH3.5 overexpression resulted in approximately twofold lower IAA and up to sixfold higher IAA-Asp than in wild type (Fig. 4 A and B). More striking results were observed with PAA and PAA-Asp (Fig. 4 C and D). Decreased levels of PAA (up to fivefold) and 15- to 70-fold higher accumulation of PAA-Asp than in wild-type plants were observed. In vitro assays of AtGH3.5 indicate that the enzyme is also active on SA and BA but favors BA (Table S2). As with IAA and PAA, overexpression of AtGH3.5 alters the levels of benzoates. SA levels varied in the overexpression lines but were generally lower than in wild-type plants (Fig. 4E). These changes corresponded with varied levels of SA-Asp in the overexpressing lines (Fig. 4F). One line (OE1) showed 30-fold elevation in SA-Asp, but the other lines displayed approximately threefold increases in the conjugate. Levels of BA were moderately reduced (Fig. 4G) but showed up to 60-fold higher BA-Asp (Fig. 4H).
Fig. 4.
Levels of free acyl acids and acyl acid conjugates in wild-type and AtGH3.5-overexpressing lines of Arabidopsis thaliana. Levels of IAA (A), IAA-Asp (B), PAA (C), PAA-Asp (D), SA (E), SA-Asp (F), BA (G), and BA-Asp (H) from 1-mo-old leaf tissue of wild-type A. thaliana and four independently transformed AtGH3.5-overexpressing lines (OE1–4). Metabolite levels are in nanograms per mg fresh weight (FW) leaf tissue with mean ± SD (n = 12–18). *P < 0.01, **P < 0.001, ***P < 0.0001 versus wild type.
Table S5.
Free acyl acids and acyl acid conjugate levels in wild-type and AtGH3.5-overexpressing lines of A. thaliana
| Metabolite | WT | OE1 | OE2 | OE3 | OE4 |
| IAA | 4.41 ± 0.52 | 2.67 ± 0.51 | 3.16 ± 0.90 | 1.23 ± 0.75 | 2.66 ± 0.95 |
| IAA-Asp | 0.32 ± 0.14 | 1.29 ± 0.26 | 0.43 ± 0.15 | 0.82 ± 0.24 | 1.50 ± 0.57 |
| PAA | 4.82 ± 1.02 | 1.64 ± 0.48 | 2.71 ± 0.34 | 1.60 ± 0.86 | 0.91 ± 0.04 |
| PAA-Asp | 1.84 ± 0.35 | 67.7 ± 12.5 | 17.7 ± 1.1 | 49.0 ± 6.1 | 29.9 ± 17.2 |
| SA | 118 ± 27 | 83.2 ± 20.0 | 75.6 ± 28 | 45.4 ± 6.3 | 30.4 ± 4.0 |
| SA-Asp | 0.98 ± 0.08 | 33.1 ± 15.3 | 2.62 ± 0.34 | 2.37 ± 0.20 | 2.55 ± 0.96 |
| BA | 39.8 ± 13.0 | 26.8 ± 2.9 | 21.5 ± 5.6 | 16.1 ± 4.1 | 8.94 ± 0.70 |
| BA-Asp | 0.49 ± 0.14 | 62.1 ± 10.1 | 6.12 ± 1.60 | 22.9 ± 6.3 | 45.5 ± 12.7 |
Metabolite levels from 1-mo-old leaf tissue of wild-type A. thaliana and four independently transformed AtGH3.5-overexpressing lines (OE1–4) are reported as nanograms per milligram fresh weight (FW) leaf tissue and are shown as mean ± SD (n = 12–18).
Fig. S4.
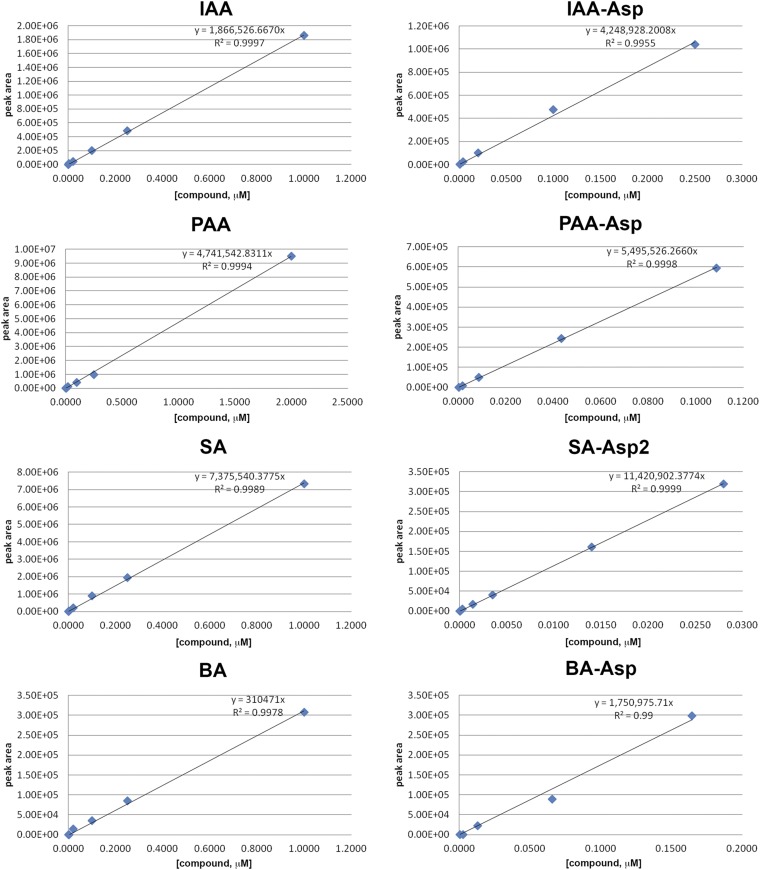
Standard curves for mass spectrometry of free and conjugated acyl acids. Using the first MRM transitions listed in Table S4 a standard curve was generated for all compounds but SA-Asp. All compounds showed a linear response. SA-Asp had a better standard curve with the second MRM transition.
Table S6.
Optimized MRM parameters for analysis of free and conjugated acyl acids
| Compound | Q1, m/z | Q3, m/z | DT, ms | DP | CE | CXP |
| IAA1 | 174 | 130 | 30 | −35 | −15 | −9 |
| IAA2 | 174 | 128 | 30 | −35 | −28 | −9 |
| IAA‐Asp1 | 289 | 132 | 30 | −55 | −24 | −10 |
| IAA‐Asp2 | 289 | 88 | 30 | −55 | −35 | −10 |
| PAA1 | 135 | 91 | 30 | −23 | −12 | −5 |
| PAA2 | 135 | 45 | 30 | −23 | −16 | −5 |
| PAA‐Asp1 | 250 | 132 | 30 | −48 | −23 | −9 |
| PAA‐Asp2 | 250 | 88 | 30 | −48 | −34 | −9 |
| SA1 | 137 | 93 | 30 | −45 | −25 | −8 |
| SA2 | 137 | 65 | 30 | −45 | −40 | −8 |
| SA‐Asp1 | 252 | 215.4 | 30 | −31 | −13 | −10 |
| SA‐Asp2 | 252 | 136 | 30 | −45 | −24 | −8 |
| BA1 | 121 | 77 | 30 | −40 | −16 | −4 |
| BA2 | 121 | 93 | 30 | −40 | −20 | −4 |
| BA‐Asp1 | 236 | 120 | 30 | −40 | −25 | −4 |
| BA‐Asp2 | 236 | 192 | 30 | −40 | −20 | −4 |
CE, collision energy; CXP, collision cell exit potential; DP, declustering potential of turbo ion spray source; DT, dwell time monitoring each MRM transition; Q1, selected m/z of the first quadrupole; Q3, selected m/z of the third quadrupole.
Discussion
GH3 proteins catalyze the conjugation of amino acids with acyl acids to regulate levels of active and inactive forms of JA and IAA (2, 3, 7, 8). Investigations of the biological action of GH3 proteins from a variety of plants have focused largely on JA and IAA; however, biochemical and structural studies suggest that the functional diversity of the GH3 protein family may be wider than previously understood. Of the 19 GH3 proteins in A. thaliana, one is required for JA-Ile biosynthesis, six are associated with conjugation and inactivation of IAA, and the rest either lack a physiological substrate or remain to be characterized (3, 7, 15–20). Of these proteins, AtGH3.5 was proposed to have a dual role in IAA and SA homeostasis (16, 17). Our analyses of AtGH3.5 support this hypothesis but also suggest an even broader role for this enzyme in the metabolism of auxins and benzoates.
To understand better the molecular basis for the substrate preference of AtGH3.5, the X-ray crystal structure was determined for comparison with other structurally characterized GH3 acyl acid amido synthetases. The structure of AtGH3.5 (Fig. 1B) shares a common ANL superfamily scaffold and a nucleotide binding site nearly invariant with other GH3 proteins (Fig. 2). The conservation of this site, which includes residues with catalytic roles in the adenylation and transferase reactions (9), indicates a common reaction mechanism for the GH3 proteins.
The structure of AtGH3.5 highlights the differences in the acyl acid binding site compared with other structurally resolved GH3 proteins (Fig. S1). Early sequence comparisons led to the proposal that GH3 proteins form three subfamilies with substrate preferences for JA, IAA, and “other” (3, 7, 24). Later, structural studies of AtGH3.11 and AtGH3.12 and comparison of residues forming the acyl acid binding site suggested up to eight distinct subfamilies, many of which include proteins that remain to be characterized (9). There are clear differences in the size and shape of the phytohormone binding sites in AtGH3.5 and VvGH3.1, both of which conjugate IAA, versus AtGH3.11 from JA-Ile biosynthesis and AtGH3.12, which is implicated in SA action through an unidentified substrate (Fig. S1) (9, 25). Additional studies will elucidate the biochemical and physiological roles of other GH3 proteins with distinct acyl acid binding sites and determine whether they also exhibit broader specificity.
Protein crystallography of AtGH3.5 and VvGH3.1 (13) revealed the structural conservation of the IAA-conjugating GH3 proteins (Fig. 3). The C-terminal domain in each structure adopts the closed active-site conformation that opens the amino acid binding site to allow the transferase step of the reaction sequence. The orientation of IAA in the AtGH3.5 structure differs slightly from the adenylated intermediate analog AIEP in the VvGH3.1 structure (Fig. 3 A and B). During the reaction catalyzed by AtGH3.5, IAA likely binds deeper in the acyl site in a position analogous to the indole-ring of AIEP and with the carboxylate oriented toward the phosphates of ATP for the chemistry of the first half-reaction. A Mg2+ ion, which is catalytically essential but crystallographically unresolved, in the active site would provide the counter ion for the adenylation reaction (9).
Within the AtGH3.5 active site, binding of AMP and IAA identifies residues forming the acyl acid binding site, which accepts a range of substrates (Table S2), including the natural auxins IAA and PAA, along with the synthetic auxin NAA. SA is a substrate for AtGH3.5 (7) but is a poor one in comparison with the auxins and BA. Mutagenesis of residues in the AtGH3.5 acyl acid binding site and kinetic analysis (Table S3) suggest that the benzoate substrates are accommodated similar to the auxins, because most mutants altered kinetic parameters for both IAA and BA (Table S3). The steady-state kinetic parameters of the mutants also suggests that the larger substrates fit into the pocket defined by Met337 and Ala339 (Fig. 3C), because the M337L and A339V substitutions did not change the kinetics with BA significantly but altered IAA preference (Table S3). These studies provide a starting point for comparisons with other documented IAA-conjugated GH3 proteins and GH3 proteins that modify other phytohormones and will help resolve the mystery of IAA specificity versus the dual IAA/BA specificity exhibited by AtGH3.5.
The residues contacting either IAA in the AtGH3.5 structure or the indole moiety of AIEP in the VvGH3.1 structure are identical and also are nearly invariant in other IAA-conjugating GH3 proteins (Fig. 3 A and C). Biochemical comparison of AtGH3.5, AtGH3.1, AtGH3.2, and AtGH3.17 reveal commonalities, but also key differences, in substrate preferences (Table S3). Although all these enzymes prefer IAA, PAA is not as efficient a substrate with AtGH3.1 as the other enzymes. Only AtGH3.5 shows comparable kcat/Km values for both IAA and BA; the rest distinctly prefer IAA. Also, AtGH3.5 was the only GH3 tested that was active with SA. Given the common acyl acid binding site, what accounts for substrate preferences of IAA-conjugating GH3 proteins?
One possibility is that differences in residues of α5 alter the orientation of residues in the acyl acid binding site (Fig. 3C); however, with the exception of AtGH3.17, changes in this motif are conservative. The second option is that the ping-pong kinetic mechanism (12), which involves multiple steps in the reaction sequence, may account for variations in steady-state parameters. As described for other adenylating enzymes (26–28), the conformational transition between two distinct half-reactions provides a proofreading mechanism. The structural change can lead to the release of noncognate adenylated intermediates into solution or their hydrolysis within the active site before the transferase step (26–28). Differences in the kinetics of the reaction sequence, in particular movement of the C-terminal domain, of the IAA-conjugating GH3 proteins may affect substrate preference, even though these enzymes retain conserved acyl acid binding sites.
Previous work suggests that AtGH3.5, unlike other IAA-conjugating GH3 proteins in A. thaliana, works in both IAA and SA homeostasis (7, 16, 17, 21–23). Examination of the Arabidopsis gain-of-function mutants wes1-D and gh3.5-1D revealed dwarf phenotypes consistent with lower IAA levels and increased AtGH3.5 activity. A similar phenotype was observed in the 35S-driven overexpressing lines here (Fig. S3). The wes1-D and gh3.5-1D plants displayed enhanced pathogen resistance, which was attributed to the SA-conjugating role of AtGH3.5 and the lower IAA levels that activate pathogen responses (16, 17). Based on the biochemical analysis of AtGH3.5 (Table S2) and earlier in planta studies focused on IAA, IAA-amino acid conjugates, and SA levels (16, 17, 21–23), we decided to examine the effect of AtGH3.5 overexpression on IAA, PAA, SA, and BA levels in Arabidopsis (Fig. 4). The in vivo data indicate that AtGH3.5 modifies metabolites in auxin and SA-related pathways.
As described for the AtGH3.5 gain-of-function mutants and overexpression of other IAA-conjugating GH3 proteins in Arabidopsis and rice (16, 17, 20, 29–31), AtGH3.5 overexpression decreased IAA and increased IAA-Asp levels (Fig. 4 A and B). Consistent with the AtGH3.5 substrate profile, overexpression altered PAA and PAA-Asp levels (Fig. 4 C and D). A recent report describes the formation of PAA-Asp and PAA-Glu in plants overexpressing either AtGH3.6 or AtGH3.9 (31). Although IAA is considered the predominant auxin in plants, PAA functions as a growth-regulating molecule (1, 32, 33).
PAA is present at levels similar to that of IAA in plants, it functions through the same TIR1/AFB auxin receptor-mediated pathway as IAA, and it regulates the same auxin-responsive genes as IAA (32). The two auxins show distinct distribution patterns, and, unlike IAA, PAA is not actively transported to form polar gradients in plant cells (32). Given that multiple IAA-conjugating GH3 proteins (7, 12, 32) as well as AtGH3.5 display significant activity with PAA, the physiological role of these enzymes as mediators of auxin responses likely extends to PAA. The accumulation of PAA-Asp versus IAA-Asp in the Arabidopsis AtGH3.5-overexpressing lines also suggests that the subsequent metabolism of these molecules may differ. Conjugation of IAA with acidic amino acids is believed to lead to the inactivation and degradation of IAA (6). The low levels of IAA-Asp in the AtGH3.5-overexpressing lines (Fig. 4 A and B) are consistent with rapid turnover through the degradation pathway. In contrast, overexpression of AtGH3.5 results in a striking accumulation of PAA-Asp (Fig. 4 C and D), suggesting either that degradation of this conjugate may not be as efficient as that of IAA-Asp or that the amino acid conjugate serves as a storage form for PAA. Although the results reported here and by others (32, 33) highlight the parallels and differences in IAA and PAA metabolism in plants, further investigation is required to understand the roles of PAA and its conjugates in plant growth and development; however, it is likely that AtGH3.5 plays a key role in IAA/PAA homeostasis.
The analysis of AtGH3.5 indicates, as described previously (7), that this protein can conjugate SA but is more efficient using BA as a substrate. In plants, SA is a major phytohormone that mediates plant disease resistance and abiotic stress responses (34). The overexpression of AtGH3.5 in Arabidopsis led to changes in SA levels in some overexpressing lines but not in others; however, SA-Asp increased in all lines (Fig. 4 E and F). Analysis of the wes1-D and gh3.5-1D gain-of-function mutants indicated elevated SA content, although not to the levels as observed during pathogen infection (16, 17). SA-Asp levels were not analyzed in the gain-of-function mutants, although total SA conjugates, including glycosylated forms, increased (16, 17). AtGH3.5 may serve to attenuate SA activity by conjugation in vivo. Given that other Arabidopsis GH3 proteins do not accept SA as a substrate, this SA conjugation appears to be a specialized role for AtGH3.5.
Overexpression of AtGH3.5 resulted in even greater changes in BA and BA-Asp (Fig. 4 G and H). The biological role for altered BA content is unclear at this time, but it is possible that BA-Asp provides a precursor pool for SA during pathogen challenge. Two routes have been proposed for SA synthesis, one through isochorismate and another through metabolism of benzoic acids (35). Plant benzoic acids can be modified and converted to other compounds, including SA (35). Metabolically, BA-2-hydroxylase, a soluble P450 monooxygenase, which has been partially purified and characterized, can convert BA to SA (35, 36). Studies in tobacco suggest that BA conjugates, in particular BA-glucose, may contribute to SA synthesis in plant defense responses (37). For BA-Asp to function as an SA precursor, a hydrolase would be needed to release the free acid from the conjugate. The lack of BA-Asp accumulation in wild-type plants compared with the overexpressing lines suggests that hydrolysis of BA-Asp may occur slowly and that increased AtGH3.5 causes a bottleneck in this process.
Extensive studies of the wes1-D and gh3.5-1D gain-of-function mutants in A. thaliana indicate that these plants have higher SA levels than wild-type plants during pathogen challenge (16, 17). It is possible that the increased levels of SA are derived from BA-Asp, which may provide a precursor pool for SA synthesis, in addition to induction of the isochorismate route (35). AtGH3.5 can inactivate SA by conjugation but does so less efficiently than with BA. The increased disease resistance in the wes1-D and gh3.5-1D gain-of-function mutants is linked to the lower IAA levels but also may be tied to reduced PAA levels and a pool of BA-Asp for mobilization to SA. Additional studies on benzoate metabolism and SA biosynthesis are required to untangle these hypotheses. Overall, structural and biochemical studies of the GH3 acyl acid amido synthetases help define their roles in plant hormone responses.
Methods
Protein Expression, Protein Purification, and Site-Directed Mutagenesis.
The coding region for AtGH3.5 was PCR-amplified from an ORF clone inserted in pENTR223 [The Arabidopsis Information Resource (TAIR) accession no G18499] and was ligated into pET-28a (Novagen). The construct was transformed into E. coli Rosetta (DE3) cells and was grown in Terrific broth containing 50 μg/mL kanamycin and 34 μg/mL chloramphenicol to A600nm ∼0.8. Protein expression was induced with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) overnight at 18 °C. Recombinant protein was purified using Ni2+-affinity chromatography (9, 11, 12). For protein crystallization, the His-tagged protein was incubated with thrombin in 50 mM Tris (pH 8), 500 mM NaCl, 20 mM imidazole, 10% (vol/vol) glycerol overnight at 4 °C and then was passed over a mixed nickel/benzamidine Sepharose column to remove uncut protein and the protease. The flow-through was purified by size-exclusion chromatography on a Superdex-200 26/60 HiLoad FPLC column equilibrated with 25 mM Hepes (pH 7.5), 100 mM NaCl, and 1 mM MgCl2. Protein concentration was determined by Bradford assay with BSA as the standard. Point mutants of AtGH3.5 were generated using the QuikChange PCR method with proteins expressed and purified as above. Constructs for AtGH3.1 (TAIR accession no. AT2G14960), AtGH3.2 (TAIR accession no. AT4G37390), AtGH3.6 (TAIR accession no. AT5G54510), AtGH3.9 (TAIR accession no. AT2G47750), and AtGH3.17 (TAIR accession no. AT1G28130) were generated as above.
Protein Crystallography.
AtGH3.5 was crystallized using the hanging-drop, vapor-diffusion method at 4 °C. A 1:1 ratio of protein (6 mg/mL) and 20% (vol/vol) PEG-8000, 0.1 M 2-Cyclohexylaminoethanesulfonic acid (CHES) (pH 9.5), 2 mM MgCl2, 5 mM Tris(2-carboxyethyl)phosphine (TCEP), 5 mM IAA, and 5 mM AMP was used. Crystals were frozen in liquid nitrogen with mother liquor supplemented with 25% (vol/vol) glycerol as a cryoprotectant. Diffraction data were collected at beamline ID23-2 of the European Synchrotron Radiation Facility, indexed and integrated with XDS, and scaled with XSCALE (38). Molecular replacement was performed with PHASER (39) using a homology model of AtGH3.5 generated in SWISS-MODEL (40) with the AtGH3.12 structure (9) (PDB ID code 4EQL; 50% identity) as template. The initial structure was refined with BUSTER (41) and COOT (42) used for model building. Subsequent refinement was performed in PHENIX (43). Crystallographic statistics are summarized in Table S1.
Enzyme Assays.
Assays of GH3 protein activity used a coupled-enzyme assay with a standard reaction buffer of 50 mM Tris (pH 8.0) and 1 mM MgCl2, as previously described (12). For the amino acid screen, IAA and ATP were held constant (1 mM) with each amino acid at 5 mM. Kinetic analysis of acyl acid substrates used ATP (1 mM) and aspartate (5 mM) with varied concentrations of acyl acid (up to 10 mM). The data then were fit to the Michaelis–Menten equation using SigmaPlot.
Arabidopsis AtGH3.5-Overexpressing Lines.
A. thaliana ecotype Col-0 seeds were obtained from the Arabidopsis Biological Research Center, Ohio State University, Columbus, OH. For Arabidopsis transformation, the coding region was PCR-amplified from pET28a-AtGH3.5 and cloned into the pENTR/d-TOPO vector (Invitrogen). The coding region then was transferred into the pEARLY GATE-202 (35S promoter, N-terminal FLAG tag) vector (44) using a recombinase kit (Invitrogen). The resulting vector was electroporated into Agrobacterium tumefaciens LBA4404. Plants were transformed by floral dip and grown to maturity. After T1 seeds were selected and harvested, multiple herbicide-resistant lines were isolated for the generation of T2 seeds. T2 seeds exhibiting a 3:1 segregation ratio on plates with glufosinate ammonium (50 mg/L) were used for isolation of independent homozygous lines whose identity was confirmed by immunoblot analysis for expression of FLAG-tagged AtGH3.5. For protein analysis, seedlings were ground in liquid nitrogen and extracted in 50 mM potassium phosphate buffer (pH 8). Protein amount was quantified by Bradford assay. Proteins were separated by SDS/PAGE and electrotransferred to nitrocellulose membranes for immunoblot analysis with anti-FLAG M2 antibody (Sigma-Aldrich) with immunoreactive bands visualized by enhanced chemiluminescence (Amersham).
Mass Spectrometry of Free and Conjugated Acyl Acids.
For analysis of free and conjugated acyl acids, standard curves were generated for IAA, PAA, SA, and BA and their respective conjugates. For the commercially available molecules, 1-mM solutions in 50 mM Tris (pH 8) were used. Enzymatic conversion of PAA, SA, and BA to PAA-Asp, SA-Asp, and BA-Asp, respectively, used purified AtGH3.5. Reactions contained 1 mM acyl acid, 1 mM ATP, 1 mM MgCl2, 5 mM aspartate, 50 mM Tris (pH 8.0) (15 min at 25 °C). Analysis used a protocol similar to previous work on IAA conjugates (11). Metabolites were analyzed on a Shimadzu LC with an Applied Biosystems 4000 Q-TRAP mass spectrometer equipped with an electrospray ion source and with a C18 column (Onyx; 4.6 mm × 100 mm; Phenomenex) using a gradient of 40% solvent A (0.1% acetic acid) held for 2 min to 100% solvent B [90% (vol/vol) acetonitrile and 0.1% acetic acid] for 8 min at a rate of 1 mL/min. Acyl acid forms were used to select multiple reaction monitoring (MRM) transitions and to optimize the compound-dependent parameters (Table S4). Reaction mixtures then were analyzed using the disappearance of the free acid to calculate conjugate. Standard curves were generated for each compound (Fig. S4).
For analysis of free and conjugated acyl acids, leaf tissue was collected from 1-mo-old A. thaliana Col-0 and four independent AtGH3.5-overexpressing lines (OE1–4). For each line, 50–150 mg of plant tissue was collected in triplicate from four to six plants per line. Tissues were frozen in liquid nitrogen and ground for 30 s with ice-cold methanol/acetonitrile (1:1; 900 μL) added to each sample before the sample was homogenized again for 2 min. Samples were centrifuged (16,000 × g; 5 min; 4 °C), and the supernatant was collected and transferred to fresh tubes. The pellet was resuspended in methanol/acetonitrile (1:1; 500 μL), homogenized for 2 min, and centrifuged. The supernatant was combined with previous supernatant and then was dried. Samples were dissolved in 30% (vol/vol) methanol (200 μL) and were analyzed by LC-MS/MS, as described.
Acknowledgments
Portions of this research were carried out at the European Synchrotron Radiation Facility and the Donald Danforth Plant Science Center Proteomics and Mass Spectrometry Facility. This work was supported by National Science Foundation (NSF) Grant MCB-1614539 (to J.M.J.). C.S.W. was a US Department of Agriculture Predoctoral Fellow supported by Grant MOW-2010-05240. A.M.S. was an NSF Graduate Research Fellow supported by Grant DGE-1143954.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Crystallography, atomic coordinates, and structure factors reported in this work have been deposited in the Protein Data Bank (PDB ID code 5KOD).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1612635113/-/DCSupplemental.
References
- 1.Korasick DA, Enders TA, Strader LC. Auxin biosynthesis and storage forms. J Exp Bot. 2013;64(9):2541–2555. doi: 10.1093/jxb/ert080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Westfall CS, Muehler AM, Jez JM. Enzyme action in the regulation of plant hormone responses. J Biol Chem. 2013;288(27):19304–19311. doi: 10.1074/jbc.R113.475160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Staswick PE, Tiryaki I, Rowe ML. Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell. 2002;14(6):1405–1415. doi: 10.1105/tpc.000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fonseca S, et al. (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat Chem Biol. 2009;5(5):344–350. doi: 10.1038/nchembio.161. [DOI] [PubMed] [Google Scholar]
- 5.Meesters C, et al. A chemical inhibitor of jasmonate signaling targets JAR1 in Arabidopsis thaliana. Nat Chem Biol. 2014;10(10):830–836. doi: 10.1038/nchembio.1591. [DOI] [PubMed] [Google Scholar]
- 6.LeClere S, Tellez R, Rampey RA, Matsuda SP, Bartel B. Characterization of a family of IAA-amino acid conjugate hydrolases from Arabidopsis. J Biol Chem. 2002;277(23):20446–20452. doi: 10.1074/jbc.M111955200. [DOI] [PubMed] [Google Scholar]
- 7.Staswick PE, et al. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell. 2005;17(2):616–627. doi: 10.1105/tpc.104.026690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Westfall CS, Herrmann J, Chen Q, Wang S, Jez JM. Modulating plant hormones by enzyme action: The GH3 family of acyl acid amido synthetases. Plant Signal Behav. 2010;5(12):1607–1612. doi: 10.4161/psb.5.12.13941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Westfall CS, et al. Structural basis for prereceptor modulation of plant hormones by GH3 proteins. Science. 2012;336(6089):1708–1711. doi: 10.1126/science.1221863. [DOI] [PubMed] [Google Scholar]
- 10.Gulick AM. Conformational dynamics in the Acyl-CoA synthetases, adenylation domains of non-ribosomal peptide synthetases, and firefly luciferase. ACS Chem Biol. 2009;4(10):811–827. doi: 10.1021/cb900156h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Q, Zhang B, Hicks LM, Wang S, Jez JM. A liquid chromatography-tandem mass spectrometry-based assay for indole-3-acetic acid-amido synthetase. Anal Biochem. 2009;390(2):149–154. doi: 10.1016/j.ab.2009.04.027. [DOI] [PubMed] [Google Scholar]
- 12.Chen Q, Westfall CS, Hicks LM, Wang S, Jez JM. Kinetic basis for the conjugation of auxin by a GH3 family indole-acetic acid-amido synthetase. J Biol Chem. 2010;285(39):29780–29786. doi: 10.1074/jbc.M110.146431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peat TS, et al. Crystal structure of an indole-3-acetic acid amido synthetase from grapevine involved in auxin homeostasis. Plant Cell. 2012;24(11):4525–4538. doi: 10.1105/tpc.112.102921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Round A, et al. Determination of the GH3.12 protein conformation through HPLC-integrated SAXS measurements combined with X-ray crystallography. Acta Crystallogr D Biol Crystallogr. 2013;69(Pt 10):2072–2080. doi: 10.1107/S0907444913019276. [DOI] [PubMed] [Google Scholar]
- 15.Takase T, et al. ydk1-D, an auxin-responsive GH3 mutant that is involved in hypocotyl and root elongation. Plant J. 2004;37(4):471–483. doi: 10.1046/j.1365-313x.2003.01973.x. [DOI] [PubMed] [Google Scholar]
- 16.Park JE, et al. GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J Biol Chem. 2007;282(13):10036–10046. doi: 10.1074/jbc.M610524200. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Z, et al. Dual regulation role of GH3.5 in salicylic acid and auxin signaling during Arabidopsis-Pseudomonas syringae interaction. Plant Physiol. 2007;145(2):450–464. doi: 10.1104/pp.107.106021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakazawa M, et al. DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J. 2001;25(2):213–221. doi: 10.1046/j.1365-313x.2001.00957.x. [DOI] [PubMed] [Google Scholar]
- 19.Khan S, Stone JM. Arabidopsis thaliana GH3.9 influences primary root growth. Planta. 2007;226(1):21–34. doi: 10.1007/s00425-006-0462-2. [DOI] [PubMed] [Google Scholar]
- 20.Zheng Z, et al. Local auxin metabolism regulates environment-induced hypocotyl elongation. Nat Plants. 2016;2:16025. doi: 10.1038/nplants.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park JE, et al. An Arabidopsis GH3 gene, encoding an auxin-conjugating enzyme, mediates phytochrome B-regulated light signals in hypocotyl growth. Plant Cell Physiol. 2007;48(8):1236–1241. doi: 10.1093/pcp/pcm086. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Z, Wang M, Li Z, Li Q, He Z. Arabidopsis GH3.5 regulates salicylic acid-dependent and both NPR1-dependent and independent defense responses. Plant Signal Behav. 2008;3(8):537–542. doi: 10.4161/psb.3.8.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y, Shen H, Wang M, Li Q, He Z. Salicyloyl-aspartate synthesized by the acetyl-amido synthetase GH3.5 is a potential activator of plant immunity in Arabidopsis. Acta Biochim Biophys Sin (Shanghai) 2013;45(10):827–836. doi: 10.1093/abbs/gmt078. [DOI] [PubMed] [Google Scholar]
- 24.Terol J, Domingo C, Talón M. The GH3 family in plants: Genome wide analysis in rice and evolutionary history based on EST analysis. Gene. 2006;371(2):279–290. doi: 10.1016/j.gene.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 25.Okrent RA, Brooks MD, Wildermuth MC. Arabidopsis GH3.12 (PBS3) conjugates amino acids to 4-substituted benzoates and is inhibited by salicylate. J Biol Chem. 2009;284(15):9742–9754. doi: 10.1074/jbc.M806662200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reger AS, Carney JM, Gulick AM. Biochemical and crystallographic analysis of substrate binding and conformational changes in acetyl-CoA synthetase. Biochemistry. 2007;46(22):6536–6546. doi: 10.1021/bi6026506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yadavalli SS, Ibba M. Quality control in aminoacyl-tRNA synthesis its role in translational fidelity. Adv Protein Chem Struct Biol. 2012;86:1–43. doi: 10.1016/B978-0-12-386497-0.00001-3. [DOI] [PubMed] [Google Scholar]
- 28.Manandhar M, Cronan JE. Proofreading of noncognate acyl adenylates by an acyl-coenzyme a ligase. Chem Biol. 2013;20(12):1441–1446. doi: 10.1016/j.chembiol.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ding X, et al. Activation of the indole-3-acetic acid-amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. Plant Cell. 2008;20(1):228–240. doi: 10.1105/tpc.107.055657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Domingo C, Andrés F, Tharreau D, Iglesias DJ, Talón M. Constitutive expression of OsGH3.1 reduces auxin content and enhances defense response and resistance to a fungal pathogen in rice. Mol Plant Microbe Interact. 2009;22(2):201–210. doi: 10.1094/MPMI-22-2-0201. [DOI] [PubMed] [Google Scholar]
- 31.Zhang SW, et al. Altered architecture and enhanced drought tolerance in rice via the down-regulation of indole-3-acetic acid by TLD1/OsGH3.13 activation. Plant Physiol. 2009;151(4):1889–1901. doi: 10.1104/pp.109.146803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sugawara S, et al. Distinct characteristics of indole-3-acetic acid and phenylacetic acid, two common auxins in plants. Plant Cell Physiol. 2015;56(8):1641–1654. doi: 10.1093/pcp/pcv088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cook SD, et al. Auxin biosynthesis: Are the indole-3-acetic acid and phenylacetic acid biosynthesis pathways mirror images? Plant Physiol. 2016;171(2):1230–1241. doi: 10.1104/pp.16.00454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan S, Dong X. Perception of the plant immune signal salicylic acid. Curr Opin Plant Biol. 2014;20:64–68. doi: 10.1016/j.pbi.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Widhalm JR, Dudareva N. A familiar ring to it: Biosynthesis of plant benzoic acids. Mol Plant. 2015;8(1):83–97. doi: 10.1016/j.molp.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 36.León J, Shulaev V, Yalpani N, Lawton MA, Raskin I. Benzoic acid 2-hydroxylase, a soluble oxygenase from tobacco, catalyzes salicylic acid biosynthesis. Proc Natl Acad Sci USA. 1995;92(22):10413–10417. doi: 10.1073/pnas.92.22.10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chong J, et al. Free and conjugated benzoic acid in tobacco plants and cell cultures. Induced accumulation upon elicitation of defense responses and role as salicylic acid precursors. Plant Physiol. 2001;125(1):318–328. doi: 10.1104/pp.125.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de la Fortelle E, Bricogne G. Maximum-likelihood heavy-atom parameter refinement for the multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 40.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics. 2006;22(2):195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 41.Blanc E, et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- 42.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 43.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Earley KW, et al. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45(4):616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]