Significance
The small lectin-like protein ZG16 (zymogen granulae protein 16) aggregates bacteria and by that works together with the inner colon mucus layer to maintain bacteria at a safe distance from the epithelial cell surface. In the absence of Zg16, more bacteria penetrate the epithelium, enter the regional lymph nodes and spleen, trigger the immune system, and cause abdominal fat pad mass increase. ZG16 is important for a safe normal host-bacteria symbiosis as it does not kill commensal bacteria, but limit bacterial translocation into the host.
Keywords: mucin, colon, inflammation, obesity, peptidoglycan
Abstract
The distal colon functions as a bioreactor and harbors an enormous amount of bacteria in a mutualistic relationship with the host. The microbiota have to be kept at a safe distance to prevent inflammation, something that is achieved by a dense inner mucus layer that lines the epithelial cells. The large polymeric nets made up by the heavily O-glycosylated MUC2 mucin forms this physical barrier. Proteomic analyses of mucus have identified the lectin-like protein ZG16 (zymogen granulae protein 16) as an abundant mucus component. To elucidate the function of ZG16, we generated recombinant ZG16 and studied Zg16−/− mice. ZG16 bound to and aggregated Gram-positive bacteria via binding to the bacterial cell wall peptidoglycan. Zg16−/− mice have a distal colon mucus layer with normal thickness, but with bacteria closer to the epithelium. Using distal colon explants mounted in a horizontal perfusion chamber we demonstrated that treatment of bacteria with recombinant ZG16 hindered bacterial penetration into the mucus. The inner colon mucus of Zg16−/− animals had a higher load of Gram-positive bacteria and showed bacteria with higher motility in the mucus close to the host epithelium compared with cohoused littermate Zg16+/+. The more penetrable Zg16−/− mucus allowed Gram-positive bacteria to translocate to systemic tissues. Viable bacteria were found in spleen and were associated with increased abdominal fat pad mass in Zg16−/− animals. The function of ZG16 reveals a mechanism for keeping bacteria further away from the host colon epithelium.
The intestinal mucus limits the interaction between the host and the 1013–1014 luminal bacteria in the intestine. The organization of the mucus differs between the small and the large intestine (1–3). The small intestine has a single mucus layer that is easily removed and penetrable to bacteria. The large intestine has a two-layered mucus system with an inner laminated and dense layer attached to the epithelium that acts as a filter to exclude bacteria. This filter function is made possible by the macromolecular structure of the large and glycosylated MUC2 mucin, which by its disulfide-bonded polymeric nature forms large net-like sheets (4). At about 50 µm from the epithelium in mice (200 µm in humans), the inner mucus layer is released from its attachment and slowly starts to expand in a host-controlled process (2). The expanded outer mucus layer is penetrable and colonized by commensal bacteria, a process which probably involves bacterial mucin-specific adhesins, and provides an enormous glycan food source for the bacteria (5). Some commensal bacteria are equipped with large repertoires of genes, usually organized in clusters, for the degradation and utilization of nondegraded food and mucin glycans (6). The rapid turnover of mucus in the colon (∼1 h) ensures that this bacterial food source is constantly regenerated (7). Bacteria do not use all this energy for themselves, as a substantial amount is sent back to the host in the form of short fatty acids (6).
Proteomic studies of the colonic mucus revealed additional highly abundant proteins (2). One of these, ZG16 (zymogen granulae protein 16), a 16-kDa protein with a Jacalin-type lectin domain, caught our attention. Carbohydrate array studies performed by us (Center Functional Glycomics consortium) and others suggested binding to polymannose structures (8, 9). However, as ZG16 did not bind several types of yeast in our hands, we instead turned our attention to the most abundant glycan in bacteria, peptidoglycan. We can now show that ZG16 binds to peptidoglycan, resulting in bacterial aggregation and inhibition of mucus penetration, thus moving bacteria further away from the host epithelium.
Results
ZG16 Binds Peptidoglycan and Gram-Positive Bacteria.
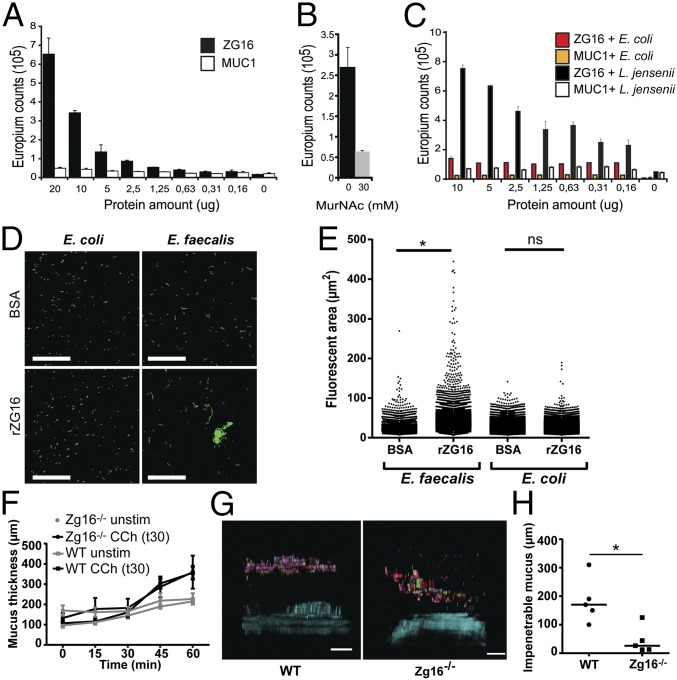
Pure recombinant ZG16-Fc (SI Appendix, Fig. S1A), but not an irrelevant Ig-fusion protein, was found to bind insoluble peptidoglycan in a concentration-dependent fashion (Fig. 1A). This binding was inhibited by one of the sugar building blocks of peptidoglycan, N-Acetylmuraminic acid (MurNAc, NAM), although at a relatively high concentration (Fig. 1B). Gram-positive bacteria, such as Lactobacillus jensenii, have peptidoglycan as their outer surface and were shown to bind ZG16, but no binding to Gram-negative Eschericia coli was detected (Fig. 1C). As the ZG16 structure has been determined, we made in silico docking studies of the interaction with peptidoglycan (SI Appendix, Fig. S1 B–D) (10, 11). The ZG16 structure is barrel-shaped with the carbohydrate-binding pocket on one side. ZG16 was found to dock well with a terminal NAM unit (SI Appendix, Fig. S1 B–D). Such terminal epitopes appear relatively frequently in peptidoglycan, although the amount differs depending on the bacterial species (12). The in silico docking suggests that ZG16 could bind to peptidoglycan both by glycan and amino acid moieties.
Fig. 1.
ZG16 binds peptidoglycan and Gram-positive bacteria. Binding experiments were performed using a DELFIA-based assay. (A) Different concentrations of recombinant ZG16-Fc were applied to wells containing insoluble peptidoglycan. ZG16-Fc (filled bars) bound to the peptidoglycan after extensive washing compared with the control MUC1-Fc (open bars). Error bars represent SEM (n = 3). (B) Preincubation with 30 mM MurNAc partially inhibited binding of ZG16-Fc to peptidoglycan. (C) ZG16-Fc binding to the Gram-positive bacteria L. jensenii and Gram-negative bacteria E. coli. The assay was performed in a similar way as for the peptidoglycan binding. (D) Syto9-stained E. faecalis and E. coli cultures were incubated with BSA or rZG16, spread on microscopy slides, and imaged; images show representative field views of BSA (Top) or rZG16 (Bottom) treated bacteria. (Scale bars, 50 µm.) (E) Fluorescent areas from the microscopy images calculated using Imaris software; statistical significance calculated using Tukey’s multiple comparison test (ns, not significant; *P < 0.001). Data representative of n = 3 independent experiments. (F) Mucus measurement of WT (n = 9) and Zg16−/− mice (n = 6). Distal colon tissue was mounted in a horizontal chamber, charcoal was added to visualize the mucus, and the distance to the epithelial cells was measured. The secretagogue carbachol was added after 30 min. (G) Penetrability measurements of WT (n = 5) and Zg16−/− (n = 5) mice. Distal colon tissue was mounted in a horizontal chamber, and fluorescent bacterial-size beads were added on the explant 20 min after mounting. Beads were allowed to sediment for 40 min before the distribution was visualized using confocal microscopy. Images are representative confocal z-stack projections obtained from the experiments. (Scale bars, 100 µm.) (H) Scatter plot of the mean distance of the 20 fluorescent beads closest to the epithelium; lines represent median. *P = 0.016 using Mann–Whitney u test.
ZG16 Is Not Directly Bactericidal but Aggregates Gram-Positive Bacteria.
To determine if ZG16 affected the viability of Gram-positive (Enterococcus faecalis, Bacillus subtilis) or Gram-negative bacteria (Bacteroides fragilis, E. coli), bacterial viability was monitored after the addition of untagged recombinant ZG16 (rZG16). Whereas treatment with lysozyme had a clear bactericidal effect, the relative number of bacteria was the same with or without added rZG16 (SI Appendix, Fig. S2). Human β-defensin 1 was recently shown to only have antimicrobial activity under reducing conditions (13). ZG16 contains two C-terminal cysteines proposed to form an intramolecular disulfide bond that might need to be reduced for bactericidal effects (14). However, reduced rZG16 also did not show any antibacterial effect (SI Appendix, Fig. S2B).
To address if ZG16 had any effect other than just binding, stained bacteria were treated with rZG16 and imaged (Fig. 1D). Significantly larger Gram-positive bacterial aggregates were observed after rZG16 treatment in contrast to BSA-treated control bacteria (Fig. 1E). To assess the effect of bacterial aggregation in a system that is similar to the polymeric mucus network, the motile Gram-positive bacterium B. subtilis was applied to low-density agar plates impregnated with either BSA or rZG16. Bacteria applied to plates containing rZG16 showed a higher degree of aggregation and decreased spreading in the agar in contrast to BSA-treated controls (SI Appendix, Fig. S3). Together these results show that whereas ZG16 does not affect the viability of bacteria, it causes aggregation of Gram-positive cells, which limits their ability to penetrate polymeric networks.
Characterization of Zg16−/− Murine Mucus.
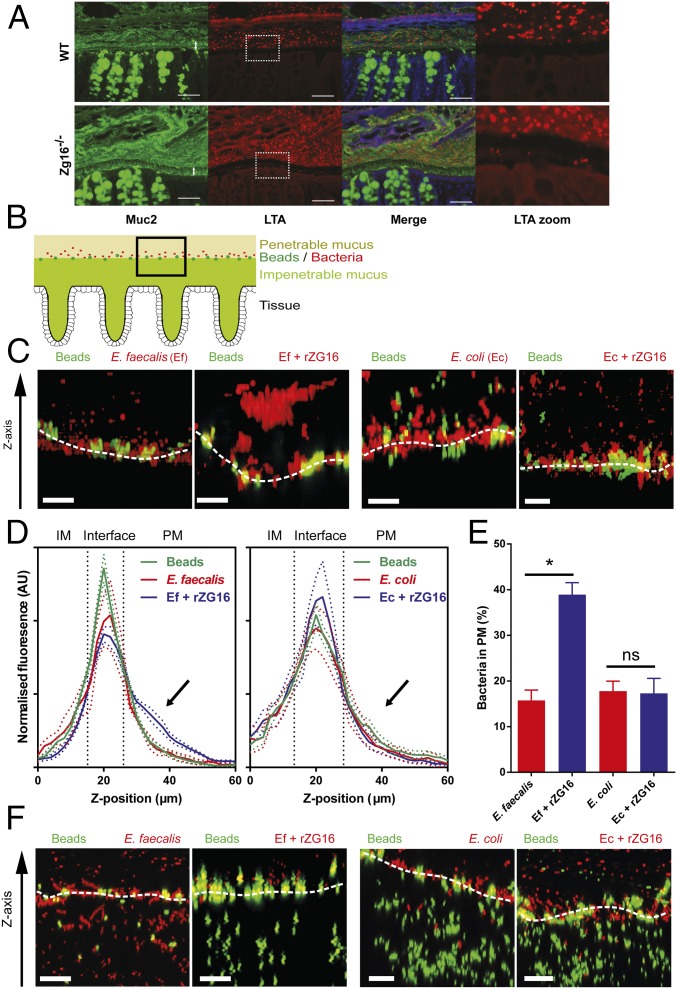
When the distal colon mucus growth in the Zg16−/− mouse was measured ex vivo and compared with WT over 60 min with or without the addition of the cholinergic secretagogue carbachol, no difference was observed (Fig. 1F). When the quality of the mucus was analyzed by bacterial-size bead penetration of the WT mice, a spatial separation of at least 100 µm from the epithelium was observed. The Zg16−/− mice had more beads closer to the epithelium (Fig. 1 G and H). Carnoy-fixed distal colon sections were H&E stained and showed no signs of inflammation such as crypt elongation or neutrophil infiltration. Alcian blue and Periodic Acid–Schiff stains were also similar (SI Appendix, Fig. S4). Colon tissue sections were costained with an anti-Muc2 antiserum and an antibody that reacts with lipoteichoic acid (LTA) present on Gram-positive bacteria (Fig. 2A). The Zg16−/− inner mucus layer showed more Gram-positive bacteria in the inner mucus layer. Combined, these results indicated that Zg16−/− mice had a normally thick inner mucus layer, but a larger degree of bacterial penetration into the inner mucus layer.
Fig. 2.
ZG16 alters distal colonic mucus penetration by Gram-positive bacteria in vivo and in vitro. (A) Carnoy-fixed distal colon immunohistochemistry of the mucus layer (anti-MUC2, green) and Gram-positive bacteria (anti-LTA antibody, red) (SI Appendix, Fig. S5). (Scale bars, 50 µm.) (B–F) C57BL/6 WT or Zg16−/− distal colon tissues were mounted in an imaging chamber; Fluorescent beads were applied apically to visualize the interface between the impenetrable (IM) and penetrable (PM) mucus layers; 108 cfu E. faecalis (Ef) or E. coli (Ec) were stained with BacLight Red then treated with 10 µg rZG16 and applied apically to mucus; 3D z-stacks of the mucus surface were acquired by confocal microscopy. (B) Schematic representation of data acquisition region. (C) Confocal z-stacks of WT mucus surface exposed to untreated Ef/Ec (Left) or Ef/Ec treated with rZG16 (Right); White dashed line indicates IM/PM interface. (D) Distribution of beads and untreated or rZG16-treated Ef (Left) or Ec (Right) along the z-stack z axis at the WT IM/PM interface. Colored dashed lines represent SEM from n = 6 mice; black dashed lines separate IM, interface, and PM regions of the z-stack. (E) Quantification of control and rZG16-treated Ef and Ec from the PM z-stack region indicated by the arrows in D. Error bars represent SEM from n = 6 mice. (F) Confocal z-stacks of Zg16−/− mucus surface exposed to untreated Ef/Ec (Left) or Ef/Ec treated with rZG16 (Right). White dashed line indicates IM/PM interface. All images are representative of n = 6 mice. Statistical significance calculated with Tukey’s multiple comparison test (ns, not significant; *P < 0.05). (Scale bars, 20 µm.)
Recombinant ZG16 Alters Distal Colonic Mucus Penetration.
Distal colon WT tissue was mounted in a confocal imaging chamber, and 2-µm fluorescent beads were apically applied to mark the interface between the impenetrable (IM) and penetrable (PM) mucus layers. Gram-positive E. faecalis and Gram-negative E. coli were fluorescently labeled, treated with rZG16 or BSA, and applied to the colonic mucus, and confocal z-stacks were acquired over the IM/PM interface (Fig. 2B). Under BSA-treated conditions the majority of both Gram-positive and negative bacterial cells settled at the IM/PM interface of the WT mucus, but rZG16-treated bacteria aggregated Gram-positive cells, and the cells did not fully penetrate the PM layer (Fig. 2C). This reproducible effect shifted the rZG16-treated bacteria significantly out into the PM layer compared with BSA-treated bacteria (arrow, Fig. 2 D and E), a shift not observed in the rZG16-treated E. coli. The same method was applied to Zg16−/− colons where patches of the IM layer were more penetrable to both beads and bacteria. However, adding rZG16 to the Gram-positive bacteria rendered them unable to penetrate the IM layer, while Gram-negative bacteria were unaffected (Fig. 2F). Interestingly, the inhibition of Gram-positive bacterial IM layer penetration by rZG16 was not limited to aggregated bacteria as individual cells also failed to penetrate the IM layer despite its remaining penetrability to beads of similar size, suggesting that ZG16 can exclude bacteria from the IM layer by aggregation-independent mechanisms.
ZG16 Alters the Distribution, Composition, and Mobility of the Mucus-Associated Intestinal Microbiota.
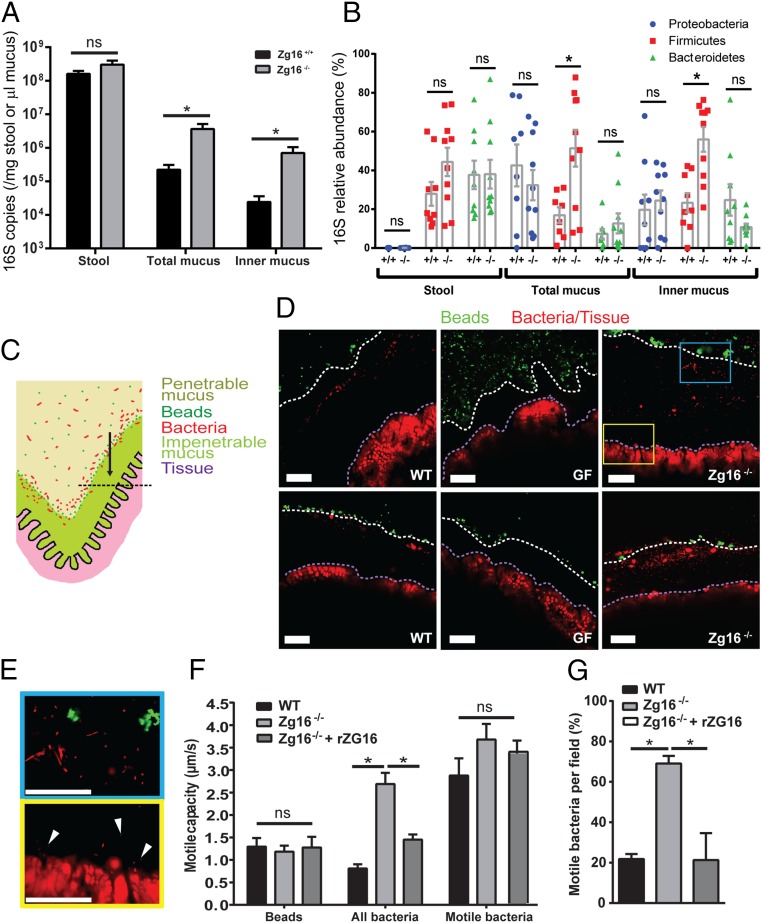
DNA was extracted from stool and biopsy punch-collected tissues from unflushed (total mucus) or flushed (inner mucus) distal colon from littermate Zg16+/+ and Zg16−/− mice. Bacterial 16S was quantified by qPCR and normalized to stool mass or mucus thickness (SI Appendix, Fig. S6). The amount of bacteria per microliter of mucus was significantly higher in the Zg16−/− total and inner mucus compared with Zg16+/+ (Fig. 3A). However, no differences were observed in the stool. Analysis of the relative abundance of 16S rRNA from the phyla Proteobacteria, Firmicutes, and Bacteroidetes by qualitative qPCR showed significant increases in the Gram-positive Firmicutes in the mucus (Fig. 3B). A deeper relative analysis by 16S rRNA gene sequencing of mucus samples from these cohoused littermate animals showed a more complex alteration in their bacterial ecology, including a decrease in Deferribacteraceae and increase in the Clostridiales (SI Appendix, Fig. S7). No differences were observed in the stool, suggesting a selection effect of Zg16 in mucus only (Fig. 3B and SI Appendix, Fig. S7),
Fig. 3.
ZG16 alters the mucus-associated intestinal microbiota as well as the distribution and motile capacity of bacteria within the mucus. (A) Zg16+/+ and Zg16−/− littermate bacterial 16S copy number detected in stool and unflushed (total mucus) or flushed (inner mucus) colonic tissue samples by qPCR. Data were normalized to stool mass or mucus volume (SI Appendix, Fig. S6). (B) Group-specific qPCR showing an increased abundance of Gram-positive Firmicutes in the total and inner mucus of the Zg16−/− in relation to littermate Zg16+/+ mice. (C–E) Conventionally raised C57BL/6 (WT), germ-free (GF), and Zg16−/− distal colon tissues were mounted in an imaging chamber; fluorescent beads were applied apically to visualize the interface between the impenetrable (IM) and penetrable (PM) mucus layers; and bacteria and tissues were visualized in situ using the nucleic acid-binding dye, Syto9. Mucus, bacteria, and tissues were imaged. (C) Schematic representation of chamber-mounted colonic tissues with IM and PM layers, beads, and bacteria indicated. Black dashed line represents the focal plane where confocal optical sections were acquired. (D) Confocal micrographs through WT, GF, and Zg16−/− colonic mucus. White dashed lines indicate the IM/PM interface; purple dashed lines indicate the edge of the colonic tissue. (E) Magnified confocal micrographs from the Inset blue and yellow boxes in D. (Top, blue border) Morphologically distinctive bacteria in the mucus. (Bottom, yellow border) Bacteria (white arrows) near the tissue surface. (F and G) Bacterial motility in WT, Zg16−/−, and Zg16−/− + rZG16 mucus was assessed by recording 1-min time series of optical sections through the colonic mucus as pictured in E (Movies S1–S7). (F) Quantification of motile capacity (maximum velocity) of beads at the IM/PM interface, all bacteria within the mucus, and only bacteria classed as motile within the mucus. (G) Proportion of bacteria determined as motile per imaging field. All error bars are SEM from n = 9 Zg16+/+ and n = 10 Zg16−/−animals. Statistical significances calculated with Mann–Whitney u test (A, B) or Sidak’s (F) or Dunn’s (G) multiple comparison tests (ns, not significant; *P < 0.05). (All scale bars, 50 µm.)
To investigate the effect of ZG16 on the distribution of the microbiota within the mucus in vivo, unflushed distal colon tissue was mounted in an imaging chamber with green fluorescent beads to mark the IM/PM interface (Fig. 3C). Syto9 nucleic acid-binding dye (red) was used to stain the host tissue and microbiota in situ. Confocal microscopy revealed a band of Syto9-stained objects in the mucus of WT (conventionally raised) mice that resembled morphologically diverse bacterial cells, and these objects were absent in GF (germ-free) mucus (Fig. 3 D and E). In WT mucus the microbiota were typically found in a discrete zone close to the IM/PM interface with limited bacteria deeper within the IM (Fig. 3D). The Zg16−/− IM mucus contained notably more bacterial cells than the WT mucus with bacterial cells observed at the tissue surface (Fig. 3 D and E and Movie S1).
As it was noted that mucus-associated bacteria in the Zg16−/− mucus appeared more motile, the motile capacity (MC; the maximum speed observed for an individual bacterium) of all detected bacteria was analyzed by live imaging of bacteria in the mucus. The WT microbiota were mostly static, with a mean MC equal to the background movement of beads in the mucus, whereas the mean MC of the Zg16−/− microbiota was significantly higher (Fig. 3F and Movies S2–S5). Analysis of only motile bacteria in WT and Zg16−/− mucus demonstrated that the MC difference was due to an increase in the proportion of motile bacteria in the Zg16−/− mucus rather than overall velocity (Fig. 3 F and G). Incubation of Zg16−/− colonic mucus with rZG16 restored the mean bacterial MC and proportion of motile bacteria to WT levels (Fig. 3 F and G and Movies S6 and S7). This suggests that ZG16 works together with the mucin network and forms an enhanced barrier to Gram-positive colonization of the IM layer. Trapping of bacteria and reduced motility in the mucus may be the mechanistic explanation of ZG16’s contribution to colonic mucus barrier.
Zg16−/− Mice Have Increased Host Systemic Bacterial Load and Serum Cytokine Levels.
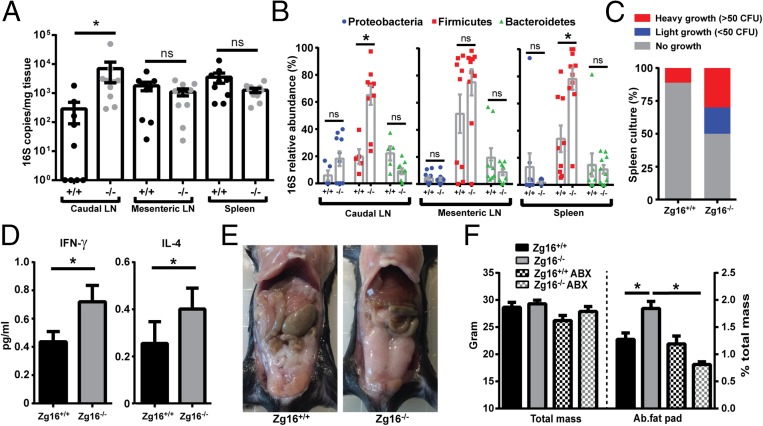
As bacterial contact with the colonic epithelium has been shown to result in bacterial translocation and inflammation (2, 15), lymph nodes (LN) and spleen were sampled from littermate Zg16+/+ and Zg16−/− mice and their bacterial load quantified by 16S qPCR. The 16S load was significantly higher in the distal colon, draining caudal LN of Zg16−/− (Fig. 4A). Qualitative analysis of LNs and spleen 16S DNA showed that the dominant taxonomic group was the Gram-positive phylum Firmicutes in the caudal LN and spleen of the Zg16−/− compared with Zg16+/+ mice (Fig. 4B). Viable bacteria were possible to culture from 50% Zg16−/− spleens, but only one of the Zg16+/+ spleens had viable bacteria (Fig. 4C and SI Appendix, Fig. S8A). Gram-staining of the bacterial isolates demonstrated that all were Gram-positive, and 16S gene sequencing identified almost all isolates as either species of Staphylococcus or Enterococcus faecium (SI Appendix, Fig. S8B). Screening DNA extractions using primers targeting genes specific to these bacteria demonstrated their presence in the animals from which viable splenic bacteria had been identified, making it unlikely that cultured bacteria were contaminants (SI Appendix, Fig. S8C). Staphylococcal and Enterococcal species are ubiquitous, and many are considered opportunistic pathogens due to their well-characterized ability to survive in the systemic environment and cause infections. This may explain why viable spleen bacteria were exclusively from this group. These results indicated that viable Gram-positive bacteria had penetrated the systemic tissues of the Zg16−/− mice, but these experiments do not exclude other bacteria. The systemic presence of viable bacteria may elicit an immune response, and, in fact, IFNγ and IL-4 showed significantly increased levels in serum (Fig. 4D); also, other analyzed cytokines showed a tendency to be increased (SI Appendix, Fig. S9). Increased intestinal bacterial penetration and low-grade inflammation can correlate with increased fat and body mass (16, 17). Indeed, increased fat was typically observed directly when the abdomen was opened (Fig. 4E). Comparison of cohoused littermate Zg16−/− and Zg16+/+ mice showed a significant increase in abdominal fat pad mass, but not overall body mass (Fig. 4F). Administration of broad-spectrum antibiotics in the drinking water lowered the intestinal bacterial load and reverted the increased Zg16−/− abdominal fat pad mass to that of Zg16+/+ (Fig. 4F and SI Appendix, Fig. S10). This indicated that the increased fat pad mass was dependent on bacteria. The Zg16−/− phenotype was emphasized when comparing age-matched WT and Zg16−/− mice from the same animal facility where the Zg16−/− had been separately bred for several generations. The Zg16−/− mice were then ∼15% heavier and had significantly increased levels of the TNFα, KC/GRO, and IL6 cytokines (SI Appendix, Fig. S11). Consequently, mice lacking Zg16 show several features associated with intestinal bacterial barrier dysfunction, further suggesting a crucial role for ZG16 in maintaining a functional colonic mucus barrier.
Fig. 4.
Zg16−/− mice have an increased systemic bacterial load of viable Gram-positive bacteria and increased abdominal fat pad mass. (A–C) Caudal and mesenteric lymph nodes (LN) and spleen were acquired from littermate Zg16+/+ and Zg16−/− mice. Tissues were weighed and homogenized, and DNA was extracted. (A) Quantification of bacterial 16S copy number in LN and spleen tissue by qPCR. Error bars are SEM from n = 9 Zg16+/+ and n = 10 Zg16−/− mice. (B) Estimation of the relative abundance of different bacterial taxonomic groups (Bacteroidetes, Firmicutes and Proteobacteria) using qPCR in the tissues examined in A. (C) Growth of tissue homogenates from spleen were cultivated anaerobically on brain heart infusion-supplemented agar plates. Isolated bacterial colonies were identified by 16S rDNA sequencing and confirmed to be present in the stool, mucus, and lymphatic samples from the same individual animals (SI Appendix, Fig. S6). (D) Significant differences of IFN-γ and IL-4 in the serum of littermate n = 9 Zg16+/+ and n = 10 Zg16−/− mice. (E) Opened abdomen of 12-wk ZG16+/+ and ZG16−/− mice. (F) Total mass and abdominal fat pads of 12-wk-old male littermate Zg16+/+ (n = 9) and Zg16−/− (n = 10) as well as Zg16+/+ (n = 4) and Zg16−/− (n = 5) after 3 wk of antibiotic treatment. Error bars are SEM. Statistical significance calculated with Mann–Whitney u test (A–D) or unpaired t test with similar variance (F) (ns, not significant; *P < 0.05).
Discussion
The mucus in the distal colon is organized in a two-layered fashion where the inner mucus is a dense MUC2 mucin polymeric network and the outer is nonattached and more penetrable (2). The process of converting the inner colon mucus layer to the outer nonattached mucus layer is fast and reflected in the border observed in tissue sections, whereas the expansion in pore size is more gradual as observed in our ex vivo explants where the border between impenetrable (IM) and penetrable (PM) mucus, as defined by fluorescent beads, is more diffuse and located further from the inner–outer mucus layer interphase. When ZG16 aggregates bacteria, these larger complexes were trapped further away in the PM. Our original assumption that the exclusion of bacteria of the inner impenetrable mucus layer was caused solely by the stacked polymeric MUC2 sheets is, as shown here, an oversimplification (2, 4). ZG16, with its capacity to aggregate Gram-positive bacteria, is also an essential component of the mucus barrier, as it is partially compromised in its absence. There is an increase in the number of bacteria in the normally impenetrable mucus layer of the Zg16−/− mice and a higher rate of viable Gram-positive bacteria translocation to systemic tissues. Defects in or absence of the mucus layer allow bacteria to come in close contact with the epithelium and cause an increased bacterial uptake and local inflammation (2, 15, 18). Despite the more penetrable mucus layer found in the Zg16−/− mice, no obvious signs of local inflammation could be detected in histology sections. This is analogous to the IL10−/− mice harbored in the same animal facility that also show limited inflammation compared with genetically identical animals stored at other facilities (15, 19). Despite the lack of local inflammation, an increased 16S caudal LN load and viable splenic bacteria, as well as higher levels of serum proinflammatory cytokines, were likely caused by the increased levels of systemic live bacteria. Surprisingly, binding of ZG16 to Gram-positive bacteria did not result in any bactericidal activity as described for another peptidoglycan-binding lectin present in the gut, the antimicrobial protein Reg IIIγ found in the small intestine (20). Zg16 is, in contrast to Reg IIIγ, primarily found in the colon (21). Having a bacterial aggregator like ZG16, rather than bactericidal proteins, in the colon might be beneficial for the host as this protective mechanism will not eliminate metabolically important bacteria in the mucus; instead, ZG16 works together with the MUC2 polymeric network to maintain bacteria at a safe distance from the epithelial cell surface. The role of ZG16 in the colon mucus further emphasizes that a physical separation of commensal bacteria and the host epithelium is the primary protective principle for the colon.
Materials and Methods
Binding to Peptidoglycan.
Binding experiments were performed as a Dissociation-Enhanced Lanthanide Fluorescent Immunoassay (DELFIA) based method in V-shaped NUNC microtiter plates as follows. Insoluble peptidoglycan from Bacillus subtilis (Sigma), Lactobacillus jensenii, or Eschericia coli bacteria were incubated with different concentrations of ZG16-IgG or MUC1-IgG at +4 °C overnight. Peptidoglycan or bacteria were pelleted by centrifugation at 1,200 × g for 5 min. Unspecific binding was prevented by incubating the plates for 1 h at RT in DELFIA-blocking solution (22). Europium-labeled donkey anti-mouse antibody diluted 1:200 in DELFIA assay solution was added to the wells. Each well was washed 5 times with DELFIA wash solution before 200 μL DELFIA enhancement solution was added. Fluorescent signal was read in VICTOR2 plate reader (Wallac). Each assay was performed in triplicate. For inhibition experiments, recombinant protein was preincubated with MurNAc (Sigma) before addition to the wells.
Bacterial Aggregation Assay.
Bacterial cultures were grown to OD600 0.1 in LB. One milliliter of culture was centrifuged at 5,000 × g for 5 min, and cells were washed with PBS. Washed cells were recentrifuged and resuspended in PBS supplemented with 10 µM Syto9 (Life Technologies) with 10 µg BSA or 10 µg rZG16 and incubated for 30 min at room temperature. Cell suspensions were transferred to microscope slides, overlaid with a coverslip, and imaged using a confocal microscope (Zeiss LSM 700) using a 488-nm laser and a Plan-Apochromat 20×/0.8 M27 objective lens. Multiple 2.5 mm2 tile scans were acquired from each slide. Image files were exported to Imaris (Bitplane), and the areas of individual Syto9-stained objects were quantified.
Animals.
The Zg16−/− mice were generated by Taconic (accession number: TF0327), by deletion of all three Zg16 exons by the insertion of a resistance gene. Mice were backcrossed >10 generations into a C57BL/6 background. Heterozygous-bred Zg16+/− littermate mice were cohoused until used in experiments as Zg16+/+ or Zg16−/−. WT mice (C57BL/6) were used for backcross and as control animals in some experiments. Male mice at the age between 10–14 wk were used. Mice were treated with ampicillin (1 g/L) (Meda), vancomycin (500 mg/L) (Hospira), neomycin (1 g/L, Sigma) and metronidazole (1 g/L, Sigma) in the drinking water for 3 wk. All animal experiments were approved and performed according to Gothenburg laboratory animal ethics committee guidelines. Animals were killed by isoflurane and cervical dislocation.
Mucus Measurements.
Mucus measurements on Zg16−/− and WT were performed as previously described (23). For measurement of total and inner mucus before DNA extraction, colon tissue without any fecal content was collected and either not flushed (total mucus) or flushed (inner mucus) with sterile Krebs buffer. Tissue was opened and pinned flat on sterile silicone gels, and the mucus was visualized using sterile 10 µm black polystyrene beads (Polysciences) applied to the mucosal surface. Mucus thickness was measured using a micropipette as before.
Mucus Penetrability Measurements.
The penetrability of the distal colon mucus was measured as previously described (23). Z-stacks using a LSM700 microscope (Carl Zeiss) were taken, and the distribution of the beads in the mucus was analyzed. Acquired results were analyzed in Velocity (Perkin–Elmer).
Immunohistochemistry.
Distal colon with fecal material was fixed in Carnoy solution (MethaCarn) to preserve the mucus (2). Fixed tissue was paraffin-embedded, sectioned, dewaxed, and stained with Haematoxylin–Eosin (Htx) or Alcian blue/periodate acid Schiff (AB/PAS). For fluorescent microscopy, dewaxed sections were stained with the anti-MUC2C3 antiserum (2) followed by anti-rabbit IgG Alexa 488 (Life Technologies) and with Gram-positive LTA mouse monoclonal antibody (Thermo Scientific) followed by anti-mouse IgG1 Alexa 555 (Life Technologies). DNA was counterstained with Hoescht 35258 (Molecular Probes) and mounted with ProLong Gold (Life Technologies). Images were obtained using a Nikon E-1000 fluorescent microscope (Nikon).
Bacterial ex Vivo Mucus Penetration.
Colonic tissue was mounted in a horizontal perfusion imaging chamber, and 2-µm Fluosphere beads were applied apically to visualize the IM/PM interface. One milliliter OD600 0.1 bacterial cultures were centrifuged at 5,000 × g for 5 min and resuspended in PBS. Cells were labeled using 0.2 µM BacLight Red stain in the dark at RT for 15 min. Cells were then centrifuged at 5,000 × g for 5 min and resuspended in Krebs-mannitol buffer with 10 µg rZG16 or 10 µg BSA and incubated at RT for 30 min. Labeled bacteria were added to colonic explant mucus after addition of beads and incubated at 37 °C for 15 min. Bacteria and beads were imaged using a confocal microscope (Zeiss LSM 700) using 488/555-nm lasers and a Plan-Apochromat 20×/1.0DIC water objective lens. Confocal z-stacks covering the IM/PM interface were acquired, and fluorescence values for beads and labeled bacteria at each focal plane were recorded. Fluorescence values for each stack were normalized to total fluorescence and used to determine the distribution of beads and bacteria along the z axis of the stack. Normalized data from multiple z-stacks were aligned using the IM/PM interface (focal plane with maximum normalized bead fluorescence) to allow comparison of bacterial distribution in the mucus between different treatment groups.
Visualization and Tracking of Mucus-Associated Bacteria.
Colonic tissue was mounted in a horizontal perfusion imaging chamber, and 2 µm Fluosphere beads were applied apically to visualize the IM/PM interface. Mucus-associated bacteria and colonic tissues were stained with 10 µM Syto9 in Krebs-mannitol for 30 min at RT. For experiments where tissue was treated with ZG16, the recombinant protein was added to the staining solution at a concentration of 350 µg/mL. The staining solution was replaced with fresh Krebs-mannitol buffer. Bacteria, tissue, and beads were imaged using a confocal microscope (Zeiss LSM 700) using 488/555-nm lasers and a Plan-Apochromat 20×/1.0DIC water objective lens. Images and 1.5-min time series were acquired. Time series were exported to Imaris software (Bitplane) to track the movement of bacteria within the mucus and beads at the IM/PM interface. The motile capacity (MC) of bacteria and beads was determined by recording the maximum speed of individual detected tracks. Bacteria were classed as motile if their MC exceeded the mean MC of the beads tracked in each time series or were classed as nonmotile if their MC equaled or was less than this value.
DNA Extraction from Stool, Mucus, and Systemic Tissues.
Mucus and underlying tissue were acquired from opened and pinned colonic tissue using sterile 2-mm biopsy punches (Miltex). Stool and systemic tissues were acquired by dissection. All samples were transferred to 1-mL Tris-EDTA buffer with 0.5% SDS (wt/vol) and 200 µg/mL proteinase K in Lysing Matrix E tubes (MP Biomedicals). Tubes were incubated at 55 °C for 1 h, after which DNA was isolated using mechanical disruption and phenol:chloroform:isoamylalcohol extraction as previously described (24). Extracted DNA was precipitated using ethanol and 0.2M NaCl. DNA was pelleted by centrifugation at 20,000 × g for 20 min and washed with 70% ethanol. DNA pellets were rehydrated in 100 µL Tris-EDTA buffer and stored at ‒20 °C until used. Blank sample collection tubes were prepared and exposed to the dissection equipment, buffers, etc., at the time of sampling to control for potential contamination.
The 16S qPCR Analysis.
DNA extractions were quantified using a Nanodrop spectrophotometer (Thermo). To allow quantification of low-copy number bacterial 16S DNA by qPCR the ratio of 16S DNA to total DNA was increased by limited cycle number (LCN) PCRs amplifying the whole 16S gene (20). Fifty microliters LCN PCRs were prepared using HotStar Taq Plus PCR Mastermix (Qiagen), 0.2 µM universal forward primer 27F (AGAGTTTGATCMTGGCTCAG), 0.2 µM universal reverse primer 1492R (CGGTTACCTTGTTACGACTT), and 500 ng template DNA. Thermocycling conditions were: 95 °C for 5 min, 16 cycles of 94 °C for 1 min, 55 °C for 1 min, 72 °C for 1.5 min, and 72 °C for 10 min. The 16S standards (quantified E. coli 16S DNA), contamination controls, and no-template controls were amplified at the same time as samples. Amplified samples, standards, and controls were then analyzed by qPCR to determine the total number of 16S copies and the relative proportions of total 16S belonging to three taxonomic groups of bacteria (Bacteroidetes, Firmicutes, and Proteobacteria) as previously described (25). qPCRs were run on a CFX96 instrument (Bio-Rad) using the previously described thermocycling conditions (26). Quantification cycle (Cq) values and calibration curves were constructed from 16S standard data. Calibration curves were used to calculate the total original 16S copy number from mucus and systemic tissue samples. The 16S data obtained from samples were initially normalized to data from contamination controls and then normalized to mucus volume or tissue mass to allow comparison between different experimental groups.
Additional materials and methods are in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We acknowledge the assistance of Valentina Tremaroli, Rozita Akrami, and the Mammalian Protein Expression and Centre Cellular Imaging Core Facilities. This work was supported by the Swedish Research Council, The Swedish Cancer Foundation, The Knut and Alice Wallenberg Foundation, IngaBritt and Arne Lundberg Foundation, Sahlgren's University Hospital (LUA-ALF), Wilhelm and Martina Lundgren’s Foundation, Wiberg Foundation, Torsten Söderbergs Stiftelse, The Sahlgrenska Academy, National Institute of Allergy and Infectious Diseases (U01AI095473), The Swedish Foundation for Strategic Research–The Mucus-Bacteria-Colitis Center of the Innate Immunity Program, European Union Marie Curie, and Human Frontiers (B.O.S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Microbiota 16S rDNA sequencing data have been deposited to the ENA sequence read archive, www.ebi.ac.uk/ena/data/view (accession no. PRJEB15635).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1611400113/-/DCSupplemental.
References
- 1.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307(5717):1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 2.Johansson MEV, et al. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci USA. 2008;105(39):15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ermund A, Schütte A, Johansson MEV, Gustafsson JK, Hansson GC. Studies of mucus in mouse stomach, small intestine, and colon. I. Gastrointestinal mucus layers have different properties depending on location as well as over the Peyer’s patches. Am J Physiol Gastrointest Liver Physiol. 2013;305(5):G341–G347. doi: 10.1152/ajpgi.00046.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ambort D, et al. Calcium and pH-dependent packing and release of the gel-forming MUC2 mucin. Proc Natl Acad Sci USA. 2012;109(15):5645–5650. doi: 10.1073/pnas.1120269109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johansson MEV, Larsson JM, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4659–4665. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol. 2012;10(5):323–335. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johansson MEV. Fast renewal of the distal colonic mucus layers by the surface goblet cells as measured by in vivo labeling of mucin glycoproteins. PLoS One. 2012;7(7):e41009. doi: 10.1371/journal.pone.0041009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tateno H, et al. Human ZG16p recognizes pathogenic fungi through non-self polyvalent mannose in the digestive system. Glycobiology. 2012;22(2):210–220. doi: 10.1093/glycob/cwr130. [DOI] [PubMed] [Google Scholar]
- 9.Kumazawa-Inoue K, et al. ZG16p, an animal homolog of β-prism fold plant lectins, interacts with heparan sulfate proteoglycans in pancreatic zymogen granules. Glycobiology. 2012;22(2):258–266. doi: 10.1093/glycob/cwr145. [DOI] [PubMed] [Google Scholar]
- 10.Kanagawa M, et al. Structural basis for multiple sugar recognition of Jacalin-related human ZG16p lectin. J Biol Chem. 2014;289(24):16954–16965. doi: 10.1074/jbc.M113.539114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meroueh SO, et al. Three-dimensional structure of the bacterial cell wall peptidoglycan. Proc Natl Acad Sci USA. 2006;103(12):4404–4409. doi: 10.1073/pnas.0510182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim SJ, Chang J, Singh M. Peptidoglycan architecture of Gram-positive bacteria by solid-state NMR. Biochim Biophys Acta. 2014;1848(1 Pt B):350–62. doi: 10.1016/j.bbamem.2014.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schroeder BO, et al. Reduction of disulphide bonds unmasks potent antimicrobial activity of human β-defensin 1. Nature. 2011;469(7330):419–423. doi: 10.1038/nature09674. [DOI] [PubMed] [Google Scholar]
- 14.Kanagawa M, et al. Crystal structures of human secretory proteins ZG16p and ZG16b reveal a Jacalin-related β-prism fold. Biochem Biophys Res Commun. 2011;404(1):201–205. doi: 10.1016/j.bbrc.2010.11.093. [DOI] [PubMed] [Google Scholar]
- 15.Johansson MEV, et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut. 2014;63(2):281–291. doi: 10.1136/gutjnl-2012-303207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mathis D. Immunological goings-on in visceral adipose tissue. Cell Metab. 2013;17(6):851–859. doi: 10.1016/j.cmet.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444(7121):860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 18.Van der Sluis M, et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology. 2006;131(1):117–129. doi: 10.1053/j.gastro.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 19.Kühn R, Löhler J, Rennick D, Rajewsky K, Müller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75(2):263–274. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- 20.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313(5790):1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodríguez-Piñeiro AM, et al. Studies of mucus in mouse stomach, small intestine, and colon. II. Gastrointestinal mucus proteome reveals Muc2 and Muc5ac accompanied by a set of core proteins. Am J Physiol Gastrointest Liver Physiol. 2013;305(5):G348–G356. doi: 10.1152/ajpgi.00047.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baeckström D, Karlsson N, Hansson GC. Purification and characterization of sialyl-Le(a)-carrying mucins of human bile; evidence for the presence of MUC1 and MUC3 apoproteins. J Biol Chem. 1994;269(20):14430–14437. [PubMed] [Google Scholar]
- 23.Gustafsson JK, et al. An ex vivo method for studying mucus formation, properties, and thickness in human colonic biopsies and mouse small and large intestinal explants. Am J Physiol Gastrointest Liver Physiol. 2012;302(4):G430–G438. doi: 10.1152/ajpgi.00405.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zoetendal EG, et al. Isolation of DNA from bacterial samples of the human gastrointestinal tract. Nat Protoc. 2006;1(2):870–873. doi: 10.1038/nprot.2006.142. [DOI] [PubMed] [Google Scholar]
- 25.Bacchetti De Gregoris T, Aldred N, Clare AS, Burgess JG. Improvement of phylum- and class-specific primers for real-time PCR quantification of bacterial taxa. J Microbiol Methods. 2011;86(3):351–356. doi: 10.1016/j.mimet.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 26.Teo IA, Choi JW, Morlese J, Taylor G, Shaunak S. LightCycler qPCR optimisation for low copy number target DNA. J Immunol Methods. 2002;270(1):119–133. doi: 10.1016/s0022-1759(02)00218-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.