Significance
Endogenous circadian rhythms have been demonstrated in several model systems, including mammals, insects, and fungi, among many others. Cycles in behavior, physiology and gene expression have also been reported in the nematode Caenorhabditis elegans, although limited by experimental conditions. Here we report the application of a luciferase-based reporter to investigate circadian regulation in C. elegans. Our study demonstrates entrainable, endogenous, and temperature-dependent circadian rhythms in gene expression as well as part of the pathway for synchronization. Our results represent an innovative approach for the study of long-term gene expression in real time in this system, opening the way for novel research in neuroscience and molecular pathways in general, including the precise determination of its elusive circadian clock.
Keywords: C. elegans, circadian rhythms, luminescence, temperature, light
Abstract
Circadian rhythms are based on endogenous clocks that allow organisms to adjust their physiology and behavior by entrainment to the solar day and, in turn, to select the optimal times for most biological variables. Diverse model systems—including mice, flies, fungi, plants, and bacteria—have provided important insights into the mechanisms of circadian rhythmicity. However, the general principles that govern the circadian clock of Caenorhabditis elegans have remained largely elusive. Here we report robust molecular circadian rhythms in C. elegans recorded with a bioluminescence assay in vivo and demonstrate the main features of the circadian system of the nematode. By constructing a luciferase-based reporter coupled to the promoter of the suppressor of activated let-60 Ras (sur-5) gene, we show in both population and single-nematode assays that C. elegans expresses ∼24-h rhythms that can be entrained by light/dark and temperature cycles. We provide evidence that these rhythms are temperature-compensated and can be re-entrained after phase changes of the synchronizing agents. In addition, we demonstrate that light and temperature sensing requires the photoreceptors LITE and GUR-3, and the cyclic nucleotide-gated channel subunit TAX-2. Our results shed light on C. elegans circadian biology and demonstrate evolutionarily conserved features in the circadian system of the nematode.
Circadian rhythms are ubiquitously found in nature, and the underlying molecular architecture of circadian clocks appears to be relatively conserved (1, 2). This mechanism is based on a transcriptional-translational feedback loop (TTFL) in which transcription factors induce the expression of clock genes that then act to regulate their own transcription negatively, creating oscillating patterns of gene expression with a period close to 24 h (3). Additionally, this molecular circuitry can receive input cues from the environment and can convey temporal information to regulate overt rhythmic processes through output pathways. The most important synchronizers (zeitgebers) of biological rhythms are light and temperature (4). In addition to the TTFL, multiple levels of posttranscriptional controls are needed to fine-tune the molecular clock by adjusting the period of the oscillations, maintaining a robust amplitude and buffering the clock mechanisms against abrupt environmental changes (5).
Caenorhabditis elegans is a free-living soil nematode widely used as a model organism in neuroscience, genetics, and developmental biology, whose natural history is beginning to be uncovered (6). In its niche of soil and rotting vegetable matter, C. elegans encounters a complex biological environment subjected to daily temperature fluctuations as a consequence of solar energy (7) but also might be directly exposed to light on the soil surface or when it displays nictation behavior (6). Indeed, temperature and humidity vary with time of day, distance from the surface, and across the seasons. Soil temperature varies daily as a propagating sine wave whose amplitude decays exponentially with depth, with a maximum of 10 °C at the surface but less than 2 °C at a depth of 15–20 cm (7, 8), where C. elegans has been isolated (9). As the depth increases, the maximum of the sine wave is delayed from noon to the late afternoon, resulting in a greater phase delay between the light and temperature cues. This particular combination of environmental factors might be of relevance for C. elegans timekeeping (7).
C. elegans is a powerful and attractive model with which to study chronobiology because it is an organism with a completely sequenced genome, a compact nervous system, and a fully known connectome and is especially amenable to genetic modifications. However, the study of circadian rhythms in this model is far from complete, and the mechanism underlying the molecular basis of the central pacemaker remains elusive. Numerous circadian cycles have been described in the nematode at different levels, including behavior (locomotor activity) (10–12), physiology (defecation rate, pharyngeal pumping rate, and olfaction) (13, 14), metabolism (abiotic and biotic stress tolerance, food and oxygen consumption) (15, 16), protein activity and regulation (14, 17, 18), and gene expression (19). Recent exhaustive bioinformatics searches have identified proteins of relatively high homology to clock components from other species (20). However, these homolog genes play a key role in the developmental program of C. elegans (21–23), and their function in the circadian clock of the nematode is not clear.
Most attempts to identify the C. elegans clock have consisted mainly of a description of circadian outputs, applying classical in vitro methods or behavioral recordings, which have limitations for long-term studies in terms of sensitivity and accuracy (24, 25). Here we report the application of a luciferase-based reporter to investigate the circadian regulation of the suppressor of activated let-60 Ras (sur-5) gene in C. elegans, a strategy that has been very successful in other systems (26). We found that the sur-5 promoter drives the rhythmic expression of the luciferase reporter under a paradigm of light and temperature cues resembling the nematode’s natural conditions and that these rhythms showed the fundamental properties fulfilled by all biological clocks described so far.
The assay is noninvasive, highly sensitive, and has real-time resolution, which, combined with a fully annotated genome and numerous existing mutants, offers an exceptional opportunity for the molecular study of the circadian biology of C. elegans.
Results
In Vivo Bioluminescence Monitoring of sur-5 Circadian Expression in Nematode Populations.
The ubiquitous sur-5 promoter is widely used in C. elegans because it exhibits strong and constitutive expression throughout all developmental stages (27). Motivated in part by these features and because of the previously reported successful expression of the Photinus pyralis luciferase (luc) under this promoter in the nematode (28, 29), we chose it to test the reporter system in long-term recordings. We generated transgenic strains carrying the firefly luciferase cDNA fused by a short linker to the ORF of the green fluorescent protein (gfp) (28, 29) under the control of 1,052 bp of the sur-5 gene promoter (Fig. 1A). The fusion with gfp increases luciferase expression and generates high luminescence, whereas the light emitted by the unfused protein encoded by the sur-5::luc construct cannot be detected. Because the luc sequence was cloned in frame with the gfp S65C variant, which has three synthetic introns, we believe that these introns may act in cis to generate alternative splicing of the premature luc::gfp RNA, thus improving the luciferase expression and increasing luminescence emission (Fig. S1). This strategy allows bioluminescence monitoring of a small number of living adult nematodes in liquid medium for several days. Furthermore, the GFP fluorescence also enables studies of the pattern of spatiotemporal expression of the gene of interest and the selection of transgenic animals with the highest level of transgene expression.
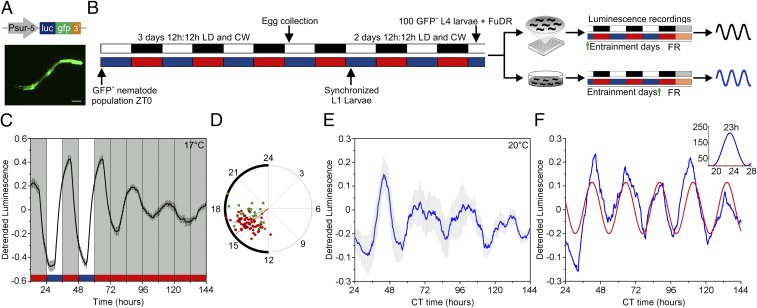
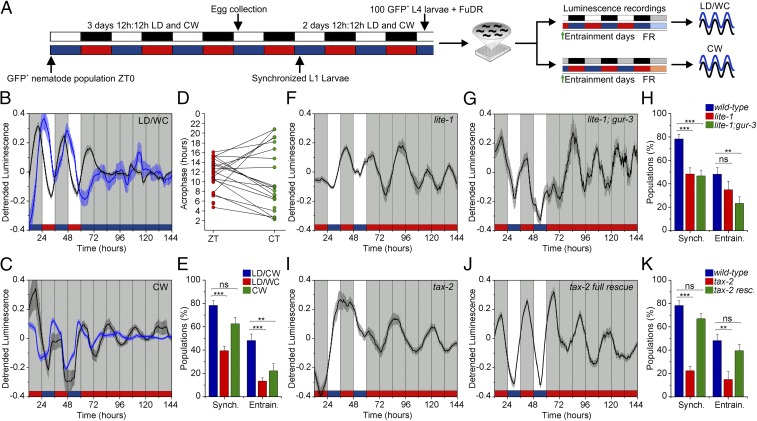
Fig. 1.
sur-5–driven luminescence is rhythmic under entrained and FR conditions. (A) Diagram of the reporter construct and fluorescent microscopy of an adult transgenic nematode illustrating the strong and ubiquitous expression of GFP. (Scale bar, 50 µm.) (B) Schematic of the assay strategy and photo/thermo-periodic conditions. Black/white bars indicate dark/light periods; blue/red bars indicate cold/warm periods; gray and orange bars indicate subjective night and subjective warm temperature, respectively. Nematodes on NGM plates with a lawn of E. coli were entrained under combined environmental 12-h/12-h cycles of LD (400/0 lx)/CW (18.5/20 °C) (ZT0, lights on and the onset of the cold-temperature phase). On the sixth day, 100 L4 transgenic nematodes expressing GFP were selected manually and transferred to the liquid luminescence medium. Luminescence was assayed under entrainment (LD/CW, 15.5/17 °C; multiwell setup) followed by FR (DD/WW, 20 °C) conditions (black waveform) or under FR conditions (dish plate setup; blue waveform). The green arrow indicates the start of recording. Black or blue line colors in C, E, and F indicate the type of assay performed. (C) Average reporter activity of entrained adult populations under LD/CW and FR conditions (n = 28). (D) Rayleigh plot showing the acrophases of luminescence emission in LD/CW (n = 58, red dots) and FR (n = 28, green dots) conditions. The remaining 30 populations were arrhythmic or showed masking under FR conditions (Rayleigh test, P < 0.001, multisample test for equal median directions, LD/CW median phase vs. FR, ns). (E) Average reporter activity of adult populations under FR conditions (DD/WW, 20 °C; n = 12). (F) Representative luminescence rhythm of one selected population from E. The red line represents the best-fitting curve (Cosinor analysis; R2 = 0.61). (Inset) The LS periodogram (P < 0.05). Luminescence signals are shown as mean ± SEM. Each population consisted of 100 adult nematodes and was considered an independent biological replicate.
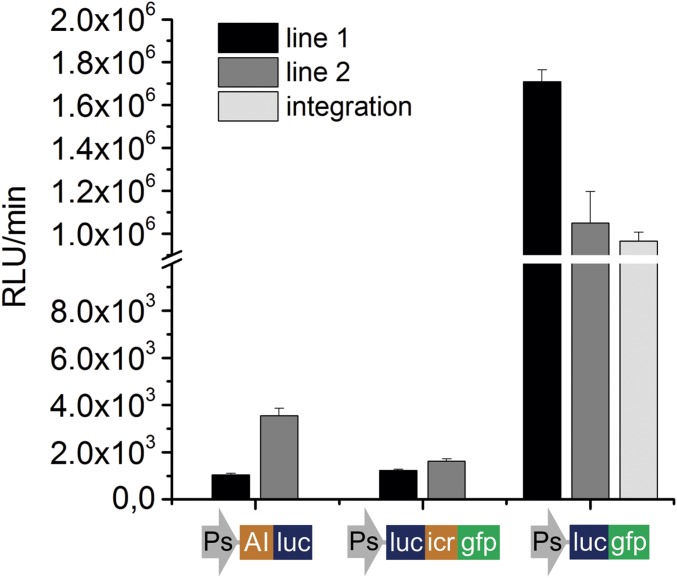
Fig. S1.
The luc::gfp fusion enhances reporter luminescence activity. Average relative luminescence units (RLU) per minute are shown for transgenic nematode populations with three different luciferase constructs: transgenic line 1, black bars; transgenic line 2, dark gray bars; and integrated line, light gray bars (n = 8 for each population). Below each set of bars a schematic of each construct is shown representing a vector with a fusion between an artificial intron (AI) and the luc CDS, a bicistronic vector containing the luc and the gfp S65C variant CDS (with three artificial introns) separated by an icr region [from Lee et al. (58)], and a vector containing a translational fusion between the luc and the gfp S65C variant CDS. All constructs contained the same sur-5 promoter (Ps) and the 3′ UTR region of pPD158.87. Under the same conditions, the luminescence signal from nematodes carrying Psur-5::luc alone was undetectable.
We used cycling conditions that resemble those found in the nematodes’ natural niche, i.e., an entrainment schedule that combines light and temperature cues: Light onset coincides with the start of the cold phase, and the temperature variations are small and therefore mimic the conditions found at C. elegans’ usual soil depth. We grew age-synchronized animals of transgenic lines carrying sur-5::luc::gfp under an environmental 12-h/12-h cycle of light/dark (LD) and cold/warm (CW) temperature (Δ = 1.5 ± 0.125 °C) (Fig. 1B) and assayed bioluminescence using a microplate luminometer inside an incubator.
We found that luminescence of nematode populations expressing sur-5::luc::gfp showed a clear circadian oscillation that entrained to the LD/CW cycle (15.5/17 °C). Luminescence increased before the onset of darkness and warm temperature, indicating anticipation of the zeitgeber transition, a signature feature of circadian synchronization (Fig. 1C). There was a clear phase correlation when the nematodes were left in constant darkness (DD) and warm temperature (WW), indicating that entrainment, rather than masking by environmental signals, occurred [the bioluminescence peaks occurred at zeitgeber time (ZT)15.6 ± 0.3 h and circadian time (CT) 16.8 ± 0.7 h] (Fig. 1D). The average period of the oscillation under constant conditions of darkness and 17 °C was 25 ± 0.4 h.
We also tested the ability of the nematodes to sustain endogenous rhythms in the absence of environmental stimuli, a fundamental property of circadian rhythms that reflects the internal regulation of the clock. Populations consisting of 100 adult nematodes in the luminescence medium were entrained under the LD/CW cycle for 3 d, and their bioluminescence was recorded in conditions of DD/WW (20 °C). We found robust circadian oscillations with an average period of 23.9 ± 0.5 h and a peak phase at CT18.8 ± 0.9 h (with CT12 derived from the previous ZT12 phase). The oscillations were sustained for at least 4 d at 20 °C, indicating that individual nematodes within each population were in synchrony with each other for several days (Fig. 1 E and F).
sur-5 Luminescence Rhythms Are Temperature-Compensated and Respond to Phase Changes in the Environmental Cues.
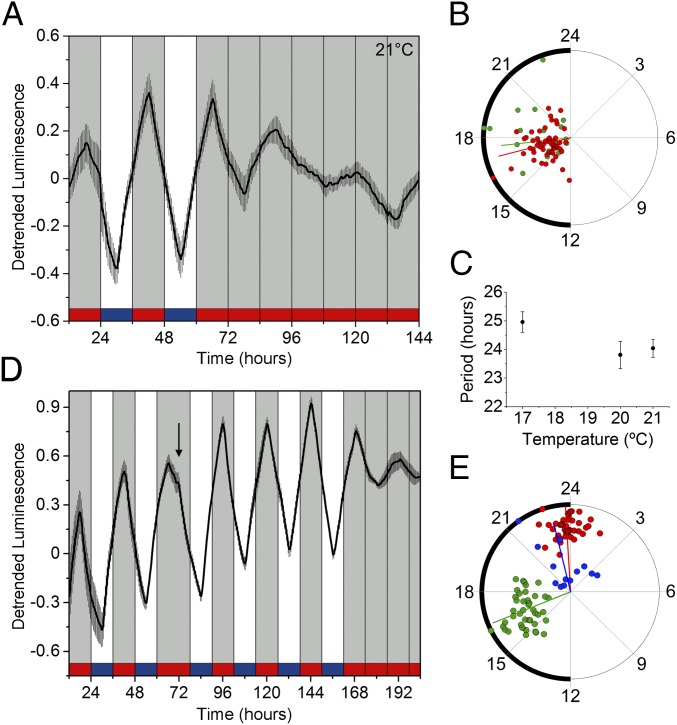
For the sur-5 luminescence rhythms to be considered circadian, they must fulfill two additional fundamental properties: (i) temperature compensation, which means that the period under free-running (FR) conditions must be roughly consistent across a range of different temperatures within the organism’s physiological range, and (ii) phase resetting after changes in environmental cues. As shown in Fig. 2 A and B, we found a strong temperature compensation of the circadian period between 17 °C and 21 °C (25 ± 0.4 h and 24 ± 0.3 h, respectively) with a Q10, the factor by which the rate of a reaction increases with the rise in the temperature, of 1.1 (Fig. 2C). Also, there was a small difference in acrophase at 21 °C (ZT16.9 ± 0.5 h) and 17 °C (ZT15.6 ± 0.3 h) (Fig. 2B).
Fig. 2.
Luminescence rhythms exhibit temperature compensation and respond to a phase shift in the photic and thermal periodic conditions. (A) Average reporter activity of adult populations under LD/CW and FR conditions (21 °C, n = 21). (B) Rayleigh plot showing the acrophases of the luminescence signal in LD/CW (red dots, n = 61) and FR (green dots, n = 21) conditions. The remaining 40 populations were arrhythmic or were masked under FR conditions (Rayleigh test, P < 0.001, multisample test for equal median directions, LD/CW median phase vs. FR, ns). (C) The circadian period did not change significantly when recorded at 17, 20, or 21 °C (one-way ANOVA with Tukey’s post hoc test, ns). (D) Average population luminescence rhythms after a 6-h phase shift (n = 13). The arrow indicates the time of the phase shift. When placed in FR conditions, acrophases were similar to those after re-entrainment (LD/CW 1: ZT16.3 ± 0.3 h; LD/CW 2: ZT23.8 ± 0.2 h; FR: CT22.6 ± 0.6 h). (E) Rayleigh plots of LD/CW 1 (green dots; n = 43), LD/CW 2 (red dots; n = 43), and FR (blue dots; n = 13) (Rayleigh test, P < 0.001; one-way ANOVA with Tukey’s post hoc test, LD/CW 1 median phase vs. LD/CW 2, P < 0.0001; LD/CW 2 median phase vs. FR, ns). Luminescence signals are shown as mean ± SEM. Each population consisted of 100 adult nematodes and was considered an independent biological replicate.
To determine if the rhythms could be reset by the zeitgebers, we applied a classical phase-shift approach in which transgenic nematode populations were first entrained to an LD/CW cycle for 4 d and then were subjected to a phase change caused by a 6-h night extension (lights on and warm temperature onset were shifted from 9:00 AM to 3:00 PM). C. elegans populations re-entrained rapidly after the 6-h phase delay and gradually adjusted to the new schedule. The response represented true re-entrainment rather than masking, because the phase peak was maintained under constant conditions for the populations that were still luminescent at the end of the experiment (peak during LD/CW 1: ZT1 6.3 ± 0.3 h; peak during LD/CW 2: ZT2 3.8 ± 0.2 h; peak during DD/WW: CT22.6 ± 0.6 h) (Fig. 2 D and E and Fig. S2).
Fig. S2.
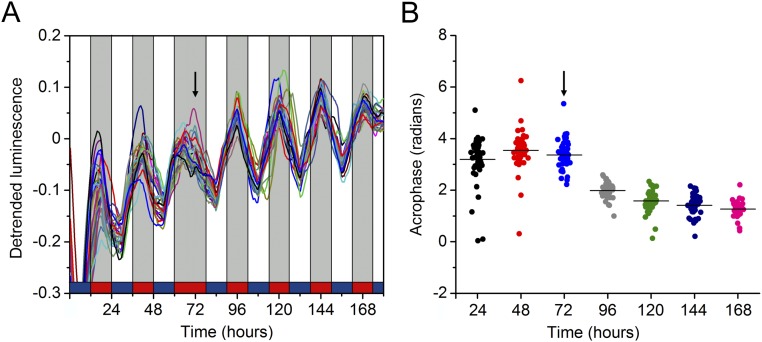
Bioluminescent rhythms resynchronize after a phase shift. (A) Individual luminescence plots of worm populations during a 6-h phase-shift experiment (n = 43). The arrow indicates the time of the phase shift. Black/white bars indicate dark/light, respectively; blue/red bars indicate cold/warm, respectively. (B) Representation of acrophase changes throughout the entrainment days showing the gradual phase adjustment in the days after the shift. For comparison purposes, the x axis corresponds to the temporal scale shown in A. Each population consisted of 100 adult nematodes and was considered an independent biological replicate.
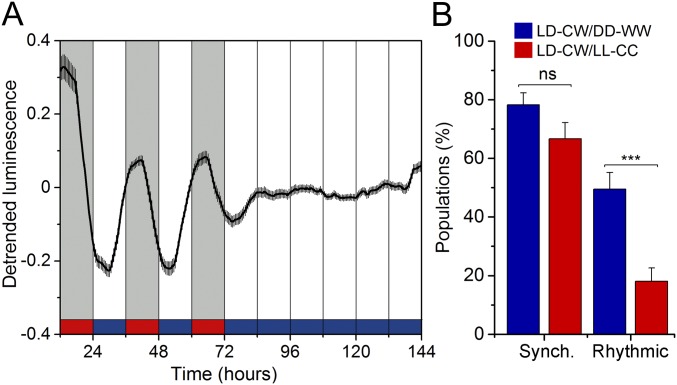
Constant light (LL) can induce behavioral and physiological arrhythmicity, for example by desynchronizing clock neurons in the central master clock (SCN) in mammals (30). We sought to determine if a similar response could be found in nematodes entrained under a LD/CW cycle followed by LL and constant cold temperature (CC) conditions (LL/CC). We found a significant decrease in rhythmicity, as revealed by the proportion of bioluminescent populations compared with those in the LD/CW followed by DD/WW conditions (18 vs. 49%, respectively) (Fig. S3).
Fig. S3.
Constant light conditions severely affect luminescence rhythms. (A) Average reporter activity of adult populations under LD/CW and then LL/CC conditions (15.5 °C, n = 36). (B) Proportion of synchronized (i.e., with their phase set by the LD/CW zeitgeber) and rhythmic (circadian under constant conditions) populations. Constant light and cold temperature increased the proportion of arrhythmic populations (LD/CW–DD/WW, n = 101; LD/CW–LL/CC n = 72; ***P < 0.001, two-tailed Fisher’s exact test). Luminescence signals are shown as mean ± SEM. Each population consisted of 100 adult nematodes and was considered an independent biological replicate.
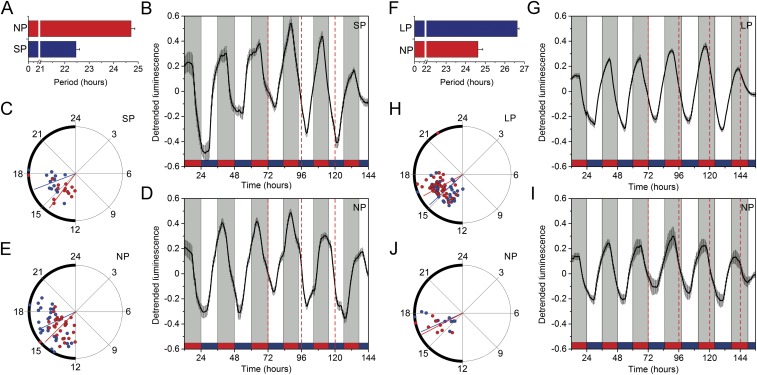
To test the entrainment capabilities of C. elegans further, we subjected nematode populations to short (22 h; T22) or long (26 h; T26) LD/CW cycles. As shown in Fig. S4, in both cases we found two well-defined groups of nematode populations, one which tended to entrain to the cycle and the other one which free-ran with a circadian period close to 24 h. Moreover, under T22 cycles nematodes exhibited a phase delay, whereas under T26 cycles a phase advance was evident, compared with regular (T24) cycles. These results suggest that 22- and 26-h cycles are close to the limits of entrainment for C. elegans.
Fig. S4.
Luminescence rhythms can be entrained to T22 and T26 cycles. Nematode populations were kept for 4 d under normal 12-h:12-h LD/CW (15.5/17 °C) conditions and for the following 4 d under a short (11-h:11-h) LD/CW (15.5/17 °C) or a long (13-h:13-h) LD/CW (15.5/17 °C) cycle. (A) Period length of nematode populations kept under T22 cycles. The reporter activity of populations (n = 60) exhibited either a short period (SP; 22.5 ± 0.1 h; blue bar, n = 14) or a normal period (NP, 24.7 ± 0.1 h; red bar, n = 29). The remaining 17 populations were arrhythmic. (B and C) Average luminescence signal (B) and Rayleigh plot (C) showing the acrophases of luminescence emission of the SP group under T24 (red dots) and T22 (blue dots) conditions (Rayleigh test, P < 0.001; LD/CW 24 h ZT14.5 ± 0.4 h vs. LD/CW 22 h ZT16.7 ± 0.3 h, P < 0.001; multisample test for equal median directions). (D and E) Average luminescence signal (D) and Rayleigh plot (E) showing the acrophases of luminescence emission of the NP group under T24 (red dots) and T22 (blue dots) conditions (Rayleigh test, P < 0.001; LD/CW 24 h ZT15 ± 0.4 h vs. LD/CW 22 h ZT16.3 ± 0.5 h, ns; multisample test for equal median directions). (F) Period length of nematodes kept under T26 cycles. The reporter activity of populations (n = 60) exhibited either a long period (LP, 26.6 ± 0.1, blue bar, n = 37) or a normal period (NP, 24.6 ± 0.2 h, red bar, n = 10). The remaining 13 populations were arrhythmic. (G and H) Average luminescence signal (G) and Rayleigh plot (H) showing the acrophases of luminescence emission of the LP group under T24 (red dots) and T26 (blue dots) conditions (Rayleigh test, P < 0.01; LD/CW 24 h ZT16.3 ± 0.4 h vs. LD/CW 26 h ZT15.5 ± 0.4 h, P < 0.05; multisample test for equal median directions). (I and J) Average luminescence signal (I) and Rayleigh plot (J) showing the acrophases of luminescence emission of the NP group under T24 (red dots) and T26 (blue dots) cycles (Rayleigh test P < 0.001; LD/CW 24 h ZT16.3 ± 0.3 h vs. LD/CW 26 h ZT16.5 ± 0.3 h, ns; multisample test for equal median directions). Luminescence signals are shown as mean ± SEM. Each population consisted of 100 adult nematodes and was considered an independent biological replicate. Red dashed lines in B, D, G, and I indicate a time lapse of 24 h in the T22 and T26 cycles.
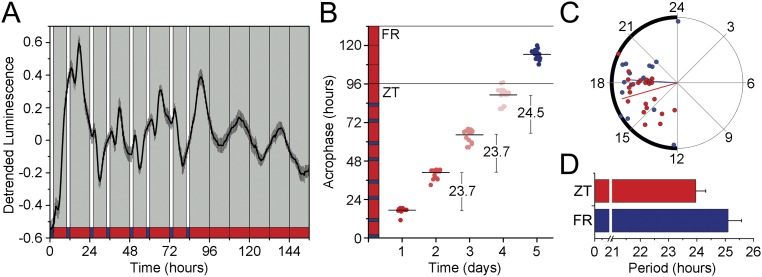
Finally, we also entrained the nematodes to a classical skeleton photo- and thermoperiod of two pulses (2 h of light and cold conditions) at the beginning and at the end of the circadian day (ZT0–2 and ZT10–12). As shown in Fig. S5, about 50% of the populations were clearly able to synchronize to the skeleton cycles with an average period of 23.9 ± 0.3 h. Interestingly, most of the populations also exhibited some masking in response to the 2-h light/cold pulses. After the nematode populations were released into constant conditions, clear and well-defined circadian activity was found with a period of 25.1 ± 0.5 h [Lomb–Scargle (LS) periodogram] in ∼40% of the populations.
Fig. S5.
Luminescence rhythms can be entrained to skeleton photo/thermo-cycles. Nematode populations (n = 48) were entrained to a classical skeleton photo/thermo-cycle for 4 d [ZT, 2-h L/C pulses (light and 15.5 °C) at ZT0–2 and ZT10–12) followed by 3 d of FR conditions (DD/WW, 17 °C). (A) Average reporter activity of the entrained populations (n = 15). (B) Dot plot of daily acrophases under ZT (daily periods were calculated as a mean of the distance between the acrophases with a moving average of 24 h are shown) and the first day of FR conditions (n = 15). (C) Rayleigh plot showing the acrophases of luminescence emission of populations under ZT (red dots) and FR (blue dots) conditions (Rayleigh test, P < 0.001; ZT16.9 ± 0.5 h vs. CT18.24 ± 0.7 h, ns; multisample test for equal median directions). (D) Average period of the luminescence activity under ZT conditions (red bar, n = 24; ZT LS period 23.9 h ± 0.3 h) or FR conditions (blue bar, n = 19; LS period 25.1 h ± 0.5 h). The remaining populations were arrhythmic. Luminescence signals are shown as mean ± SEM. Each population consisted of 100 adult nematodes and was considered an independent biological replicate.
Single-Nematode Bioluminescence Rhythms.
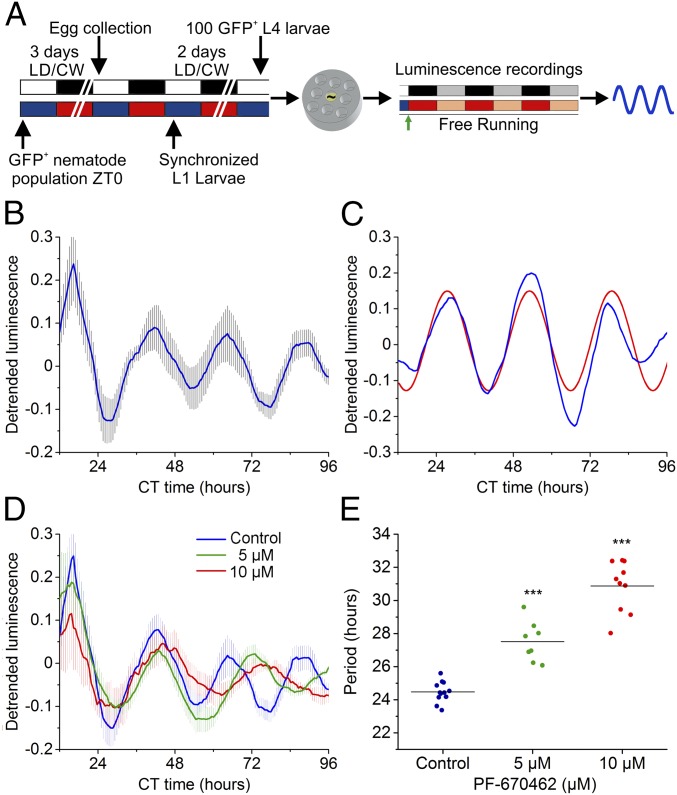
Once we were able to record and analyze robust luminescence rhythms at the population level, we sought to record luminescence from individual nematodes. It should be noted that single-nematode recordings are quite challenging to perform because of the small size of the nematodes and their high motility rate and because the far lower luminescence signal emitted by single nematodes compared with that of a population of 100 individuals greatly reduces the signal-to-noise ratio.
By adjusting the entrainment and data-acquisition protocols, we recorded clear single-nematode luminescence under FR conditions (Fig. 3A). These rhythms are detectable for at least 4 d at 20 °C, with an average period of 24.5 ± 0.2 h and a nocturnal peak phase close to that found in the nematode populations (Fig. 3B). Because single-nematode signals decrease more rapidly, nematodes were transferred immediately to FR conditions after the medium-change step of the protocol (compare Figs. 1B and 3A). The lack of this entrainment step could explain, to some extent, the loss of synchrony evidenced in some nematodes that exhibited rhythms with diurnal peak phases (Fig. 3C).
Fig. 3.
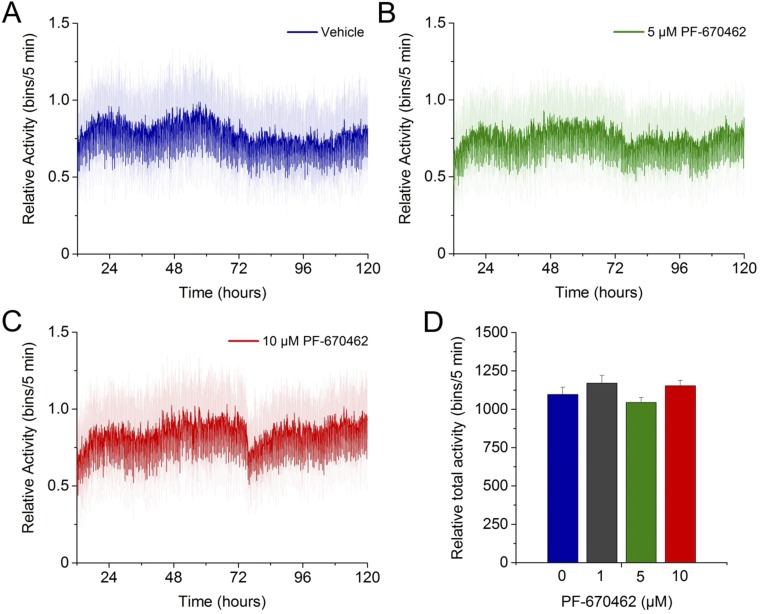
Single-nematode luminescence assays reveal robust FR rhythms that can be modulated pharmacologically by the application of a CK1ε/δ inhibitor. (A) Schematic of the assay strategy and photo/thermo-periodic conditions. Black/white bars indicate dark/light periods; blue/red bars indicate cold/warm periods; gray and orange bars indicate subjective night and subjective warm temperature, respectively. Nematodes on NGM plates with a lawn of E. coli were entrained under combined 12-h/12-h LD (400/0 lx)/CW (18.5/20 °C; ZT0, lights on and onset of the cold-temperature phase) environmental cycles. On the sixth day, individual L4 transgenic nematodes expressing GFP were selected manually and were transferred to the liquid luminescence medium. The signal was monitored under FR conditions (DD, 20 °C) for several days. (B) Average rhythms of reporter activity in single-nematode assays (n = 12). (C) Plot of representative single-nematode reporter activity with a diurnal phase. The red line represents the best-fitting curve (cosinor analysis, R2 = 0.72, LS period 24.4 h acrophase CT1.76 h). (D and E) Treatment with PF-670462 lengthens the circadian luminescence period in vivo in a concentration-dependent manner. (D) Average luciferase activity rhythms of single-nematode assays under FR conditions with treatment with vehicle (water; blue line; n = 12), 5 µM of PF-670462 (green line; n = 8), or 10 µM of PF-670462 (red line; n = 10). (E) Average periods of reporter activity under control (blue dots; 24.5 ± 0.2 h; n = 12) or treatment (green dots; 27.5 ± 0.4 h, n = 8; red dots, 30.9 ± 0.5 h, n = 10) conditions. PF-670462 significantly lengthens the circadian period (***P < 0.001 vs. vehicle, one-way ANOVA with Dunnett's post hoc test). Luminescence signals are shown as mean ± SEM.
Treatment with a Casein Kinase 1 ε/δ Inhibitor Lengthens the Period of the Luminescent Rhythms.
Casein kinase 1 (CK1) plays an important role in the posttranslational modification of core clock proteins, modulating protein–protein interactions, nuclear entry and export, and degradation (31). A dominant role for CK1-mediated protein phosphorylation in circadian timing has been described in mammals (32), Drosophila melanogaster (33), Neurospora crassa (34), and Ostreococcus tauri (35), even when the putative target proteins of CK1 in their TTFL systems are not conserved. C. elegans has only one protein, encoded by the protein kinase gene kin-20 (20), that is a homolog to the mammalian casein kinases 1ε/δ. To test whether PF-670462, a CK1ε and δ inhibitor and a common pharmacological modulator of the circadian period in other species, could also influence the sur-5 luminescent rhythms, we recorded the luminescence of single nematodes exposed to different concentrations of the drug. Treatment with PF-670462 induced significant period lengthening under FR conditions in a dose-dependent manner (control, 24.5 ± 0.2 h; 5 µM PF-670462, 27.5 ± 0.4 h; 10 µM PF-670462, 30.9 ± 0.5 h) (Fig. 3 D and E). A similar effect on circadian period length was reported previously in other systems (35, 36). It is important to note that no toxic effects were found with these PF-670462 concentrations in a locomotor activity toxicity test (Fig. S6). These results strongly suggest the presence of a conserved posttranslational mechanism of clock modulation in C. elegans.
Fig. S6.
PF-670462 is not toxic at the concentrations used in the luminescence assays. (A–D) Results from a toxicity text that recorded the global motility of populations of wild-type adult nematodes (N2) over time under different concentrations of the PF-670462, using an infrared tracking device (WMicroTracker) previously reported for toxicity screenings (59). Plots show the mean relative activity pattern ± SEM of nematodes treated with vehicle (A) or with 5 µM (B) or 10 µM of PF-670462 (C) under DD/WW (18 °C) conditions (n = 24 populations per treatment, 50 nematodes per well). The assay was performed for a period equivalent to the duration of a typical single-nematode luminescence assay. (D) No significant differences were found in the total activity of the control group treated with vehicle (water) and that of groups treated with different concentrations of PF-670462 (1 µM, 5 µM, and 10 µM) (one-way ANOVA with Dunnett's post hoc test, ns).
Rhythmic Expression of sur-5 Underlies Luminescence Rhythms.
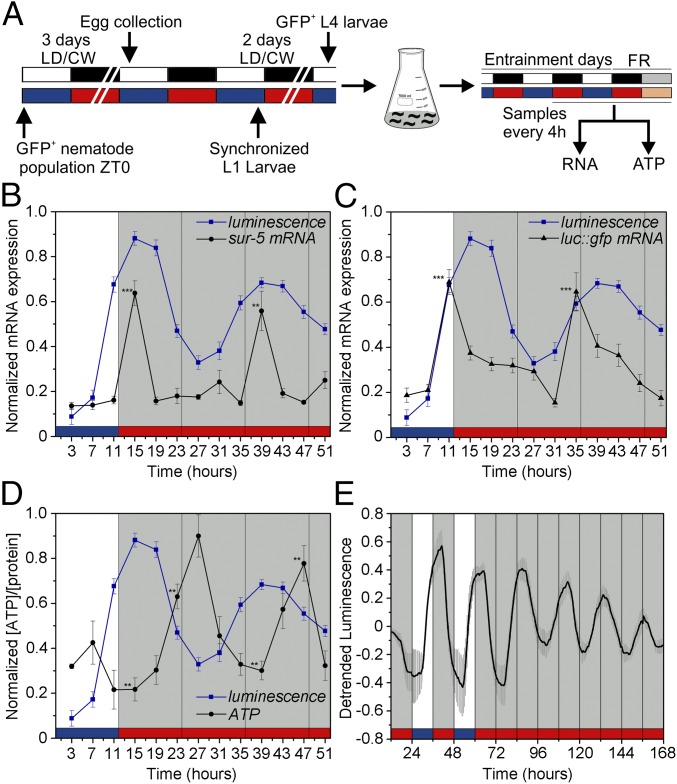
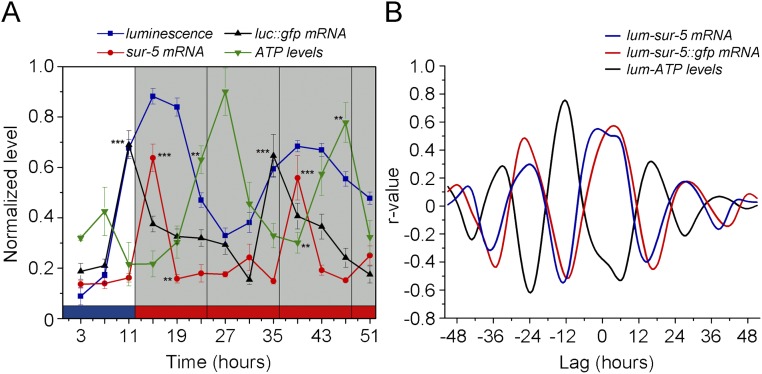
After demonstrating that sur-5–driven luminescence shows a clear oscillation with defining features of circadian rhythmicity, we next sought to explore whether these cycles could be explained by changes in mRNA levels. We performed quantitative real-time PCR experiments under light and temperature cycles (Fig. 4A) and observed a significant rhythmic expression of the sur-5 gene with a peak close to ZT15 that is sustained even in the absence of cyclic environmental cues, indicating a circadian transcriptional regulation of the gene (Fig. 4B). Accordingly, the luc::gfp reporter transcription also was rhythmic and phase-advanced with respect to bioluminescence (Fig. 4C). Taken together, the direct correlation between the reporter transcript levels and in vivo luminescence and the rhythmic expression pattern of the sur-5 mRNA (Fig. S7) strongly support the notion that circadian transcriptional regulation of sur-5 expression underlies the oscillations in luminescence.
Fig. 4.
Rhythms in reporter luminescence reflect transcriptional activity of the sur-5 gene. (A) Schematic of the assay strategy and photo/thermo-periodic conditions. Black/white bars indicate dark/light periods; blue/red bars indicate cold/warm periods; gray and orange bars indicate subjective night and subjective warm temperature, respectively. Nematodes on NGM plates with a lawn of E. coli were grown under combined environmental 12-h/12-h LD (400/0 lx)/CW (18.5/20 °C; ZT0, lights on and the onset of the cold-temperature phase) cycles. On the sixth day, L4 transgenic nematodes expressing GFP were transferred to flasks with liquid luminescence medium, entrained under the LD/CW (15.5/17 °C) condition for three more days, and released into FR conditions (DD, 17 °C). Samples were collected every 4 h starting at ZT3 during the last 3 d for population real-time PCR and ATP analysis. (B and C) mRNA oscillations of sur-5 (B) and the luc::gfp reporter (C) in the LD/CW condition (n = 4; ***P < 0.001 vs. peak; one-way ANOVA with Dunnett's post hoc test) and in the FR condition (DD, 17 °C) (n = 4; ***P < 0.001, **P < 0.01 vs. peak, one-way ANOVA with Dunnett's post hoc test) (D) Endogenous normalized ATP levels measured in vitro in the LD/CW condition (n = 4; **P < 0.01 ZT15 vs. ZT23, one-way ANOVA with Dunnett's post hoc test) and the FR condition (DD,17 °C (n = 4; **P < 0.01 ZT39 vs. ZT47, one-way ANOVA with Dunnett's post hoc test). The blue line represents the in vivo average luminescence of worm populations (100 nematodes in each population) obtained under the same conditions (n = 26). (E) Bioluminescent reporter activity of worm populations with the exogenous addition of 1 mM ATP (n = 8, mean ± SEM). Real-time PCR (B and C) and ATP levels (D) plots show the mean ± SEM of four independent biological replicates.
Fig. S7.
Luminescence rhythms correlate with transcriptional activity but do not reflect endogenous ATP levels. (A) The sur-5 and luc::gfp mRNA circadian patterns are in phase with the average luminescence pattern of nematode populations, whereas the daily variations in total ATP level are in antiphase with bioluminescence. (B) Cross-correlation analysis shows that the acrophase of the luminescence rhythms exhibits a 3-h phase advance with respect to the sur-5 expression pattern (cross-correlation test; luminescence vs. sur-5 mRNA, blue line; n = 26 and n = 4, respectively), a 3-h phase delay with respect to luc::gfp reporter mRNA (cross-correlation test; luminescence vs. luc::gfp mRNA, red line; n = 26 and n = 4, respectively), and an 11-h phase delay with respect to total ATP levels (cross-correlation test; luminescence vs. ATP, black line; n = 26 and n = 4, respectively).
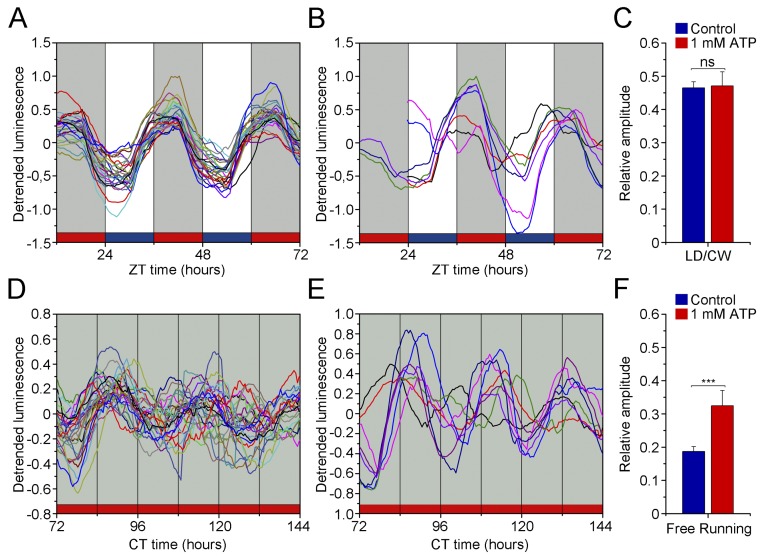
Reporter activity cycles also could reflect a basic circadian regulation of metabolic variables that also are considered a robust output of the clock in several models (37). The luciferase encoded by the reporter catalyzes the oxidation of d-luciferin in a reaction that uses ATP as substrate (38). We set out to test if ATP levels also are under circadian regulation by determining the endogenous ATP levels of samples of transgenic populations without d-luciferin taken at different time points under the LD/CW cycle. We found a clear and significant circadian rhythm in the endogenous ATP levels, in antiphase to the bioluminescence rhythm (Fig. 4D and Fig. S7). To confirm that the luciferase gene activity does not directly reflect the endogenous ATP level (29), we repeated the same luminescence recordings of transgenic worm populations in vivo but with an excess of exogenous ATP (1 mM) added to the liquid culture medium. No differences were found in the rhythmic pattern of sur-5 expression, although the circadian rhythms under FR conditions appeared to have higher amplitude with excess ATP than with normal ATP levels (Fig. 4E and Fig. S8). The clear daily variation in endogenous ATP levels indicates a circadian regulation of the metabolism.
Fig. S8.
The addition of exogenous ATP improves the amplitude of the general pattern of the luminescence oscillations under FR conditions. (A and B) Individual luminescence plots of nematode populations under LD/WC conditions without ATP treatment (A, n = 26) and with 1 mM ATP (B, n = 8). (C) The exogenous addition of 1 mM ATP does not change the general pattern of luminescence oscillations under LD/CW (ns, t test). (D and E) Individual luminescence plots of nematode populations under FR conditions without ATP treatment (D, n = 26) and with 1 mM ATP (E, n = 8). (F) Treatment with 1 mM ATP generates a significantly higher amplitude in circadian oscillations with respect to basal conditions only in the FR condition (***P < 0.001, t test). Each population consisted of 100 adult nematodes and was considered an independent biological replicate.
Light and Temperature Entrainment of Bioluminescent Rhythms Requires the Photoreceptors LITE and GUR-3, and the Cyclic Nucleotide-Gated Channel Subunit TAX-2.
One unanswered question is how the nematodes sense zeitgebers to regulate gene expression. The nematode lacks eyes, has a unique and noncanonical photoreception system different from all studied model organisms (39), and shows a high sensitivity to temperature (40). We explored the underlying mechanisms that control synchronization of the circadian clock by analyzing how light and temperature cues influence oscillation patterns.
To study if the phase relationship between each zeitgeber is important for synchronizing the nematodes, we maintained the light schedule and inverted the temperature cycle. In this inverted LD/WC paradigm, ZT0 indicates lights on and the onset of the warm phase (Fig. 5A). Remarkably, when transgenic nematode populations were subjected to these new conditions, only 39% were synchronized, and most (85%) showed masking evidenced by an arrhythmic pattern of the luminescence recordings under constant conditions (Fig. 5 B and E). Luminescence rhythms showed two well-defined groups with different peak phases, one close to the late dark phase of the cycle and the cold temperature condition (ZT19.9 ± 0.6 h; Rayleigh test; P < 0.001; black line in Fig. 5B) and the other related to the transition from the dark/cold to the light/warm condition (ZT2.30 ± 0.4 h; Rayleigh test; P < 0.001; blue line in Fig. 5B). These results suggest that only a precise combination of both environmental cues is capable of better entraining the nematodes (Fig. 5E).
Fig. 5.
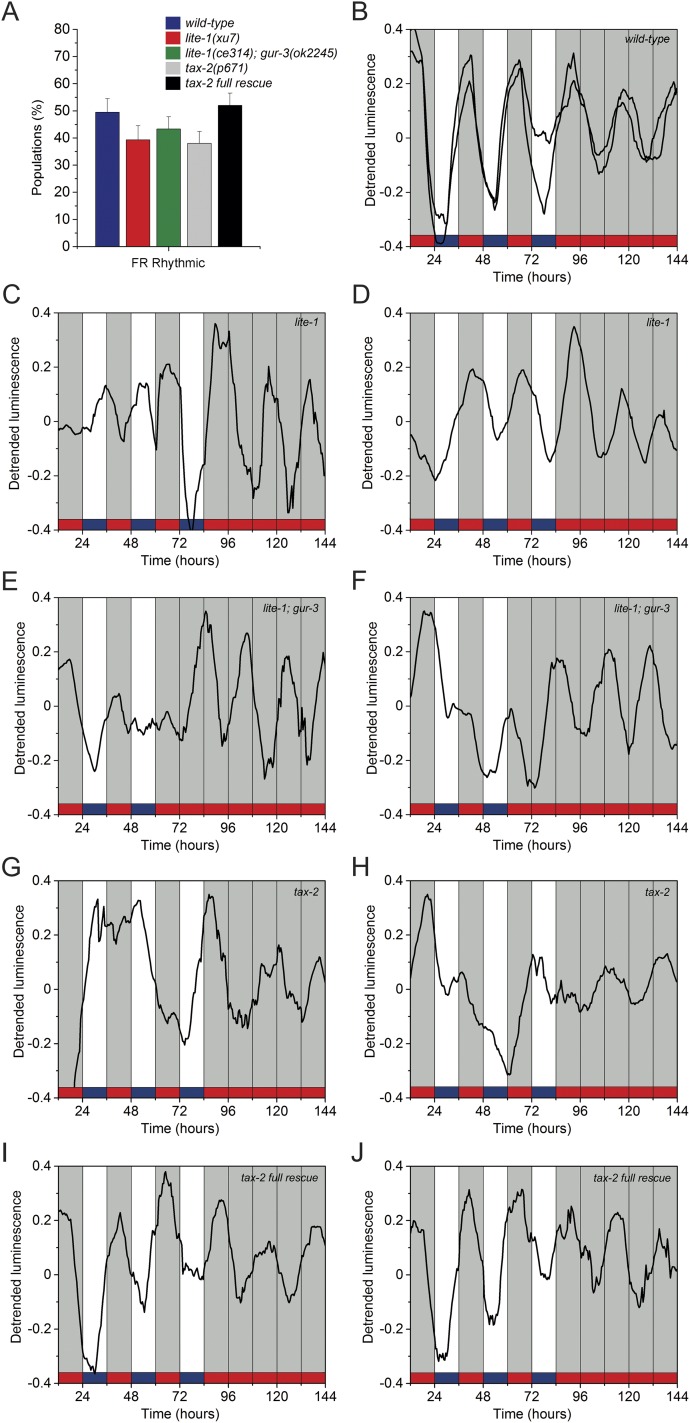
Photic and temperature entrainment pathways for luminescence rhythms. (A) Schematic of the assay strategy and photo/thermo-periodic conditions for the experiments in B and C. Black/white bars indicate dark/light periods; blue/red bars indicate cold/warm periods; gray bars indicate subjective night, and blue-red/orange bars indicate subjective cold/warm temperatures, respectively. (B and C) Nematodes on NGM plates with a lawn of E. coli were entrained under combined 12-h/12-h environmental LD (400/0 lx)/CW (18.5/20 °C; ZT0, lights on and onset of the cold-temperature phase) cycles. On the sixth day, 100 L4 transgenic nematodes expressing GFP were selected manually and were transferred to the liquid luminescence medium. Luminescence was assayed in a multiwell setup either under a different paradigm of LD/WC (17/15.5 °C; ZT0, lights on and onset of the warm-temperature phase) and DD/CC conditions (B) or under a 12-h/12-h DD/CW (15.5/17 °C) cycle (C). The green arrow indicates the start of recording. The percentage of synchronized populations is defined as the number of populations with a period and phase set by the zeitgeber conditions over the total number of populations tested; the percentage of entrained populations is the relative number of populations that retained their circadian phase when placed under constant conditions. (B) Bioluminescent reporter activity of nematode populations masked under the LD/WC cycle showed two well-defined groups with different acrophases (ZT19.9 ± 0.6 h, black line, n = 39; and ZT2.30 ± 0.4, blue line, n = 17; Rayleigh test, P < 0.001). (C) Bioluminescent reporter activity of nematode populations with entrained (black line, n = 10) and masked (blue line, n = 35) under the CW cycle. (D) Linear Rayleigh plot of the acrophases under CW (n = 45; Rayleigh test, P < 0.01; mean phase ZT: 13.2 ± 0.7 h) and WW conditions (n = 24; Rayleigh test, ns). The remaining 21 populations were arrhythmic under the FR condition. (E) Proportion of synchronized and entrained populations under different environmental conditions (LD/CW, n = 101; LD/WC, n = 165; CW, n = 72; ***P < 0.001, **P < 0.01, two-tailed Fisher’s exact test). (F and G) Representative luciferase rhythms of lite-1(xu7) mutants (F) and lite-1(ce314); gur-3(ok2245) mutants (G) (n = 30) under LD/CW and FR (DD/WW) cycles. (H) Percentage of entrained and synchronized populations of wild-type N2 (n = 101), lite-1(xu7) (n = 89), and lite-1(ce314); gur-3(ok2245) mutants (n = 120) under the assayed conditions; ***P < 0.001; **P < 0.01, two-tailed Fisher’s exact test. (I and J) Representative rhythms of luciferase activity of tax-2(p671) mutants (I) and tax-2 (p671) full rescue worms (J) (n = 30) under the classical LD/CW and FR (DD/WW) cycles. (K) Percentage of entrained and synchronized wild-type N2 (n = 101), tax-2(p671) (n = 121), and tax-2(p671) full rescue (n = 121) populations under the assayed conditions. ***P < 0.001, **P < 0.01, two-tailed Fisher’s exact test. Luminescence signals are shown as mean ± SEM. Each population consisted of 100 adult nematodes and was considered an independent biological replicate.
To understand the influence of the temperature component in circadian entrainment further, we tested the nematode populations under a 12-h/12-h CW temperature cycle (Δ = 1.5 ± 0.125 °C; ZT0 corresponds to the start of the 15.5 °C cold phase) and DD conditions (Fig. 5A). Although this thermal variation was sufficient to entrain some populations in the absence of light cues, the peak phase of bioluminescence was slightly different from that seen with the dual zeitgeber entrainment (DD/CW: ZT13.2 ± 0.7 h; LD/CW: ZT15.6 ± 0.3 h) (Fig. 5C). Furthermore, temperature alone tended to mask rather than entrain circadian rhythms: Most populations lost their circadian phase or even rhythmicity when placed under constant conditions (Fig. 5 D and E). Taken together, these results suggest that light and temperature are jointly required to obtain entrained and robust circadian rhythms.
To understand further how the clock senses environmental cues, we analyzed the expression of the bioluminescent reporter in different mutants deficient in thermo- and/or photo-reception. Nematodes have an extremely sensitive thermosensory system capable of discriminating temperature changes as small as 0.05 °C by means of the AFD and AWC sensory neurons, through the activation of guanylate cyclases and the TAX-2/TAX-4 cyclic nucleotide-gated channel (40–42). Regarding light sensing, at least two noncanonical photoreceptors of the GPCR gustatory receptor family, LITE-1 and GUR-3, were reported to sense short-length UV light directly or indirectly through a phototransduction pathway in which TAX-2 and TAX-4 also participate (39, 43, 44). Although photoreceptors are expressed in many neurons in the nervous system of the nematode, the ASJ, AWB, ASK, and ASH neurons have been reported to play a major role as photoreceptor cells for phototaxis in a redundant manner (45).
Although ∼78% of the wild-type luminescent populations were synchronized to the LD/CW cycle, the number decreased to 48% in the lite-1(xu7) photoreceptor mutant populations (Fig. 5 F and H and Fig. S9). Although LITE-1 plays a major role in the response to blue-violet light, the residual photic response could be explained by the relatively subtle phenotype of all LITE-1 mutants and the intact function of the remaining photoreceptor GUR-3 (39). In fact, although the double-mutant strain lite-1(ce314); gur-3(ok2245) showed a similar trend (around 46% synchronized populations under the same conditions), they were less able than either the wild-type population or the lite-1 single mutants to entrain to the zeitgebers (23 versus 48% and 35%, respectively; Fig. 5 G and H and Fig. S9). These results are consistent with the notion of a cooperative response between different photoreceptors.
Fig. S9.
Light and temperature sensing requires LITE-1, GUR-3, and TAX-2 proteins. (A) photoreception- and thermoreception-mutant nematode populations exhibit an FR phenotype similar to the wild-type strain. Average percentage of rhythmic nematode populations under FR conditions [wild-type, n = 101; lite-1(xu7), n = 89; lite-1(ce314); gur-3(ok2245), n = 120; tax-2 (p671), n = 121; tax-2 (p671) full rescue, n = 121; two-tailed Fisher’s exact test, all vs. wild-type, ns]. (B) Representative luciferase activity rhythms of two individual wild-type populations under LD/CW and DD/WW conditions from the data recorded in Fig. 5E. (C–J) Representative luciferase activity rhythms of two individual mutant populations under LD/CW and DD/WW conditions from the data recorded in Fig. 5 F–K. (C and D) lite-1(xu7) mutants. (E and F) lite-1(ce314) mutants; gur-3(ok2245) mutants. (G and H) tax-2 (p671) mutants. (I and J) The tax-2 (p671) full rescue strain. Each population consisted of 100 adult nematodes and was considered an independent biological replicate.
Synchronization to the LD/CW cycle decreased to 22% in the tax-2(p671) CNG channel mutant populations; moreover, luciferase expression did not maintain an entrained phase upon transfer to constant conditions (Fig. 5 I and K and Fig. S9), suggesting that the TAX-2/TAX-4 CNG channel has a major role in circadian entrainment. The apparent loss the average population rhythm under constant conditions results from a phase difference among individual populations. In this sense, the proportion of individual nematode populations that were rhythmic in constant conditions was similar among the mutants and the wild-type strain (Fig. S9). It is important to point out that the defective entrainment phenotype shown by tax-2(p671) mutants was completely recovered by a wild-type copy of the tax-2 gene (Fig. 5 J and K and Fig. S9). Consistent with these results, the severe loss of rhythmicity under entrainment conditions in tax-2(p671) mutants further supports the notion of dual temperature and light entrainment of the clock, because the TAX-2/TAX-4 CNG channel is a common component of both the photic and thermal transduction cascades.
Discussion
Much of the current understanding of the C. elegans circadian clock has been limited to the description of output behaviors, based on variable and nonrobust readouts that are not applicable for high-throughput screening studies. In this work, we applied a noninvasive and highly sensitive luciferase-based approach that revealed robust in vivo transcriptional luminescence rhythms at the population and single-nematode levels and demonstrated the fundamental properties of circadian rhythmicity in this powerful genetic model.
Population bioluminescence experiments were particularly useful for analyzing circadian properties and the effects of mutations in components of the entrainment pathway, properties that require several days of recording to allow accurate data analysis. Future forward genetic or RNAi screenings will be instrumental in uncovering the molecular components of the central clock of C. elegans. Additionally, population assays will allow the study of the factors involved in social synchronization and the effect of interindividual interactions on circadian rhythms. Population recordings could be followed for at least up to 4 d in constant conditions, suggesting that individual nematodes were still synchronized with each other. Without such synchronization, bioluminescence rhythms would dampen quickly or would not be discernable at all because of an averaging effect of each individual of the population.
On the other hand, single-nematode assays proved useful for revealing robust circadian rhythms in short-term studies and, more importantly, allowed us to avoid the averaging effect that results in the loss of information caused by individual desynchrony within a population. However, single-nematode assays showed more variability and required a larger number of replicates. Among the advantages of single-nematode recordings are the ability to measure responses to environmental stimuli and pharmacological treatments more precisely and to study the effect on the clock of single neuronal impairment through laser or genetic ablation, a promising approach to elucidate which neurons are involved in light- and temperature-sensing networks and the central pacemaker of the C. elegans’ clock.
Through integrative in vivo luminescence recordings and in vitro analyses, our study demonstrates circadian regulation of C. elegans’ metabolism and reveals a clock-controlled gene, sur-5, that had not been associated with the nematode circadian system by using classical methods. Consistent with this finding, by bioinformatics analysis we have found three putative E-box elements in the sur-5 promoter region: E1–E2 (−345), CAACTG(7N)CACGTT; the canonical E-box (−526), CACGTG; and the noncanonical E-box (−742), AACGTG). Interestingly, one of these elements, the E1−E2 tandem element, comprises a direct repeat of E-box–like elements conserved in many organisms and has been determined to be the minimal required sequence for the generation of cell-autonomous transcriptional oscillation of clock and clock-controlled genes in insects and mammals (46, 47). However, determining if these E-box elements play a similar role in C. elegans or if the nematodes have a distinct mechanism of transcriptional regulation, as do fungi and plants, requires further in-depth studies.
The sur-5 promoter is a common driver used to express transgenes constitutively and ubiquitously throughout development, but no previous studies have analyzed the expression pattern of sur-5 under a LD/CW circadian scheme in adult nematodes. Although the biochemical functions of sur-5 are still unknown, this gene encodes a putative evolutionarily conserved protein that is homologous to the Homo sapiens Acetyl-CoA synthetase (BlastP, 88% coverage, 40% identity, E = 2 × 10−162) (27), which is involved in lipid metabolism by catalyzing the generation of acetyl-CoA, a carbon source for the synthesis and elongation of fatty acids, in an ATP- and acetate-dependent manner. Interestingly, it recently has been reported that, although the mRNA levels do not oscillate, the activity of the mammalian acetyl-CoA Synthetase 1 (AceCS1) is under circadian modulation by acetylation through the action of the deacetylase SIRT1 (48). The cyclic acetylation of AceCS1 generates rhythmic acetyl-CoA levels and the consequent rhythmicity in the fatty acid elongation both in vivo and in cultured cells. Consistent with this report, we found a robust circadian rhythm in the endogenous ATP level in the nematode that was in antiphase to the mRNA levels of sur-5 transcript. It is noteworthy that the antiphase pattern of the ATP circadian rhythm with respect to the bioluminescence output demonstrates that the reporter does not reflect the metabolic state in vivo in our assay conditions, as was previously shown (29), but rather responds to sur-5–driven transcription. In line with this finding, the exogenous addition of an excess of 1 mM ATP did not alter the bioluminescence rhythmic pattern, supporting the notion that the amount of enzyme is the limiting factor in the luminescence reaction.
The circadian expression of sur-5 and the pharmacological sensitivity of the luminescence rhythms to the CK1ε/δ inhibitor support the notion of an evolutionarily conserved mechanism of transcriptional and posttranslational regulation of the circadian molecular machinery in this nematode. Several proteins have been described as putative homologs to the insect/mammalian core clock genes, notably the aryl hydrocarbon receptor associated protein (AHA-1), the abnormal cell lineage 42 protein isoforms b/c (LIN-42b/c), or timeless related protein (TIM-1), which show different degrees of homology to CLOCK, PERIOD, and TIMELESS, respectively (20, 49). However, these genes thus far have been reported to play developmental roles in the nematode (21–23), and none has been demonstrated to show circadian rhythmicity at the mRNA level (14, 19). To construct a classical TTFL (5) with a negative feedback loop, at least one of these genes should cycle. Although there is no evidence from whole-nematode experiments showing that the PER homolog lin-42 is rhythmic at the mRNA level under LD or WC cycles (7, 15), it remains possible that the gene cycles only in a subset of cells or that it could be regulated posttranslationally, as are peroxiredoxins, a conserved circadian marker in C. elegans and many other model organisms (7). Additionally, complex cytoplasmic regulations that occur by alternating species-specific heterodimerization partners and that modulate clock proteins function could be involved in the regulation of LIN-42 activity. For example, as has been shown in mice and Drosophila (50), the interaction of PER protein with a specific protein partner determines its different subcellular localizations during the day and its sensitivity to CK1ε. Also, certain mutations in Drosophila are able to affect patterns of cytoplasmic heterodimerization of PER and TIM specifically, altering or blocking the rate of nuclear transportation or even the rate of disassembly of the PER/TIM dimer to change the period length (50).
It is worth noting that the expressions of the translational reporter fusion constructs plin-42::gfp and pkin-20::gfp overlap perfectly, suggesting that they may interact in vivo (51). In addition, our bioinformatics analysis showed that KIN-20 has a high degree of homology with its counterpart in arthropods and mammals, with a complete conservation of all regulatory and functional sites (20); thus KIN-20 is a likely target of the PF-670642 selective casein kinase 1ε and δ inhibitor. Conservation of the regulatory sites between LIN-42 and PER and between CK1ε/δ and KIN-20 suggests the possibility of interactions similar to those found in other species. The precise role of LIN-42 and KIN-20 in the circadian molecular clock of C. elegans remains to be determined.
The circadian expression of core clock genes demonstrates different phases among tissues and might suffer quick dampening in the absence of environmental cues, i.e., upon transfer to constant light or constant temperature. It remains possible that masking caused by noncycling tissues or tissues with different phases could be responsible, at least in part, for the inability to find mRNA cycles of the putative clock genes in whole-nematode experiments. Additionally, samples containing large number of nematodes per time point might lead to false negatives because of the effect of phase averaging or the loss of valuable information between time points. Therefore our luciferase-based approach, which allows the recording of single-nematode molecular rhythms in real time and in intact animals, is a more sensitive and reliable method for revealing transcriptional rhythms. This fact is particularly relevant when working with C. elegans, in which circadian outputs do not appear to be particularly robust and there is a marked variability among nematodes (10–12, 52).
In addition to efforts to understand the nematode biological clock, there has been a growing interest in studying how this blind organism is able to sense light and temperature. Briefly, both pathways are based on G protein-coupled transmembrane receptors able to sense either light or temperature. These receptors activate a G protein that turns on a series of pathway-specific guanylate cyclases, increasing cGMP concentration and activating the TAX-4/TAX-2 cyclic nucleotide-gated channel (39, 41–44). Although the mechanisms underlying light and temperature sensing are well described, and most of the molecular components of the cascades have been reported, very little is known about the role of these pathways in the synchronization of the nematode clock.
Our findings provide evidence that both light and temperature, in a particular combination, are synergistic zeitgebers needed to entrain the robust luminescence rhythms observed for sur-5. In C. elegans’ natural environment, and in particular beneath the soil, the temperature increase is phase-delayed with respect to sunrise, reaching its maximum in the late afternoon (8). In this context, the hours with higher temperature are concentrated toward the end of the day, when solar radiation is minimal but the soil has absorbed most of it. Thus, we hypothesize that the combination of a cold onset with the “start” of the day in the LD/CW cycle would resemble natural conditions more accurately than a warm onset at the beginning of the day (LD/WC). Interestingly, D. melanogaster exhibits a strong preference for a temperature rhythm in which the temperature is lower in the morning and higher at dusk: The preferred temperature increases gradually from morning (ZT1–3) to peak in the evening (ZT10–12) (53). A similar synergistic effect was described in flies, in which a combination of light and temperature zeitgebers reinforced entrainment at the molecular and behavioral level (54). In our current study, the phase relationship to the LD/CW cycle showed a marked dependency on temperature, reflecting some adaptive aspects of the rhythm related to the natural environment of the nematode. A strong temperature dependency of the acrophase was also observed previously in circadian stress tolerance in C. elegans (52). However, with a Q10 of about 1.1, the circadian period is well compensated over the temperature range in which the bioluminescence could be recorded.
Additionally, our bioluminescent reporter revealed that LITE-1, GUR-3, and TAX-2 proteins are components involved in the photic and temperature synchronization pathways of the central oscillator. Similar results regarding tax-2 involvement were described by van der Linden et al. (19), who reported that a mutation in TAX-2 abolished the circadian expression of different transcripts entrained under either LD or WC conditions, indicating that TAX-2 is required for the transduction of both zeitgebers. In addition, tax-2 also was recently shown to be required for the transduction of thermal cues to the C. elegans circadian clock in locomotor activity rhythms (12). However, no evident role for lite-1 in light-entrained transcripts has been found previously (19), and to our knowledge there are no studies linking gur-3 to the input pathways of the clock. Our results, which do not show complete arrhythmicity in the mutant populations, indicate that more components probably are involved in the entrainment pathway, and follow-up experiments will be required for a complete understanding of the entrainment mechanism of the clock.
In summary, our study reveals key features of the circadian clock of C. elegans and opens the door for the identification of the central genes and neuronal circuits in this model through genetic screens that might provide fundamental information about its elusive molecular circadian clock and contribute to the understanding of general principles of circadian biology.
Materials and Methods
Experimental Conditions.
Nematode stocks were maintained on nematode growth medium (NGM) plates seeded as previously described (55) with HB101 Escherichia coli strain under 12-h/12-h LD/CW cycle (400/0 lx and CW (18.5/20 °C, Δ = 1.5 ± 0.125 °C) environmental cycles. Light and temperature are known to be the main zeitgebers, and in this schedule ZT0 (9:00 AM) indicates the time of lights on and the onset of the cold-temperature phase. CT refers to a particular time in the FR (i.e., DD/WW) cycle. Photo and thermal conditions were controlled with an I-291PF incubator (INGELAB), and temperature was monitored using DS1921H-F5 Thermochron iButtons (Maxim Integrated). Four Philips Daylight TL-D 18W/54-765 1SL fluorescent tubes were used as a light source [color temperature = 6200K and color-rendering index (CRI) = 72 Ra8].
Nematode Strains.
C. elegans strains N2 (Bristol strain, wild-type), TQ1101 lite-1(xu7), MT21793 lite-1(ce314);gur-3(ok2245), and PR671 tax-2(p671) were provided by the Caenorhabditis Genetics Center, University of Minnesota (cbs.umn.edu/cgc/home). Transgenic animals were generated by standard microinjection techniques (56). The Psur-5::luc::gfp construct was injected at 50 or 100 ng/μL with the pRF4 marker (100 ng/μL) in each genetic background. For the PR671 tax-2(p671) full rescue, we injected 10 ng/μL of Ptax-2::tax-2(genomic DNA)::yfp with 100 ng/μL of Psur-5::luc::gfp. All bioluminescent recordings were performed with strains carrying the extrachromosomal arrays in the different backgrounds. A spontaneously integrated line (100% transmission rate of both Psur-5::luc::gfp and pRF4) in the N2 (wild-type) background was selected for real-time PCR and ATP measurements. Microscopy was performed with an AxioScope.A1 (Zeiss) equipped with a light source X-Cite 120Q System (EXFO, Canada) and an Axiocam 506 mono camera (Zeiss).
Molecular Constructs.
The Psur-5::luc::gfp vector was constructed by ligation of a 1,659-bp PCR-amplified P. pyralis luciferase ORF from the pGL3-Basic Vector (Promega), with the last STOP codon removed, between the BamHI and KpnI sites of pPD158.87 (Addgene), in frame with gfp to generate a fusion protein. The luciferase ORF was amplified using the primers forward, 5′-GCGGATCCATGGAAGACGCCAAAAAC-3′ and reverse, 5′-CGGGGTACCACGGCGATCTTTCCGCC-3′. More details are provided in SI Materials and Methods.
Bioluminescence Recordings and Treatments.
Transgenic Psur-5::luc::gfp nematode populations were synchronized to the same developmental stage by the chlorine method (57). The harvested eggs were cultured overnight in a 50-mL Erlenmeyer flask with 3.5 mL of M9 buffer, 1× antibiotic-antimycotic (Thermo Fisher Scientific), and 10 µg/mL of tobramycin (Tobrabiotic; Denver Farma) at 110 rpm with a Vicking M23 shaker, in LD/CW (400/0 lx; 18.5/20 °C, Δ = 1.5 °C ± 0.125 °C) conditions. The following day, L1 larvae were transferred to NGM plates at ZT1 and were grown for 48 h to the L4 stage under the same LD/CW cycle. Starting at ZT1 (1 h post lights on), the most fluorescent nematodes were selected manually under a SMZ100 stereomicroscope equipped with an epi-fluorescence attachment (Nikon) with a cool Multi-TK-LED light source (Tolket) to avoid warming the plate. The picking was performed in a room kept at a constant temperature of 18 °C (with the white lights in the stereomicroscope also on at low intensity to mimic the cold temperature of the light phase of the entrainment in the incubator). Each plate was exposed to the blue light for only 15 min to prevent a long-term effect on entrainment or viability. The entrainment of the LD/CW cycle was performed at 18.5/20 °C to accelerate the growth of the nematodes and to get L4 larvae after 48 h so they could be selected during the day phase. Luminescence was assayed either under FR conditions (DD/WW, 20 °C; dish plate setup) or under entrainment followed by FR conditions (LD/CW, 15.5/17 °C or other temperatures; multiwell setup). The luminescence medium contained Leibovitz’s L-15 medium without phenol red (Thermo Fisher Scientific) supplemented with 1× antibiotic–antimycotic (Thermo Fisher Scientific), 40 μM of 5-fluoro-2′-deoxyuridine (FUdR) to avoid new eclosions, 5 mg/mL cholesterol, 10 µg/mL tobramycin (Tobrabiotic; Denver Farma), 1 mM d-luciferin (Gold Biotechnology), and 0.05% Triton X-100 to increase cuticle permeabilization. All chemical compounds were purchased from Sigma-Aldrich unless otherwise specified. More details are provided in SI Materials and Methods.
Quantitative Real-Time PCR.
Synchronized transgenic Psur-5::luc::gfp nematode populations were grown under a 12-h/12-h LD/CW cycle (18.5/20 °C; ZT0 indicates lights on and the onset of the cold-temperature phase) for 48 h and were collected from the NGM plates at the L4 stage at ZT3, washed, and transferred to four 1.5-L flasks with 170 mL of luminescence medium without d-luciferin (5 nematodes/10 μL). The nematodes then were cultured under LD/CW (15.5/17 °C) conditions with agitation at 100 rpm for three additional days. Then they were released in FR conditions (DD/WW; 17 °C) for the two remaining days of the assay. Four independent biological replicates (n = 4; 4,000 adult nematodes in each replicate) were collected every 4 h, starting at ZT3 in the LD/CW cycle during the last day of entrainment and during the following 2 d of the FR condition. Nematodes were centrifuged and flash frozen at −80 °C in TRIzol (Thermo Fisher Scientific). Total RNA was extracted from the samples using the TRIzol method according to the manufacturer’s instructions. Three micrograms of total RNA and poly-T primers (20 nt long) (Thermo Fisher Scientific) were used for cDNA synthesis using SuperScript II Reverse Transcriptase (Thermo Fisher Scientific). Quantitative PCR analyses were performed with SYBR Green PCR Master Mix (Applied Biosystems) using a StepOne Real-Time PCR thermal cycler (Applied Biosystems) following the manufacturer’s instructions. More details are provided in SI Materials and Methods.
In Vitro ATP Measurements.
Synchronized transgenic Psur-5::luc::gfp nematode populations were grown and entrained under the same environmental conditions used for the quantitative real-time PCR experiments. Four independent biological replicates of 3,500 adult nematodes each were collected every 4 h, starting at ZT3 under the LD/CW condition during the last day of entrainment and during the last 2 d of the FR condition. Nematodes were centrifuged, washed twice, and flash frozen at −80 °C in the same luminescence medium without d-luciferin. Nematodes were disrupted by sonication using a VCX 130PB Vibra-Cell ultrasonic processor (Sonics) in two cycles of 15 s at 40% power and then were centrifuged to pellet the debris. This step releases the endogenous ATP and completely disrupts the luciferase reporter activity of the transgenic nematodes. The total amount of ATP was quantified using 30 µL of nematode extract incubated with 6.2 µL of a mix containing 9 µg/mL of purified commercial Luciferase from P. pyralis (Sigma), 0.8 mg/mL CoA (Sigma), 1.6 mM d-luciferin, and 5% (wt/vol) BSA. Luminescence kinetics was measured with a Berthold Centro LB 960 microplate luminometer in a white, flat-bottomed, 96-well plate at 20 °C for a total of 3 min with 1 s of integration time, and the total amount of ATP was calculated using a calibration curve with different ATP standards. The total amount of ATP was normalized to the total amount of protein, as determined by the Bradford method, and was normalized to the sample with the highest ATP/protein level. All chemical compounds were purchased from Sigma-Aldrich unless otherwise specified.
Data Analysis and Statistics.
Raw luminescence data were analyzed using custom scripts written in MATLAB. For raw data from the AB-2550 Kronos Dio luminometer experiments, the waiting time background was subtracted in all cases, and the first and the last 12 h of raw data were discarded (except in the single-nematode assays). The data then were trend-corrected by dividing by a fixed moving-average window of 24 h and were smoothed for 12 h. Afterwards, the data were normalized to the maximum level of luminescence of all of the biological replicates along the entire temporal series on each experiment and were plotted in Origin 2016 Software (OriginLab). Data are presented as mean ± SEM. For the luminescence signals, the mean corresponds to nematode populations or to single-nematode recordings, as appropriate in each assay. If not stated otherwise, statistical significance was calculated by one-way ANOVA and Dunnett’s multiple comparisons test, with P < 0.05 considered statistically significant. Statistical significance levels are denoted as follows: ***P < 0.001; **P < 0.01; *P < 0.05. All statistical tests were two-tailed where applicable. No statistical methods were used to predetermine sample size. More details are provided in SI Materials and Methods.
Additional experimental details are provided in SI Materials and Methods.
SI Materials and Methods
Molecular Constructs.
Psur-5::luc was constructed by ligation of a 1,673-bp PCR-amplified P. pyralis luciferase ORF from the pGL3-Basic Vector (Promega) between the KpnI and EcoRI sites of pPD158.87 (Addgene), replacing the gfp coding sequence (CDS). The luciferase ORF was amplified using the primers forward, 5′-CATCGGGGTACCATGGAAGACGCCAAAAAC-3′ and reverse, 5′- GGAATTCTACACGGCGATCTTTCCGCCCTTC-3′.
Psur-5::luc was constructed by ligation of a 1,673-bp PCR-amplified P. pyralis luciferase ORF from the pGL3-Basic Vector (Promega) between the KpnI and EcoRI sites of pPD158.87 (Addgene), replacing the gfp CDS. The luciferase ORF was amplified using the primers forward, 5′-CATCGGGGTACCATGGAAGACGCCAAAAAC-3′ and reverse, 5′- GGAATTCTACACGGCGATCTTTCCGCCCTTC-3′.
Psur-5::AI::luc was constructed by ligation of a 1,673-bp PCR-amplified P. pyralis luciferase ORF from the pGL3-Basic Vector (Promega) between the KpnI and EcoRI sites of pPD95.75 (Addgene), replacing the gfp CDS. Then, the AI::luc was cut with BamHI and EcoRI and cloned into pPD158.87 (Addgene) under the sur-5 promoter. The luciferase ORF was amplified using the primers used for the Psur-5::luc.
For the Psur-5::luc::icr::gfp, we first synthetized the intercistronic region (icr) sequence described by Lee et al. (58) in GenScript with KpnI and BglII sites in the 5′ and XhoI and EcoRI in the 3′ extreme (pGEM-T Easy Vector; Promega). Then we amplified the luciferase ORF from the pGL3-Basic Vector (Promega) and ligated the 1,679-bp fragment between the KpnI and BglII sites of pGEMT::icr with primers forward, 5′-CATCGGGGTACCATGGAAGACGCCAAAAAC-3′ and reverse, 5′- CATGAGAAGATCTTTTACACGGCGATCTTTCCGC-3′. Next, we amplified a 900-bp fragment of the gfp ORF from the pPD158.87 (Addgene) and cloned it into the pGEMT::luc::icr between SalI (XhoI compatible end) and EcoRI sites using the primers forward, 5′-CAACGCGTCGACATGAGTAAAGGAGAAGAAC-3′ and reverse, 5′- CAGCGCGGATCCGAATTCCTATTTGTATAGTTCATCC-3′. Finally, we cut and ligated the luc::icr::gfp construct between KpnI and EcoRI with pPD158.87 (Addgene) under the sur-5 promoter.
Bioluminescence Recordings and Treatments.
Assays of FR luminescence (dish plate setup).
For experiments in the FR conditions, eight populations of 100 L4-stage nematodes (each population was considered an independent biological replicate) were selected as described previously, washed twice with M9 buffer to remove all traces of bacteria, and transferred to a 35-mm plate dish (Greiner CELLSTAR) with 1 mL of the luminescence medium; then the dish was sealed. Nematodes were placed in the incubator at ZT2 and were left there under entrainment conditions for 2 d. Then, they were transferred at ZT12 to an AB-2550 Kronos Dio luminometer (ATTO) in which luminescence was monitored for 7 d in DD/WW (20 °C, which is the minimal temperature allowed in this luminometer). The signal was integrated for 1 min, and readings were taken every 10 min.
For single-nematode measurements, Psur-5::luc::gfp nematodes at the L4 stage were selected as before but starting at ZT10, at a constant temperature of 18 °C, washed, and then transferred directly to the liquid luminescence medium. A small, square piece of a transparent 96-well plate containing three × three wells was inserted inside a 35-mm dish plate to reduce the total volume and limit the nematode movement to the center of the plate. One nematode was placed in the center well with 200 μL of the luminescence medium, and water was placed in the other wells to avoid evaporation. The plate was sealed with a Microseal ‘B’ Adhesive Seal (Bio-Rad) to avoid evaporation and contamination, and the seal over each well was perforated twice to avoid condensation. At ZT12, the plates were transferred to an AB-2550 Kronos Dio luminometer, and luminescence was monitored in DD/WW (20 °C) for 7 d. Single-nematode recordings were taken every 37 min with an integration time of 4 min.
For treatment with the CKIδ/ε inhibitor PF-670462 (Abcam), 5 or 10 µM of drug dissolved in water was added to the medium just before the start of the luminescence recording at ZT12, and the same volume of vehicle was used as control. Single-nematode recordings were taken every 37 min with an integration time of 4 min.
Assay CKI Ɛ/δ inhibitor toxicity.
The toxicity of PF-670467 was tested by recording the global motility of populations of wild-type adult nematodes (N2) over time under different concentrations of PF-670462, using an infrared tracking device (WMicroTracker, PhylumTech) previously used for toxicity screenings (59). We tested wild-type nematodes instead of the transgenic Psur-5::luc::gfp lines, because the transgenic lines have a roller marker and are not able to swim correctly. Nematodes were grown on an LD/CW cycle as described above. At the L4 stage, synchronized populations (40 nematodes each) were transferred by pipetting to a flat-bottomed, 96-well plate (Greiner) according to the previously described protocol (10, 11) but using the luminescence assay medium without d-luciferin. Plates were covered with an optic film to avoid evaporation, and the film over each well was perforated twice to avoid condensation. Nematode population recordings are preferred to single-worm recordings because of the higher amplitude of activity resulting from a higher number of infrared beam crosses. Individual recordings exhibit a low signal-to-noise ratio and therefore are not suitable for accurate recordings of long-term activity. The assay was performed under 17 °C and constant darkness in an I-291PF incubator (Ingelab), and temperature was monitored using DS1921H-F5 Thermochron iButtons (Maxim Integrated). The activity was recorded every min and binned in 5-min blocks. The experiment was performed for a period equivalent to the duration of a typical luminescence assay.
Assays of luminescence in entrainment followed by FR conditions (multiwell setup).
For these experiments, 48 populations each comprised of 100 stage-L4 larvae (each population was considered an independent biological replicate) were selected manually as described above starting at ZT1, washed twice with M9 buffer to remove all traces of bacteria, and resuspended in 200 µL of luminescence medium. Nematodes were transferred to a white, flat-bottomed, 96-well plate (Greiner). The plate was sealed with optic film with two small perforations per well to prevent condensation. Performing the manual selection required around 5 h. The nematodes were left in the new LD/CW cycle until ZT10, and luminescence was registered using a Berthold Centro LB 960 microplate luminometer (Berthold Technologies) stationed inside an E-30B incubator (Percival) to allow tight control of the light and temperature in each experiment. Microwin 2000 software version 4.43 (Mikrotek-Laborsysteme) was programed to leave the plate outside the luminometer after each recording was performed to expose nematodes to the environmental cues. Only 48 of the 96 wells were used to avoid background contamination between contiguous wells; water was placed in the empty wells to avoid evaporation. The luminescence of each well was integrated for 10 s every 30 min. Every experiment was repeated at least twice.
The general entrainment conditions used for most experiments were 3.5 d at a 12-h/12-h LD/CW cycle (400/0 lx; 15.5/17 °C; ZT0, lights on and onset of the cold-temperature phase) and 4 d at the FR condition (DD/WW, 17 °C). This protocol was performed to analyze synchronization of the N2 (wild-type) strain under LD/CW, LD/WC, and CW conditions, temperature compensation, phase-shift assays, and ATP treatment and to analyze the rhythms of the mutants strains TQ1101 lite-1(xu7), MT21793 lite-1(ce314);gur-3(ok2245), PR671 tax-2(p671), and the PR671 full-rescue strain. We choose the LD/CW (15.5/17 °C) cycle for most recordings because 15.5 °C is the minimal temperature allowed in the Berthold Centro LB 960 microplate luminometer and because the populations have a longer life span under this condition than at 18.5/20 °C, resulting in a sustained luminescence signal for more days than at higher temperatures.
For the in vivo ATP treatment in the N2 strain, 1 mM of filter-sterilized ATP (GE Healthcare) dissolved in water was added to the medium at ZT10. Vehicle was used as control.
For temperature-compensation assays, the results of the general entrainment protocol were compared with the results of a similar assay performed for 3.5 d in LD/CW (400/0 lx; 19.5/21 °C) and for 4 d at DD/WW (21 °C). The period of the luminescence rhythms at 20 °C under DD was obtained from the AB-2550 Kronos Dio luminometer experiments as explained before.
For phase-shift assays, nematode populations were entrained for 3.5 d under a 12-h/12-h LD/CW cycle (400/0 lx; 15.5 °C/17 °C) and then were subjected to a phase shift caused by a 6-h night extension. After four more days, the nematodes were released into FR conditions (DD/WW, 17 °C) for 2 d.
For inverted zeitgeber cycle assays, nematode populations were entrained for 3.5 d under a 12-h/12-h regime of LD/WC (400/0 lx; 17/15.5 °C) and then were released into FR conditions (DD/CC, 15.5 °C) for 4 d.
For temperature-entrainment assays, nematode populations were entrained for 3.5 d in a 12-h/12-h DD/CW (15.5/17 °C) cycle and then were released into FR conditions with constant warm temperature (WW, 17 °C) for 4 d.
Finally, for the constant-light assays, nematode populations were entrained for 3.5 d under a 12-h/12-h LD/CW cycle (400/0 lx; 15.5/C/17 °C) and then were released into constant light and cold temperature (LL/CC, 15.5 °C) for 4 d.
Quantitative Real-Time PCR.
The results for sur-5 and luc::gfp expression were normalized to the mRNA levels of three selected references genes: cdc-42, pmp-3, and Y45F10D.4 (60) by the standard curve method and were normalized to the sample with the highest mRNA level. Primers for amplifying each target were sur-5 forward, 5′-CACCCCAAGGTTTTGTTCAC-3′; sur-5 reverse, 5′-TGAAGGTGTCGGATACAACG-3′; luc::gfp forward, 5′-CTAGCCGGCCATACAAGTAATC-3′; luc::gfp reverse, 5′-CGGAATACGAATTGGGAGAC-3′; cdc-42 forward, 5′-CTGCTGGACAGGAAGATTACG-3′; cdc-42 reverse, 5′-CTCGGACATTCTCGAATGAAG-3′; Y45F10D.4 forward, 5′-GTCGCTTCAAATCAGTTCAGC-3′; Y45F10D.4 reverse, 5′-GTTCTTGTCAAGTGATCCGACA-3′; pmp-3 forward, 5′-GTTCCCGTGTTCATCACTCAT-3′; pmp-3 reverse, 5′-ACACCGTCGAGAAGCTGTAGA-3′. All primers were purchased from Thermo Fisher Scientific.
Data Analysis and Statistics.
The circadian period was calculated by autocorrelation, fast Fourier nonlinear least square algorithm (FFT-NLLS), Maximum Entropy Spectral Analysis (MESA) (61) and the LS periodogram (62), evaluated inside a period range between 20 and 32 h. We chose to inform only the results of the LS algorithm because it produced the most accurate fits and also because it was reported to be the best method for period determination in short-term circadian recordings (63). With the period obtained from the LS algorithm we then estimated phase and amplitude using a cosinor algorithm, and a least-square regression fit using these parameters was calculated to evaluate the R2 of the adjustment. Any luminescent signal of nematode populations with a period of 24 h and an R2 least-square fit ≥0.7 was considered to be synchronized under the entrainment conditions. For FR conditions, any luminescent signal of a nematode population or of a single-nematode assay with a period close to 24 h and an R2 less-square fit ≥0.5 was considered to be rhythmic. We considered entrained populations to be those whose acrophases in FR conditions had a difference of less than 3 h with respect to that of the entrainment conditions. To avoid confusion, the following nomenclature is used in this work: “percentage of synchronized” means the number of populations with a period and phase set by the zeitgeber conditions over the total number of populations tested; “percentage of entrained” means the number of populations that retain their circadian phase when placed under constant conditions over the number of populations set by the zeitgeber conditions; and “percentage of rhythmic under FR” means the number of rhythmic populations under FR over the total number of populations tested.
Rayleigh tests were used to determine the statistical significance of the mean phases, and a multisample test for equal median directions (a circular analog to the Kruskal–Wallis test) was performed to compare the mean phases under entrainment and FR conditions. In the phase-shift assay, Rayleigh tests followed by one-way ANOVA and Tukey’s multiple comparisons test were used to assess significant differences between mean phases under the LD/CW condition before and after the phase shift and to compare the mean phases of the second entrainment condition (LD/CW2) with the FR condition.
Fisher’s exact test was performed to compare the proportions of synchronized, entrained, and rhythmic populations in all experiments. One-way ANOVA was performed to identify statistically significant differences between control and PF-670462–treated samples, and an unpaired Student’s t test was performed to compare the amplitude of the rhythms in ATP-addition experiments.
A cross-correlation test was applied to evaluate the phase differences between the average rhythmic patterns of luminescence, mRNA expression data, and endogenous ATP levels.
Bioinformatics Analysis.
The sur-5 minimal promoter portion of the Psur-5::luc::gfp construct was analyzed to search for the circadian regulatory elements with the next International Union of Pure and Applied Chemistry syntaxis: E-box (CACGTG) and E1–E2 box (CACGTG-[7- to 12-nt]-CAAGTG) (46, 64, 65). Analysis of regulatory elements was performed with the jPREdictor v1.23 with default parameters and one error allowed (66).
Acknowledgments
We thank S. Simonetta for initial discussions; P. Schwarzbaum and C. Alvarez for protocols and reagents; H. de la Iglesia for critical discussions and reading of the manuscript; M. Yanovsky for advice, reading of the manuscript, and technical support; L. Larrondo for discussions about the project and suggestions of experiments; A. Thackeray, W. Joyce, and M. Lamberti for critical technical assistance; S. Xu for the Ptax-2::tax-2::YFP plasmid; D. Banerjee for advice with kin-20 mutants; and the Caenorhabditis Genetics Center funded by NIH Office of Research Infrastructure Programs Grant P40 OD010440 for strains. This work was supported by grants from the Agencia Nacional de Promoción Científica y Tecnológica, Consejo Nacional de Investigaciones Científicas y Técnicas de Argentina (CONICET) and Universidad Nacional de Quilmes (Argentina) (to D.A.G.), and by NIH Grant R01-AG041870-01 (to C.Y.B.). M.E.G. and C.S.C. are PhD CONICET Fellows, and A.R. is a Bunge y Born Foundation Fellow.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1605769113/-/DCSupplemental.
References
- 1.Rosbash M. The implications of multiple circadian clock origins. PLoS Biol. 2009;7(3):e62. doi: 10.1371/journal.pbio.1000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell-Pedersen D, et al. Circadian rhythms from multiple oscillators: Lessons from diverse organisms. Nat Rev Genet. 2005;6(7):544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15(Spec No 2):R271–R277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- 4.Daan S, Aschoff J. The entrainment of circadian systems. In: Takahashi JS, Turek FW, Moore RY, editors. Circadian Clocks. Kluwer Academic; Plenum, New York: 2011. pp. 7–34. [Google Scholar]
- 5.Harmer SL, Panda S, Kay SA. Molecular bases of circadian rhythms. Annu Rev Cell Dev Biol. 2001;17:215–253. doi: 10.1146/annurev.cellbio.17.1.215. [DOI] [PubMed] [Google Scholar]
- 6.Frezal L, Felix MA. The natural history of model organisms: C. elegans outside the petri dish. eLife. 2015;4:e05849. doi: 10.7554/eLife.05849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robinson AF. Movement of five nematode species through sand subjected to natural temperature gradient fluctuations. J Nematol. 1994;26(1):46–58. [PMC free article] [PubMed] [Google Scholar]
- 8.Monteith JL, Unsworth MH. Transient Heat Balance. Principles of Environmental Physics: Plants, Animals, and the Atmosphere. Elsevier/Academic; Amsterdam: 2013. pp. 273–287. [Google Scholar]
- 9.Petersen C, Dirksen P, Prahl S, Strathmann EA, Schulenburg H. The prevalence of Caenorhabditis elegans across 1.5 years in selected North German locations: The importance of substrate type, abiotic parameters, and Caenorhabditis competitors. BMC Ecol. 2014;14:4. doi: 10.1186/1472-6785-14-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herrero A, et al. Pigment-dispersing factor signaling in the circadian system of Caenorhabditis elegans. Genes Brain Behav. 2015;14(6):493–501. doi: 10.1111/gbb.12231. [DOI] [PubMed] [Google Scholar]
- 11.Simonetta SH, Migliori ML, Romanowski A, Golombek DA. Timing of locomotor activity circadian rhythms in Caenorhabditis elegans. PLoS One. 2009;4(10):e7571. doi: 10.1371/journal.pone.0007571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Winbush A, Gruner M, Hennig GW, van der Linden AM. Long-term imaging of circadian locomotor rhythms of a freely crawling C. elegans population. J Neurosci Methods. 2015;249:66–74. doi: 10.1016/j.jneumeth.2015.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saigusa T, et al. Circadian behavioural rhythm in Caenorhabditis elegans. Curr Biol. 2002;12(2):R46–R47. doi: 10.1016/s0960-9822(01)00669-8. [DOI] [PubMed] [Google Scholar]
- 14.Olmedo M, et al. Circadian regulation of olfaction and an evolutionarily conserved, nontranscriptional marker in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2012;109(50):20479–20484. doi: 10.1073/pnas.1211705109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Migliori ML, Simonetta SH, Romanowski A, Golombek DA. Circadian rhythms in metabolic variables in Caenorhabditis elegans. Physiol Behav. 2011;103(3-4):315–320. doi: 10.1016/j.physbeh.2011.01.026. [DOI] [PubMed] [Google Scholar]
- 16.Simonetta SH, Romanowski A, Minniti AN, Inestrosa NC, Golombek DA. Circadian stress tolerance in adult Caenorhabditis elegans. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2008;194(9):821–828. doi: 10.1007/s00359-008-0353-z. [DOI] [PubMed] [Google Scholar]
- 17.Edgar RS, et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature. 2012;485(7399):459–464. doi: 10.1038/nature11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Migliori ML, et al. Daily variation in melatonin synthesis and arylalkylamine N-acetyltransferase activity in the nematode Caenorhabditis elegans. J Pineal Res. 2012;53(1):38–46. doi: 10.1111/j.1600-079X.2011.00969.x. [DOI] [PubMed] [Google Scholar]
- 19.van der Linden AM, et al. Genome-wide analysis of light- and temperature-entrained circadian transcripts in Caenorhabditis elegans. PLoS Biol. 2010;8(10):e1000503. doi: 10.1371/journal.pbio.1000503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romanowski A, Garavaglia MJ, Goya ME, Ghiringhelli PD, Golombek DA. Potential conservation of circadian clock proteins in the phylum Nematoda as revealed by bioinformatic searches. PLoS One. 2014;9(11):e112871. doi: 10.1371/journal.pone.0112871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tennessen JM, Gardner HF, Volk ML, Rougvie AE. Novel heterochronic functions of the Caenorhabditis elegans period-related protein LIN-42. Dev Biol. 2006;289(1):30–43. doi: 10.1016/j.ydbio.2005.09.044. [DOI] [PubMed] [Google Scholar]
- 22.Qin H, Powell-Coffman JA. The Caenorhabditis elegans aryl hydrocarbon receptor, AHR-1, regulates neuronal development. Dev Biol. 2004;270(1):64–75. doi: 10.1016/j.ydbio.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 23.Chan RC, et al. Chromosome cohesion is regulated by a clock gene paralogue TIM-1. Nature. 2003;423(6943):1002–1009. doi: 10.1038/nature01697. [DOI] [PubMed] [Google Scholar]
- 24.Wood KV. Marker proteins for gene expression. Curr Opin Biotechnol. 1995;6(1):50–58. doi: 10.1016/0958-1669(95)80009-3. [DOI] [PubMed] [Google Scholar]
- 25.Yu W, Hardin PE. Use of firefly luciferase activity assays to monitor circadian molecular rhythms in vivo and in vitro. Methods Mol Biol. 2007;362:465–480. doi: 10.1007/978-1-59745-257-1_38. [DOI] [PubMed] [Google Scholar]
- 26.Welsh DK, Imaizumi T, Kay SA. Real-time reporting of circadian-regulated gene expression by luciferase imaging in plants and mammalian cells. Methods Enzymol. 2005;393:269–288. doi: 10.1016/S0076-6879(05)93011-5. [DOI] [PubMed] [Google Scholar]
- 27.Gu T, Orita S, Han M. Caenorhabditis elegans SUR-5, a novel but conserved protein, negatively regulates LET-60 Ras activity during vulval induction. Mol Cell Biol. 1998;18(8):4556–4564. doi: 10.1128/mcb.18.8.4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olmedo M, Geibel M, Artal-Sanz M, Merrow M. A high-throughput method for the analysis of larval developmental phenotypes in Caenorhabditis elegans. Genetics. 2015;201(2):443–448. doi: 10.1534/genetics.115.179242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lagido C, Pettitt J, Flett A, Glover LA. Bridging the phenotypic gap: Real-time assessment of mitochondrial function and metabolism of the nematode Caenorhabditis elegans. BMC Physiol. 2008;8:7. doi: 10.1186/1472-6793-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohta H, Yamazaki S, McMahon DG. Constant light desynchronizes mammalian clock neurons. Nat Neurosci. 2005;8(3):267–269. doi: 10.1038/nn1395. [DOI] [PubMed] [Google Scholar]
- 31.Mehra A, Baker CL, Loros JJ, Dunlap JC. Post-translational modifications in circadian rhythms. Trends Biochem Sci. 2009;34(10):483–490. doi: 10.1016/j.tibs.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee H, Chen R, Lee Y, Yoo S, Lee C. Essential roles of CKIdelta and CKIepsilon in the mammalian circadian clock. Proc Natl Acad Sci USA. 2009;106(50):21359–21364. doi: 10.1073/pnas.0906651106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Price JL, et al. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94(1):83–95. doi: 10.1016/s0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- 34.Querfurth C, et al. Circadian conformational change of the Neurospora clock protein FREQUENCY triggered by clustered hyperphosphorylation of a basic domain. Mol Cell. 2011;43(5):713–722. doi: 10.1016/j.molcel.2011.06.033. [DOI] [PubMed] [Google Scholar]
- 35.van Ooijen G, et al. Functional analysis of casein kinase 1 in a minimal circadian system. PLoS One. 2013;8(7):e70021. doi: 10.1371/journal.pone.0070021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meng QJ, et al. Entrainment of disrupted circadian behavior through inhibition of casein kinase 1 (CK1) enzymes. Proc Natl Acad Sci USA. 2010;107(34):15240–15245. doi: 10.1073/pnas.1005101107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bass J. Circadian topology of metabolism. Nature. 2012;491(7424):348–356. doi: 10.1038/nature11704. [DOI] [PubMed] [Google Scholar]
- 38.de Wet JR, Wood KV, DeLuca M, Helinski DR, Subramani S. Firefly luciferase gene: Structure and expression in mammalian cells. Mol Cell Biol. 1987;7(2):725–737. doi: 10.1128/mcb.7.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edwards SL, et al. A novel molecular solution for ultraviolet light detection in Caenorhabditis elegans. PLoS Biol. 2008;6(8):e198. doi: 10.1371/journal.pbio.0060198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clark DA, Biron D, Sengupta P, Samuel AD. The AFD sensory neurons encode multiple functions underlying thermotactic behavior in Caenorhabditis elegans. J Neurosci. 2006;26(28):7444–7451. doi: 10.1523/JNEUROSCI.1137-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kimura KD, Miyawaki A, Matsumoto K, Mori I. The C. elegans thermosensory neuron AFD responds to warming. Curr Biol. 2004;14(14):1291–1295. doi: 10.1016/j.cub.2004.06.060. [DOI] [PubMed] [Google Scholar]
- 42.Ramot D, MacInnis BL, Lee HC, Goodman MB. Thermotaxis is a robust mechanism for thermoregulation in Caenorhabditis elegans nematodes. J Neurosci. 2008;28(47):12546–12557. doi: 10.1523/JNEUROSCI.2857-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu J, et al. C. elegans phototransduction requires a G protein-dependent cGMP pathway and a taste receptor homolog. Nat Neurosci. 2010;13(6):715–722. doi: 10.1038/nn.2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bhatla N, Horvitz HR. Light and hydrogen peroxide inhibit C. elegans Feeding through gustatory receptor orthologs and pharyngeal neurons. Neuron. 2015;85(4):804–818. doi: 10.1016/j.neuron.2014.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ward A, Liu J, Feng Z, Xu XZ. Light-sensitive neurons and channels mediate phototaxis in C. elegans. Nat Neurosci. 2008;11(8):916–922. doi: 10.1038/nn.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakahata Y, et al. A direct repeat of E-box-like elements is required for cell-autonomous circadian rhythm of clock genes. BMC Mol Biol. 2008;9:1. doi: 10.1186/1471-2199-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paquet ER, Rey G, Naef F. Modeling an evolutionary conserved circadian cis-element. PLOS Comput Biol. 2008;4(2):e38. doi: 10.1371/journal.pcbi.0040038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sahar S, et al. Circadian control of fatty acid elongation by SIRT1 protein-mediated deacetylation of acetyl-coenzyme A synthetase 1. J Biol Chem. 2014;289(9):6091–6097. doi: 10.1074/jbc.M113.537191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hasegawa K, Saigusa T, Tamai Y. Caenorhabditis elegans opens up new insights into circadian clock mechanisms. Chronobiol Int. 2005;22(1):1–19. doi: 10.1081/cbi-200038149. [DOI] [PubMed] [Google Scholar]
- 50.Young MW, Kay SA. Time zones: A comparative genetics of circadian clocks. Nat Rev Genet. 2001;2(9):702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- 51.Banerjee D, Kwok A, Lin SY, Slack FJ. Developmental timing in C. elegans is regulated by kin-20 and tim-1, homologs of core circadian clock genes. Dev Cell. 2005;8(2):287–295. doi: 10.1016/j.devcel.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 52.Kippert F, Saunders DS, Blaxter ML. Caenorhabditis elegans has a circadian clock. Curr Biol. 2002;12(2):R47–R49. doi: 10.1016/s0960-9822(01)00670-4. [DOI] [PubMed] [Google Scholar]
- 53.Kaneko H, et al. Circadian rhythm of temperature preference and its neural control in Drosophila. Curr Biol. 2012;22(19):1851–1857. doi: 10.1016/j.cub.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoshii T, Vanin S, Costa R, Helfrich-Förster C. Synergic entrainment of Drosophila’s circadian clock by light and temperature. J Biol Rhythms. 2009;24(6):452–464. doi: 10.1177/0748730409348551. [DOI] [PubMed] [Google Scholar]
- 55.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- 57.Lewis JA, Fleming JT. Basic Culture Methods. Caenorhabditis elegans: Modern Biological Analysis of an Organism. Vol 48. Academic; San Diego: 1995. pp. 3–29. [PubMed] [Google Scholar]
- 58.Lee LW, Lo HW, Lo SJ. Vectors for co-expression of two genes in Caenorhabditis elegans. Gene. 2010;455(1-2):16–21. doi: 10.1016/j.gene.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 59.Bianchi JI, Stockert JC, Buzzi LI, Blázquez-Castro A, Simonetta SH. Reliable screening of dye phototoxicity by using a Caenorhabditis elegans fast bioassay. PLoS One. 2015;10(6):e0128898. doi: 10.1371/journal.pone.0128898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoogewijs D, Houthoofd K, Matthijssens F, Vandesompele J, Vanfleteren JR. Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol Biol. 2008;9:9. doi: 10.1186/1471-2199-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dowse HB. Maximum entropy spectral analysis for circadian rhythms: Theory, history and practice. J Circadian Rhythms. 2013;11(1):6. doi: 10.1186/1740-3391-11-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Press WH, Flannery BP, Teukolsky SA, Vetterling WT. Numerical Recipes in FORTRAN: The Art of Scientific Computing. 2nd Ed. Cambridge Univ Press; New York: 1992. p. 1010. [Google Scholar]
- 63.Zielinski T, Moore AM, Troup E, Halliday KJ, Millar AJ. Strengths and limitations of period estimation methods for circadian data. PLoS One. 2014;9(5):e96462. doi: 10.1371/journal.pone.0096462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kumaki Y, et al. Analysis and synthesis of high-amplitude Cis-elements in the mammalian circadian clock. Proc Natl Acad Sci USA. 2008;105(39):14946–14951. doi: 10.1073/pnas.0802636105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ukai-Tadenuma M, Kasukawa T, Ueda HR. Proof-by-synthesis of the transcriptional logic of mammalian circadian clocks. Nat Cell Biol. 2008;10(10):1154–1163. doi: 10.1038/ncb1775. [DOI] [PubMed] [Google Scholar]
- 66.Fiedler T, Rehmsmeier M. jPREdictor: A versatile tool for the prediction of cis-regulatory elements. Nucleic Acids Res. 2006;34(Web Server issue):W546–550. doi: 10.1093/nar/gkl250. [DOI] [PMC free article] [PubMed] [Google Scholar]