Significance
Until it was demonstrated that the Aedes aegypti mosquito transmitted yellow fever, the disease was possibly the most feared pestilence in the western hemisphere. This finding, by Walter Reed’s Yellow Fever Commission, is credited with eradication of the disease in many areas, through sanitation programs designed to eliminate the vector. Since this discovery, the mosquito has been widely believed to be complicit in the transmission of viral diseases, earning the enmity of humans everywhere. However, we show here that yellow fever virus encodes a protein that blocks the mosquito’s immune response, suggesting the pathogen’s continued existence in nature depends on staying one step ahead of the vector’s antiviral defense.
Keywords: yellow fever virus, flavivirus, capsid, Zika virus, RNA interference
Abstract
Mosquito-borne flaviviruses, including yellow fever virus (YFV), Zika virus (ZIKV), and West Nile virus (WNV), profoundly affect human health. The successful transmission of these viruses to a human host depends on the pathogen’s ability to overcome a potentially sterilizing immune response in the vector mosquito. Similar to other invertebrate animals and plants, the mosquito’s RNA silencing pathway comprises its primary antiviral defense. Although a diverse range of plant and insect viruses has been found to encode suppressors of RNA silencing, the mechanisms by which flaviviruses antagonize antiviral small RNA pathways in disease vectors are unknown. Here we describe a viral suppressor of RNA silencing (VSR) encoded by the prototype flavivirus, YFV. We show that the YFV capsid (YFC) protein inhibits RNA silencing in the mosquito Aedes aegypti by interfering with Dicer. This VSR activity appears to be broadly conserved in the C proteins of other medically important flaviviruses, including that of ZIKV. These results suggest that a molecular “arms race” between vector and pathogen underlies the continued existence of flaviviruses in nature.
Arthropod-borne viruses (arboviruses) are maintained in nature through transmission cycles that involve alternating replication in hematophageous insect vectors and vertebrate hosts. The Flavivirus genus of the family Flaviviridae contains a number of important mosquito-borne viruses responsible for large epidemics of human disease. These include Zika virus (ZIKV), West Nile virus (WNV), dengue virus (DENV; serotypes 1–4), Japanese encephalitis virus (JEV), and the type virus for the family, yellow fever virus (YFV). The YFV genome is a single-stranded RNA of positive polarity that is ∼11 kb in length. The 5′-capped RNA encodes the structural proteins (C-prM-E) and nonstructural proteins (NS1-NS2A-NS2B-NS3-NS4A-2K-NS4B-NS5) (1). Nonstructural components are assembled, with host factors, into membrane-bound replication complexes, where the various enzymatic functions of these proteins (e.g., RNA-dependent RNA polymerase, helicase, capping machinery) replicate the genome. The relatively smooth surface of the flavivirus virion is decorated with the envelope (E) and membrane (M) proteins, anchored into the host-derived lipid bilayer via C-terminal transmembrane domains. Within the virion is a nucleocapsid core comprising a single copy of the RNA genome in complex with multiple capsid (C) proteins (2–4). In an urban transmission cycle, the yellow fever mosquito, Aedes aegypti, acquires the virus after feeding on an infected human host. After a brief incubation period, the mosquito may transmit the virus to other susceptible humans. Although infection of the vertebrate human host is often associated with pathology and disease, a persistent nonlethal infection is established in the insect vector.
When vector mosquitoes become infected with arboviral pathogens, antiviral immunity is essential to the survival of the insect (5). Although a sufficiently robust immune response may be sterilizing, the antiviral defenses of the mosquito are not always an effective barrier to the transmission of viral pathogens. In plant and invertebrate organisms, double-stranded RNA (dsRNA) replicative intermediates (RIs) produced during viral infection activate an antiviral defense based on RNA silencing (6). In flies, the ribonuclease Dicer-2 (Dcr-2) recognizes dsRNA (7). Cleavage of dsRNA RIs by Dcr-2 generates viral small interfering RNAs (vsiRNAs) ∼21 nt in length. These siRNA duplexes are incorporated into the RNA-induced silencing complex (RISC). RISC maturation involves loading a duplex siRNA, choosing and retaining a guide strand, and ejecting the antiparallel passenger strand (8–11). The guide strand directs Argonaute 2 (Ago-2), an essential RISC component possessing endonuclease activity, to complementary RNAs in the cell, leading to their sequence-specific degradation. When infected with RNA viruses, Drosophila melanogaster dcr-2 or ago-2 loss-of-function mutants exhibit an “enhanced disease phenotype,” characterized by elevated levels of virus replication and increased mortality (12–14).
In an evolutionary “arms race,” the genomes of numerous plant and insect viruses have evolved to encode one or more viral suppressors of RNA silencing (VSRs) (15, 16). For example, the flock house virus (FHV) B2 is a well-characterized dsRNA-binding protein that shields the RIs produced during viral infection from processing by Dcr-2 and interferes with the incorporation of vsiRNA duplexes into the RISC (17–20). Well-documented examples of plant and insect virus proteins that interfere with small RNA pathways suggest that arboviruses may encode similar proteins; however, the mechanisms by which medically important arboviruses modulate small RNA pathways to productively infect disease vectors remain unknown. Elucidating these mechanisms would fundamentally alter our understanding of pathogen transmission by arthropod vectors.
We have previously shown that an antiviral response directed by vsiRNAs is essential to limiting the pathogenesis of arbovirus infections in mosquitoes (5). Suppressing the accumulation of vsiRNAs in Ae. aegypti by infection with a recombinant Sindbis virus (SINV) expressing the FHV B2 resulted in elevated levels of virus replication and increased mortality, i.e., a disease phenotype. In the present study, we show that this phenotype can be used to reliably identify VSRs that interfere with RNA silencing by diverse mechanisms. Thus, to investigate the possible presence of VSRs encoded in the genomes of flaviviruses and other mosquito-borne pathogens, we infected Ae. aegypti with recombinant SINVs (Alphavirus genus) expressing heterologous virus sequences. Here we report suppression of RNA silencing in Ae. aegypti mosquitoes infected with a recombinant SINV expressing the yellow fever virus capsid (YFC). Furthermore, through a series of biochemical assays, we demonstrate that the observed antagonism of RNA silencing is mediated by the binding of YFC to long dsRNAs, interfering with the production of vsiRNAs by Dicer. Finally, we provide evidence that this VSR activity is broadly conserved in the C proteins of other medically important flaviviruses, including that of ZIKV.
Results
Studies identifying VSRs have often been guided by previous experimental evidence implicating a particular viral protein or sequence as a virulence factor. Infection of an invertebrate host with arboviruses tends to be avirulent, however, making the identification of VSR activity in these viruses more challenging. We previously showed that expression of FHV B2 protein from recombinant alphaviruses dramatically increases the virulence of infection in the mosquito host, resulting in a disease phenotype (5). We hypothesized that this phenotype would be predictive of VSR functions in other viral proteins and sequences.
As an initial test of this hypothesis, we created recombinant SINVs expressing well-characterized VSR proteins from a duplicated subgenomic promoter (Fig. 1A). Ae. aegypti mosquitoes were infected with SINVs expressing the Cricket paralysis virus 1A protein (SINV/CrPV-1A) (12, 21), D. melanogaster C virus 1A protein (SINV/DCV-1A) (14), Carnation Italian ringspot tombusvirus P19 protein (SINV/CIRV-P19) (22–24), or the influenza A virus NS1 protein (SINV/H1N1-NS1) (25–27). Control viruses were engineered to contain nontranslatable versions of these proteins expressed from the second subgenomic promoter. We previously showed that the survival of Ae. aegypti infected with recombinant SINV is indistinguishable from that of uninfected mosquitoes (5). In the present experiment, however, mortality was significantly higher in Ae. aegypti infected with SINV/CrPV-1A, SINV/DCV-1A, SINV/CIRV-P19, or SINV/H1N1-NS1 compared with mosquitoes infected with SINV or control viruses expressing nontranslatable transcripts (Fig. 1B). These results suggest a strong correlation between the expression of heterologous VSR proteins and a disease phenotype in mosquitoes infected with SINV, even under circumstances in which the heterologous VSRs are derived from a diverse range of viruses employing different mechanisms of RNA interference (RNAi) suppression.
Fig. 1.
A disease phenotype in Ae. aegypti infected with recombinant SINVs expressing heterologous VSR proteins. (A) Schematic of a recombinant SINV expressing a VSR from an engineered subgenomic promoter (SG2). (B) Survival of mosquitoes after infection with recombinant SINVs expressing well-known VSR proteins (red triangles) or with viruses containing nontranslatable versions of the VSR sequences (black circles). Survival curves represent cohorts of ≥40 adult female mosquitoes injected with 500 pfu of virus. Significance was determined using the Mantel–Cox log-rank test.
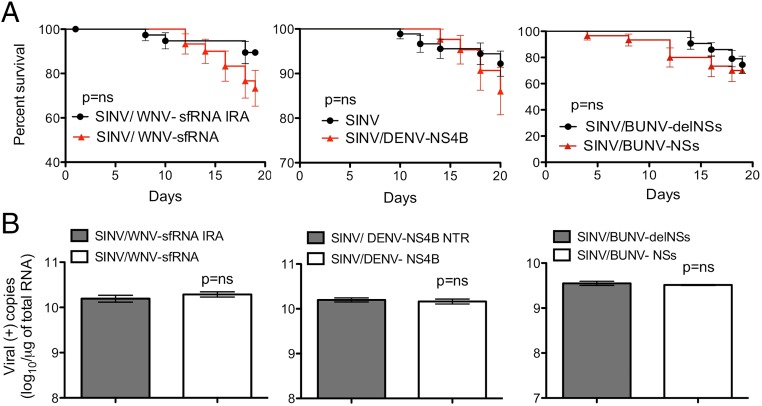
We next tested whether the expression of candidate VSRs, previously identified in the genomes of WNV, DENV, and Bunyamwera virus (BUNV; Orthobunyavirus genus), correlated with a disease phenotype in mosquitoes infected with SINV. Ae. aegypti were infected with viruses expressing the WNV sfRNA (SINV/WNV-sfRNA) (28), DENV NS4B protein (SINV/DENV-NS4B) (29), or BUNV NSs protein (SINV/BUNV-NSs) (30). Control viruses expressed inactive mutant versions of the WNV sfRNA or BUNV NSs protein, containing the previously described deletions sfRNA IRA (31) or delNSs (30), respectively. In contrast to the results described above, there was no significant difference between the survival of mosquitoes infected with control viruses and those infected with SINV/WNV-sfRNA, SINV/DENV-NS4B, or SINV/BUNV-NSs (Fig. 2A). Real-time PCR (RT-PCR) analysis of SINV RNA in the infected mosquitoes corroborated the survival results, indicating no significant increase in levels of virus replication in mosquitoes infected with SINV/WNV-sfRNA, SINV/DENV-NS4B, or SINV/BUNV-NSs compared with mosquitoes infected with control viruses (Fig. 2B). These results suggest that the WNV sfRNA, DENV NS4B, and BUNV NSs provide little to no protection from an RNA silencing response induced by virus replication in Ae. aegypti.
Fig. 2.
Infection of Ae. aegypti with recombinant SINVs expressing heterologous sequences with suspected VSR activity. (A) Survival after infection with recombinant SINVs expressing suspected VSRs (red triangles) or with control viruses (black circles). Survival curves represent cohorts of ≥40 adult female mosquitoes injected with 500 pfu of virus. Significance was determined using the Mantel–Cox log-rank test. (B) RT-PCR results demonstrating accumulation of viral RNA in mosquitoes infected with recombinant SINVs expressing suspected VSR proteins or control viruses. Error bars indicate the SEM among three biological replicates (n = 5 per replicate).
Whereas a disease phenotype is evident in mosquitoes infected with alphaviruses expressing heterologous VSRs, similar experiments had not been performed with flaviviruses. Therefore, we assessed the replication of YFV in Ae. aegypti in which Dicer-2 has been genetically ablated (32). Our results show that the replication of YFV is significantly higher in dcr-2 null mutant mosquitoes compared with wild type (WT) mosquitoes at the same time point, suggesting that RNAi is an important antiviral mechanism targeting flaviviruses in the vector host (Fig. S1).
Fig. S1.
Accumulation of viral RNA in WT and dcr-2 null Ae. aegypti infected with YFV. The graph shows RT-PCR results for three independent biological replicates (n = 5/replicate). Error bars indicate SEM.
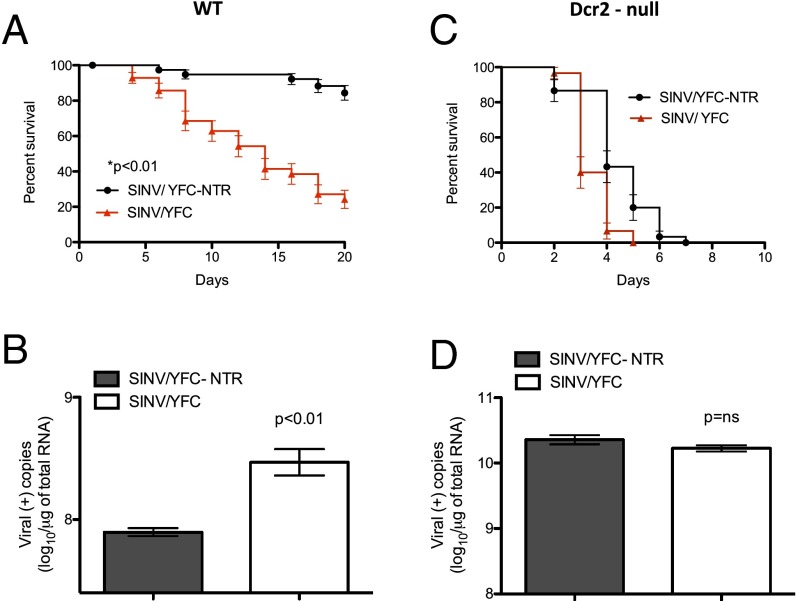
Although not a feature of all VSRs, dsRNA binding is essential to the antiviral activity of many well-characterized proteins that interfere with small RNA pathways (14, 17–19, 22–27, 33, 34). Even though a dsRNA-binding function has not previously been assigned to the flavivirus capsid, the protein has physical properties common to other VSRs that indiscriminately bind dsRNAs, shielding them from Dicer and RISC assembly; for example, evidence indicates that the flavivirus capsid protein binds RNA through nonspecific electrostatic interactions (35, 36), similar to FHV B2 (17–20) and Influenza A virus NS1 (25–27). Therefore, we assessed the potential of the YFC protein to act as an antagonist of antiviral immunity in the disease vector host. To do so, we infected Ae. aegypti with a virus expressing the YFC protein (SINV/YFC) or a control virus containing a nontranslatable version of the protein-coding sequence (SINV/YFC-NTR). Similar to the results obtained with well-known VSR proteins, mortality was significantly higher in mosquitoes infected with SINV/YFC compared with those infected with the virus containing the nontranslatable version of the protein-coding sequence (Fig. 3A). Also consistent with a disease phenotype, RT-PCR analysis indicated significantly higher levels of virus replication in mosquitoes infected with SINV/YFC compared with those infected with SINV/YFC-NTR (Fig. 3B).
Fig. 3.
The YFV capsid is a suppressor of RNA silencing. (A and C) Survival curves for Liverpool strain WT Ae. aegypti (A) and dcr-2 null mutant mosquitoes (C) infected with either recombinant SINV expressing YFC (red triangles) or virus containing a nontranslatable version of the YFC sequence (black circles). Survival curves represent cohorts of ≥30 adult female mosquitoes injected with 500 pfu of virus. (B and D) Accumulation of viral RNA in WT mosquitoes (B) and dcr-2 mutants (D) infected with recombinant SINV expressing YFC or virus containing a nontranslatable YFC sequence. Error bars for RT-PCR results indicate the SEM among three independent biological replicates (n = 5 per replicate).
To confirm that the observed disease phenotype in mosquitoes infected with SINV/YFC is due to a specific VSR activity present in the expressed YFV capsid, and not to some other function of the protein, we infected dcr-2 null mutant Ae. aegypti defective for the production of vsiRNAs with the recombinant SINVs (32). Not surprisingly, dcr-2 null mutant mosquitoes infected with the control virus also exhibited a disease phenotype, as would be expected in the absence of a functional siRNA pathway (12–14). However, in contrast to infections of the WT mosquitoes, survival curves for the dcr-2 mosquitoes infected with SINV/YFC or SINV/YFC-NTR were remarkably similar, indicating that in the absence of the primary antiviral response, the expression of YFC did not dramatically enhance virus pathogenicity (Fig. 3C). Consistent with this, the replication of SINV/YFC-NTR did not differ significantly from that of SINV/YFC in the mutant mosquitoes (Fig. 3D), indicating that the dcr-2 null genotype genetically rescued replication of the control virus. Overall, these experiments suggest VSR activity in YFC capable of suppressing RNA silencing in the natural disease vector host, Ae. aegypti.
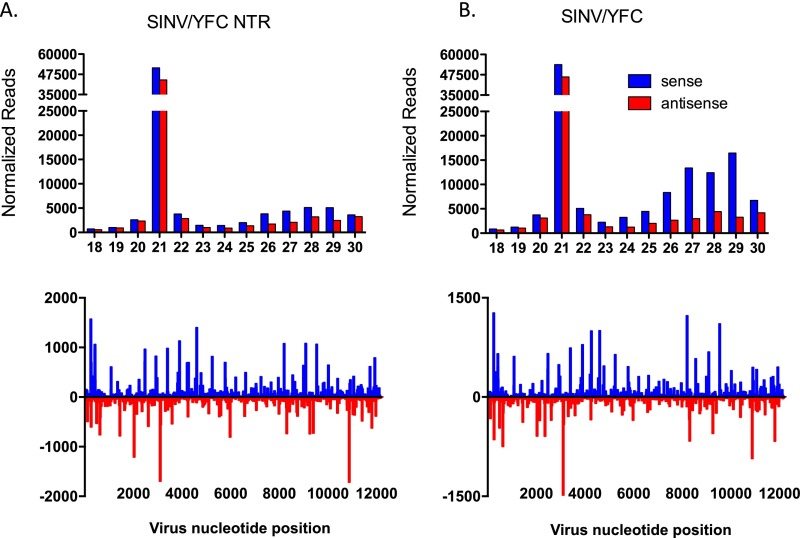
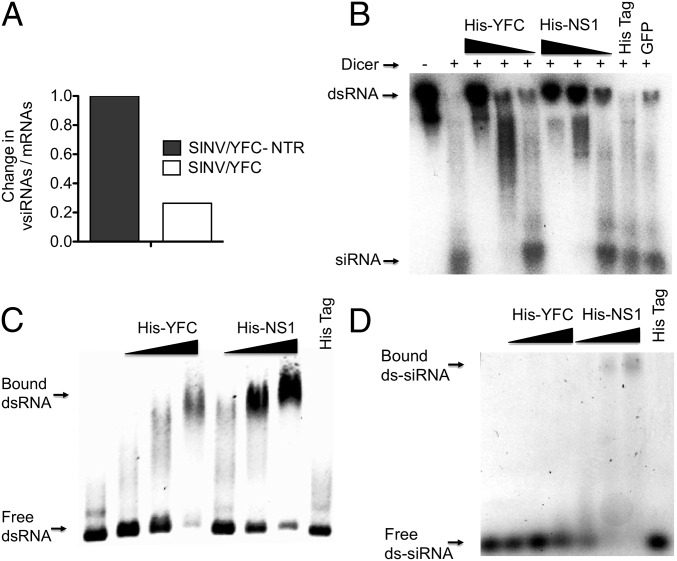
Because small RNAs ultimately serve as the effector molecules that direct the sequence-specific degradation of cognate target RNAs in the cell, we next analyzed the ratio of viral small RNAs (through Illumina-based sequencing) to viral RNAs in mosquitoes infected with recombinant SINVs (Fig. 3B and Fig. S2). The number of vsiRNAs targeting SINV/YFC decreased approximately fourfold compared with those targeting SINV/YFC-NTR in a WT genetic background (Fig. 4A), suggesting that the VSR activity of YFC is mediated by specific inhibition of Dicer. Thus, to further characterize the VSR activity of the YFV capsid, we expressed and purified a polyhistidine-tagged (His-tag) YFC protein (His-YFC) from Escherichia coli.
Fig. S2.
Small RNA reads mapped to the infecting virus genome. Shown are representative size distributions and density plots for virus-derived small RNAs in Ae. aegypti mosquitoes infected with SINV/YFC-NTR (A) and SINV/YFC (B).
Fig. 4.
The YFV capsid binds dsRNA and interferes with Dicer processing. (A) Fold change in the ratio of viral small RNAs to viral mRNAs in mosquitoes infected with recombinant SINVs, calculated from normalized small RNA reads mapping to the viral genome and strand-specific RT-PCR analysis of SINV plus-strands in 1 µg of total RNA. (B) An in vitro dicing assay with radiolabeled 700 bp dsRNA, recombinant human Dicer, and increasing concentrations of His-YFC (lanes 3–5) or His-NS1 (lanes 6–8). A His-tag peptide and GFP served as negative controls (lanes 9 and 10). Products of the dicing assay were analyzed on a 15% PAGE gel with a low molecular weight size marker. Lane 1, dsRNA with reaction buffer; lane 2, dsRNA with reaction buffer and recombinant Dicer. The concentrations of His-YFC and His-NS1 were 0.075 in lanes 3 and 6, 0.03 in lanes 4 and 7, and 0.015 nM in lanes 5 and 8; concentrations for the His-tag and GFP were 0.075 nM in lanes 9 and 10, respectively. (C) EMSA with 200-bp dsRNA (lane 1) and increasing concentrations of His-YFC protein (lanes 2–4), His-NS1 (lanes 5–7), and His-tag peptide (lane 8). RNA mobility shift was visualized with SYBR Green on a 1% agarose gel. The concentrations of His-YFC and His-NS1 used were 0.03 (lanes 2 and 5), 0.06 (lanes 3 and 6), and 0.12 nM (lanes 4 and 7). The concentration of His-tag used was 0.12 nM (lane 8). (D) EMSA with 21-bp duplex siRNA (lane 1) and increasing concentrations of His-YFC protein (lanes 2–4), His-NS1 (lanes 5–7), and His-tag peptide (lane 8). The concentrations of His-YFC and His-NS1 were 0.03 nM (lanes 2 and 5), 0.06 nM (lanes 3 and 6), and 0.12 nM (lanes 4 and 7). The concentration of His-tag was 0.12 nM (lane 8).
To assess the effect of His-YFC on the processing of a dsRNA substrate into siRNAs, we performed in vitro dicing assays. In the absence of His-YFC, dsRNA was efficiently processed into siRNAs by the recombinant Dicer enzyme (Fig. 4B, lanes 1 and 2); however, addition of His-YFC to the in vitro dicing assay inhibited the production of siRNAs in a dose-dependent manner (Fig. 4B, lanes 3–5). Similarly, His-tagged influenza A virus NS1, a well-characterized dsRNA-binding protein with previously demonstrated VSR activity (25–27), also inhibited dicing of the dsRNA substrate in this assay (Fig. 4B, lanes 6–8). In contrast, an unconjugated His-tag peptide or a green fluorescent protein (GFP) had no effect on the activity of Dicer (Fig. 4B, lanes 9 and 10). These results confirm that YFC inhibits cleavage of dsRNA substrates by Dicer.
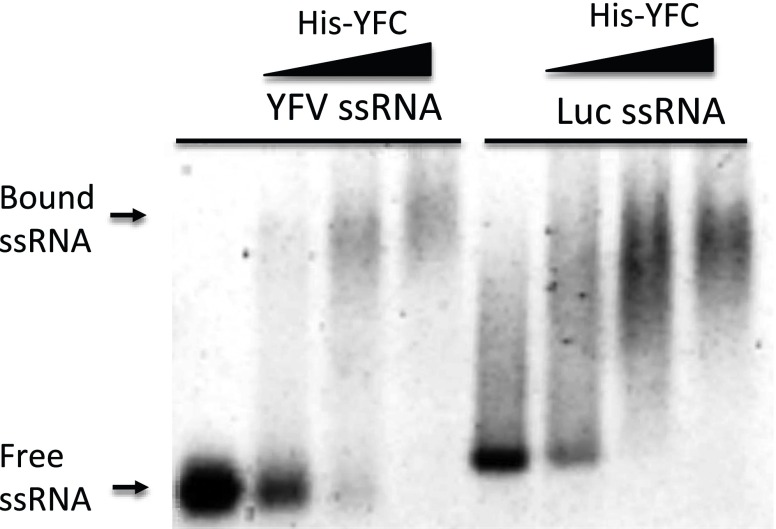
To determine whether the mechanism of Dicer inhibition by the YFC is similar to that of NS1 (i.e., nonspecific binding to dsRNAs) (25–27), we performed electrophoretic mobility shift assays (EMSAs) with both a long dsRNA and a 21-bp duplex siRNA. Although YFC was able to bind the longer dsRNA with high affinity (Fig. 4C), there was little evidence of interaction of the recombinant protein with the shorter duplex siRNA (Fig. 4D), suggesting that YFC does not interfere with loading of the RISC. Whereas there was an observable shift in the mobility of both the long dsRNA and the duplex siRNA with the recombinant NS1 protein, there was no interaction of either RNA with the unconjugated His-tag, even at the equivalent highest concentration. Finally, we also observed binding of YFC with long ssRNAs derived from YFV or luciferase sequences (Fig. S3), indicating that the recombinant protein is able to bind nonspecifically to both dsRNAs and ssRNAs. Overall, these results demonstrate that YFC has a dsRNA-binding function that interferes with the cleavage of long dsRNAs by Dicer.
Fig. S3.
The YFV capsid binds single-stranded RNA nonspecifically. EMSA with a 150-bp YFV RNA sequence (lanes 2–4), a 175-bp luciferase RNA sequence (lanes 5–7), and increasing concentrations of His-YFC protein.
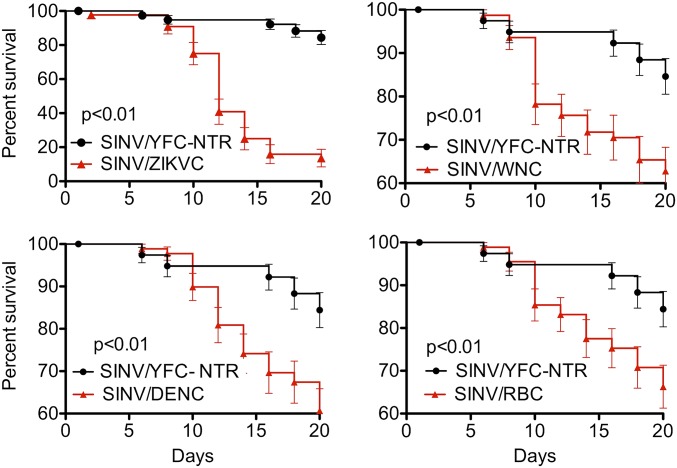
To investigate a possible correlation between the expression of other flavivirus C proteins and a disease phenotype in mosquitoes infected with SINV, we generated recombinant SINVs expressing the capsid of ZIKV (SINV/ZIKC), WNV (SINV/WNC), DENV-2 (SINV/DENC), or Rio Bravo virus (RBV; SINV/RBC). Compared with Ae. aegypti infected with SINV/YFC-NTR, mosquitoes infected with the recombinant SIN viruses expressing the heterologous C proteins exhibited evidence of viral pathogenicity (Fig. 5). Given that these experiments included the C proteins of the genetically distant ZIKV, DENV-2 (vectored by Ae. aegypti), WNV (vectored by culex species mosquitoes), and a representative of the group of flaviviruses without any known vector, RBV (isolated from bats) (37), these results strongly suggest that the VSR activity identified in the YFC is broadly conserved in the C proteins of other flaviviruses as well.
Fig. 5.
A disease phenotype in Ae. aegypti infected with recombinant SIN viruses expressing flavivirus capsid proteins. Shown are survival curves for mosquitoes infected with recombinant SIN viruses expressing flavivirus capsid proteins (red triangles). Significance was determined by comparing curves with the survival of mosquitoes infected with recombinant SINV containing a nontranslatable YFC (black circles; replicated for each panel from Fig. 3A) using the Mantel–Cox log-rank test. Survival curves represent cohorts of ≥40 adult female mosquitoes injected with 500 pfu of virus.
Discussion
Although many questions remain regarding the role of mammalian small RNA pathways in antiviral defense, it is clear that the presence of conserved antiviral silencing pathways in invertebrate organisms plays an important role in the transmission of agents of human disease. Mosquito-borne viruses are responsible for a broad spectrum of human diseases, including arthritis, hemorrhagic fever, encephalitis, and recently microcephaly and Guillain-Barré syndrome. Members of the Flavivirus genus, such as ZIKV, WNV, DENV 1–4, JEV, and YFV, are among the most important mosquito-borne pathogens because of their widespread prevalence and the severe morbidity and mortality they cause in humans. Transmission of these viruses to human or animal hosts depends on the pathogen’s ability to overcome the potentially sterilizing RNAi-based immune response of the mosquito vector; however, the specific mechanisms by which this may occur remain unknown.
There are numerous examples of VSRs encoded in the genomes of plant and insect viruses, which presumably evolved as a result of an evolutionary arms race with antiviral silencing pathways (15, 16). However, for various reasons, interrogating mosquito-borne viruses for the presence of VSR proteins has proven challenging. In this paper, we describe a simple but effective assay that is highly informative with respect to identifying VSR functions in proteins and other sequences. With this assay, we have identified a VSR encoded in the genome of YFV, the prototype flavivirus. Results obtained both in vivo and in vitro indicate that the YFC protein antagonizes RNA silencing by binding long dsRNAs with high affinity, interfering with efficient processing by Dicer. Our studies also suggest that this VSR function is broadly conserved among flavivirus C proteins.
Precedent for a VSR function in a viral structural protein can be found in the coat protein (CP) of Turnip crinkle virus, which has been shown to suppress RNA silencing through a mechanism that likely also involves the binding of dsRNAs (38). In comparison with other positive-strand viruses, the flavivirus nucleocapsid has a poorly defined and unique architecture (2, 3, 39). Reported production of noninfectious virus-like particles by overexpressing only precursor membrane (prM) and E protein indicates that the C protein is not required for formation of the virion’s spherical shell (40, 41). Indeed, there appears to be little interaction between the C protein and the E and M proteins, raising questions about how nucleocapsids are efficiently incorporated into budding membranous structures containing the flavivirus surface proteins (42). A packaging signal has not been identified in genomes of flaviviruses, and in vitro formation of nucleocapsid-like particles with positive- or negative-stranded DENV RNAs of differing lengths is consistent with the idea that these sequences do not exist (36). Rather, interactions between RNA and the C protein appear to be mediated by nonspecific electrostatic interactions (35, 36), and our results are consistent with this idea. Evidence indicates that packaging requires active synthesis of viral RNA, suggesting that replication and encapsidation are tightly coupled (43). This may explain the apparent specificity with which genomic RNA is packaged into the virion, despite a lack of organization in the way in which C proteins and viral RNA assemble (2, 3, 39). Our results, presented here, suggest that the C protein interacts with viral dsRNA RIs in the infected cell.
Similar to other positive-strand viruses, the replication complexes of flaviviruses are believed to assemble on internal cellular membranes (42). In DENV-infected cells, this process involves extensive rearrangements of the endoplasmic reticulum to form a continuous network of vesicles consisting of series of invaginations of the endoplasmic reticulum membrane (44). Similar structures have been observed in mosquito cells infected with DENV (45). Detection of dsRNA within membrane vesicles suggests that these are sites of viral replication within the infected cell (44, 45). Although evidence suggests that membrane vesicles are connected to the cytosol by pores (44, 45), it has been postulated that these structures may protect the viral RNA from the cellular components of innate immunity (46). However, the existence of VSRs encoded in the genomes of flaviviruses suggests that replication complexes are accessible to RNA silencing pathways, and possibly other antiviral responses as well.
In mammalian cells, viral infection induces the synthesis and secretion of type I IFNs (α/β). Activation of signaling pathways by the secreted IFN results in the transcription of hundreds of genes, contributing to the establishment of a general antiviral state in the cell. A number of viruses have been found to express proteins that are antagonists of the IFN α/β response. Similar to VSR proteins, viruses have evolved diverse mechanisms for inhibiting the IFN α/β response (47). Also similar to VSR proteins, dsRNA binding is a molecular mechanism by which some viral proteins inhibit the IFN α/β system. For example, the dsRNA-binding function of the influenza NS1 protein counteracts the establishment of an antiviral state in the cell by blocking virus-mediated activation of cellular transcription factors that induce synthesis of IFN α/β (26, 27, 47). Thus, a particularly intriguing possibility is that the dsRNA-binding domain of the flavivirus capsid protein evolved to counteract immune responses in both mammalian and insect hosts. Conservation of a dsRNA-binding function in the C proteins of flaviviruses without a known insect vector, as our data suggests, justifies future studies to address the possibility that flavivirus C proteins inhibit the type I IFN response.
Although flavivirus C proteins have low sequence identity, several functional elements appear to be well conserved (48), for example, the oligomeric state of the protein, which forms a dimer in solution (49, 50). The individual subunits of the dimer each comprise four alpha helices (α1–α4). The dimer conformation is stabilized through hydrophobic interactions occurring between two pairs of antiparallel helices (50). The flavivirus C protein is highly basic, with positively charged residues distributed throughout. The nonuniform distribution generates an unusually large net positive charge that makes the oligmerization of protein-only cores unlikely, which is consistent with the the role of flavivirus C protein in RNA binding and encapsidation (50). In vitro experiments have implicated the first 32 and last 26 residues of the Kunjin virus (KUNV) C protein as important in RNA binding (51); however, studies mapping the minimal functional elements of the dimeric YFC protein required for packaging and virus assembly have demonstrated remarkable functional flexibility at the N and C termini of the protein (48).
Removal of the first 40 amino acid residues from the N terminus, including the entire α1 helix, fail to abolish the C protein’s ability to package replicon RNA. Similarly, the last 27 residues of the YFC protein’s terminus, including complete deletion of the α4 helix, are also expendable for virus assembly. These findings have led to the proposal that genomic RNA is bound initially at the N terminus of the C protein, followed by binding of residues at the C terminus (48). However, the YFC protein’s C terminus appears to be sufficient for packaging of genomic RNA in the absence of binding sites located in N terminus and vice versa (48). In future studies, it will be interesting to determine whether the dsRNA-binding domain present in the YFC is independent, identical, or overlaps with domains already implicated as important to the binding of genomic RNA.
With rare exceptions, all cellular organisms have evolved a system of protection from foreign genetic material (52). In response, viruses have evolved to encode a diverse array of countermeasures, composing the majority of genes in some viral genomes, with the complete catalog constantly updated (52). Evolutionary history is replete with examples of antiviral mechanisms being co-opted by the cell for other functions (52); indeed, a similar process is likely to have occurred in the small genomes of RNA viruses, leading to the evolution of multifunctional proteins. The establishment of persistent nonlethal infections in the invertebrate vector is essential to the natural maintenance cycles of many mosquito-borne viruses. Whether mosquito-borne viral pathogens have evolved to encode VSRs, as have other viruses with plant and insect hosts, has been unclear. Small RNAs are important in modulating the pathogenesis of viral infections in disease vector mosquitoes. Thus, infection of the invertebrate host with a virus encoding a VSR could in theory result in a fitness cost that would adversely affect the probability of the virus being transmitted to a new vertebrate host. However, the results of the present study indicate that flaviviruses encode at least one protein capable of blocking RNAi in the mosquito, suggesting coevolution of these viruses in response to the strong selective pressure exerted by the RNA silencing pathways of the host, a classic “red queen hypothesis.” Thus, a molecular arms race occurring between vector and pathogen underlies the continued existence of these viruses in nature, illustrating the importance of RNA silencing pathways in the transmission of many important human diseases.
Materials and Methods
Mosquito Infections.
Ae. aegypti (Liverpool) WT and dcr-2 null strains were reared at 28 °C and 80% relative humidity, with a 14/10-h day/night light cycle. Dcr-2 null mosquitoes were obtained as described previously (32). Dcr-2 null mosquitoes were crossed with a previously described Ae. aegypti “sensor” strain that expresses an eye-specific GFP reporter only when RNAi is impaired (53). Dcr-2 null mosquitoes were screened at 6 d after emergence for the expression of eye-specific GFP. Mosquitoes were injected with 0.5 µL (106 pfu/mL) of recombinant SINV, and survival was monitored daily. The WT mosquitoes were 2 d old at injection, whereas the dcr-2 null mosquitoes were 8 d old due to the screening process.
Recombinant Virus Production.
Recombinant SIN viruses were generated by inserting heterologous sequences encoding known or suspected VSRs into a multiple cloning site, located downstream of a duplicated subgenomic promoter. Recombinant viruses were rescued as described previously (5).
RNA Isolation and Detection.
Total RNA was extracted from pools of mosquitoes with Tri Reagent RT (Molecular Research Center) according to the manufacturer’s instructions. SINV mRNA levels were determined using a strand-specific quantitative RT-PCR Taqman assay (Life Technologies) as described previously (54), with the following primers and Taqman probe: forward, 5′-ATCACAATTGGCAACGAGAAGAG-3′; reverse, 5′-CTGTGGGTTCGGAGAATAGTGG-3′; probe, 5′-CTAAAAGCAGCCGAACTC-3′. The following primers and Taqman probe were used for YFV: forward, 5′-GGTTCCATGAGCGTGGCTAT-3′; reverse, 5′-GCGCAGCAGCGTAGTAACAC-3′; probe: 5′-CAAGCTGGAAGGTAGGGTGAT-3′. A minimum of three independent biological replicates were tested, and results were analyzed using the t test to determine significance.
Small RNA Library Preparation and Analysis.
Libraries were prepared from total RNA isolated with Tri RT (Molecular Research Center) from adult female mosquitoes at 96 h after infection with recombinant SINVs. In brief, small RNAs (18–35-nt) were recovered by PAGE separation, and libraries were prepared with the Truseq Small RNA Sample Preparation Kit (Illumina) according to the manufacturer's instructions. To reduce sources of nonbiological variation, libraries were multiplexed and biological replicates were sequenced in a single lane of a HiSeq (Illumina) flow cell. Following removal of the 3′ adapters, 10 million small RNA reads were selected at random from each dataset and mapped to the SINV genome. Differential expression of vsiRNAs was determined as described previously (55). The small RNA libraries used in this study are available for download from the Gene Expression Omnibus (accession no. GSE80691).
Dicing Assays.
Processing of long dsRNA into siRNAs was analyzed with an ultra-active form of recombinant human Dicer, capable of cleaving more than 95% of dsRNA template into siRNAs within 2 h (Genlantis), in an in vitro cleavage assay. In brief, 32P-labeled dsRNA (700 bp, 30 ng) and recombinant human Dicer (0.5 U) were incubated in the buffer solutions provided by the manufacturer for 2 h at 37 °C, and then analyzed on a 15% PAGE gel. The indicated concentrations of recombinant His-YFC protein, His-NS1 protein (Bioclone), 6× His-tag peptide (Abcam), or recombinant GFP (Kerafast) were coincubated in the dicing assay.
EMSA.
EMSA was performed using the SYBR Green EMSA Kit (Life Technologies) according to the manufacturer’s instructions. In brief, a 200-bp dsRNA or 21-bp siRNA duplex was incubated with various concentrations of recombinant proteins for 20 min at room temperature, and then visualized on an agarose gel with SYBR Green.
Acknowledgments
This work was supported by National Institute for Allergy and Infectious Diseases Grants AI077726, AI103265, and AI119081. The funding agency had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. G.D. is a Guest Editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE80691).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1600544113/-/DCSupplemental.
References
- 1.Lindenbach BD, Murray CL, Thiel H-J, Rice CM. 2013. Flaviviridae: The viruses and their replication. Fields Virology, eds Knipe DM, Howley PM (Wolters Kluwer/Lippincott Williams & Wilkins Health, Philadelphia), 6th Ed.
- 2.Kuhn RJ, et al. Structure of dengue virus: Implications for flavivirus organization, maturation, and fusion. Cell. 2002;108(5):717–725. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mukhopadhyay S, Kim BS, Chipman PR, Rossmann MG, Kuhn RJ. Structure of West Nile virus. Science. 2003;302(5643):248. doi: 10.1126/science.1089316. [DOI] [PubMed] [Google Scholar]
- 4.Zhang W, et al. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat Struct Biol. 2003;10(11):907–912. doi: 10.1038/nsb990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Myles KM, Wiley MR, Morazzani EM, Adelman ZN. Alphavirus-derived small RNAs modulate pathogenesis in disease vector mosquitoes. Proc Natl Acad Sci USA. 2008;105(50):19938–19943. doi: 10.1073/pnas.0803408105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu Q, Wang X, Ding SW. Viral suppressors of RNA-based viral immunity: Host targets. Cell Host Microbe. 2010;8(1):12–15. doi: 10.1016/j.chom.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409(6818):363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 8.Tomari Y, Du T, Zamore PD. Sorting of Drosophila small silencing RNAs. Cell. 2007;130(2):299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marques JT, et al. Loqs and R2D2 act sequentially in the siRNA pathway in Drosophila. Nat Struct Mol Biol. 2010;17(1):24–30. doi: 10.1038/nsmb.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123(4):621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y, et al. C3PO, an endoribonuclease that promotes RNAi by facilitating RISC activation. Science. 2009;325(5941):750–753. doi: 10.1126/science.1176325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang XH, et al. RNA interference directs innate immunity against viruses in adult Drosophila. Science. 2006;312(5772):452–454. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL. Essential function in vivo for Dicer-2 in host defense against RNA viruses in Drosophila. Nat Immunol. 2006;7(6):590–597. doi: 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- 14.van Rij RP, et al. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 2006;20(21):2985–2995. doi: 10.1101/gad.1482006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li F, Ding SW. Virus counterdefense: Diverse strategies for evading the RNA-silencing immunity. Annu Rev Microbiol. 2006;60:503–531. doi: 10.1146/annurev.micro.60.080805.142205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Neal ST, Samuel GH, Adelman ZN, Myles KM. Mosquito-borne viruses and suppressors of invertebrate antiviral RNA silencing. Viruses. 2014;6(11):4314–4331. doi: 10.3390/v6114314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu R, et al. Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature. 2005;436(7053):1040–1043. doi: 10.1038/nature03870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chao JA, et al. Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat Struct Mol Biol. 2005;12(11):952–7. doi: 10.1038/nsmb1005. [DOI] [PubMed] [Google Scholar]
- 19.Lingel A, Simon B, Izaurralde E, Sattler M. The structure of the flock house virus B2 protein, a viral suppressor of RNA interference, shows a novel mode of double-stranded RNA recognition. EMBO Rep. 2005;6(12):1149–1155. doi: 10.1038/sj.embor.7400583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aliyari R, et al. Mechanism of induction and suppression of antiviral immunity directed by virus-derived small RNAs in Drosophila. Cell Host Microbe. 2008;4(4):387–397. doi: 10.1016/j.chom.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nayak A, et al. Cricket paralysis virus antagonizes Argonaute 2 to modulate antiviral defense in Drosophila. Nat Struct Mol Biol. 2010;17(5):547–554. doi: 10.1038/nsmb.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silhavy D, et al. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 2002;21(12):3070–3080. doi: 10.1093/emboj/cdf312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vargason JM, Szittya G, Burgyán J, Hall TM. Size-selective recognition of siRNA by an RNA silencing suppressor. Cell. 2003;115(7):799–811. doi: 10.1016/s0092-8674(03)00984-x. [DOI] [PubMed] [Google Scholar]
- 24.Qiu W, Park JW, Scholthof HB. Tombusvirus P19-mediated suppression of virus-induced gene silencing is controlled by genetic and dosage features that influence pathogenicity. Mol Plant Microbe Interact. 2002;15(3):269–280. doi: 10.1094/MPMI.2002.15.3.269. [DOI] [PubMed] [Google Scholar]
- 25.Li WX, et al. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc Natl Acad Sci USA. 2004;101(5):1350–1355. doi: 10.1073/pnas.0308308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bornholdt ZA, Prasad BV. X-ray structure of influenza virus NS1 effector domain. Nat Struct Mol Biol. 2006;13(6):559–560. doi: 10.1038/nsmb1099. [DOI] [PubMed] [Google Scholar]
- 27.Cheng A, Wong SM, Yuan YA. Structural basis for dsRNA recognition by NS1 protein of influenza A virus. Cell Res. 2009;19(2):187–195. doi: 10.1038/cr.2008.288. [DOI] [PubMed] [Google Scholar]
- 28.Schnettler E, et al. Noncoding flavivirus RNA displays RNA interference suppressor activity in insect and mammalian cells. J Virol. 2012;86(24):13486–13500. doi: 10.1128/JVI.01104-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kakumani PK, et al. Role of RNA interference (RNAi) in dengue virus replication and identification of NS4B as an RNAi suppressor. J Virol. 2013;87(16):8870–8883. doi: 10.1128/JVI.02774-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szemiel AM, Failloux AB, Elliott RM. Role of Bunyamwera Orthobunyavirus NSs protein in infection of mosquito cells. PLoS Negl Trop Dis. 2012;6(9):e1823. doi: 10.1371/journal.pntd.0001823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pijlman GP, et al. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe. 2008;4(6):579–591. doi: 10.1016/j.chom.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 32.Basu S, et al. Silencing of end-joining repair for efficient site-specific gene insertion after TALEN/CRISPR mutagenesis in Aedes aegypti. Proc Natl Acad Sci USA. 2015;112(13):4038–4043. doi: 10.1073/pnas.1502370112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Cleef KW, et al. Mosquito and Drosophila entomobirnaviruses suppress dsRNA- and siRNA-induced RNAi. Nucleic Acids Res. 2014;42(13):8732–8744. doi: 10.1093/nar/gku528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valli A, et al. The VP3 factor from viruses of Birnaviridae family suppresses RNA silencing by binding both long and small RNA duplexes. PLoS One. 2012;7(9):e45957. doi: 10.1371/journal.pone.0045957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pong WL, Huang ZS, Teoh PG, Wang CC, Wu HN. RNA binding property and RNA chaperone activity of dengue virus core protein and other viral RNA-interacting proteins. FEBS Lett. 2011;585(16):2575–2581. doi: 10.1016/j.febslet.2011.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Teoh PG, Huang ZS, Pong WL, Chen PC, Wu HN. Maintenance of dimer conformation by the dengue virus core protein α4-α4′ helix pair is critical for nucleocapsid formation and virus production. J Virol. 2014;88(14):7998–8015. doi: 10.1128/JVI.00940-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Billoir F, et al. Phylogeny of the genus flavivirus using complete coding sequences of arthropod-borne viruses and viruses with no known vector. J Gen Virol. 2000;81(Pt 9):2339. doi: 10.1099/0022-1317-81-9-2339. [DOI] [PubMed] [Google Scholar]
- 38.Qu F, Ren T, Morris TJ. The coat protein of turnip crinkle virus suppresses posttranscriptional gene silencing at an early initiation step. J Virol. 2003;77(1):511–522. doi: 10.1128/JVI.77.1.511-522.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X, et al. Cryo-EM structure of the mature dengue virus at 3.5-Å resolution. Nat Struct Mol Biol. 2013;20(1):105–110. doi: 10.1038/nsmb.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Allison SL, et al. Two distinct size classes of immature and mature subviral particles from tick-borne encephalitis virus. J Virol. 2003;77(21):11357–11366. doi: 10.1128/JVI.77.21.11357-11366.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schalich J, et al. Recombinant subviral particles from tick-borne encephalitis virus are fusogenic and provide a model system for studying flavivirus envelope glycoprotein functions. J Virol. 1996;70(7):4549–4557. doi: 10.1128/jvi.70.7.4549-4557.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Apte-Sengupta S, Sirohi D, Kuhn RJ. Coupling of replication and assembly in flaviviruses. Curr Opin Virol. 2014;9:134–142. doi: 10.1016/j.coviro.2014.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khromykh AA, Varnavski AN, Sedlak PL, Westaway EG. Coupling between replication and packaging of flavivirus RNA: Evidence derived from the use of DNA-based full-length cDNA clones of Kunjin virus. J Virol. 2001;75(10):4633–4640. doi: 10.1128/JVI.75.10.4633-4640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Welsch S, et al. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe. 2009;5(4):365–375. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Junjhon J, et al. Ultrastructural characterization and three-dimensional architecture of replication sites in dengue virus-infected mosquito cells. J Virol. 2014;88(9):4687–4697. doi: 10.1128/JVI.00118-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chatel-Chaix L, Bartenschlager R. Dengue virus- and hepatitis C virus-induced replication and assembly compartments: The enemy inside—caught in the web. J Virol. 2014;88(11):5907–5911. doi: 10.1128/JVI.03404-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.García-Sastre A. Mechanisms of inhibition of the host interferon alpha/beta-mediated antiviral responses by viruses. Microbes Infect. 2002;4(6):647–655. doi: 10.1016/s1286-4579(02)01583-6. [DOI] [PubMed] [Google Scholar]
- 48.Patkar CG, Jones CT, Chang YH, Warrier R, Kuhn RJ. Functional requirements of the yellow fever virus capsid protein. J Virol. 2007;81(12):6471–6481. doi: 10.1128/JVI.02120-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jones CT, et al. Flavivirus capsid is a dimeric alpha-helical protein. J Virol. 2003;77(12):7143–7149. doi: 10.1128/JVI.77.12.7143-7149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma L, Jones CT, Groesch TD, Kuhn RJ, Post CB. Solution structure of dengue virus capsid protein reveals another fold. Proc Natl Acad Sci USA. 2004;101(10):3414–3419. doi: 10.1073/pnas.0305892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khromykh AA, Westaway EG. RNA binding properties of core protein of the flavivirus Kunjin. Arch Virol. 1996;141(3-4):685–699. doi: 10.1007/BF01718326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koonin EV, Dolja VV. A virocentric perspective on the evolution of life. Curr Opin Virol. 2013;3(5):546–557. doi: 10.1016/j.coviro.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adelman ZN, Anderson MA, Morazzani EM, Myles KM. A transgenic sensor strain for monitoring the RNAi pathway in the yellow fever mosquito, Aedes aegypti. Insect Biochem Mol Biol. 2008;38(7):705–713. doi: 10.1016/j.ibmb.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Plaskon NE, Adelman ZN, Myles KM. Accurate strand-specific quantification of viral RNA. PLoS One. 2009;4(10):e7468. doi: 10.1371/journal.pone.0007468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morazzani EM, Wiley MR, Murreddu MG, Adelman ZN, Myles KM. Production of virus-derived ping-pong-dependent piRNA-like small RNAs in the mosquito soma. PLoS Pathog. 2012;8(1):e1002470. doi: 10.1371/journal.ppat.1002470. [DOI] [PMC free article] [PubMed] [Google Scholar]