Abstract
Autism spectrum disorders (ASD) are classified as neurodevelopmental disorders characterised by diminished social communication and interaction. Recently, evidence has accrued that a significant proportion of individuals with autism have concomitant diseases such as mitochondrial disease and abnormalities of energy generation. This has therefore led to the hypothesis that autism may be linked to mitochondrial dysfunction. We review such studies reporting decreased activity of mitochondrial electron transport chain (ETC) complexes and reduced gene expression of mitochondrial genes, in particular genes of respiratory chain complexes, in individuals with autism. Overall, the findings support the hypothesis that there is an association of ASD with impaired mitochondrial function; however, many of the studies have small sample sizes and there is variability in the techniques utilised. There is therefore a vital need to utilise novel imaging techniques, such as near-infrared spectroscopy, that will allow non-invasive measurement of metabolic markers for neuronal activity such as cytochrome c oxidase, in order to better establish the link between autism and mitochondrial dysfunction.
Keywords: Autism, Mitochondrial dysfunction, Mitochondrial health, Energy metabolism, Electron transport chain, Mitochondrial deficiency
Introduction
Autism spectrum disorders (ASD) are classified as a group of neurodevelopmental disorders which include autistic disorder, Asperger’s syndrome, pervasive developmental disorder and childhood disintegrative disorder. ASD is diagnosed on the basis of behavioural observations such as impaired social interaction, diminished verbal and non-verbal communication and repetitive behaviours [1]. ASD is characterised through certain behavioural and cognitive features which are primarily thought to be a result of atypical development of the brain itself. Over recent years however, there is increasing evidence indicating that a significant proportion of the ASD population have implicating comorbidities such as mitochondrial dysfunction, oxidative stress, gastrointestinal abnormalities and abnormalities in regulation of the immune system [2]. In this context, autism may involve, or be a result of, systemic physiological abnormalities rather than entirely being a neurodevelopmental disorder.
The initial hypothesis associating ASD with an underlying comorbidity such as mitochondrial dysfunction was made by Coleman and Blass in 1985, who hypothesised that individuals with ASD can have “abnormal carbohydrate metabolism” [3]. Later on, Lombard made a similar proposition in 1998 [4] suggesting that “autism may be a disorder of atypical mitochondrial function”. Since then, there have been an increasing number of research and clinical studies involving individuals with ASD and there is mounting evidence linking mitochondrial dysfunction to ASD. We review the major findings from studies that present evidence of mitochondrial dysfunction in ASD. Prior to this, the reader is presented with a brief overview of the mitochondria, mitochondrial genome, and the mitochondrial mechanisms of energy generation, with particular reference to those biochemical pathways discussed in the studies reviewed.
Mitochondrial Function
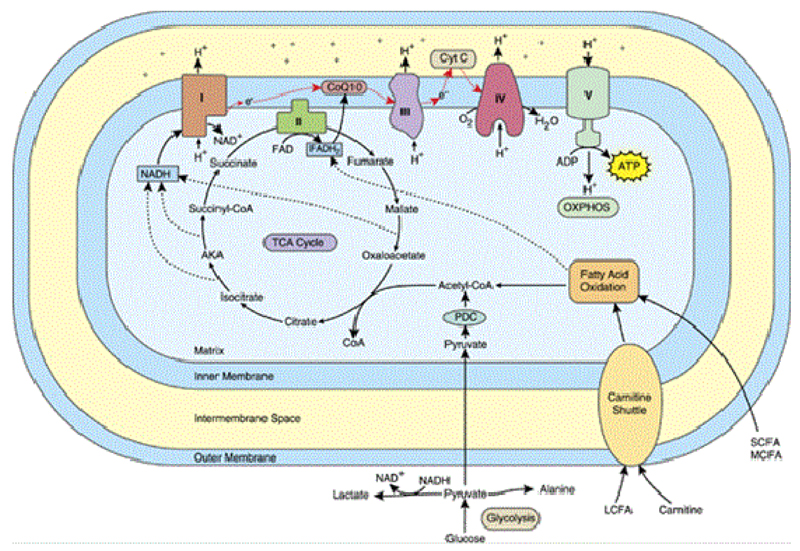
Mitochondria are cellular organelles that play a vital role in ensuring the proper functioning of cells [5]. As the site of energy generation, they are deemed the “powerhouse” of the cell and are responsible for providing cells with energy to function. Mitochondria are especially important in areas of high energy demand such as the brain where neuronal cells utilise an estimated 4.7 billion ATP per second [6]. ATP generation takes place through three major processes; glycolysis which takes place in the cytosol, tricarboxylic acid (TCA) cycle in the mitochondrial matrix and the electron transport chain (ETC) in the inner mitochondrial membrane. In the first step of glycolysis, glucose is broken down into pyruvate. Pyruvate then enters the mitochondrial matrix where it is converted to the key intermediate enzyme acetyl-CoA which is further used in the TCA cycle to reduce electron carriers nicotinamide adenine dinucleotide (NAD) or NADH in its reduced form and flavin adenine dinucleotide (FAD) or FADH2 in its reduced form. These high-energy electron carriers are then transported to the inner mitochondrial membrane for the final stage known as oxidative phosphorylation which takes place at the site of the electron transport chain (ETC), in the inner mitochondrial membrane. The respiratory chain comprises of different components, which include five enzyme complexes I-V as well as two electron carriers; ubiquinone and cytochrome c [2]. During this process, an initial donation of electrons is made by the carriers NADH and FADH2 to complexes I and II, respectively. These electrons are then passed down through the remaining ETC complexes, down electrochemical gradient to eventually produce ATP. Figure 1 shows the processes through which ATP is generated in the mitochondria.
Figure 1.
Processes involved in energy generation and the mitochondrial respiratory chain. “Abbreviations: ADP, adenosine diphosphate; AKA, α-Ketoglutarate; ATP, Adenosine Triphosphate; CoQ, Co-enzyme Q; Cyt C, Cytochrome C; e, electron; FAD, Flavin Adenine Dinucleotide; FADH2, Reduced FAD; H, Hydrogen; LCFA, Long Chain Fatty Acid; MCFA, Medium Chain Fatty Acid; NAD, Nicotinamide Adenine Dinucleotide; NADH, Reduced NAD; OXPHOS, Oxidative Phosphorylation; PDC, Pyruvate Dehydrogenase Complex; SCFA, Short Chain Fatty Acid; TCA, Tricarboxylic Acid; I, complex I; II, complex II; III, complex III; IV, complex IV; V, complex V. The red arrows denote the flow of electrons in the ETC” [2].
In addition to ATP synthesis, the mitochondria also play a number of other important roles. These include calcium homeostasis through regulation of Ca2+ signalling, regulation of apoptosis (cell death) and production of reactive oxygen species (ROS) [7]. These diverse mitochondrial functions coupled with the importance of the mitochondria in the control of cellular energy demands imply that any form of dysfunction could have serious consequences on the health of an individual and may lead to disease. Mitochondrial disease has already been linked to the pathogenesis of diseases such as and Alzheimer’s [8].
Furthermore, mitochondria possess their own genome. Although 99% of mitochondrial proteins are coded for by nuclear DNA [9], the ETC complexes contain subunits that are encoded both by mitochondrial DNA (mtDNA) and nuclear DNA (nDNA) [10]. The mitochondrial genome has a total of 37 genes; 13 of which encode for subunits of complexes I and III-V of the ETC and the rest-22 are transfer RNAs (tRNAs) and 2 ribosomal RNAs (rRNAs) [11] the machinery required for translation and transcription of the mitochondrial genes into the ETC subunits. With both the nuclear and mitochondrial genome regulating the gene expression of mitochondrial proteins, mutations in either DNA could lead impaired mitochondrial function as well as abnormalities in the ETC complexes.
Mitochondrial Dysfunction in ASD
Mitochondrial dysfunction can be classified into two types [12]; the first type (“primary dysfunction”) refers to the dysfunction that occurs as a result of a mutation in a gene that is directly involved in the ATP-generating pathway. The second type (“secondary dysfunction”) refers to the dysfunction occurring due to other genetic, biochemical or metabolic abnormalities and deficiencies that may impair the mitochondria’s ability to synthesise ATP. These biochemical markers of mitochondrial dysfunction include “lactate, pyruvate, lactate-to-pyruvate ratio, ubiquinone, alanine, alanine-to-lysine ratio and acyl-carnitine”, all of which are produced at some stage of respiration [2].
We now review the major findings from studies that present evidence of mitochondrial dysfunction in ASD. The findings from the studies include a diverse range of results, some of which include evidence of decreased activity of mitochondrial respiratory chain complexes, evidence of the presence of biomarkers of oxidative stress and mitochondrial dysfunction as well as indication of mtDNA mutations (deletions and replications). Table one summarises the major findings from the studies discussed herein.
Table 1.
Summary of major findings from studies, including methods used. This table has been adapted from [5].
| Citation | ASD Cases/Controls | Methods | Major findings |
|---|---|---|---|
| Minshew et al. [13] | 11/11 | Phosphorus-31 magnetic resonance spectroscopy | Markers of mitochondrial function were abnormal in ASD brain and correlated with ASD behaviours |
| Friedman et al. [17] | 45/28 | Echo planar spectroscopic imaging | “Reduced metabolite N-acetyl-aspartate1 (NAA) concentrations in the ASD group compared to typically developing children” |
| Palmieri et al. [36] | 6/6 | Quantification of gene expression Western blotting Protein quantification Genomic DNA sequencing |
“Higher ETC complex IV activity in ASD group” “ASD group showed higher level of oxidative damage to mitochondrial proteins in Brodmann Area (BA) BA41/422 or BA223” |
| Giulivi et al. [25] | 10/10 | Quantification of gene expression (q-PCR) Spectrophotometry Measurement of oxygen consumption using Clark-type electrodes |
“Mitochondrial dysfunction, mitochondrial DNA over replication and mitochondrial DNA deletions were more likely in ASD group” |
| Chauhan et al. [20] | 8/8 | Western blotting | “Decreased ETC complex activity in the frontal lobe, temporal lobe and cerebellum in ASD group” |
| Anitha et al. [31] | 8/10 | Quantification of gene expression (q-PCR) | “Decreased expression of mitochondrial genes in anterior cingulate gyrus, motor cortex and thalamus in ASD group” |
| Ginsberg et al. [30] | 9/9 | Whole genome gene expression analysis Genome-wide DNA methylation |
“Decreased ETC complex gene expression in cerebellum and BA194 in ASD group |
| Rose et al. [37] | 15/15 | Protein assay kit High-performance liquid chromatography Mass spectrometry | “Decreased aconitase5 activity in cerebellum and BA22 in ASD group” |
| Smith et al. [38] | 69/89 | Microarray analysis for copy number variation | “Copy number variants (CNV) in autistic patients commonly encompass genes important in mitochondrial function, ion transport and synaptic structure and function” |
| Anitha et al. [31] | 8/10 | Quantification of gene expression (q-PCR) | “Reduced expression of ETC genes (complexes I, III, IV and V) in the anterior cingulate gyrus, thalamus, motor cortex in ASD group” |
| Gu et al. [22] | 14/12 | Protein quantification mtDNA copy number analysis (q-PCR) | “Reduced ETC complex I and V activity in the frontal lobe in ASD group” “Higher mitochondrial DNA copy number compared to nuclear DNA in 3 different mitochondrial genes found in ASD group” |
| Tang et al. [21] | 20/25 | Western blotting Quantification and assessment of mtDNA (q-PCR and long-range PCR) Protein quantification |
“Reduced ETC activity complexes I and IV in BA216 in ASD group” “Higher levels of mitochondrial fission proteins and lower levels of mitochondrial fusion proteins in BA21 of ASD group” |
| Chen et al. [32] | 78/83 | mtDNA copy number analysis (q-PCR) | “Elevated mtDNA copy number in peripheral blood cells of children with autism” |
| Hardan et al. [19] | 17/17 | Proton magnetic resonance spectroscopy (1H MRS) | Decreased ratio of NAA to phosphocreatine and creatine in children with ASD |
| Goldenthal et al. [15] | 92/68 | Quantification of protein activity levels (immunocapture assays and spectrophotometry) | Significant mitochondrial respiratory complex chain deficiencies in 42% of children with ASD |
Evidence of Mitochondrial Dysfunction using Magnetic Resonance Spectroscopy
Minshew et al. [13] conducted a study in 1993 using Phosphorus-31 magnetic resonance spectroscopy (31P MRS) and reported evidence of mitochondrial energy metabolism abnormalities. The study investigated brain energy and phospholipid metabolism in the dorsal prefrontal cortex of autistic adolescents, by looking at phosphocreatine, ADP, ATP, inorganic phosphate and phosphomonoesters, phosphodiesters respectively. The study reported abnormal metabolite levels for both brain energy and phospholipid metabolism, in the group with ASD. Furthermore, these metabolite levels were compared with scores from language and intelligence tests within the same group. The study reported a positive correlation between low intelligence and language scores and abnormal metabolite levels in the autistic group, in comparison to controls.
Similar evidence has emerged for brain metabolic marker “N-acetyl-aspartate” (NAA) using the same technique. NAA is an important brain metabolite and a marker for mitochondrial dysfunction [14–16] since it is synthesised solely by neuronal mitochondria and one of its main functions includes contributing to the production of energy by mitochondria. Reduced NAA concentrations were also reported in a study conducted by Friedman et al. Friedman et al. in a group of 45 children with ASD, 13 typically developing (TD) controls and 15 children with development delay reported reduced in the ASD group in comparison to the controls [17]. Furthermore, Ipser et al. [18] conducted a study including autistic children and adults and found a significantly lower levels of NAA in the white and grey matter of the brains of the autistic children. Meanwhile across age, the study reported significantly lower levels of NAA in the grey matter of the cerebellum, the anterior cingulate cortex and parietal cortex.
Goldenthal et al. reported reduced NAA, phosphocreatine and creatine levels in the left thalamus of children with autism [16]. Phosphocreatine and creatine provide information about cellular energy metabolism that have a particularly important role in tissues with high fluctuating energy demands. Furthermore, a more recent study using proton spectroscopy conducted by the same group reports decreased ratio of NAA to phosphocreatine (PCr) and creatine (Cr) in the anterior white matter of children with autism [19].
Levels of Electron Transport Chain Complexes
As the ETC has a vital role in ATP production, many studies have investigated the respiratory chain complexes for function, gene expression levels and activity levels. A study conducted by Chauhan et al. in 2011 examined a number of different areas of the post-mortem brain tissue [20] (over a wide age range of individuals with ASD), including the frontal, temporal, parietal and occipital cortices as well as the cerebellum, to investigate the protein levels of the ETC complexes. The results of this study demonstrated decreased levels of respiratory chain complexes in the frontal and temporal cortex and the cerebellum in the ASD group of children aged 4-10 years. Through further investigation, the cerebellum was found to have decreased levels of ETC complexes III and V, while the frontal cortex was reported to have reduced levels of all respiratory chain complexes [20]. These results were not found in the ASD group aged 14-39 years, therefore suggesting brain region-specific developmental changes in the respiratory chain complexes in ASD children. It could also suggest that development of abnormalities in the respiratory chain could later on lead to mitochondrial disease and potentially contribute to the onset of autism symptoms. A similar study by Tang et al. [21] observed severe ETC impairment with reduced levels of all ETC complexes (except complex II) in children with ASD of age less than 10 years, in post-mortem temporal lobe brain tissue. The study also found lower complex I and IV enzyme activity.
Gu et al. in 2013 investigated the respiratory chain complexes as well as pyruvate dehydrogenase activity in the post-mortem frontal cortex tissue in 14 ASD individuals [22]. The study reported significantly decreased pyruvate dehydrogenase activity (35% reduction) along with reduced complex I and V activity, in the ASD group. Pyruvate dehydrogenase is a key enzyme involved in conversion of pyruvate to acetyl-CoA. Its reduced activity would result in insufficient removal of pyruvate and lactate for the TCA cycle, causing insufficient ATP generation. The results from this study support others that have reported the association of mitochondrial dysfunction, abnormal pyruvate and lactate levels [23,24], with autism by suggesting that abnormal pyruvate dehydrogenase activity may play a role in high levels of these metabolites in ASD.
A number of other studies investigated the activity of mitochondrial ETC complexes in tissue other than brain. Giulivi et al. Investigated mitochondrial dysfunction by studying lymphocytes in 10 children with ASD and 10 controls [25]. Here too, the results demonstrated impaired ETC complex I in the ASD group. They also reported on pyruvate dehydrogenase activity which was found to be significantly decreased (50%) in the ASD group. A more recent study conducted by Goldenthal et al. investigated mitochondrial enzyme deficiencies in buccal swabs of children with autism [15]. This study reports extensive ETC abnormalities in 42% of the ASD group.
Although there are inconsistencies in the brain areas examined in these studies, there is clear indication of the presence of abnormalities in respiratory chain complexes in individuals with autism and these may in fact play a role in the etiology of ASD. This can be explained by the fact that the respiratory chain complexes are vital to energy generation, particularly in an area of high energy demand such as the brain. Their deficiency could lead to severe implications such as insufficient energy metabolism and acceleration in production of free radicals, resulting in oxidative stress.
Oxidative Stress
Reactive oxygen species (ROS) or free radicals are produced during the metabolism of oxygen at the site of oxidative phosphorylation, the electron transport chain. They are a molecular species that contain an unpaired electron and are therefore highly reactive and unstable. Although a regulated level of free radicals is required for key processes such calcium homeostasis, they are capable of attacking important macromolecules such as proteins and lipids, causing cell damage and which can eventually lead to disease. Cell damage caused by ROS is implicated in a number of diseases such as cancer [26,27] and Alzheimer’s [8]. Several antioxidant defence mechanisms are employed by the human body to maintain the level of free radicals and regulate the damage they can cause. These defence mechanisms include the production of antioxidant enzymes such as glutathione peroxidase and superoxide dismutase (SOD) [5]. Mitochondrial dysfunction, particularly deficiencies in the respiratory chain complexes, could lead to an elevated level of ROS thereby causing an imbalance of ROS production/antioxidant defences, leading to oxidative stress.
Recent studies have demonstrated the co-occurrence of mitochondrial abnormalities and oxidative stress in the post-mortem brain tissue of children with ASD. As discussed earlier, the study by Chauhan et al. [20] demonstrated brain region-specific deficiencies in ETC complexes. This study additionally presented evidence of elevated levels of ROS in the same brain regions where respiratory chain complex deficiencies were observed. Similarly the study by Tang et al [21] also discussed earlier reported elevated levels of biomarkers of oxidative stress in the temporal cortex of the ASD group, the same brain region where ETC complex deficiencies were observed. Additionally, the same study reported significantly decreased activity of antioxidant enzyme superoxide dismutase2, which plays an important role in the defence mechanisms against ROS, in the group of children with ASD. A different study by Giulivi et al. [25] reported increased mitochondrial rate of production of antioxidant hydrogen peroxide in the children with autism in comparison to controls.
An earlier study by Chauhan et al. investigated the presence of markers of oxidative stress in brain tissue as well as in urine and blood in a group of individuals with ASD [28]. The study revealed significantly increased levels of these markers in the ASD group in comparison to the controls, thereby strengthening the hypothesis that oxidative stress may play a role in the etiology of autism [28].
Electron Transport Chain Gene Expression
Due to the recent implication of mitochondrial dysfunction in a number of diseases such as Alzheimer’s [8,29] and cancer [27], research groups have regained interest in field of mitochondrial genetics and epigenetics. Many processes of mitochondrial gene expression and regulation are, to date, not very well understood. Various studies have set out to explore the variation of mitochondrial gene expression in the respiratory chain enzymes, in individuals with autism. In order to investigate this, one such study by Ginsberg et al. used genome-wide genotyping and DNA methylation sequencing in post-mortem brain tissue, namely cerebellar and occipital, from a group 9 individuals with ASD and 9 controls [30]. The results of the study presented evidence of down-regulation of the genes coding for the ETC complexes I and III as well as ATP synthase.
Anitha et al. conducted a gene expression analysis study with specific focus on the 84 genes encoding the mitochondrial respiratory chain complexes [31]. The study examined three areas of post-mortem brain tissue, namely the “anterior cingulate gyrus, motor cortex and thalamus” areas in a group of 8 individuals with autism versus 10 controls. The results showed reduced gene expression in a number of the genes coding for respiratory chain enzymes in the ASD group. More specifically, in 11 genes coding for complex I, 5 of those coding for complex III and IV and 7 coding for complex V [31].
Mitochondrial DNA Mutations
More recently, studies have utilised real-time polymerase chain reaction (q-PCR) technique to look at mitochondrial DNA copy number in individuals with ASD. DNA copy number variation is a form of structural variations of DNA results in (ab)normal variation in the number of copies of one or more sections of DNA. The study conducted by Gu et al. [22], as discussed earlier, found deficient ETC complexes and decreased pyruvate dehydrogenase activity. The group additionally investigated copy number variations (CNVs) of mitochondrial respiratory chain genes. “CNVs are the most common types of structural variations in the human genome” [22]. The results demonstrated increased CNVs in 3 respiratory chain genes encoding for subunits of complex I and III a result that was also presented in the study by Giulivi et al. in the lymphocytes of ASD individuals [25]. A more recent study conducted by Chen et al. [32] shows results consistent with the study conducted by Gu et al. The study reports mtDNA copy number to be significantly elevated in in peripheral blood cells in children with autism.
Conclusion
Autism is classified as a psychiatric disorder which is characterised entirely by behaviours. Recent studies however, have presented evidence for systemic physiological abnormalities in ASD. The studies reviewed herein provide support to the idea of a link between mitochondrial dysfunction and the behavioural phenotype of autism. A number of different techniques have been employed to investigate a range of potential aspects of this link and many have reported decreased mitochondrial gene expression, decreased activity of electron transport chain complexes and abnormal levels of peripheral markers of mitochondrial function. However, due to the variation and inconsistency in techniques used, there are limitations to the studies reviewed here. These limitations make it difficult to determine whether mitochondrial dysfunction is a feature of all individuals with ASD, whether it is a cause or an effect of autism and whether such dysfunctions are localised to specific regions of the brain. Even in light of these however, there are consistent encouraging findings across the different studies and methodologies, indicating that mitochondrial disease may be commonly linked with autism and may be critical in the causal pathway that determines atypicalities in brain function and structure in autism. While further work is required, many studies have reported a stronger association between mitochondrial abnormalities and ASD in children, thereby providing indirect support for a causal role.
Majority of the studies reviewed have examined the post-mortem brain tissue of individuals with ASD. While this is an important step in investigating a potential link between mitochondrial dysfunction and autism, it does not provide the prospective insight that in vivo investigation could deliver. Near-infrared spectroscopy (NIRS) is an optical imaging technique that allows in vivo measurement of chromophore concentration changes, which can be used to derive a measure of cerebral oxygenation changes. With recent developments in the NIRS imaging technique, novel NIRS systems allow investigation of mitochondrial function in vivo by measuring optical changes in the brain resulting from oxidative phosphorylation involving mitochondrial electron transport chain enzyme cytochrome-c-oxidase [33,34]. Studies in animals and humans have demonstrated that these changes, measured following hypoxic ischemia, to be correlated with 31Phosphorus magnetic resonance spectroscopy (MRS) biomarkers of cerebral energy failure. Thereby providing evidence for the potential of measuring changes in mitochondrial function resulting from regional changes in the brain induced by neural activity [35]. Adapting these novel NIRS systems for use in children with ASD is an exciting step which can provide useful insight into the role of mitochondrial function in autism. Additionally, it could provide the opportunity to explore whether mitochondrial dysfunction is a causal effect of autism and how its development occurs in the ASD brain.
Notes
The definitions of the Brodmann areas defined below were taken from [39].
“NAA is synthesized in the mitochondria of neurons. One of its functions is to contribute to energy production from the amino acid glutamate in neuronal mitochondria” [14].
“Brodmann area is a region of the cerebral cortex, in the human or other primate brain, defined by its structure and organization of cells. BA41/42 is Brodmann areas 41 and 42 respectively that are involved in auditory processing.”
“BA22 is the Brodmann area 22 region of the human brain. On the left side of the brain this area helps with generation and understanding of individual words. On the right side of the brain it helps to discriminate pitch and sound intensity.”
“BA19 is the Brodmann area 19 region of the human brain. It is part of the occipital lobe cortex and along with area 18; it comprises the extrastriate (or peristriate) cortex. In humans with normal sight, extrastriate cortex is a visual association area, with feature extracting, shape recognition, and multimodal integrating functions.”
Aconitase is an enzyme primarily located in the mitochondria involved in the TCA cycle.
“BA21 is Brodmann area 21 and it is part of the temporal cortex in the human brain. The region encompasses most of the lateral temporal cortex, a region believed to play a part in auditory processing and language.”
Acknowledgement
This work was supported by the Biotechnology and Biological Sciences Research Council [grant number BB/J014567/1]. Johnson received support from the UK Medical Research Council (G0701484 ID: 85031 & MR/K021389/1) and from the Innovative Medicines Initiative Joint Undertaking under Grant Agreement No. 115300, resources of which are composed of financial contribution from the European Union’s Seventh Framework Programme (FP7/2007– 2013) and EFPIA companies’ in kind contribution.
References
- 1.American Psychiatric Association. Diagnostic and statistical manual of mental disorders, 5th Edition (DSM-5) Diagnostic Stat Man Ment Disord 4th Ed. TR; 2013. p. 280. [Google Scholar]
- 2.Rossignol DA, Frye RE. Mitochondrial dysfunction in autism spectrum disorders: A systematic review and meta-analysis. Mol Psychiatry. 2012;17:290–314. doi: 10.1038/mp.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coleman M, Blass JP. Autism and lactic acidosis. J Autism Dev Disord. 1985;15:1–8. doi: 10.1007/BF01837894. [DOI] [PubMed] [Google Scholar]
- 4.Lombard J. Autism: A mitochondrial disorder? Med Hypotheses. 1998;50:497–500. doi: 10.1016/s0306-9877(98)90270-5. [DOI] [PubMed] [Google Scholar]
- 5.Rossignol DA, Frye RE. Evidence linking oxidative stress, mitochondrial dysfunction and inflammation in the brain of individuals with autism. Front Physiol. 2014;5:150. doi: 10.3389/fphys.2014.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu XH, Qiao H, Du F, Xiong Q, Liu X, et al. Quantitative imaging of energy expenditure in human brain. Neuroimage. 2012;60:2107–2117. doi: 10.1016/j.neuroimage.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devall M, Roubroeks J, Mill J, Weedon M, Lunnon K. Epigenetic regulation of mitochondrial function in neurodegenerative disease: New insights from advances in genomic technologies. Neurosci Lett. 2016;625:47–55. doi: 10.1016/j.neulet.2016.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devall M, Mill J, Lunnon K. The mitochondrial epigenome: A role in Alzheimer's disease? Epigenomics. 2014;6:665–675. doi: 10.2217/epi.14.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boengler K, Heusch G, Schulz R. Nuclear-encoded mitochondrial proteins and their role in cardioprotection. Biochim Biophys Acta. 2011;1813:1286–1294. doi: 10.1016/j.bbamcr.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 10.Cotter D, Guda P, Fahy E, Subramaniam S. MitoProteome: Mitochondrial protein sequence database and annotation system. Nucleic Acids Res. 2004;32:D463–467. doi: 10.1093/nar/gkh048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DiMauro S, Schon EA. Mitochondrial respiratory-chain diseases. N Engl J Med. 2003;348:2656–2668. doi: 10.1056/NEJMra022567. [DOI] [PubMed] [Google Scholar]
- 12.Haas RH, Parikh S, Falk MJ, Saneto RP, Wolf NI, et al. Mitochondrial disease: A practical approach for primary care physicians. Pediatrics. 2007;120:1326–1333. doi: 10.1542/peds.2007-0391. [DOI] [PubMed] [Google Scholar]
- 13.Minshew NJ, Goldstein G, Dombrowski SM, Panchalingam K, Pettegrew JW. A preliminary 31P MRS study of autism: evidence for under synthesis and increased degradation of brain membranes. Biol Psychiatry. 1993;33:762–773. doi: 10.1016/0006-3223(93)90017-8. [DOI] [PubMed] [Google Scholar]
- 14.Clark JB. N-acetyl aspartate: A marker for neuronal loss or mitochondrial dysfunction. Dev Neurosci. 1998;20:271–276. doi: 10.1159/000017321. [DOI] [PubMed] [Google Scholar]
- 15.Goldenthal MJ, Damle S, Sheth S, Shah N, Melvin J, et al. Mitochondrial enzyme dysfunction in autism spectrum disorders; a novel biomarker revealed from buccal swab analysis. Biomark Med. 2015;9:957–965. doi: 10.2217/bmm.15.72. [DOI] [PubMed] [Google Scholar]
- 16.Hardan AY, Minshew NJ, Melhem NM, Srihari S, Jo B, et al. An MRI and proton spectroscopy study of the thalamus in children with autism. Psychiatry Res Neuroimaging. 2008;163:97–105. doi: 10.1016/j.pscychresns.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman SD, Shaw DW, Artru AA, Richards TL, Gardner J, et al. Regional brain chemical alterations in young children with autism spectrum disorder. Neurology. 2003;6:100–107. doi: 10.1212/wnl.60.1.100. [DOI] [PubMed] [Google Scholar]
- 18.Ipser JC, Syal S, Bentley J, Adnams CM, Steyn B, et al. 1H-MRS in autism spectrum disorders: A systematic meta-analysis. Metab Brain Dis. 2012;27:275–287. doi: 10.1007/s11011-012-9293-y. [DOI] [PubMed] [Google Scholar]
- 19.Hardan AY, Fung LK, Frazier T, Berquist SW, Minshew NJ, et al. A proton spectroscopy study of white matter in children with autism. Prog Neuro Psychopharmacology Biol Psychiatry. 2015;66:48–53. doi: 10.1016/j.pnpbp.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chauhan A, Gu F, Essa MM, Wegiel J, Kaur K, et al. Brain region-specific deficit in mitochondrial electron transport chain complexes in children with autism. J Neurochem. 2011;117:209–220. doi: 10.1111/j.1471-4159.2011.07189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang G, Rios PG, Kuo SH, Akman HO, Rosoklija G, et al. Mitochondrial abnormalities in temporal lobe of autistic brain. Neurobiol Dis. 54:349–361. doi: 10.1016/j.nbd.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu F, Chauhan V, Kaur K, Brown WT, LaFauci G, et al. Alterations in mitochondrial DNA copy number and the activities of electron transport chain complexes and pyruvate dehydrogenase in the frontal cortex from subjects with autism. Transl Psychiatry. 2013;3:e299. doi: 10.1038/tp.2013.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weissman JR, Kelley RI, Bauman ML, Cohen BH, Murray KF, et al. Mitochondrial disease in autism spectrum disorder patients: A cohort analysis. PLoS One. 2008;3:11. doi: 10.1371/journal.pone.0003815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oliveira G, Diogo L, Grazina M, Garcia P, Ataíde CA, et al. Mitochondrial dysfunction in autism spectrum disorders: A population-based study. Dev Med Child Neurol. 2005;47:185–189. doi: 10.1017/s0012162205000332. [DOI] [PubMed] [Google Scholar]
- 25.Cecilia Giulivi PY-FZ, Alicja Omanska-Klusek BS, Catherine Ross-Inta MS, Sarah Wong BS, Irva Hertz-Picciotto BS, et al. Mitochondrial dysfunction in autism. JAMA. 304:2389–2396. doi: 10.1001/jama.2010.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng S, Xiong L, Ji Z, Cheng W, Yang H. Correlation between increased ND2 expression and demethylated displacement loop of mtDNA in colorectal cancer. Mol Med Rep. 2012;6:125–130. doi: 10.3892/mmr.2012.870. [DOI] [PubMed] [Google Scholar]
- 27.Wen SL, Zhang F, Fenn S. Decreased copy number of mitochondrial DNA: A potential diagnostic criterion for gastric cancer. Oncol Lett. 2013;6:1098–1102. doi: 10.3892/ol.2013.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chauhan A, Chauhan V. Oxidative stress in autism. Pathophysiology. 2006;13:171–181. doi: 10.1016/j.pathophys.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 29.Chandrasekaran K, Giordano T, Brady DR, Stoll J, Martin LJ, et al. Impairment in mitochondrial cytochrome oxidase gene expression in Alzheimer disease. Brain Res Mol Brain Res. 1994;24:336–340. doi: 10.1016/0169-328x(94)90147-3. [DOI] [PubMed] [Google Scholar]
- 30.Ginsberg MR, Rubin RA, Falcone T, Ting AH, Natowicz MR. Brain transcriptional and epigenetic associations with autism. PLoS One. 2012;7:e44736. doi: 10.1371/journal.pone.0044736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anitha A, Nakamura K, Thanseem I, Yamada K, Iwayama Y, et al. Brain region-specific altered expression and association of mitochondria-related genes in autism. Mol Autism. 2012;3:12. doi: 10.1186/2040-2392-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen S, Li Z, He Y, Zhang F, Li H, et al. Elevated mitochondrial DNA copy number in peripheral blood cells is associated with childhood autism. BMC Psychiatry. 2015;15:50. doi: 10.1186/s12888-015-0432-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palmieri L, Persico AM. Mitochondrial dysfunction in autism spectrum disorders: Cause or effect? Biochim Biophys Acta. 2010;1797:1130–1137. doi: 10.1016/j.bbabio.2010.04.018. [DOI] [PubMed] [Google Scholar]
- 34.Bale G, Mitra S, Meek J, Robertson N, Tachtsidis I. A new broadband near-infrared spectroscopy system for in vivo measurements of cerebral cytochrome-c-oxidase changes in neonatal brain injury. Biomed Opt Express. 2014;5:3450–3466. doi: 10.1364/BOE.5.003450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitra S, Bale G, Meek J, Mathieson S, Uria C, et al. In vivo measurement of cerebral mitochondrial metabolism using broadband near infrared spectroscopy following neonatal stroke,” in oxygen transport to tissue XXXVII. Springer New York: 2016. pp. 493–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palmieri L, Papaleo V, Porcelli V, Scarcia P, Gaita L, et al. Altered calcium homeostasis in autism-spectrum disorders: Evidence from biochemical and genetic studies of the mitochondrial aspartate/glutamate carrier {AGC} 1. Mol Psychiatry. 2010;15:38–52. doi: 10.1038/mp.2008.63. [DOI] [PubMed] [Google Scholar]
- 37.Rose S, Melnyk S, Pavliv O, Bai S, Nick TG, et al. Evidence of oxidative damage and inflammation associated with low glutathione redox status in the autism brain. Transl Psychiatry. 2012;2:e134. doi: 10.1038/tp.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith M, Flodman PL, Gargus JJ, Simon MT, Verrell K, et al. Mitochondrial and ion channel gene alterations in autism. Biochim Biophys Acta. 2002;1817:1796–1802. doi: 10.1016/j.bbabio.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brodmann K. Vergleichende Lokalisationslehre der Grosshirnrinde. J Nerv Ment Dis. 1910;37L:783–784. [Google Scholar]