Abstract
BACKGROUND
The Amish and Hutterites are U.S. agricultural populations whose lifestyles are remarkably similar in many respects but whose farming practices, in particular, are distinct; the former follow traditional farming practices whereas the latter use industrialized farming practices. The populations also show striking disparities in the prevalence of asthma, and little is known about the immune responses underlying these disparities.
METHODS
We studied environmental exposures, genetic ancestry, and immune profiles among 60 Amish and Hutterite children, measuring levels of allergens and endotoxins and assessing the microbiome composition of indoor dust samples. Whole blood was collected to measure serum IgE levels, cytokine responses, and gene expression, and peripheral-blood leukocytes were phenotyped with flow cytometry. The effects of dust extracts obtained from Amish and Hutterite homes on immune and airway responses were assessed in a murine model of experimental allergic asthma.
RESULTS
Despite the similar genetic ancestries and lifestyles of Amish and Hutterite children, the prevalence of asthma and allergic sensitization was 4 and 6 times as low in the Amish, whereas median endotoxin levels in Amish house dust was 6.8 times as high. Differences in microbial composition were also observed in dust samples from Amish and Hutterite homes. Profound differences in the proportions, phenotypes, and functions of innate immune cells were also found between the two groups of children. In a mouse model of experimental allergic asthma, the intranasal instillation of dust extracts from Amish but not Hutterite homes significantly inhibited airway hyperreactivity and eosinophilia. These protective effects were abrogated in mice that were deficient in MyD88 and Trif, molecules that are critical in innate immune signaling.
CONCLUSIONS
The results of our studies in humans and mice indicate that the Amish environment provides protection against asthma by engaging and shaping the innate immune response. (Funded by the National Institutes of Health and others.)
Many genetic risk factors have been reported to modify susceptibility to asthma and allergy,1,2 but the dramatic increase in the prevalence of these conditions in westernized countries in the past half-century suggests that the environment also plays a critical role.3 The importance of environmental exposures in the development of asthma is most exquisitely illustrated by epidemiologic studies conducted in Central Europe that show significant protection from asthma and allergic disease in children raised on traditional dairy farms. In particular, children's contact with farm animals and the associated high microbial exposures4,5 have been related to the reduced risk.6,7 However, the effect of these traditional farming environments on immune responses is not well defined.
To address this gap in knowledge, we designed a study that compares two distinctive U.S. farming populations — the Amish of Indiana and the Hutterites of South Dakota — that recapitulate the differences in the prevalences of asthma and allergy observed in farmers and nonfarmers in Europe. These two particular groups of farmers originated in Europe — the Amish in Switzerland and the Hutterites in South Tyrol — during the Protestant Reformation and then emigrated to the United States in the 1700s and 1800s, respectively. Both groups have since remained reproductively isolated.8,9 Their lifestyles are similar with respect to most of the factors known to influence the risk of asthma, including large sibship size, high rates of childhood vaccination, diets rich in fat, salt, and raw milk, low rates of childhood obesity, long durations of breast-feeding, minimal exposure to tobacco smoke and air pollution, and taboos against indoor pets. However, whereas the Amish practice traditional farming, live on single-family dairy farms, and use horses for fieldwork and transportation, the Hutterites live on large, highly industrialized, communal farms. Strikingly, the prevalence of asthma in Amish versus Hutterite schoolchildren is 5.2% versus 21.3% and the prevalence of allergic sensitization is 7.2% versus 33.3%, as previously reported.10,11
METHODS
OVERVIEW
We characterized the immune profiles of Amish and Hutterite schoolchildren. Furthermore, we used mouse models of asthma to study the effect of the environment on airway responses and to create a mechanistic framework for the interpretation of our observations in humans.
STUDY PARTICIPANTS AND STUDY OVERSIGHT
In November 2012, we studied 30 Amish children 7 to 14 years of age who lived in Indiana, and in December 2012 we studied 30 Hutterite children who lived in South Dakota and were matched with the Amish children for sex and for age within 1 year. (For information on the characteristics of the children see Table 1, and the Methods section in the Supplementary Appendix, available with the full text of this article at NEJM.org). Written informed consent was obtained from the parents and written assent was obtained from the children. One parent of each child responded to a questionnaire on asthma symptoms and previous diagnoses. The study was approved by the institutional review boards at the University of Chicago and at St. Vincent Hospital in Indianapolis.
Table 1.
Demographic and Clinical Characteristics of the Study Populations.*
| Characteristic | Amish (N=30) | Hutterite (N=30) |
|---|---|---|
| Age (yr) | ||
| Median | 11 | 12 |
| Range | 8–14 | 7–14 |
| Girls (no.) | 10 | 10 |
| Sibships (no.) | 15 | 14 |
| Children with asthma (no.) | 0 | 6 |
| Positivity for allergen-specific IgE (no.) | ||
| >0.7 kUA/liter | 5 | 9 |
| >3.5 kUA/liter | 2 | 9 |
| Serum IgE (kU/liter) | ||
| Median | 21 | 64 |
| Interquartile range | 10–57 | 15–288 |
UA denotes allergen-specific unit.
BLOOD-SAMPLE COLLECTION AND ANALYSIS
Whole blood was collected in tubes that contained culture medium alone, medium plus 0.1 μg per milliliter of lipopolysaccharide, or medium plus 0.4 μg per milliliter of anti-CD3 plus 0.33 μg per milliliter of anti-CD28 monoclonal antibodies (TruCulture Blood Collection System, Myriad RBM). After incubation at 37°C for 30 hours, supernatant and cells were frozen for use in gene-expression and cytokine studies. Levels of 26 cytokines were measured with the use of the Milliplex Map Human TH17 Magnetic Bead Panel (EMD Millipore) or enzyme-linked immunoabsorbent assay (eBioscience) in the supernatant, in accordance with standard protocols. Additional blood was collected to obtain peripheral-blood leukocytes for flow cytometry and DNA isolation, and serum was collected for IgE studies (as described in the Methods section in the Supplementary Appendix).
Cryopreserved human peripheral-blood leukocytes were incubated for 10 minutes with pooled human IgG antibodies (FcX, Biolegend) to block nonspecific antibody binding before undergoing surface staining with fluorescently conjugated antibodies (see Table S1 in the Supplementary Appendix) and intracellular staining for FoxP3 (eBioscience). Flow-cytometry data were acquired on an LSRFortessa cell analyzer (BD Biosciences), and acquisition data were analyzed with FlowJo software (Tree Star).
DUST COLLECTION AND EXTRACT PREPARATION
Electrostatic dust collectors were placed in one bedroom and the living room in each of 10 Amish and 10 Hutterite homes to collect airborne house dust. All 10 Amish homes and 9 of 10 Hutterite homes housed children who participated in the study. After 1 month, dust was analyzed for endotoxin and allergen levels, and extracts were prepared for studies in mice. In addition, a vacuum was used to collect dust from the living-room floor in Amish homes and from mattresses in Amish and Hutterite homes for use in microbiome studies (as described in the Methods section in the Supplementary Appendix). Aqueous extracts of house dust from Amish and Hutterite homes were prepared as described in the Methods section in the Supplementary Appendix.
GENETIC STUDIES
RNA was extracted from thawed cells with the use of AllPrep DNA/RNA Mini Kits (Qiagen). RNA underwent complementary DNA synthesis and was then hybridized to HumanHT-12 v4 Expression BeadChip arrays (Illumina). A common set of 118,789 single-nucleotide polymorphisms (SNPs) was genotyped or imputed in the 60 children in the study (see Fig. S1 in the Supplementary Appendix).
MOUSE MODELS
We instilled 50 μl of house-dust extract intranasally every 2 to 3 days (for a total of 14 times) into 7-week old BALB/c mice (Harlan Laboratories), beginning on day 0. The mice had been sensitized intraperitoneally with 20 μg of ovalbumin (grade V, Sigma) plus alum (Pierce) on days 0 and 14 and were challenged intranasally with 50 μg of ovalbumin on days 28 and 38. Beginning 5 days before day 0, we also instilled 50 μl of dust extract from Amish homes intranasally every 2 to 3 days (for a total of 14 times) in 7-week old, C57BL6 wild-type, MyD88-deficient mice12 and in mice deficient in both MyD88 and Trif13 (Jackson Laboratories). These mice were sensitized intraperitoneally with 20 μg of ovalbumin plus alum on days 0 and 14 and challenged intranasally with 75 μg of ovalbumin on days 26, 27, and 28.
STATISTICAL ANALYSIS
Statistical analyses were performed with the use of R or Prism software (GraphPad). Differences in the distributions of median cytokine levels between Amish and Hutterite children were determined with use of a Wilcoxon signed-rank test. We used linear regression to identify differentially expressed genes in the Amish and Hutterite untreated samples of peripheral-blood leukocytes. The methods of Benjamini and Hochberg14 were used to control the false discovery rate. For flow cytometric studies and the study in mice, differences in cell populations and airway resistance were assessed with the use of an unpaired Student's t-test. Additional details on sample processing, quality control, and statistical analysis for all methods described here are provided in the Methods section in the Supplementary Appendix.
RESULTS
ASTHMA AND ALLERGIC-SENSITIZATION RATES AND GENETIC ANCESTRY
None of the Amish children and six (20%) of the Hutterite children had asthma, rates similar to those reported in earlier studies.10,11 Levels of total serum IgE and the number of children whose levels of IgE against common allergens were high (defined as more than 3.5 kUA [allergen-specific unit] per liter) were lower in the Amish group than in the Hutterite group (Table 1, and Table S2 in the Supplementary Appendix). No statistical differences were observed in levels of serum Ig isotypes other than IgE (Fig. S2 in the Supplementary Appendix).
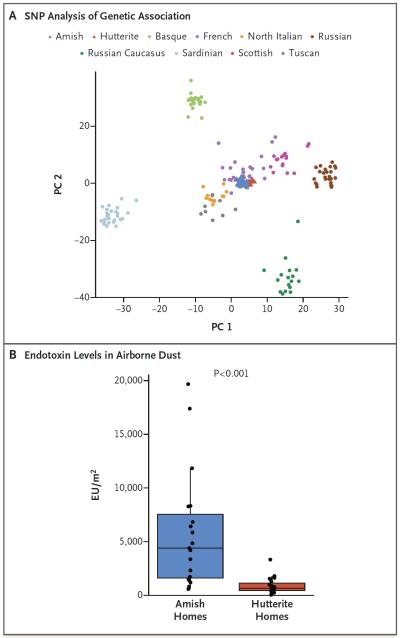
To evaluate whether the differences in the prevalence of asthma and of allergic sensitization and differences in IgE level could be attributed to population history, we assessed ancestry by conducting principal component analysis and compared allele frequencies using genomewide SNPs. These studies revealed remarkable genetic similarities between the Amish children and the Hutterite children, as compared with other European populations15 (Fig. 1A, and Fig. S3 in the Supplementary Appendix).
Figure 1. Ancestries and Environments of Amish and Hutterite Children.
Panel A shows a principal components plot of the first two principal components (PC 1 and PC 2) of the analysis of 72,034 single-nucleotide polymorphisms (SNPs). Amish and Hutterite genotypes were projected onto the sample space created by Human Genome Diversity Project (HGDP) for European populations.15 Panel B shows endotoxin levels in airborne dust from 10 Amish and 10 Hutterite homes. Box-and-whisker plots show a horizontal line indicating median value, a box representing the interquartile range, and whiskers showing the 95% confidence interval. The P value was calculated with the use of the Wilcoxon rank-sum test. EU denotes endotoxin units.
EXPOSURES TO ALLERGENS, MICROBES, AND MICROBIAL PRODUCTS
Common allergens (from cats, dogs, house-dust mites, and cockroaches) were detectable in airborne dust from 4 of 10 Amish and 1 of 10 Hutterite homes (Table S3 in the Supplementary Appendix). In contrast, endotoxin levels were measurable in airborne dust from all 20 homes, and median levels were strikingly higher (6.8 times as high) in Amish homes than in Hutterite homes (4399 endotoxin units [EU] per square meter vs. 648 EU per square meter, P <0.001) (Fig. 1B). Analysis of a single pooled sample of mattress dust from each population revealed different profiles of the relative abundance of bacteria at the family level (Fig. S4 in the Supplementary Appendix).
COMPOSITION AND PHENOTYPE OF PERIPHERAL-BLOOD LEUKOCYTES
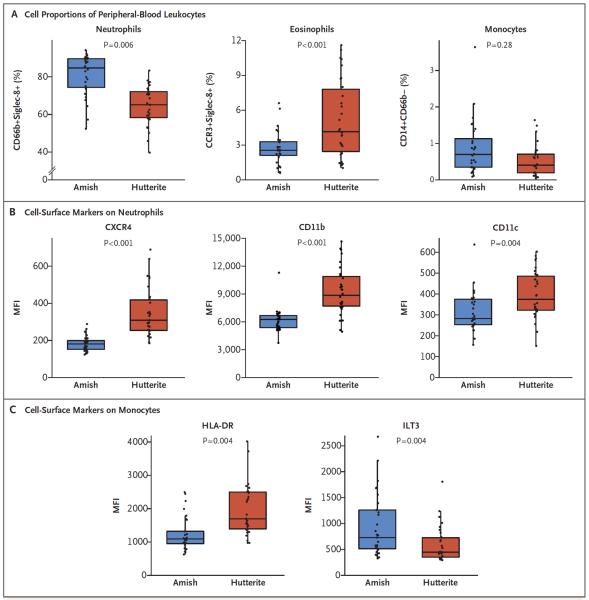
Peripheral-blood leukocytes from Amish children had increased proportions of neutrophils, decreased proportions of eosinophils, and similar proportions of monocytes as compared with samples from Hutterite children (Fig. 2A). Neutrophils from Amish children expressed lower levels of the chemokine receptor CXCR4 and the adhesion molecules CD11b and CD11c than did neutrophils from Hutterite children, suggesting that these cells may have recently emigrated from the bone marrow (Fig. 2B). Although proportions of monocytes were similar in Amish and Hutterite children, monocytes from Amish children, unlike those from Hutterite children, exhibited a suppressive phenotype characterized by lower levels of human leukocyte antigen DR (HLA-DR) and higher levels of the inhibitory molecule immunoglobulin-like transcript 3 (ILT3)16,17 (Fig. 2C). In contrast with previous studies,18,19 no significant differences in percentages of T regulatory cells (defined as CD3+, CD4+, FoxP3+, and CD127−) was observed in Amish and Hutterite children (0.056±0.054% vs. 0.079±0.081% of peripheral-blood leukocytes, P = 0.29).
Figure 2. Proportions of Peripheral-Blood Leukocytes and Cell-Surface–Marker Phenotypes in Amish and Hutterite Children.
The percentages of total peripheral-blood leukocytes (Panel A) were determined with flow cytometry for neutrophils (defined as CD66b+Siglec-8+), eosinophils (defined as CCR3+Siglec-8+), and monocytes (defined as CD14+CD66b−). Box-and-whisker plots show a line indicating median value, with the box showing the interquartile range and whiskers showing the 95% confidence interval. Neutrophils (Panel B) were characterized according to the surface expression of CXCR4, CD11b, and CD11c (shown here), along with CXCR1 and CXCR2, expressed as mean fluorescence intensity (MFI). The expression of the interleukin-8 coreceptors CXCR1 and CXCR2 was not significantly different between groups (P = 0.26 and P = 0.91, respectively). Monocytes (Panel C) were characterized for the surface expression of HLA-DR and immunoglobulin-like transcripts (ILTs), including ILT3 (shown here). There was no significant difference in the MFI of inhibitory receptors ILT2 and ILT4 between Amish and Hutterite children (P = 0.69 and P = 0.21, respectively; data not shown), whereas the surface expression of ILT5 was increased on Amish monocytes (P = 0.001; data not shown). All P values were calculated with the use of an unpaired Student's t-test. Cell proportions and phenotypes after the exclusion of children with asthma or allergic sensitization are shown in Table S4 in the Supplementary Appendix.
CYTOKINE RESPONSES TO INNATE AND ADAPTIVE STIMULATION
Cytokine levels were measured in supernatants from peripheral-blood leukocytes that were cultured for 30 hours, with or without innate stimuli (lipopolysaccharide) or adaptive stimuli (combined anti-CD3 and anti-CD28 antibodies). Twenty-three cytokines were detectable in the supernatants from peripheral-blood leukocytes treated with lipopolysaccharide. Median levels of each of these 23 cytokines were lower in the Amish children than in the Hutterite children, and these distributions were significantly different (P <0.001 by Wilcoxon signed-rank test) (see Tables S5 and S6 in the Supplementary Appendix). Results were similar after the exclusion of children who were known to have asthma or allergies (Table S7 in the Supplementary Appendix). In contrast, after adaptive stimulation, the overall distributions of median cytokine levels were not significantly different in peripheral-blood leukocytes from Amish and Hutterite children (P = 0.08 by Wilcoxon signed-rank test) (Table S8 in the Supplementary Appendix).
GENE-EXPRESSION PROFILES
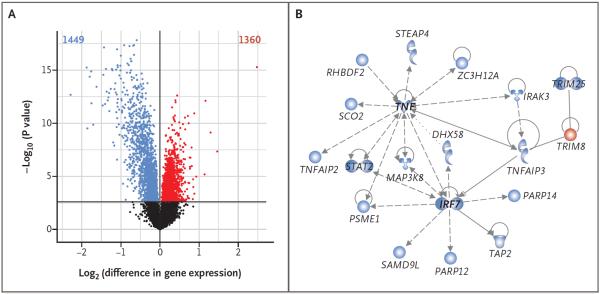
The striking differences in the proportions of peripheral-blood leukocytes observed in Amish and Hutterite children were reflected in the gene-expression profiles of these cells (Fig. S5 in the Supplementary Appendix). At a false discovery rate of 1%, 1449 genes were up-regulated in the peripheral-blood leukocytes of Amish children (blue points in Fig. 3A) as compared with 1360 genes up-regulated in the cells of Hutterite children (red points in Fig. 3A). These differentially expressed genes were organized into 15 coexpression modules with the use of the Whole Genome Co-Expression Network Analysis (Table S9 in the Supplementary Appendix). To better understand the biologic relationships within each set of genes in each module, we used Ingenuity Pathway Analysis (Qiagen) to construct unsupervised networks on the basis of prior knowledge of the physical and functional connections between the molecules encoded by the genes. The most significant network (P = 1.0×10−30 by Fisher's exact test) was in a module that contained 43 genes. This module was associated with both Amish and Hutterite status (P = 7.1×10−9) and was therefore also associated with the proportions of neutrophils (P = 1.5×10−6) and eosinophils (P = 1.0×10−3). Eighteen of the genes in this module were over-expressed in Amish peripheral-blood leukocytes, and all were clustered in a network that had as hubs tumor necrosis factor (TNF) and interferon regulatory factor 7 (IRF7), two key proteins in the innate immune response to microbial stimuli (Fig. 3B).
Figure 3. Gene-Expression Profiles in Peripheral-Blood Leukocytes from Amish and Hutterite Children.
In Panel A, a volcano plot shows differences in baseline gene expression in peripheral-blood leukocytes from Amish and Hutterite children. The x axis indicates the log2 differences in gene-expression level between groups, with larger positive values representing genes with higher expression in the Hutterites relative to the Amish (1360 genes, shown in red points) and larger negative values representing genes with higher expression in the Amish relative to the Hutterites (1449 genes, shown in blue points). The y axis shows the −log10 of the P values for each gene, with larger values indicating greater statistical significance. The solid horizontal line indicates the 1% false discovery rate. Black points represent genes from Amish and Hutterite cells for which there was no significant difference in gene expression. Differences in gene expression remain after the data for children with asthma or allergic sensitization were excluded (Figs. S6 and S7 in the Supplementary Appendix). Changes in gene expression between the two groups after correcting for differences in cell proportion are shown in Figure S4 in the Supplementary Appendix. In Panel B, a network of differentially expressed genes in untreated peripheral-blood leukocytes is shown. Genes shown in blue have increased expression in Amish children, and the gene shown in red has increased expression in Hutterite children. The gene shapes indicate the class of each gene's protein product (spirals denote enzymes, a v-shape denotes cytokines, conjoined circles denote a transcription regulator, hollow upside-down triangles denote kinases, cups denote transporters, and circles denote other products). Lines represent different biologic relationships (solid lines indicate direct interaction, dashed lines indirect interaction, arrows direction of activation, arrows with a horizontal line direction of activation and inhibition, and lines without arrows binding only).
EFFECTS OF HOUSE-DUST EXTRACTS ON EXPERIMENTAL ASTHMA
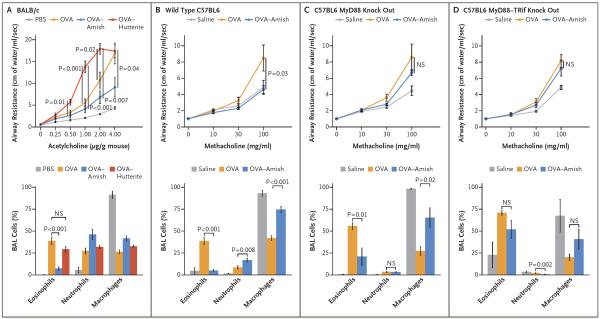
To create a framework that would help us to interpret our observations, which were pointing toward a protective role of innate immunity, we used a classic ovalbumin mouse model of allergic asthma, comparing the effects of house dust obtained from Amish and Hutterite homes by administering extracts intranasally to mice over the course of 4 to 5 weeks. Eosinophilia was observed in bronchoalveolar-lavage samples, and airway hyperresponsiveness was exacerbated in mice treated with ovalbumin and Hutterite dust extracts as compared with mice treated with ovalbumin alone, findings that were consistent with the absence of protection from asthma observed in Hutterite children (Fig. 4A). In contrast, inhalation of Amish dust extracts was sufficient to significantly inhibit ovalbumin-induced airway hyperresponsiveness, eosinophilia in the bronchoalveolar lavage, and the elevation of serum ovalbumin-specific IgE levels (Fig. 4A, and Table S10 in the Supplementary Appendix). Levels of lung T regulatory cells (defined as CD3+, CD4+, and FoxP3+) were not increased (Table S11 in the Supplementary Appendix), and all cytokines measured in bronchoalveolar-lavage samples, including interleukin-10, were suppressed in mice that received Amish dust extracts (Table S12 in the Supplementary Appendix). The inhibitory effects of these extracts in wild-type mice probably required innate immunity, because protection was strongly reduced in mice deficient in MyD88 (Fig. 4C) and completely abrogated in mice deficient in both MyD88 and Trif (Fig. 4D), two molecules that are critical to the development of multiple innate immune-signaling pathways.
Figure 4. Effects of Amish and Hutterite House-Dust Extracts on Airway Responses in Mouse Models of Allergic Asthma.
Panel A shows the effects of the intranasal instillation of 50 μl of Amish or Hutterite dust extract in 7-week-old mice (BALB/c strain) every 2 to 3 days for a total of 14 times beginning at day 0. The mice were sensitized with ovalbumin (OVA) intraperitoneally on days 0 and 14 and challenged with ovalbumin intranasally on days 28 and 38. Airway resistance (shown as centimeters of water per milliliter per second and stimulated in response to increasing doses of acetylcholine administered intravenously) and bronchoalveolar-lavage (BAL) cellularity were measured on day 39 (4 to 6 mice per group). The total amount of Amish and Hutterite dust extract administered over the course of the experiment represented the total load of airborne dust deposited on electrostatic dust collectors placed in Amish or Hutterite homes for 1 month. Statistical differences in experimental measures were assessed with the use of Student's t-test. Amish house-dust extracts (7.5 mg of dust equivalent in 50 μl) were instilled intranasally every 2 to 3 days for a total of 14 times beginning 5 days before day 0 into 7-week old wild-type mice (Panel B), mice deficient in MyD88 (Panel C), and mice deficient in MyD88 and Trif (Panel D) (all C57BL6 strains). These mice were sensitized intraperitoneally with 20 μg of ovalbumin on days 0 and 14 and were challenged intranasally with 75 μg of ovalbumin on days 26, 27, and 28. Airway resistance (shown as an increase from baseline in response to increasing doses of nebulized methacholine) and bronchoalveolar-lavage cellularity were measured on day 30 (12 mice per group for wild-type mice and 6 mice per group for those deficient in MyD88 or MyD88 and Trif). Statistical differences in experimental measures were assessed with the use of Student's t-test. I bars represent the standard errors of the data. NS denotes not significant and PBS phosphate-buffered saline.
DISCUSSION
Our studies in Amish and Hutterite schoolchildren revealed marked differences in the prevalence of asthma despite similar genetic ancestries and lifestyles. As compared with the Hutterites, the Amish, who practice traditional farming and are exposed to an environment rich in microbes, showed exceedingly low rates of asthma and distinct immune profiles that suggest profound effects on innate immunity. Data generated in an experimental model of asthma support this notion by showing that the protective effect of the Amish environment requires the activation of innate immune signaling.
Analyses of the proportions and gene-expression profiles of peripheral-blood immune cells in Amish and Hutterite children revealed differences in the cells and genes involved in innate immune responses to microbes. Indeed, neutrophils, eosinophils, and monocytes appeared to be major targets of the distinct environments to which Amish and Hutterite children are exposed because these cell types differed between the two groups in terms of their relative abundance, their phenotypes, or both. Moreover, the network most associated with these differences consisted of innate immune genes. Notable among the genes that were more highly expressed in the Amish children was TNFAIP3, which encodes A20, a ubiquitin-editing enzyme that limits the activity of multiple inflammatory pathways that depend on nuclear factor κB (NF-κB)20 and that has also been shown to mediate the protective effects of European farm-dust extracts in murine models of allergic asthma.21 IRF7, a hub in this network, regulates type I interferon transcription and is therefore essential for innate airway responses against viruses22 that are linked to susceptibility to asthma.23,24 In turn, TRIM8, the one gene in the network that was more highly expressed in the Hutterites, acts as a positive regulator of TNF-α–and interleukin-1β–induced activation of NF-κB.25 These findings suggest that in the Amish, intense and presumably sustained exposure to microbes activates innate pathways that shape and calibrate downstream immune responses.
Sustained microbial exposure was also reflected in the phenotypes of peripheral innate immune cells in the Amish. Repeated microbial stimulation can lead to reduced expression of HLA-DR on monocytes26,27 and drive immature neutrophils from the bone marrow.28–31 Indeed, Amish children had immature neutrophils bearing markers suggestive of recent emigration from the bone marrow, and they had monocytes with reduced expression of HLA-DR and increased expression of ILT3, all of which are suggestive of antiinflammatory function. Proportions of T regulatory cells and levels of interleukin-10, which typically mediate immune-balancing effects, were not increased in the Amish children. However, qualitative and functional differences in regulatory-cell populations remain to be defined.
Innate immunity has evolved to sense the environment and transduce signals that calibrate adaptive responses to exogenous antigens. The proteins MyD88 and Trif are located at the convergence of multiple innate signaling pathways,32 and deletion of these molecules virtually disables innate immune responses, thereby also dysregulating adaptive immunity. The fact that the loss of protection was more marked in mice deficient in both MyD88 and Trif than in mice deficient only in MyD88 points to the involvement of multiple innate pathways. The concordance between findings from studies in humans and in mice was remarkable: in both studies protection was accompanied by lower levels of eosinophils, higher levels of neutrophils, generally suppressed cytokine responses, and no increase in levels of T regulatory cells or interleukin-10. Thus, the finding that these features were largely dependent on innate immune pathways in mice suggests that innate immune signaling may also be the primary target of protection in the Amish children, in whom downstream adaptive immune responses may also be modulated.
Our study has several limitations. First, we were unable to include children younger than 6 years of age, we collected samples at a single time point, and the numbers of Amish and Hutterite children in our study were relatively small. As a result, we may have missed important windows of immune development or lacked the ability to detect early, subtle shifts in cell composition, response, or phenotype that are critical for immune maturation. Second, our microbiome assessments were limited, since only pooled dust samples from a limited number of homes were available for the studies in which we assessed bacterial composition. Therefore, we cannot further dissect microbial composition and identify potentially protective microbes to target. However, the striking differences found in endotoxin levels support the notion that the Amish indoor environment is much richer in microbial exposures than the Hutterite environment. Third, the strategy used for sampling the Hutterite children enriched selection for those with asthma, although the prevalence of asthma in our sample was similar to that reported in previous population-based studies.11 Moreover, the exclusion of children with asthma or allergic sensitization from our analyses of gene expression, cell composition, and immune phenotypes did not affect the outcomes.
Our study in a small number of children was sufficient to show significant differences in the prevalence of asthma and in immune profiles, suggesting that very strong environmental factors must account for these differences. Indeed, we showed that there are remarkable genetic similarities between Amish and Hutterite children. Although we interrogated only common variants, other variants that occur at very low frequency in these populations are unlikely to account for the observed large differences in the prevalence of asthma. In the end, the novelty of our work lies in the identification of innate immunity as the primary target of the protective Amish environment, a finding supported by results obtained in both humans and mice. Conversely, our work suggests that susceptibility to asthma may be increased when innate immune stimulation is weak. A deeper understanding of the relevant stimuli and the innate immune pathways they engage may ultimately pave the way for the development of effective strategies for the prevention of asthma.
Supplementary Material
Acknowledgments
Supported by the National Institutes of Health, St. Vincent Foundation, and the American Academy of Allergy, Asthma, and Immunology Foundation.
We thank the Hutterite and Amish volunteers and their families for participating in this study; Gorka Alkorta-Aranburu, Maitane Arrubarrena Orbegozo, Kathleen Bailey, Christine Billstrand, Kelly Blaine, Daniel Cook, Donna Decker, Mohammad Jaffery, Courtney Burrows, Katherine Naughton, Raluca Nicolae, Rob Stanaker, Meghan Sullivan, and Emma Thompson for assistance on field trips and with sample processing; Peace Ezeh, Amanda Herrell, Ashley Horner, Kenneth Addison, Dominik Schenten, and Shane Snyder for their contributions to the studies in mice; and Minal Çalışkan, Yoav Gilad, Jessie Nicodemus-Johnson, John Novembre, and Matthew Stephens for helpful comments and statistical advice.
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
REFERENCES
- 1.Ober C, Yao TC. The genetics of asthma and allergic disease: a 21st century perspective. Immunol Rev. 2011;242:10–30. doi: 10.1111/j.1600-065X.2011.01029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyers DA, Bleecker ER, Holloway JW, Holgate ST. Asthma genetics and personalised medicine. Lancet Respir Med. 2014;2:405–15. doi: 10.1016/S2213-2600(14)70012-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bach JF. The effect of infections on susceptibility to autoimmune and allergic diseases. N Engl J Med. 2002;347:911–20. doi: 10.1056/NEJMra020100. [DOI] [PubMed] [Google Scholar]
- 4.Braun-Fahrländer C, Riedler J, Herz U, et al. Environmental exposure to endotoxin and its relation to asthma in school-age children. N Engl J Med. 2002;347:869–77. doi: 10.1056/NEJMoa020057. [DOI] [PubMed] [Google Scholar]
- 5.Ege MJ, Mayer M, Normand AC, et al. Exposure to environmental microorganisms and childhood asthma. N Engl J Med. 2011;364:701–9. doi: 10.1056/NEJMoa1007302. [DOI] [PubMed] [Google Scholar]
- 6.Ege MJ, Frei R, Bieli C, et al. Not all farming environments protect against the development of asthma and wheeze in children. J Allergy Clin Immunol. 2007;119:1140–7. doi: 10.1016/j.jaci.2007.01.037. [DOI] [PubMed] [Google Scholar]
- 7.Ege MJ, Strachan DP, Cookson WO, et al. Gene-environment interaction for childhood asthma and exposure to farming in Central Europe. J Allergy Clin Immunol. 2011;127:138–44. doi: 10.1016/j.jaci.2010.09.041. [DOI] [PubMed] [Google Scholar]
- 8.Hostetler JA. Hutterite society. Johns Hopkins University Press; Baltimore: 1974. [Google Scholar]
- 9.Hostetler JA. Amish society. 4th ed Johns Hopkins University Press; Baltimore: 1993. [Google Scholar]
- 10.Holbreich M, Genuneit J, Weber J, Braun-Fahrländer C, Waser M, von Mutius E. Amish children living in northern Indiana have a very low prevalence of allergic sensitization. J Allergy Clin Immunol. 2012;129:1671–3. doi: 10.1016/j.jaci.2012.03.016. [DOI] [PubMed] [Google Scholar]
- 11.Motika CA, Papachristou C, Abney M, Lester LA, Ober C. Rising prevalence of asthma is sex-specific in a US farming population. J Allergy Clin Immunol. 2011;128:774–9. doi: 10.1016/j.jaci.2011.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adachi O, Kawai T, Takeda K, et al. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity. 1998;9:143–50. doi: 10.1016/s1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
- 13.Yamamoto M, Sato S, Hemmi H, et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301:640–3. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 14.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 15.Cann HM, de Toma C, Cazes L, et al. A human genome diversity cell line panel. Science. 2002;296:261–2. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- 16.Kim-Schulze S, Scotto L, Vlad G, et al. Recombinant Ig-like transcript 3-Fc modulates T cell responses via induction of Th anergy and differentiation of CD8+ T suppressor cells. J Immunol. 2006;176:2790–8. doi: 10.4049/jimmunol.176.5.2790. [DOI] [PubMed] [Google Scholar]
- 17.Velten FW, Duperrier K, Bohlender J, Metharom P, Goerdt S. A gene signature of inhibitory MHC receptors identifies a BDCA3(+) subset of IL-10-induced dendritic cells with reduced allostimulatory capacity in vitro. Eur J Immunol. 2004;34:2800–11. doi: 10.1002/eji.200324732. [DOI] [PubMed] [Google Scholar]
- 18.Lluis A, Depner M, Gaugler B, et al. Increased regulatory T-cell numbers are associated with farm milk exposure and lower atopic sensitization and asthma in childhood. J Allergy Clin Immunol. 2014;133:551–9. doi: 10.1016/j.jaci.2013.06.034. [DOI] [PubMed] [Google Scholar]
- 19.Schaub B, Liu J, Höppler S, et al. Maternal farm exposure modulates neonatal immune mechanisms through regulatory T cells. J Allergy Clin Immunol. 2009;123(4):774–82.e5. doi: 10.1016/j.jaci.2009.01.056. [DOI] [PubMed] [Google Scholar]
- 20.Ma A, Malynn BA. A20: linking a complex regulator of ubiquitylation to immunity and human disease. Nat Rev Immunol. 2012;12:774–85. doi: 10.1038/nri3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schuijs MJ, Willart MA, Vergote K, et al. Farm dust and endotoxin protect against allergy through A20 induction in lung epithelial cells. Science. 2015;349:1106–10. doi: 10.1126/science.aac6623. [DOI] [PubMed] [Google Scholar]
- 22.Bosco A, Ehteshami S, Panyala S, Martinez FD. Interferon regulatory factor 7 is a major hub connecting interferon-mediated responses in virus-induced asthma exacerbations in vivo. J Allergy Clin Immunol. 2012;129:88–94. doi: 10.1016/j.jaci.2011.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loss GJ, Depner M, Hose AJ, et al. The early development of wheeze: environmental determinants and genetic susceptibility at 17q21. Am J Respir Crit Care Med. 2016;193:889–97. doi: 10.1164/rccm.201507-1493OC. [DOI] [PubMed] [Google Scholar]
- 24.Calışkan M, Bochkov YA, Kreiner-Møller E, et al. Rhinovirus wheezing illness and genetic risk of childhood-onset asthma. N Engl J Med. 2013;368:1398–407. doi: 10.1056/NEJMoa1211592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Q, Yan J, Mao AP, et al. Tripartite motif 8 (TRIM8) modulates TNFα- and IL-1β-triggered NF-κB activation by targeting TAK1 for K63-linked polyubiquitination. Proc Natl Acad Sci U S A. 2011;108:19341–6. doi: 10.1073/pnas.1110946108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meisel C, Schefold JC, Pschowski R, et al. Granulocyte-macrophage colony-stimulating factor to reverse sepsis-associated immunosuppression: a double-blind, randomized, placebo-controlled multi-center trial. Am J Respir Crit Care Med. 2009;180:640–8. doi: 10.1164/rccm.200903-0363OC. [DOI] [PubMed] [Google Scholar]
- 27.Gomez HG, Gonzalez SM, Londoño JM, et al. Immunological characterization of compensatory anti-inflammatory response syndrome in patients with severe sepsis: a longitudinal study. Crit Care Med. 2014;42:771–80. doi: 10.1097/CCM.0000000000000100. [DOI] [PubMed] [Google Scholar]
- 28.Fortunati E, Kazemier KM, Grutters JC, Koenderman L, Van den Bosch J. Human neutrophils switch to an activated phenotype after homing to the lung irrespective of inflammatory disease. Clin Exp Immunol. 2009;155:559–66. doi: 10.1111/j.1365-2249.2008.03791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orden S, De Pablo C, Rios-Navarro C, et al. Efavirenz induces interactions between leucocytes and endothelium through the activation of Mac-1 and gp150,95. J Antimicrob Chemother. 2014;69:995–1004. doi: 10.1093/jac/dkt468. [DOI] [PubMed] [Google Scholar]
- 30.Devi S, Wang Y, Chew WK, et al. Neutrophil mobilization via plerixafor-mediated CXCR4 inhibition arises from lung demargination and blockade of neutrophil homing to the bone marrow. J Exp Med. 2013;210:2321–36. doi: 10.1084/jem.20130056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamada M, Kubo H, Kobayashi S, et al. The increase in surface CXCR4 expression on lung extravascular neutrophils and its effects on neutrophils during endotoxin-induced lung injury. Cell Mol Immunol. 2011;8:305–14. doi: 10.1038/cmi.2011.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Netea MG, Wijmenga C, O'Neill LAJ. Genetic variation in toll-like receptors and disease susceptibility. Nat Immunol. 2012;13:535–42. doi: 10.1038/ni.2284. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.