Synopsis
The molecular characterization of lung cancer has considerably changed the classification and treatment of these tumors, becoming an essential component of pathologic diagnosis and oncologic therapy decisions. Through the recognition of novel biomarkers, such as epidermal growth factor receptor mutations and anaplastic lymphoma kinase translocations, it has been possible to identify subsets of patients who benefit from targeted molecular therapies. The success of targeted anticancer therapies and new immunotherapy approaches has created a new paradigm of personalized therapy and has also led to accelerated development of new drugs for lung cancer treatment. This review focuses on clinically relevant cancer biomarkers as targets for therapy, as well as potential new targets for drug development.
Keywords: lung cancer, genotyping, biomarkers, molecular targets
1. Introduction
Lung cancer has shown a decrease in incidence and mortality in recent decades; however, it remains one of the cancers with the highest incidence and ranks first in cancer-related deaths in the United States (1). An estimated 221,200 new cases and 158,040 deaths are expected to occur in 2015, representing approximately 13% of all cancers diagnosed and 27% of all cancer deaths (2). Despite advances in early detection and standard treatment, most patients are diagnosed at an advanced stage and have a poor prognosis, with an overall 5-year survival rate of 10% to 15% (3). Lung cancer is a heterogeneous disease comprising several subtypes with pathologic and clinical relevance (4). The recognition of histologic subtypes of non-small cell lung carcinoma (NSCLC), namely adenocarcinoma, squamous cell carcinoma, and large cell lung carcinoma as the most frequent subtypes, has become important as a determinant of therapy in this disease (5). In addition, in recent years, the identification of molecular abnormalities in a large proportion of patients with lung cancer has allowed the emergence of personalized targeted therapies and has opened new horizons and created new expectations for these patients (6). The use of predictive biomarkers to identify tumors that could respond to targeted therapies has meant a change in the paradigm of lung cancer diagnosis (5).
This paradigm change affects all stakeholders in the fight against lung cancer including pathologists. Currently, several multiplex genotyping platforms for the detection of oncogene mutations, gene amplifications, and rearrangement are moving to the clinical setting. Genome-wide molecular investigations using next-generation sequencing (NGS) technologies have been evaluated in the research setting, with promising results. Further investigations in NSCLC are required for a better understanding of the implications of intratumor heterogeneity and the roles of tumor suppressor genes and epigenetic events with no known driver mutations. NGS in the clinical setting will provide comprehensive information cheaper and faster by using small amounts of tissue. Pathologists should be able to precisely handle tissue adequacy in terms of quantity and quality and maintaining tumor cells for detection of molecular alterations. The recent clinical successes of immunotherapy approaches to lung cancer have posed additional challenges to the scientific community and pathologists to develop predictive biomarkers of response to these therapies and have highlighted the need for proper procurement and processing of tissue specimens from patients with lung cancer.
In this review, we will focus on the major predictive biomarkers in NSCLC, with special emphasis on their clinical and molecular importance, as well as the current status of molecular testing for these biomarkers.
2. Histologic Subtyping of NSCLC
The advent of molecular profiling and targeted therapy has renewed interest in the classification of NSCLC into major subtypes such as adenocarcinoma, squamous cell carcinoma, and large cell lung carcinoma (7). Other subtypes, including sarcomatoid carcinoma, neuroendocrine large cell carcinoma, and others, represent a very minor proportion of the total NSCLC cases (7). The most recent histologic classification of lung cancer published by the World Health Organization in 2015 incorporates relevant genetics and immunohistochemistry (IHC) aspects of different tumor subtypes (Figure 1) (7). Lung cancers are increasingly diagnosed and staged by transthoracic core needle biopsy and fine-needle aspiration (FNA), transbronchial needle aspiration, endobronchial ultrasound-guided transbronchial needle aspiration, and endoscopic ultrasound-guided FNA. It is well established that poorly differentiated adenocarcinoma and squamous cell carcinoma of the lung can appear indistinguishable by routine microscopy, particularly in small biopsy and cytology specimens. In these small specimens, particularly in poorly differentiated tumors, we need to integrate morphology with IHC analysis to make a precise diagnosis. This includes the examination of IHC expression of thyroid transcription factor (TTF1) and the novel aspartic proteinase of the pepsin family A (napsin A) for adenocarcinoma and p40 and cytokeratin 5/6 for squamous cell carcinoma (8). In addition, histochemical staining of mucin is useful for the diagnosis of adenocarcinoma histology. The correct histologic diagnosis of these specimens is important, but it is also imperative to exercise judicial use of the tissue to maximize the yield for molecular testing (Table 1).
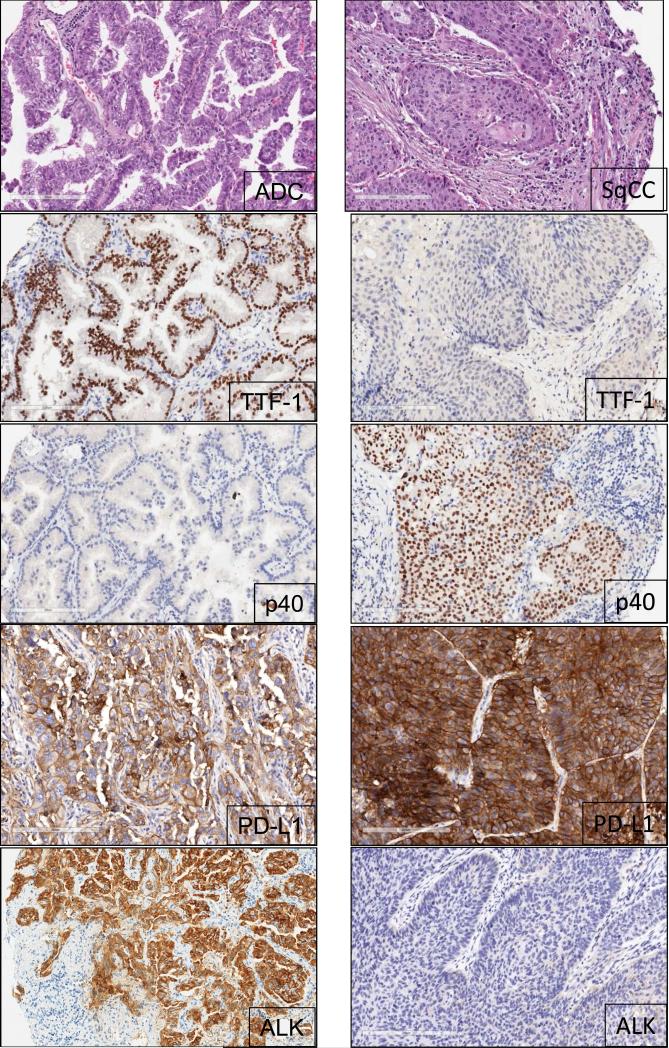
Figure 1.
Representative examples of histology of lung adenocarcinoma (ADC) and squamous cell carcinoma (SqCC) histology and biomarker analysis using IHC. ADC tumor tissue shows positive expression for TTF-1 (nuclear), PD-L1 (membrane and cytoplasm), and ALK (cytoplasm); p40 shows negative expression. SqCC shows positive expression for p40 (nuclear) and PD-L1 (membrane and cytoplasm); p40 and ALK expression are negative (20x magnification).
Table 1.
Frequency of the main molecular alterations in lung adenocarcinoma and squamous cell carcinoma
| Gene | Alteration | Adenocarcinoma | Squamous Cell Carcinoma |
|---|---|---|---|
| Frequency | |||
| EGFR | Mutation | 10% | 3% |
| ALK | Rearrangement | 4-7% | None |
| ROS | Rearrangement | 1-2% | None |
| KRAS | Mutation | 25-35% | 5% |
| MET | Mutation | 8% | 3% |
| MET | Amplification | 4% | 1% |
| NTRK1 | Rearrangement | 3% | None |
| FGFR | Amplification | 3% | 20% |
| HER2 | Mutation | 1.6-4% | None |
| BRAF | Mutation | 1-3% | 0.3% |
| PIK3CA | Mutation | 2% | 7% |
| RET | Rearrangement | 1-2% | None |
| DDR2 | Mutation | 0.5% | 3-4% |
| PTEN | Deletion | - | 16% |
3. Genomic Biomarkers in NSCLC
Advances in elucidating the molecular biology of lung cancer have led to the identification of a number of potential biomarkers that could be relevant in the clinical management of patients with NSCLC.
3.1. Epidermal growth factor receptor (EGFR)
The epidermal growth factor receptor (EGFR) is a tyrosine kinase receptor member of the ERBB family. The EGFR gene is located on the short arm of chromosome 7 at position 12 (9). When the extracellular ligand binds to EGFR, it generates homo- or heterodimerization of the receptor, leading to phosphorylation of sites in the cytoplasmic tyrosine kinase and activation of various intracellular pathways, including the phosphatidylinositol 3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) and RAS/RAF/mitogen-activated protein kinase (MAPK) pathways, which lead to cell proliferation, metastasis, and prevention of apoptosis (10). EGFR is overexpressed in 62% of NSCLCs, and its expression has been associated with poor prognosis (11). Approximately 10% of patients with adenocarcinoma of the lung in the United States and 30% to 50% in East Asia have lung tumors associated with EGFR mutations (11). These mutations occur within exons 18-21, which encode for a portion of the EGFR kinase domain (10). Approximately 90% of EGFR mutations occur as in-frame deletions in exon 19 or as missense mutations in exon 21 (44% and 41% of all mutations, respectively) (4). Activating mutations in the kinase domain of EGFR trigger ligand-independent tyrosine kinase activation, leading to hyperactivation of downstream antiapoptotic signaling pathways (10). EGFR mutations are found more often in adenocarcinomas with lepidic features from female never smokers (10). The high response rates (55%-78%) to treatment with tyrosine kinase inhibitors (TKIs), such as gefitinib, erlotinib and afatinib, in patients with EGFR-mutant tumors, and the significantly greater progression-free survival (PFS) of these patients, have made EGFR TKIs the standard treatment for patients with these mutations (6). However, most of these patients develop resistance and relapse in a short time, owing to the occurrence of a new mutation (T790M) in exon 20 of the EGFR kinase domain (50%), amplification of the MET oncogene (21%), or mutations of PI3KCA (4).
EGFR mutations are identified mostly with the use of gene sequencing methodologies and real-time polymerase chain reaction (PCR)-based assays. Both methods have been reported to have high performance and sensitivity in the detection of these mutations in formalin-fixed and paraffin-embedded tissues (4). Detection of EGFR mutations by an IHC-based approach with specific antibodies against mutant proteins has been attempted but showed variable sensitivity and significant variability between studies (12).
3.2. Anaplastic lymphoma kinase (ALK)
Anaplastic lymphoma kinase (ALK) is a tyrosine kinase receptor member of the insulin receptor superfamily. The ALK gene is located on the short arm of chromosome 2 at position 23 (13). ALK gene rearrangement was originally identified in anaplastic large cell lymphoma (14) and was subsequently described in a subset of NSCLC tumors harboring a fusion of ALK and echinoderm microtubule-associated protein-like 4 (EML4) genes (4). This rearrangement encodes for a chimeric protein with constitutive kinase activity, which promotes malignant growth and proliferation (14). The EML4-ALK fusion has been detected in 3.7% to 7% of NSCLCs (10, 14), usually in adenocarcinomas with signet-ring cells or cribriform histology features, and is more common in young patients who have never smoked (14). There are several EML4-ALK rearrangement variants and also ALK fusion with other less frequent partners, such as kinesin family member 5B (KIF5B), TRK-fused gene (TFG), kinesin light chain 1 (KLC1), and huntingtin-interacting protein 1 (HIP1) genes, resulting in oncogenic transformation (13, 15). It has been shown that EGFR, Kirsten rat sarcoma viral oncogene homolog gene (KRAS), and ALK molecular alterations are mutually exclusive events (4); nevertheless, they have been described in up to 2.7% of lung adenocarcinoma cases with concurrent molecular alterations (16). The ALK fusion defines a distinct subpopulation of patients with lung adenocarcinoma who are highly responsive (57%-74%) to ALK inhibitors such as crizotinib. Patients treated with crizotinib demonstrated significantly better median PFS and response rate compared to patients who received chemotherapy (17). As a result, testing for ALK rearrangements in patients with advanced lung adenocarcinoma is recommended in current clinical practice guidelines (5, 10). However, despite initial responses, a fraction of the patients develop acquired resistance to crizotinib, owing to secondary mutations within the kinase domain of EML4-ALK; these include L1196M, C1156Y, and F1174L, among others (18, 19). Several second-generation ALK inhibitors that target ALK-positive NSCLC, such as alectinib, ceritinib, and AP26133, have been developed and are currently under evaluation in clinical trials (20).
Current diagnostic approaches to detect ALK fusion genes and their results include break-apart fluorescence in situ hybridization (FISH), IHC, and reverse-transcription PCR (RT-PCR) (21). Break-apart FISH has been established in clinical trials as the standard method for confirmation of ALK status (22). FISH and IHC have shown high concordance in several reports, especially with the development of IHC antibodies (clones 5A4 and D5F3) with better sensitivities and specificities (83%-100%) for the detection of ALK rearrangement (23, 24). As a result, IHC detection of the ALK protein is being considered as an adequate screening tool to test NSCLC samples for ALK rearrangements or as a tool to evaluate cases that are not interpretable by FISH (25, 26). Nevertheless, other studies reveal major discordances, suggesting the need to combine both tests to optimize detection (22). Other methods, such as RT-PCR and NGS, are in use but have not been examined systematically compared to FISH as a predictor of response to ALK inhibitors (27, 28).
3.3. Kirsten rat sarcoma viral oncogene homolog (KRAS)
KRAS is an oncogene located on the long arm of chromosome 12 at position 12.1 (29). It is a member of the RAS family of membrane-associated G proteins and encodes for a protein with intrinsic GTPase activity, which is involved in a variety of cellular responses including proliferation, cytoskeletal reorganization, and survival (30). KRAS acts downstream of a number of tyrosine kinases receptors, including EGFR, and is associated with activation of the RAS/RAF/MAP kinase kinase (MEK)/extracellular signal-regulated kinase (ERK) and RAS/MAPK signaling pathways (10). KRAS mutations occur in 25% to 35% of patients with NSCLC, principally adenocarcinomas with a solid pattern (31), and are found more often in white patients compared to Asians, in former or current smokers, but without sex predilection (32). Mutations in the form of single-nucleotide missense variants are found in codons 12 and 13 in approximately 95% of cases (4, 32). In never smokers, the most common KRAS mutations are G12D and G12V, whereas G12C is the most common mutation associated with smoking (31, 32). The presence of KRAS mutation may be associated with unfavorable outcome (33) and could be a negative predictor of responsiveness to chemotherapy (34). In addition, it is associated with an increased likelihood of having a second primary tumor (35) and is a predictor of resistance to targeted therapy with EGFR-TKIs, such as gefitinib or erlotinib, in patients with NSCLC (36).
Because KRAS, EGFR, and ALK molecular alterations are mutually exclusive, it has been suggested that KRAS testing could be a surrogate assay to exclude EGFR- and ALK-positive cases (10); however, this approach is not currently recommended. Although there are no targeted therapies approved for patients with lung cancer and KRAS mutation, several clinical trials aimed at downstream signaling targets are under way. Different phase 2 trials have reported improvements in both PFS and response rate with the combination of selumetinib (MEK1/MEK2 inhibitor) and docetaxel compared to docetaxel alone (37) and promising results with sorafenib (RAS/RAF pathway inhibitor), with a disease control rate of approximately 50% (38). Conversely, trametinib (MEK1/MEK2 inhibitor) did not show advantages over docetaxel in patients with NSCLC (39).
3.4. ROS proto-oncogene 1, receptor tyrosine kinase (ROS1)
ROS proto-oncogene 1, receptor tyrosine kinase (ROS1) is a tyrosine kinase receptor member of the insulin receptor family and is located on the long arm of chromosome 6 at position 22. ROS1 plays a role in epithelial cell differentiation during the development of a variety of organs, but no ligand for this receptor has been identified (40). ROS1 rearrangements were originally described in glioblastoma and have also been reported in cholangiocarcinoma and ovarian cancer (41). Approximately 1% to 2% of NSCLCs harbor ROS1 rearrangements (41), and several fusion partners, including CD74, solute carrier family 34, member 2 (SLC34A2), leucine-rich repeats and immunoglobulin-like domains 3 (LRIG3), ezrin (EZR), syndecan 4 (SDC4), tropomyosin 3 (TPM3), and FIG, have been reported in these tumors. All of these fusions result in a chimeric protein that has been reported to be oncogenic (40, 41). ROS1-rearranged NSCLC typically occurs in young, female, never smokers with a histologic diagnosis of adenocarcinoma (40, 41) and is usually mutually exclusive with other oncogenic drivers (EGFR, KRAS, ALK) (41). Clinical trials have reported that patients with advanced NSCLC harboring ROS1 rearrangement have benefited from crizotinib treatment, showing response rates up to 80% (40, 42). Ongoing phase 1 and 2 studies are investigating the activity of crizotinib and ceritinib (ALK inhibitor) in ROS1-rearranged NSCLC (43, 44). ROS1 testing is indispensable for identifying patients who could benefit from crizotinib treatment. The National Comprehensive Cancer Network 2014 guidelines recommend that all patients with advanced triple-negative (EGFR, ALK, and KRAS) lung adenocarcinoma be tested for other molecular markers including ROS1 (5).
There is not a gold standard method, but currently available diagnostic methods include FISH, RT-PCR, and IHC (45). FISH is the only method approved by the US Food and Drug Administration (FDA) to detect ALK-rearranged NSCLC and has been used in clinical trials as the standard method for confirmation of ROS1 rearrangement; nevertheless, it is an expensive and laborious technique. Because ROS1-rearranged lung cancer is rare, assessment of ROS1 protein expression by IHC may be used as a screening tool for the identification of candidates suitable for ROS1-targeted therapy. In fact, studies have found that ROS1 IHC (D4D6 clone) has a high sensitivity (100%) and specificity (92%-97%) for ROS1 rearrangements compared to FISH (45).
3.5. Human epidermal growth factor receptor 2 (HER2)
The human epidermal growth factor receptor 2 gene HER2 (ERBB2) is a proto-oncogene located on chromosome 17 at position 12 (46). It encodes for a tyrosine kinase receptor member of the ERBB receptor family (47). HER2 lacks a specific ligand. Nevertheless, it can be combined with other ERBB receptors to form a heterodimer (48). This allows for the activation of important signal transduction pathways, including the MAPK and PI3K pathways, involved in cell proliferation, differentiation, and migration (47). HER2 expression and/or amplification is found in many cancers including breast and gastric cancer (48). Overexpression of HER2 has been reported in 7% to 34.9% of NSCLCs and has been associated with poor prognosis in patients with these tumors (47). Activating mutations of HER2 have been found in 1.6% to 4% of lung cancers (47, 49). These mutations occur in the 4 exons of the tyrosine kinase domain (exons 18-21) and are found more often in adenocarcinomas in female, Asian, never or light smokers. HER2 mutations are almost always mutually exclusive with other driver oncogene alterations in lung cancer described above (49). Different studies reinforce the importance of screening lung adenocarcinomas for HER2 mutation as a method to select patients who could benefit from HER2-targeted therapies (afatinib and trastuzumab), which have shown response rates of approximately 50% (50). Several clinical trials of targeted agents, such as trastuzumab, neratinib and pyrotinib, among others, are being conducted in patients with HER2 mutation (47). HER2 mutations are usually assessed via sequencing approaches.
3.6. RET proto-oncogene
The RET proto-oncogene is located on the long arm of chromosome 10 at position 11.2. It encodes for a tyrosine kinase receptor for the glial cell line–derived neurotrophic factor family of ligands and is involved in cell proliferation, migration, and differentiation, as well as neuronal navigation (51). RET chromosomal rearrangements were originally described in papillary thyroid carcinoma (51). Approximately 1% to 2% of NSCLCs harbor RET fusions, and several fusion partners, including kinesin family member 5B (KIF5B) (90%), coiled-coil domain containing 6 (CCDC6), nuclear receptor coactivator 4 (NCOA4), and tripartite motif-containing 33 (TRIM33), have been described (52, 53). RET-rearranged NSCLC typically occurs in adenocarcinomas with more poorly differentiated solid features in young never smokers, and it is mutually exclusive with known driver oncogenes (52, 54). In vitro studies showed that RET fusions lead to oncogenic transformation, which can be inhibited by multitargeted kinase inhibitors such as vandetanib, sorafenib, and sunitinib (54). Preliminary studies with cabozantinib (MET and vascular endothelial growth factor receptor 2 inhibitor) in RET-rearranged lung adenocarcinoma are promising (53).
FISH is currently the standard diagnostic assay for detection of RET chromosomal rearrangements. RT-PCR is usually insufficient for the detection of new partners or isoforms, and RET IHC has shown low sensitivity and specificity for RET rearrangements (52, 54). Sequencing approaches, including NGS methodologies, are also frequently used to detect RET translocations.
3.7. MET proto-oncogene
The MET gene is located on the long arm of chromosome 7 at position 31 (55). This oncogene encodes for a tyrosine kinase receptor (hepatocyte growth factor receptor), which activates multiple signaling pathways that play fundamental roles in cell proliferation, survival, motility, and invasion (4). Pathologic activation of MET includes mutation, gene amplification, and protein overexpression (56). MET alterations were first reported in patients with renal papillary carcinoma and mutations in the MET kinase domain leading to constitutive activation of the receptor (57). In lung cancer, MET mutations are found in the extracellular semaphorin and juxtamembrane domains, occurring in 3% of squamous cell lung cancers and 8% of lung adenocarcinomas (56). MET amplifications are found in 4% of lung adenocarcinomas and 1% of squamous cell lung cancers and are associated with sensitivity to MET inhibitors (56). In NSCLC, MET and hepatocyte growth factor protein expression, along with high MET gene copy number, have been described as poor prognosis factors (58, 59). Activating point mutations affecting splice sites of exon 14 of the MET gene (METex14), which occur in 4% of lung adenocarcinomas, represent a possible oncogenic driver and identify a subset of patients who may benefit from MET inhibitors such as capmatinib and crizotinib (56). This novel alteration is usually assayed by NGS methodology.
3.8. B-RAF proto-oncogene, serine/threonine kinase (BRAF)
The B-RAF proto-oncogene, serine/threonine kinase (BRAF) oncogene is located on the long arm of chromosome 7 at position 34. It encodes for a serine/threonine kinase, which is involved in the RAS/RAF/MEK/ERK signaling pathway (60). When activated by oncogenic mutations, BRAF phosphorylates MEK and promotes cell growth, proliferation, and survival (60). The highest incidence of BRAF mutation is in malignant melanoma (27%-70%), followed by papillary thyroid cancer, colorectal cancer, and serous ovarian cancer (61). BRAF mutations have also been reported in 1% to 3% of NSCLCs (60). In contrast to melanoma, only half of BRAF mutations in NSCLC are V600E mutations. Other non-V600E mutations reported in NSCLC include G469A (~35%) and D594G (~10%). All BRAF mutations are mutually exclusive with other driver alterations such as those of EGFR, KRAS, and ALK (60, 62). BRAF-mutated NSCLC has been reported to be mostly adenocarcinoma, and in contrast to patients with EGFR mutations or ALK rearrangements who are mostly never smokers, patients with BRAF mutations are mostly current or former smokers (62). Nevertheless, patients with NSCLC and BRAF V600E mutations have a worse prognosis and lower response to platinum-based chemotherapy than patients with wild-type BRAF. These patients have benefited from treatment with BRAF and MEK inhibitors (63). BRAF inhibitors, such as vemurafenib and dabrafenib, have high and selective activity against the V600E-mutant BRAF kinase, with overall responses rates from 33% to 42% (63, 64). BRAF and MEK inhibitors targeting BRAF mutation–positive NSCLC, such as trametinib, selumetinib, and dasatinib, among others, are currently under evaluation in clinical trials.
3.9. Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA)
PI3Ks are heterodimeric lipid kinases composed of catalytic and regulatory subunits and are part of several downstream pathways involved in cell growth, transformation, adhesion, apoptosis, survival, and motility (65). The PIK3CA gene is located on the long arm of chromosome 3 at position 26.3. It encodes for the catalytic subunit p110 alpha of P13Ks (66). PKI3CA amplifications, deletions, and somatic missense mutations have been reported in many tumors including lung cancers. In fact, PIK3CA is one of the most commonly mutated oncogenes, along with KRAS, in human cancers (67). Mutations are found in 1% to 4% of patients with NSCLC, usually affecting exons 9 and 20 (80%) (4, 65, 67-69). These mutations are not mutually exclusive with other driver alterations and have been reported more frequently in lung squamous cell carcinoma compared to adenocarcinoma (6.5% vs 1.5%) (69). However, PIK3CA mutations have not shown association with any clinicopathologic features (65, 68, 69). Squamous cell carcinomas with PIK3CA gains are not accompanied by other genetic alterations, suggesting that this gene may play a important role in the pathogenesis of squamous cell cancers (65). Studies have shown that PIK3CA mutations in EGFR-mutated lung cancer confer resistance to EGFRTKIs and are a negative prognostic predictor in patients with NSCLC treated with EGFR-TKIs (70). PI3KCA alterations and their downstream effectors, such as phosphatase and tensin homolog (PTEN), mTOR, and AKT, are potential therapeutic targets for NSCLC therapy and are being evaluated in clinical trials for lung cancer (71). Alterations in PI3KCA are detected using sequencing approaches, mostly NGS assays.
3.10. Neurotrophic receptor tyrosine kinase 1(NTRK1)
The neurotrophic receptor tyrosine kinase 1 (NTRK1) proto-oncogene is located on chromosome 1q21-22 and encodes for a receptor tyrosine kinase, also known as tropomyosin-related kinase (TRK) A, belonging to the TRK superfamily of receptor tyrosine kinases (117). NTRK1 is involved in the regulation of cell growth and differentiation via activation of several signal transduction pathways including MAPK, PI3K, and phospholipase C-gamma (72). NTRK1 rearrangements have been found in colon cancer, thyroid cancer, and glioblastoma multiforme (72). In lung cancer, approximately 3% of adenocarcinomas harbor NTRK1 fusions, and some fusion partners, including myosin phosphatase RHO-interacting protein (MPRIP)-NTRK1 and CD74-NTRK1, have been reported (73). All of these fusions result in constitutive TRKA kinase activity, which has been reported to be oncogenic (73). In early phase 1 studies, NTRK inhibitors, such as entrectinib and LOXO-101, have shown promising results in patients with solid tumors harboring NTRK fusions (74).
3.11. Fibroblast growth factor receptor (FGFR)
The fibroblast growth factor receptor (FGFR) gene is located on chromosome 8 at position 12 and encodes for a tyrosine kinase receptor belonging to the FGFR family. The FGFR family includes 4 receptor tyrosine kinases (FGFRs 1-4). When ligand-receptor binding occurs, FGFR dimerizes and phosphorylates FGFR substrate 2-alpha (FRS2α), leading to activation of different pathways, including the RAS/MAPK and PI3K/AKT/mTOR pathways, promoting cell survival, motility, invasiveness, and proliferation (75, 76). In cancer, FGFR gene amplifications, somatic missense mutations, and chromosomal translocations are the most frequent mechanisms of activation (76). FGFR has been identified as an oncogenic driver in breast, gastric, endometrial, urothelial, and brain tumors, among others (76). In lung cancer, the incidence of FGFR1 amplification is significantly higher in squamous cell carcinoma (20%) compared to adenocarcinoma (3%) and is more frequent in current smokers compared to former and never smokers. Other specific clinic-demographic features also correlate with FGFR1 amplification (75, 77). Some studies have recognized FGFR amplification as an independent negative prognostic factor in patients with NSCLC (78), whereas other studies have shown the opposite (75). In addition, FGFR amplifications may be found in concurrence with other tumor genetic alterations including TP53 and PIK3CA mutation and platelet-derived growth factor receptor A (PDGFRA) amplification (77). Somatic FGFR mutations in lung tumors usually occur in FGFR2 and FGFR3 and have been detected in 6% of lung squamous cell carcinomas (79). Multiple FGFR inhibitors, such as ponatinib, a multitargeted kinase inhibitor that displays potent pan–anti-FGFR activity, are in development, with promising results in cell lines and xenograft models (80). Phase 1 and 2 clinical trials of FGFR inhibitors (dovitinib, nintedanib, ponatinib, and AZD4547, among others) are ongoing in patients with NSCLC (81). FGFR gene copy number is usually assayed by FISH; however, members of this family are frequently part of NGS testing panels.
3.12. Discoidin domain receptor tyrosine kinase 2 (DDR2)
The discoidin domain receptor tyrosine kinase 2 gene (DDR2) is located on the long arm of chromosome 1 at position 23.3 and encodes for a tyrosine kinase receptor that is expressed in mesenchymal tissues and which binds fibrillar collagen as ligand. DDR2 activates important signaling pathways including SRC, SRC homology domain-containing (SHC), Janus kinase (JAK), ERK1/2, and PI3K and promotes cell migration, proliferation, and survival (82). In cancer, DDR2 mutations have been reported in melanoma as well as uterine, gastric, bladder, and colorectal cancers (83). In lung cancer, DDR2 mutations occur in 3% to 4% of lung squamous cell carcinomas (84) compared to 0.5% of adenocarcinomas (85) and are only present in smokers (86). No other significant association with clinicopathologic status has been found (84). At least 11 different DDR2 mutations have been identified (84) distributed throughout the gene and include the extracellular-binding discoidin domain and the cytoplasmic kinase domain (82, 84). DDR2 mutations have been associated with response to dasatinib (a multitargeted kinase inhibitor) in preclinical models and early phase clinical trials. Phase 2 clinical trials of dasatinib in patients with lung squamous cell carcinoma are under way (82, 84).
4. Immunotherapy Markers in Lung Cancer
Historically, lung cancer has not been considered very immunogenic because of several failed attempts with cytokines and vaccines. Nevertheless, over the past few years, immunotherapy has re-emerged strongly with the development of checkpoint inhibitors as treatments for NSCLC. Immune checkpoints are inhibitory pathways with the functions of maintaining self-tolerance and modulating immune responses (87). Immune checkpoint proteins that have been studied more comprehensively in many types of cancer, including lung cancer, are cytotoxic T-lymphocyte-associated antigen 4 (CTLA-4) and the programmed death-ligand 1 receptor (PD-1), which are expressed mainly on T cells, and programmed death-ligand 1 (PD-L1), which is expressed on tumor cells and tumor inflammatory infiltrate including macrophages, dendritic cells, and T cells (88). Many other checkpoint molecules, such as T-cell immunoglobulin and mucin domain-containing protein 3 (TIM3), B- and T-lymphocyte-associated protein (BTLA), V-domain Ig suppressor of T-cell activation (VISTA), and lymphocyte-activation gene 3 (LAG3), have been identified and are currently being evaluated as potential targets for cancer immunotherapy (89).
4.1. Cytotoxic T-lymphocyte-associated antigen 4 (CTLA-4)
Monoclonal antibodies that inhibit CTLA-4, such as ipilimumab, are available to prevent the binding of CTLA-4 with its ligands (CD80/CD86), leading to reactivation of the antitumor inmune response mediated by specific T cells (88). A phase 2 study of ipilimumab in combination with chemotherapy in patients with advanced NSCLC showed very promising results, with a significant improvement in PFS versus a control group treated with chemotherapy alone (90). A phase 3 trial of ipilimumab in combination with chemotherapy in patients with squamous histology NSCLC is ongoing (91). Currently, there is no biomarker to predict response to CTLA-4 therapy.
4.2. Programmed death-ligand 1 receptor (PD-1)
Several monoclonal antibodies targeting the interaction between PD-1 and its ligands PD-L1 and PD-L2 are available. There are different ways to block the PD-1 pathway; one is to use antibodies directed against PD-1 or by blocking its ligand PD-L1 (92). Clinical trials in NSCLC have shown sustained responses in approximately 20% of unselected patients to treatment with monoclonal antibodies against PD-1, such as nivolumab and pembrolizumab, and with antibodies against PD-L1 such as MPDL3280A. The FDA has approved the use of nivolumab in advanced NSCLC on or after platinum-based chemotherapy and pembrolizumab as second-line treatment for NSCLC after chemotherapy (93, 94). A recent study has reported that greater nonsynonymous mutation burden is associated with improved objective response, durable clinical benefit, and PFS in patients with NSCLC treated with pembrolizumab (95). Furthermore, IHC PD-L1 positivity in NSCLC has been identified as a potential predictor of response to anti– PD-1 and anti–PD-L1 monoclonal antibody therapy (96) and also as a prognostic biomarker (97). Other studies reported that PD-L1 overexpression cannot be currently considered a robust predictive biomarker for response to immunotherapy or a prognostic biomarker (98). These discrepancies may be due to assay variability and interpretive subjectivity differences for the evaluation of PD-L1 expression, including differences in detection methods, IHC antibody clones, and cut-off values for determining PD-L1 positivity, and heterogeneity in PD-L1 expression and site of PD-L1 expression (tumor cells and tumor inmune cells) (99, 100). Further studies are needed to compare different assays and to clarify and standardize testing protocols to confirm the suitability of PD-L1 expression as a biomarker.
Key points.
The molecular characterization of lung cancer has changed the classification and treatment of these tumors, becoming an essential component of pathologic diagnosis and therapy decisions.
The success of targeted therapies and new immunotherapy approaches has created a new paradigm of personalized therapy in lung cancer.
Pathologists should be able to precisely handle tissue adequacy in terms of quantity and quality and maintaining tumor cells for detection of molecular alterations.
This review focuses on clinically relevant cancer biomarkers as targets for therapy, as well as potential new targets for drug development.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure statement: The Authors have nothing to disclose.
Contributor Information
Pamela Villalobos, Department of Translational Molecular Pathology, The University of Texas MD Anderson Cancer Center, Houston, TX 77019..
Ignacio I. Wistuba, Department of Translational Molecular Pathology, The University of Texas MD Anderson Cancer Center, Houston, TX 77019.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.American Cancer Society [May 25, 2016];Cancer Facts & Figures 2015. Available at: http://www.cancer.org/research/cancerfactsstatistics/cancerfactsfigures2015/index.
- 3.Cagle PT, Allen TC, Olsen RJ. Lung cancer biomarkers: present status and future developments. Arch Pathol Lab Med. 2013;137(9):1191–1198. doi: 10.5858/arpa.2013-0319-CR. [DOI] [PubMed] [Google Scholar]
- 4.Fujimoto J, Wistuba II. Current concepts on the molecular pathology of non-small cell lung carcinoma. Semin Diagn Pathol. 2014;31(4):306–313. doi: 10.1053/j.semdp.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kerr KM, Bubendorf L, Edelman MJ, et al. Panel Members. Second ESMO consensus conference on lung cancer: pathology and molecular biomarkers for non-small-cell lung cancer. Ann Oncol. 2014;25(9):1681–1690. doi: 10.1093/annonc/mdu145. [DOI] [PubMed] [Google Scholar]
- 6.Mok TS. Personalized medicine in lung cancer: what we need to know. Nat Rev Clin Oncol. 2011;8(11):661–668. doi: 10.1038/nrclinonc.2011.126. [DOI] [PubMed] [Google Scholar]
- 7.Travis WD, Bambrilla E, Burke AP, Marx A, Nicholson AG, editors. WHO Classification of Tumours of the Lung, Pleura, Thymus and Heart (IARC WHO Classification of Tumours) 4th Edition. World Health Organization; Geneva: 2015. [DOI] [PubMed] [Google Scholar]
- 8.Travis WD, Brambilla E, Riely GJ. New pathologic classification of lung cancer: relevance for clinical practice and clinical trials. J Clin Oncol. 2013;31(8):992–1001. doi: 10.1200/JCO.2012.46.9270. [DOI] [PubMed] [Google Scholar]
- 9.Khalil FK, Altiok S. Advances in EGFR as a predictive marker in lung adenocarcinoma. Cancer Control. 2015;22(2):193–199. doi: 10.1177/107327481502200210. [DOI] [PubMed] [Google Scholar]
- 10.Sholl LM. Biomarkers in lung adenocarcinoma: a decade of progress. Arch Pathol Lab Med. 2015;139(4):469–480. doi: 10.5858/arpa.2014-0128-RA. [DOI] [PubMed] [Google Scholar]
- 11.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7(3):169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 12.Fan X, Liu B, Xu H, et al. Immunostaining with EGFR mutation-specific antibodies: a reliable screening method for lung adenocarcinomas harboring EGFR mutation in biopsy and resection samples. Hum Pathol. 2013;44(8):1499–1507. doi: 10.1016/j.humpath.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 13.Zhao Z, Verma V, Zhang M. Anaplastic lymphoma kinase: role in cancer and therapy perspective. Cancer Biol Ther. 2015;16(12):1691–1701. doi: 10.1080/15384047.2015.1095407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chatziandreou I, Tsioli P, Sakellariou S, et al. Comprehensive molecular analysis of NSCLC; clinicopathological associations. PLoS One. 2015;10(7):e0133859. doi: 10.1371/journal.pone.0133859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang DD, Zhang B, Gu Q, et al. HIP1-ALK, a novel ALK fusion variant that responds to crizotinib. J Thorac Oncol. 2014;9(3):285–294. doi: 10.1097/JTO.0000000000000087. [DOI] [PubMed] [Google Scholar]
- 16.Sholl LM, Aisner DL, Varella-Garcia M, et al. LCMC Investigators. Multi-institutional oncogenic driver mutation analysis in lung adenocarcinoma: the Lung Cancer Mutation Consortium experience. J Thorac Oncol. 2015;10(5):768–777. doi: 10.1097/JTO.0000000000000516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Solomon BJ, Mok T, Kim DW, et al. PROFILE 1014 Investigators. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med. 2014;371(23):2167–2177. doi: 10.1056/NEJMoa1408440. [DOI] [PubMed] [Google Scholar]
- 18.Choi YL, Soda M, Yamashita Y, et al. ALK Lung Cancer Study Group. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N Engl J Med. 2010;363(18):1734–1739. doi: 10.1056/NEJMoa1007478. [DOI] [PubMed] [Google Scholar]
- 19.Sasaki T, Okuda K, Zheng W, et al. The neuroblastoma-associated F1174L ALK mutation causes resistance to an ALK kinase inhibitor in ALK-translocated cancers. Cancer Res. 2010;70(24):10038–10043. doi: 10.1158/0008-5472.CAN-10-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sullivan I, Planchard D. ALK inhibitors in non-small cell lung cancer: the latest evidence and developments. Ther Adv Med Oncol. 2016;8(1):32–47. doi: 10.1177/1758834015617355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toyokawa G, Seto T. Anaplastic lymphoma kinase rearrangement in lung cancer: its biological and clinical significance. Respir Investig. 2014;52(6):330–338. doi: 10.1016/j.resinv.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 22.Cabillic F, Gros A, Dugay F, et al. Parallel FISH and immunohistochemical studies of ALK status in 3244 non-small-cell lung cancers reveal major discordances. J Thorac Oncol. 2014;9(3):295–306. doi: 10.1097/JTO.0000000000000072. [DOI] [PubMed] [Google Scholar]
- 23.Martinez P, Hernández-Losa J, Montero MÁ, et al. Fluorescence in situ hybridization and immunohistochemistry as diagnostic methods for ALK positive non-small cell lung cancer patients. PLoS One. 2013;8(1):e52261. doi: 10.1371/journal.pone.0052261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sholl LM, Weremowicz S, Gray SW, et al. Combined use of ALK immunohistochemistry and FISH for optimal detection of ALK-rearranged lung adenocarcinomas. J Thorac Oncol. 2013;8(3):322–328. doi: 10.1097/JTO.0b013e31827db604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yi ES, Boland JM, Maleszewski JJ, et al. Correlation of IHC and FISH for ALK gene rearrangement in non-small cell lung carcinoma: IHC score algorithm for FISH. J Thorac Oncol. 2011;6(3):459–465. doi: 10.1097/JTO.0b013e318209edb9. [DOI] [PubMed] [Google Scholar]
- 26.Paik JH, Choe G, Kim H, et al. Screening of anaplastic lymphoma kinase rearrangement by immunohistochemistry in non-small cell lung cancer: correlation with fluorescence in situ hybridization. J Thorac Oncol. 2011;6(3):466–472. doi: 10.1097/JTO.0b013e31820b82e8. [DOI] [PubMed] [Google Scholar]
- 27.Wallander ML, Geiersbach KB, Tripp SR, Layfield LJ. Comparison of reverse transcription-polymerase chain reaction, immunohistochemistry, and fluorescence in situ hybridization methodologies for detection of echinoderm microtubule-associated proteinlike 4-anaplastic lymphoma kinase fusion-positive non-small cell lung carcinoma: implications for optimal clinical testing. Arch Pathol Lab Med. 2012;136(7):796–803. doi: 10.5858/arpa.2011-0321-OA. [DOI] [PubMed] [Google Scholar]
- 28.Pekar-Zlotin M, Hirsch FR, Soussan-Gutman L, et al. Fluorescence in situ hybridization, immunohistochemistry, and next-generation sequencing for detection of EML4-ALK rearrangement in lung cancer. Oncologist. 2015;20(3):316–322. doi: 10.1634/theoncologist.2014-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McBride OW, Swan DC, Tronick SR, et al. Regional chromosomal localization of N-ras, K-ras-1, K-ras-2 and myb oncogenes in human cells. Nucleic Acids Res. 1983;11(23):8221–8236. doi: 10.1093/nar/11.23.8221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edkins S, O'Meara S, Parker A, et al. Recurrent KRAS codon 146 mutations in human colorectal cancer. Cancer Biol Ther. 2006;5(8):928–932. doi: 10.4161/cbt.5.8.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kempf E, Rousseau B, Besse B, Paz-Ares L. KRAS oncogene in lung cancer: focus on molecularly driven clinical trials. Eur Respir Rev. 2016;25(139):71–76. doi: 10.1183/16000617.0071-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dogan S, Shen R, Ang DC, et al. Molecular epidemiology of EGFR and KRAS mutations in 3,026 lung adenocarcinomas: higher susceptibility of women to smoking-related KRAS-mutant cancers. Clin Cancer Res. 2012;18(22):6169–6177. doi: 10.1158/1078-0432.CCR-11-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ying M, Zhu XX, Zhao Y, Li DH, Chen LH. KRAS mutation as a biomarker for survival in patients with non-small cell lung cancer, a meta-analysis of 12 randomized trials. Asian Pac J Cancer Prev. 2015;16(10):4439–4445. doi: 10.7314/apjcp.2015.16.10.4439. [DOI] [PubMed] [Google Scholar]
- 34.Macerelli M, Caramella C, Faivre L, et al. Does KRAS mutational status predict chemoresistance in advanced non-small cell lung cancer (NSCLC)? Lung Cancer. 2014;83(3):383–388. doi: 10.1016/j.lungcan.2013.12.013. [DOI] [PubMed] [Google Scholar]
- 35.Shepherd FA, Domerg C, Hainaut P, et al. Pooled analysis of the prognostic and predictive effects of KRAS mutation status and KRAS mutation subtype in early-stage resected non-small-cell lung cancer in four trials of adjuvant chemotherapy. J Clin Oncol. 2013;31(17):2173–2181. doi: 10.1200/JCO.2012.48.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pao W, Wang TY, Riely GJ, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2(1):e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jänne PA, Shaw AT, Pereira JR, et al. Selumetinib plus docetaxel for KRAS-mutant advanced non-small-cell lung cancer: a randomised, multicentre, placebo-controlled, phase 2 study. Lancet Oncol. 2013;14(1):38–47. doi: 10.1016/S1470-2045(12)70489-8. [DOI] [PubMed] [Google Scholar]
- 38.Dingemans AM, Mellema WW, Groen HJ, et al. A phase II study of sorafenib in patients with platinum-pretreated, advanced (Stage IIIb or IV) non-small cell lung cancer with a KRAS mutation. Clin Cancer Res. 2013;19(3):743–751. doi: 10.1158/1078-0432.CCR-12-1779. [DOI] [PubMed] [Google Scholar]
- 39.Blumenschein GR, Jr, Smit EF, Planchard D, et al. A randomized phase II study of the MEK1/MEK2 inhibitor trametinib (GSK1120212) compared with docetaxel in KRAS-mutant advanced non-small-cell lung cancer (NSCLC)†. Ann Oncol. 2015;26(5):894–901. doi: 10.1093/annonc/mdv072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bergethon K, Shaw AT, Ou SH, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol. 2012;30(8):863–870. doi: 10.1200/JCO.2011.35.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshida A, Kohno T, Tsuta K, et al. ROS1-rearranged lung cancer: a clinicopathologic and molecular study of 15 surgical cases. Am J Surg Pathol. 2013;37(4):554–562. doi: 10.1097/PAS.0b013e3182758fe6. [DOI] [PubMed] [Google Scholar]
- 42.Mazières J, Zalcman G, Crinò L, et al. Crizotinib therapy for advanced lung adenocarcinoma and a ROS1 rearrangement: results from the EUROS1 cohort. J Clin Oncol. 2015;33(9):992–999. doi: 10.1200/JCO.2014.58.3302. [DOI] [PubMed] [Google Scholar]
- 43.ClinicalTrials.gov # NCT01964157 [May 25, 2016];An Open-label, Multicenter, Phase II Study of LDK378 in Patients With Non-small Cell Lung Cancer Harboring ROS1 Rearrangement. Available at: https://clinicaltrials.gov/ct2/show/NCT01964157?term=An+Open-label%2C+Multicenter%2C+Phase+II+Study+of+LDK378+in+Patients+With+Non-small+Cell+Lung+Cancer+Harboring+ROS1+Rearrangement&rank=1.
- 44.ClinicalTrials.gov # NCT02183870 [May 25, 2016];EUCROSS: European Trial on Crizotinib in ROS1 Translocated Lung Cancer (EUCROSS) Available at: https://clinicaltrials.gov/ct2/show/NCT02183870?term=EUCROSS%3A+European+Trial+on+Crizotinib+in+ROS1+Translocated+Lung+Cancer&rank=1.
- 45.Cao B, Wei P, Liu Z, et al. Detection of lung adenocarcinoma with ROS1 rearrangement by IHC, FISH, and RT-PCR and analysis of its clinicopathologic features. Onco Targets Ther. 2015;9:131–138. doi: 10.2147/OTT.S94997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Popescu NC, King CR, Kraus MH. Localization of the human erbB-2 gene on normal and rearranged chromosomes 17 to bands q12-21.32. Genomics. 1989;4(3):362–366. doi: 10.1016/0888-7543(89)90343-1. [DOI] [PubMed] [Google Scholar]
- 47.Ricciardi GR, Russo A, Franchina T, et al. NSCLC and HER2: between lights and shadows. J Thorac Oncol. 2014;9(12):1750–1762. doi: 10.1097/JTO.0000000000000379. [DOI] [PubMed] [Google Scholar]
- 48.Bu S, Wang R, Pan Y, et al. Clinicopathologic characteristics of patients with HER2 insertions in non-small cell lung cancer. Ann Surg Oncol. 2016 Mar 23; doi: 10.1245/s10434-015-5044-8. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 49.Shigematsu H, Takahashi T, Nomura M, et al. Somatic mutations of the HER2 kinase domain in lung adenocarcinomas. Cancer Res. 2005;65(5):1642–1646. doi: 10.1158/0008-5472.CAN-04-4235. [DOI] [PubMed] [Google Scholar]
- 50.Mazières J, Peters S, Lepage B, et al. Lung cancer that harbors an HER2 mutation: epidemiologic characteristics and therapeutic perspectives. J Clin Oncol. 2013;31(16):1997–2003. doi: 10.1200/JCO.2012.45.6095. [DOI] [PubMed] [Google Scholar]
- 51.Knowles PP, Murray-Rust J, Kjaer S, et al. Structure and chemical inhibition of the RET tyrosine kinase domain. J Biol Chem. 2006;281(44):33577–33587. doi: 10.1074/jbc.M605604200. [DOI] [PubMed] [Google Scholar]
- 52.Wang R, Hu H, Pan Y, et al. RET fusions define a unique molecular and clinicopathologic subtype of non-small-cell lung cancer. J Clin Oncol. 2012;30(35):4352–4359. doi: 10.1200/JCO.2012.44.1477. [DOI] [PubMed] [Google Scholar]
- 53.Drilon A, Wang L, Hasanovic A, et al. Response to Cabozantinib in patients with RET fusion-positive lung adenocarcinomas. Cancer Discov. 2013;3(6):630–635. doi: 10.1158/2159-8290.CD-13-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lipson D, Capelletti M, Yelensky R, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med. 2012;18(3):382–384. doi: 10.1038/nm.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhen Z, Giordano S, Longati P, Medico E, Campiglio M, Comoglio PM. Structural and functional domains critical for constitutive activation of the HGF-receptor (Met). Oncogene. 1994;9(6):1691–1697. [PubMed] [Google Scholar]
- 56.Paik PK, Drilon A, Fan PD, et al. Response to MET inhibitors in patients with stage IV lung adenocarcinomas harboring MET mutations causing exon 14 skipping. Cancer Discov. 2015;5(8):842–849. doi: 10.1158/2159-8290.CD-14-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmidt L, Duh FM, Chen F, et al. Germline and somatic mutations in the tyrosine kinase domain of the MET proto-oncogene in papillary renal carcinomas. Nat Genet. 1997;16(1):68–73. doi: 10.1038/ng0597-68. [DOI] [PubMed] [Google Scholar]
- 58.Masuya D, Huang C, Liu D, et al. The tumour-stromal interaction between intratumoral c-Met and stromal hepatocyte growth factor associated with tumour growth and prognosis in non-small-cell lung cancer patients. Br J Cancer. 2004;90(8):1555–1562. doi: 10.1038/sj.bjc.6601718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beau-Faller M, Ruppert AM, Voegeli AC, et al. MET gene copy number in non-small cell lung cancer: molecular analysis in a targeted tyrosine kinase inhibitor naïve cohort. J Thorac Oncol. 2008;3(4):331–339. doi: 10.1097/JTO.0b013e318168d9d4. [DOI] [PubMed] [Google Scholar]
- 60.Cardarella S, Ogino A, Nishino M, et al. Clinical, pathologic, and biologic features associated with BRAF mutations in non-small cell lung cancer. Clin Cancer Res. 2013;19(16):4532–4540. doi: 10.1158/1078-0432.CCR-13-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garnett MJ, Marais R. Guilty as charged: B-RAF is a human oncogene. Cancer Cell. 2004;6(4):313–319. doi: 10.1016/j.ccr.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 62.Paik PK, Arcila ME, Fara M, et al. Clinical characteristics of patients with lung adenocarcinomas harboring BRAF mutations. J Clin Oncol. 2011;29(15):2046–2051. doi: 10.1200/JCO.2010.33.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Planchard D, Kim TM, Mazieres J, et al. Dabrafenib in patients with BRAFV600E-positive advanced non-small-cell lung cancer: a single-arm, multicentre, open-label, phase 2 trial. Lancet Oncol. 2016 Apr 11; doi: 10.1016/S1470-2045(16)00077-2. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hyman DM, Puzanov I, Subbiah V, et al. Vemurafenib in multiple nonmelanoma cancers with BRAF V600 mutations. N Engl J Med. 2015;373(8):726–736. doi: 10.1056/NEJMoa1502309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamamoto H, Shigematsu H, Nomura M, et al. PIK3CA mutations and copy number gains in human lung cancers. Cancer Res. 2008;68(17):6913–6921. doi: 10.1158/0008-5472.CAN-07-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Karakas B, Bachman KE, Park BH. Mutation of the PIK3CA oncogene in human cancers. Br J Cancer. 2006;94(4):455–459. doi: 10.1038/sj.bjc.6602970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Samuels Y, Velculescu VE. Oncogenic mutations of PIK3CA in human cancers. Cell Cycle. 2004;3(10):1221–1224. doi: 10.4161/cc.3.10.1164. [DOI] [PubMed] [Google Scholar]
- 68.Endoh H, Yatabe Y, Kosaka T, Kuwano H, Mitsudomi T. PTEN and PIK3CA expression is associated with prolonged survival after gefitinib treatment in EGFR-mutated lung cancer patients. J Thorac Oncol. 2006;1(7):629–634. [PubMed] [Google Scholar]
- 69.Kawano O, Sasaki H, Endo K, et al. PIK3CA mutation status in Japanese lung cancer patients. Lung Cancer. 2006;54(2):209–215. doi: 10.1016/j.lungcan.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 70.Chen JY, Cheng YN, Han L, et al. Predictive value of K-ras and PIK3CA in non-small cell lung cancer patients treated with EGFR-TKIs: a systemic review and meta-analysis. Cancer Biol Med. 2015;12(2):126–139. doi: 10.7497/j.issn.2095-3941.2015.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thomas A, Rajan A, Lopez-Chavez A, Wang Y, Giaccone G. From targets to targeted therapies and molecular profiling in non-small cell lung carcinoma. Ann Oncol. 2013;24(3):577–585. doi: 10.1093/annonc/mds478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alberti L, Carniti C, Miranda C, Roccato E, Pierotti MA. RET and NTRK1 protooncogenes in human diseases. J Cell Physiol. 2003;195(2):168–186. doi: 10.1002/jcp.10252. [DOI] [PubMed] [Google Scholar]
- 73.Vaishnavi A, Capelletti M, Le AT, et al. Oncogenic and drug-sensitive NTRK1 rearrangements in lung cancer. Nat Med. 2013;19(11):1469–1472. doi: 10.1038/nm.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Passiglia F, Caparica R, Giovannetti E, et al. The potential of neurotrophic tyrosine kinase (NTRK) inhibitors for treating lung cancer. Expert Opin Investig Drugs. 2016;25(4):385–392. doi: 10.1517/13543784.2016.1152261. [DOI] [PubMed] [Google Scholar]
- 75.Jiang T, Gao G, Fan G, Li M, Zhou C. FGFR1 amplification in lung squamous cell carcinoma: a systematic review with meta-analysis. Lung Cancer. 2015;87(1):1–7. doi: 10.1016/j.lungcan.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 76.Dienstmann R, Rodon J, Prat A, et al. Genomic aberrations in the FGFR pathway: opportunities for targeted therapies in solid tumors. Ann Oncol. 2014;25(3):552–563. doi: 10.1093/annonc/mdt419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Weiss J, Sos ML, Seidel D, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med. 2010;2(62):62ra93. doi: 10.1126/scitranslmed.3001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Seo AN, Jin Y, Lee HJ, et al. FGFR1 amplification is associated with poor prognosis and smoking in non-small-cell lung cancer. Virchows Arch. 2014;465(5):547–558. doi: 10.1007/s00428-014-1634-2. [DOI] [PubMed] [Google Scholar]
- 79.Liao RG, Jung J, Tchaicha J, et al. Inhibitor-sensitive FGFR2 and FGFR3 mutations in lung squamous cell carcinoma. Cancer Res. 2013;73(16):5195–5205. doi: 10.1158/0008-5472.CAN-12-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gozgit JM, Wong MJ, Moran L, et al. Ponatinib (AP24534), a multitargeted pan-FGFR inhibitor with activity in multiple FGFR-amplified or mutated cancer models. Mol Cancer Ther. 2012;11(3):690–699. doi: 10.1158/1535-7163.MCT-11-0450. [DOI] [PubMed] [Google Scholar]
- 81.Tiseo M, Gelsomino F, Alfieri R, et al. FGFR as potential target in the treatment of squamous non small cell lung cancer. Cancer Treat Rev. 2015;41(6):527–539. doi: 10.1016/j.ctrv.2015.04.011. [DOI] [PubMed] [Google Scholar]
- 82.Payne LS, Huang PH. Discoidin domain receptor 2 signaling networks and therapy in lung cancer. J Thorac Oncol. 2014;9(6):900–904. doi: 10.1097/JTO.0000000000000164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Beauchamp EM, Woods BA, Dulak AM, et al. Acquired resistance to dasatinib in lung cancer cell lines conferred by DDR2 gatekeeper mutation and NF1 loss. Mol Cancer Ther. 2014;13(2):475–482. doi: 10.1158/1535-7163.MCT-13-0817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hammerman PS, Sos ML, Ramos AH, et al. Mutations in the DDR2 kinase gene identify a novel therapeutic target in squamous cell lung cancer. Cancer Discov. 2011;1(1):78–89. doi: 10.1158/2159-8274.CD-11-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455(7216):1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.An SJ, Chen ZH, Su J, et al. Identification of enriched driver gene alterations in subgroups of non-small cell lung cancer patients based on histology and smoking status. PLoS One. 2012;7(6):e40109. doi: 10.1371/journal.pone.0040109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12(4):252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Seetharamu N, Budman DR, Sullivan KM. Immune checkpoint inhibitors in lung cancer: past, present and future. Future Oncol. 2016;12(9):1151–1163. doi: 10.2217/fon.16.20. [DOI] [PubMed] [Google Scholar]
- 89.Swatler J, Kozlowska E. [Immune checkpoint-targeted cancer immunotherapies]. Postepy Hig Med Dosw. 2016;70:25–42. doi: 10.5604/17322693.1192926. (Abstract in English; Article in Polish) [DOI] [PubMed] [Google Scholar]
- 90.Lynch TJ, Bondarenko I, Luft A, et al. Ipilimumab in combination with paclitaxel and carboplatin as first-line treatment in stage IIIB/IV non-small-cell lung cancer: results from a randomized, double-blind, multicenter phase II study. J Clin Oncol. 2012;30(17):2046–2054. doi: 10.1200/JCO.2011.38.4032. [DOI] [PubMed] [Google Scholar]
- 91.Carrizosa DR, Gold KA. New strategies in immunotherapy for non-small cell lung cancer. Transl Lung Cancer Res. 2015;4(5):553–559. doi: 10.3978/j.issn.2218-6751.2015.06.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Brahmer JR. Immune checkpoint blockade: the hope for immunotherapy as a treatment of lung cancer? Semin Oncol. 2014;41(1):126–132. doi: 10.1053/j.seminoncol.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Garon EB, Rizvi NA, Hui R, et al. KEYNOTE-001 Investigators. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372(21):2018–2028. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 94.Gettinger SN, Horn L, Gandhi L, et al. Overall survival and long-term safety of nivolumab (anti-programmed death 1 antibody, BMS-936558, ONO-4538) in patients with previously treated advanced non-small-cell lung cancer. J Clin Oncol. 2015;33(18):2004–2012. doi: 10.1200/JCO.2014.58.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med. 2015;373(17):1627–1639. doi: 10.1056/NEJMoa1507643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sun JM, Zhou W, Choi YL, et al. Prognostic significance of programmed cell death ligand 1 in patients with non-small-cell lung cancer: a large cohort study of surgically resected cases. J Thorac Oncol. 2016 Apr 18; doi: 10.1016/j.jtho.2016.04.007. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 98.Sorensen SF, Zhou W, Dolled-Filhart M, et al. PD-L1 expression and survival among patients with advanced non-small cell lung cancer treated with chemotherapy. Transl Oncol. 2016;9(1):64–69. doi: 10.1016/j.tranon.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Velcheti V, Schalper KA, Carvajal DE, et al. Programmed death ligand-1 expression in non-small cell lung cancer. Lab Invest. 2014;94(1):107–116. doi: 10.1038/labinvest.2013.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Taube JM, Anders RA, Young GD, et al. Colocalization of inflammatory response with B7-h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci Transl Med. 2012;4(127):127ra37. doi: 10.1126/scitranslmed.3003689. [DOI] [PMC free article] [PubMed] [Google Scholar]