Abstract
MicroRNAs (miRNAs) play an important role in cancer development and progression, altering several biological functions by affecting targets through either degradation of mRNAs or suppression of protein translation. One such miRNA, miR-1247, is downregulated in various cancers, but its biological role in non-small-cell lung cancer (NSCLC) is unknown. This study found that the expression of miR-1247 was significantly reduced in NSCLC cell lines and tumor tissues compared with matched normal lung tissues and cell lines as a result of DNA hypermethylation. Overexpression of miR-1247 or demethylation by 5-azacytidine (5-Aza) treatment dramatically inhibited cell growth, migration, invasion, and cell cycle progression. Furthermore, Stathmin 1 (STMN1) was found to be an immediate and functional target of miR-1247. The expression of STMN1 was significantly increased in NSCLC cell lines but was decreased by 5-Aza treatment. In addition, miR-1247 upregulation partially inhibited STMN1-induced promotion of migration and invasion of A549 and H1299 cells. The results suggest that miR-1247 was silenced by DNA methylation. MiR-1247 and its downstream target gene STMN1 may therefore be a future target for the treatment of NSCLC.
Keywords: stathmin 1, DNA methylation, biomarker, miRNAs, gene regulation, NSCLC
Introduction
Non-small-cell lung cancer (NSCLC) is one of the leading causes of cancer deaths and is the predominant form of lung cancer worldwide.1,2 Although there has been great progress in the diagnosis and treatment of NSCLC, many patients still have poor prognosis with <5 years of overall survival.3–5 Therefore, urgent exploration of NSCLC tumorigenesis is vital to improve diagnosis and treatment.
MicroRNAs (miRNAs) are a class of short (~22 nt), non-coding, regulatory RNAs involved in multiple biological processes, including cell invasion, metastasis, angiogenesis, and apoptosis.6 Accumulating evidence has shown that >50% of miRNA genes are situated in fragile sites and are involved in tumor pathogenesis, including NSCLC.7–9 MiR-1247 has previously been identified to be downregulated in prostate cancer, pancreatic cancer, and osteosarcoma.10–13 Upregulation of miR-1247 inhibited cell proliferation and metastasis in pancreatic cancer and was therefore recognized as a tumor suppressor.10 Moreover, the expression of miR-1247 has been found to be decreased in osteosarcoma cancer stem cells and influences human osteosarcoma oncogenesis by the regulation of MAP3K9.11 Furthermore, miR-1247 was found to be downregulated in lung adenocarcinoma and squamous carcinoma cancer tissue compared with normal tissues.14 Yet the function and mechanism of miR-1247 in NSCLC has rarely been explored. Stathmin 1 (STMN1), a major microtubule-depolymerizing protein, serves as a prognostic marker for multiple cancers, including NSCLC,15 gallbladder carcinoma,16 and colon cancer.17 STMN1 has been found to destabilize microtubules and plays an important role in the regulation of cell cycle progression and tumor metastasis.18,19
DNA methylation silences the expression of some miRNAs and is thought to be another form of diagnostic biomarker and target for cancer therapy.20–23 For example, overexpression of miRNA-503 mediated by DNA methylation inhibited the expression of FANCA, which regulates the resistance of NSCLC cells to cisplatin.22 MiR-148 was found to be downregulated by DNA methylation in skin cancer, which promoted metastasis by targeting TGIF2.23 Methylation of miR-1247 was reported to be related to epithelial–mesenchymal transition (EMT) in ulcerative colitis.24 Given these findings, it was speculated that miR-1247 may have some function in NSCLC through DNA methylation, a previously uninvestigated question. Therefore, this study was designed to investigate DNA methylation in relation to miR-1247 in NSCLC tissues and cell lines. In addition, this study analyzed changes in apoptosis, migration, and invasion ability after miR-1247 overexpression or demethylation with 5-azacytidine (5-Aza) in NSCLC cell lines. Furthermore, the relationship between miR-1247 and STMN1 in NSCLC was investigated. Finally, miR-1247 was examined for its involvement in inhibiting the accelerating effects of STMN1-induced regulation. This research may suggest a novel approach for the treatment of NSCLC in the future.
Methods
Cell culture and transfection
HBE, A549, H460, and H1299 cell lines, kindly provided by Cell Bank, Chinese Academy of Sciences (Shanghai, People’s Republic of China), were maintained in RPMI-1640 medium (Thermo Fisher Scientific, Waltham, MA, USA) with 10% fetal bovine serum (Thermo Fisher Scientific) in humidified 5% CO2 at 37°C.
The cells were cultured in 6-well plates (5×105 cells/well) for 24 h and then exposed to 5 μmol/L 5-Aza (Sigma, St Louis, MO, USA) for 72 h. The cells were collected for quantitative real-time polymerase chain reaction (qRT-PCR).
H1299 and A549 cells were transfected with negative control (NC) or miR-1247 mimics (HmiR0610, Funeng, Guangzhou, People’s Republic of China) using Lip2000 (Thermo Fisher Scientific) for 48 h. A group of cells (5-Aza group) were treated with 5 μmol/L of 5-Aza for 48 h. All the cells were examined for the ability to invade and metastasize by Western blot and methylation-specific PCR (MSP).
Tissues
Human NSCLC and normal lung tissues were acquired from a cancer hospital. The samples were fresh-frozen and stored in liquid nitrogen after surgery. Written informed consent was obtained for use of the tissues. The study was approved by the Central Southern University Ethics Committee.
qRT-PCR
Trizol reagent (Thermo Fisher Scientific) was used to isolate total RNA. RNA was then reverse-transcribed to cDNA using a Reverse Transcriptase Kit (Funeng). MiRNA was tested by qRT-PCR using an ABI 7300 Sequence Detector (Thermo Fisher Scientific) with SYBR Green PCR Master Mix (Bio-Rad, Hercules, CA, USA). PCR parameters were as follows: 95°C for 3 min, 40 cycles of 95°C for 10 s, and 58°C for 30 s. The relative expression level was calculated using the delta delta Ct method. Reverse-transcribed primer and real-time primers for miR-1247 and U6 were obtained from Funeng (HmiRQP3407 and HmiRQP9001). The real-time primer sequences were as follows: STMN1 (sense: 5′-GAGAAACGAGAGCACGAGAAAG-3′, antisense: 5′-GTTCTTCCGCACTTCTTCAATG-3′), β-actin (sense: 5′-AGGGGCCGGACTCGTCATACT-3′, antisense: 5′-GGCGGCACCACCATGTACCCT-3′).
Western blot
The Bradford protein assay kit (P200B; Auragene Company, Changsha, People’s Republic of China) was used to confirm the protein concentration. Proteins were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis before being transferred onto polyvinylfluoride membranes. Then the membranes were incubated with anti-STMN1 antibody (1:500, SC-55531; Santa Cruz Biotechnology Inc., Dallas, TX, USA) at 4°C overnight and then incubated in the corresponding secondary antibody preparation for 1 h at room temperature. Bands were detected with the ECL detection kit (P001WB-1; Auragene). The blots were reported with β-actin.
Proliferation and clonogenic assays
Each well in 96-well plates was seeded with 5,000 cells. At 24, 48, and 72 h after culture, cell proliferation was detected by MTT assays using a microplate reader at 570 nm wavelength.
Six-well plates had 300 cells per well seeded in adherent conditions. After 14 days of culture, colonies were fixed for 15 min with 4% buffered formalin and stained with GIEMSA (G8220-1; Solarbio, Beijing, People’s Republic of China) for 20 min. The number of colonies per well was determined.
Cell-cycle analysis
Cells were fixed in 70% ethanol for 1 h (or until analysis) at 4°C. Phosphate-buffered saline was then used to wash the fixed cells. The Cell Cycle Detection Kit (KeyGEN, Nanjing, People’s Republic of China) was used for cell-cycle analysis according to the manufacturer’s instructions.
Invasion assay
Transwell chambers (8-μm pore size; Costar, Cambridge, MA, USA) were used to measure cell invasion ability. First, the top chamber was plated with Matrigel (Becton, Dickinson and Company, Franklin Lakes, NJ, USA) and incubated at 37°C for 4 h. Then cells (5×105 per well, 200 μL) were added into the top chamber and complete medium into the bottom chambers. After 48 h of incubation, cells on the upper surface were removed by a wet cotton swab. Cells on the lower surface of the membrane were fixed and stained with crystal violet and examined under a microscope. The membranes were dissolved with 10% acetic acid, and absorbance values were detected by a microplate reader at 570 nm wavelength. Each assay was independently performed three times.
Wound healing assay
For this assay, 5×105 cells were seeded into six-well plates and incubated for 24 h in serum-free RPMI-1640 before wounding. A 10 μL pipette tip was used to create a wound across the diameter of the well. Cell migration across the wound surface was then assessed by microscopy after 48 h. Images were captured under ×200 magnification (AE31, Motic). Photoshop CS3 was used to measure the distance between the edges of the wound.
Dual luciferase reporter gene assay
To assay the transcriptional activity of the STMN1 gene with miR-1247, 24,000 cells were seeded in 24-well plates. The plasmid STMN1-3′UTR-psi-CHECK2/Mut STMN1-3′UTR-psi-CHECK2 (Auragene Company) with or without miR-1247 mimics/inhibitor/NC (Funeng) were transfected with Lipofectamine 2000 (Thermo Fisher Scientific). Luciferase activity was measured and quantified using a luminometer with the DualLuciferase Reporter Assay System (E1910, Promega Corporation, Fitchburg, WI, USA). The experiments were performed in triplicate. The results are expressed as the means of the ratio between the firefly and Renilla luciferase activities.
Methylation-specific PCR
EZ DNA Methylation-Gold™ Kit (Zymo Research, Irvine, CA, USA) was used to modify genomic DNA, and MSP was used to detect the methylation level of miR-1247 in cells and tissues. PCR amplification was performed with HotStar Taq Polymerase (Qiagen, Hilden, Germany) and consisted of initial incubation at 94°C for 4 min, followed by 34 cycles at 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s, followed by one cycle at 72°C for 5 min. PCR products were electrophoresed in 3% agarose gels and then visualized by ultraviolet illumination. The miR-1247 MSp primers were (forward) 5′-TTGTTTTTTATTTCGGGAACGTCGA and (reverse) 5′-ATACGCACTTAACGCGTCCGAACG. The miR-1247 unMSp primers were (forward) 5′-GTTGTTTTTTATTTTGGGAATGTTGA and (reverse) 5′-AAAAATACACACTTAACACATCCAAACACC.
Statistical analysis
All statistical analyses were performed using SPSS 17.0 software. The data were presented as mean ± standard deviation (SD) of three independent experiments and compared using a Student’s t-test and one-way analysis of variance. A P-value of <0.05 was considered to be statistically significant and indicated by *.
Results
MiR-1247 is epigenetically downregulated by DNA methylation in NSCLC tissues and cell lines
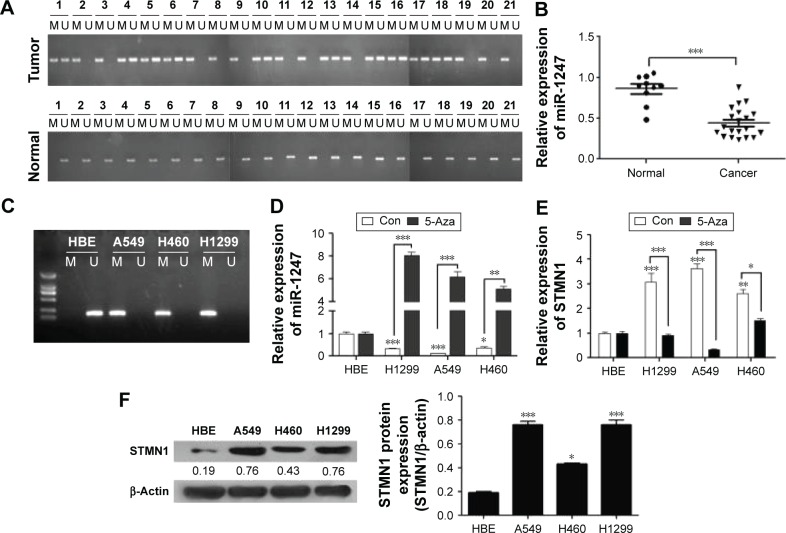
To examine the correlation between DNA methylation level and miR-1247 expression, MSP was used to assess the level of methylation of miR-1247, and qRT-PCR was used to assess the expression of miR-1247 in NSCLC tissues and cell lines. It was found that miR-1247 in NSCLC tissues was more highly methylated, while in normal tissues it was unmethylated (Figure 1A). MiR-1247 was more dramatically silenced in NSCLC tissues than in adjacent tissues (Figure 1B), indicating that DNA methylation was negatively correlated with the expression of miR-1247. The same result was found in NSCLC cell lines. MiR-1247 in H1299, H460, and A549 cell lines was hypermethylated compared with HBE cells, which were not methylated (Figure 1C). Moreover, the expression of miR-1247 in NSCLC cells (A549, H460, and H1299) was dramatically lower compared with normal cells (HBE), an effect that was reversed by 5-Aza treatment (Figure 1D). These results confirm that miR-1247 was decreased by DNA methylation in NSCLC tissues and cell lines.
Figure 1.
MiR-1247 is deregulated by methylation in non-small lung cancer tissues and cell lines.
Notes: (A) The DNA methylation level of miR-1247 in 21 pairs of NSCLC and adjacent tissue samples were analyzed by MSP. (B) MiR-1247 expression levels were detected by qRT-PCR in 21 NSCLC and 21 normal tissue samples. (C) MSP analysis of the DNA methylation level of miR-1247 in HBE, A549, H460, and H1299 cell lines. (D) MiR-1247 levels of HBE, A549, H1299, H460 cells, performed by qRT-PCR. (E) STMN1 levels. (F) The STMN1 protein expression was analyzed by Western blot. Bars represent mean values ± standard deviation. ***P<0.001; **P<0.01; *P<0.05, N=3.
Abbreviations: 5-Aza, 5′-Aza-deoxycytidine; M, methylated; NSCLC, non-small cell lung cancer; qRT-PCR, quantitative real-time polymerase chain reaction; U, nonmethylated; MSP, methylation-specific PCR; Con, control.
Next, the relationship between DNA methylation and expression of STMN1 in NSCLC cells was determined by using qRT-PCR assay. The STMN1 mRNA levels were upregulated in NSCLC cells compared with normal HBE cells, while the levels were significantly restored by 5-Aza (Figure 1E). STMN1 protein levels were measured by Western blot in NSCLC, and normal cells were similarly found to have dramatically higher expression in NSCLC cells than in HBE cells (Figure 1F). In summary, miR-1247 was downregulated in NSCLC, while STMN1 was upregulated. DNA hypermethylation inhibited the expression of miR-1247 and resulted in high expression of STMN1, an effect that could be reversed by 5-Aza.
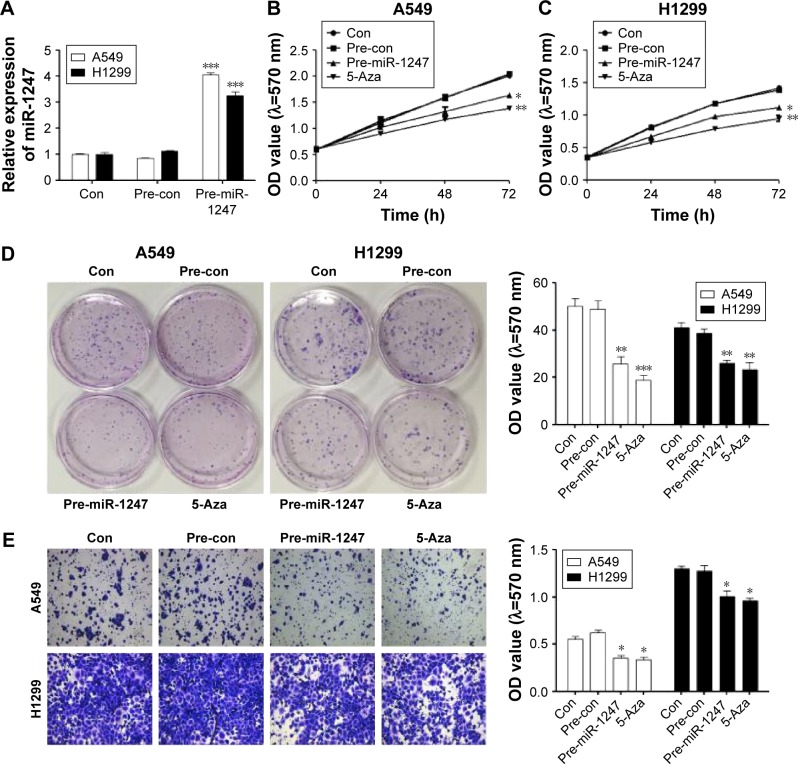
Upregulation of miR-1247 inhibited cell proliferation and invasion in A549 and H1299 cells
MiR-1247 mimics were used to establish gain-of-function models, and the overexpression efficiency was confirmed by qRT-PCR assay (Figure 2A). Additionally, the effects on cell proliferation of A549 and H1299 cells of miR-1247 upregulation either by transfection or by demethylation treatment were examined by MTT and clone formation assays. MTT results showed that overexpression of miR-1247 resulted in a significant decrease in cell growth of NSCLC cell lines (A549 and H1299), and demethylation treatment also led to a significant decrease in the proliferation of A549 (Figure 2B) and H1299 cells (Figure 2C). From the colony formation assay, it was found that overexpression of miR-1247 and demethylation significantly inhibited the viability of A549 and H1299 cells, which formed fewer and smaller clones (Figure 2D). These findings suggest that 5-Aza dramatically increased the expression of miR-1247, which then suppressed the proliferation of NSCLC cells in vitro.
Figure 2.
Upregulation of miR-1247 expression impairs cell proliferation and invasion in non-small-cell lung cancer.
Notes: (A) qRT-PCR assay detected the miR-1247 level in A549 and H1299 cells with miR-1247 overexpression. MTT analysis of cell proliferation in A549 cells (B) and in H1299 cells (C) treated with miR-1247 mimics/mimicsNC/5-Aza. (D) Cell proliferation ability was used to detect cell proliferation in A549 and H1299 cells with miR-1247 overexpression or exposure to 5-Aza. (E) Transwell migration assay with miR-1247 overexpression or exposure to 5-Aza. Bars represent mean values ± standard deviation. ***P<0.001; **P<0.01; *P<0.05, N=3.
Abbreviations: qRT-PCR, quantitative real-time polymerase chain reaction; h, hours; OD, optical density; Con, control.
The effect of miR-1247 on cell invasion was detected by a transwell assay (Figure 2E). The invasion ability of cells was dramatically decreased when miR-1247 was overexpressed and demethylated compared with control groups. These data indicate that miR-1247 has a vital role in reducing the growth and invasion of A549 and H1299 cells and is regulated by DNA methylation.
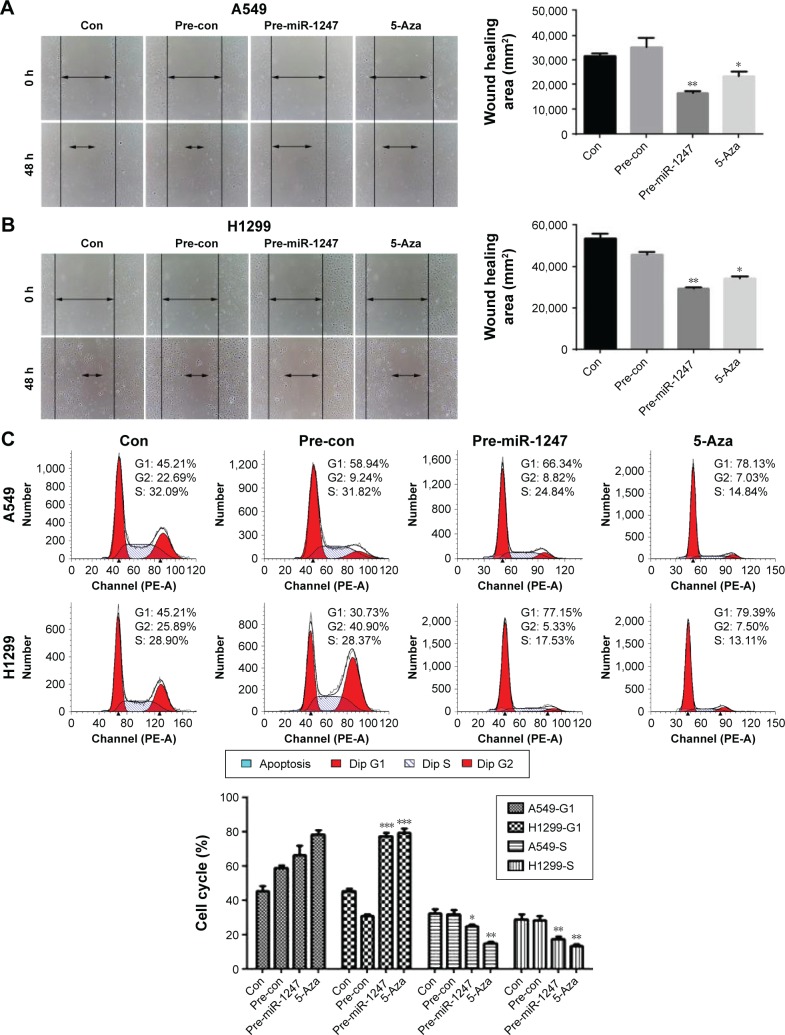
Overexpression of miR-1247 inhibited cell migration and promoted G1/S phase cell-cycle arrest in A549 and H1299 cells
To test whether miR-1247 had a functional effect on cell migration, a wound healing assay was performed. Upregulation of miR-1247 markedly reduced the migration of A549 (Figure 3A) and H1299 (Figure 3B) cells. Similar results were obtained when cells were treated with 5-Aza, demonstrating that miR-1247 inhibited effective migration.
Figure 3.
MiR-1247 impairs cell migration and cell cycle of non-small-cell lung cancer.
Notes: Wound scratch assays were performed to analyze the migration (arrows) of A549 (A) or H1299 (B) cells with miR-1247 overexpression or exposure to 5-Aza. (C) Overexpression of miR-1247 and exposure to 5-Aza arrested the cell cycle at the G1/S phase. Bars represent mean values ± standard deviation. ***P<0.001; **P<0.01; *P<0.05, N=3.
Abbreviation: Con, control.
Furthermore, to measure whether miR-1247 had a functional effect on cell-cycle arrest, a flow cytometry assay was performed. MiR-1247 overexpression significantly accelerated cell-cycle arrest at the G1/S phase in A549 and H1299 cells. Similar results were obtained when cells were treated with 5-AZa (Figure 3C). These data also demonstrate that upregulation of miR-1247 by demethylation treatment inhibited cell migration and promoted G1/S phase cell-cycle arrest in A549 and H1299 cells.
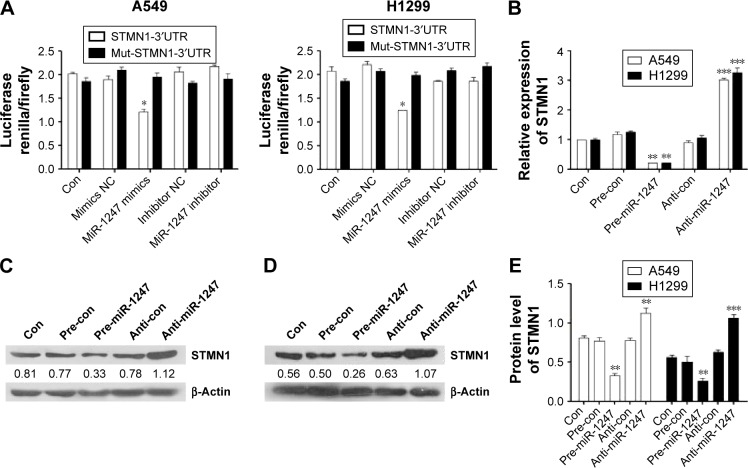
MiR-1247 directly targeted STMN1 and downregulated its expression in A549 and H1299 cells
As described earlier, it was found that the expression of STMN1 was corrected with hypomethylated miR-1247. In addition, a bioinformatics tool (microRNA.org-Targets and Expression) was used to predict the target for miR-1247 and found that STMN1 was such a target. Therefore, to confirm the regulatory role of miR-1247 on STMN1, a luciferase assay was utilized to detect the interactions between miR-1247 and the putative miR-1247-binding sites on 3′UTR of the STMN1 gene. Overexpression of miR-1247 inhibited luciferase activity by combining with the STMN1-3′-UTR segment in both A549 and H1299 cells (Figure 4A). No change was observed in the luciferase reporter activity when the cells were transfected with anti-miR-1247 or controls.
Figure 4.
MiR-1247 targeted STMN1 in A549 and H1299 cells.
Notes: (A) Luciferase reporter assay was used to determine whether STMN1 was the direct target of miR-1247 in A549 and H1299 cells. A normal and a mutated 3′-UTR of STMN1 were subcloned into the psi-CHECK2/Neo reporter vector. Psi-CHECK2/Neo-STMN1-3′UTR or psi-CHECK2/Neo-mut STMN1-3′UTR plus miR-1247 mimics or miR-1247 inhibitor were co-transfected. The fluorescence intensities were detected to mirror the capacity for direct binding. (B) The mRNA expression of STMN1 was detected in A549 and H1299 cells with miR-1247 upregulated and silenced with pre-miR and anti-miR. The expression of STMN1 protein was detected by Western blot in A549 (C) and H1299 (D) miR-1247-upregulated and -silenced cells. (E) Bars represent mean values ± standard deviation of (C) and (D). ***P<0.001; **P<0.01; *P<0.05, N=3.
Abbreviation: Con, control.
To further investigate the regulatory effect of miR-1247 on STMN1 expression, qRT-PCR and Western blot were performed with miR-1247 overexpressed or silenced in A549 and H1299 cells. As predicted, overexpression of miR-1247 significantly repressed STMN1 mRNA and protein levels compared with control groups (Figure 4B–E). By comparison, downregulation of miR-1247 in A549 (Figure 4B, C, and E) and H1299 (Figure 4B, D, and E) cells led to a moderate increase in the STMN1 mRNA and protein levels. This demonstrated that STMN1 was a direct target of miR-1247 in A549 and H1299 cells.
MiR-1247 inhibition accelerates the effects of STMN1 on cell invasion
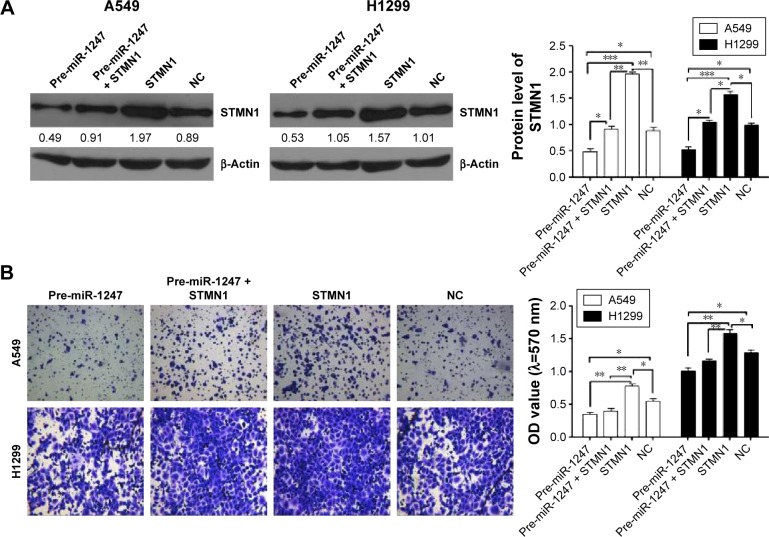
STMN1 protein expression was assessed (Figure 5) when STMN1 was overexpressed with or without miR-1247. Western blot results showed that miR-1247 inhibited STMN1 expression when both miR-1247 and STMN1 were overexpressed (Figure 5A).
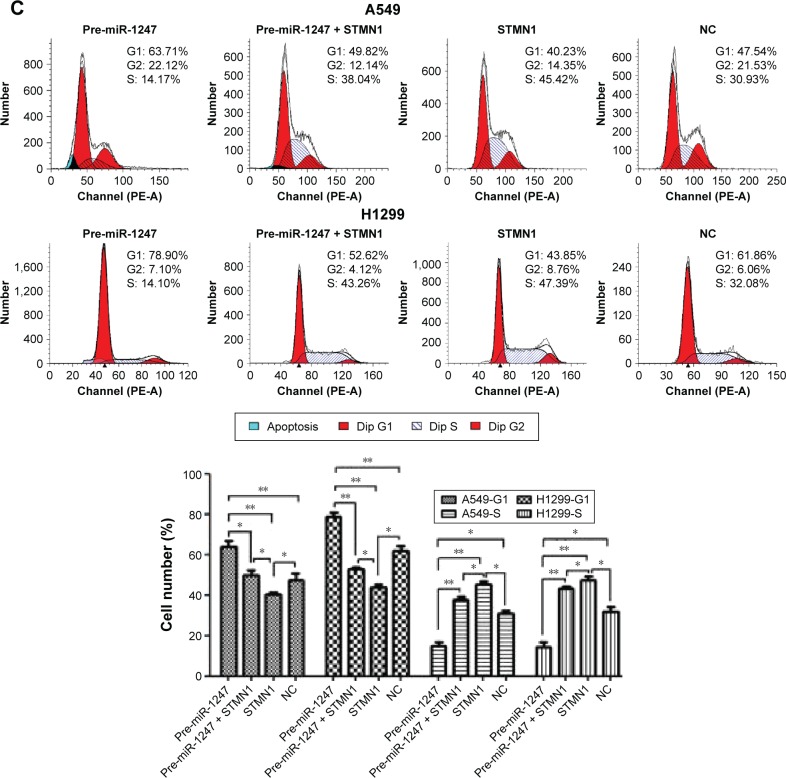
Figure 5.
MiR-1247 inhibited the effects of STMN1 that accelerated cell invasion and cell cycle.
Notes: (A) STMN1 protein expression was analyzed by Western blot. A549 and H1299 cells were co-transfected with pre-miR-1247 and pCDNA3.1-STMN1. (B) Increased miR-1247 expression reversed STMN1-induced cell invasion. (C) Overexpression of miR-1247 reversed STMN1-induced cell cycle progression at the G1/S phase in A549 and H1299 cells. Bars represent mean values ± standard deviation. ***P<0.001; **P<0.01; *P<0.05, N=3.
Abbreviation: NC, cells transfected with mimics NC and pcDNA3.1.
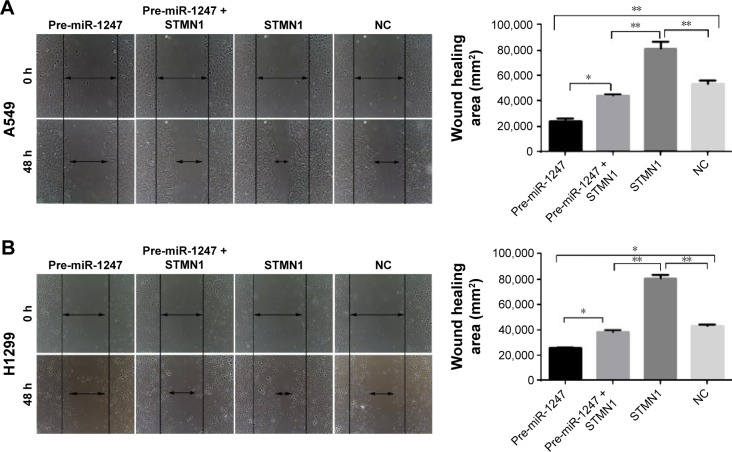
Transwell and wound scratch assays were performed to evaluate the function of STMN1 as a target gene of miR-1247 by evaluating the influence of miR-1247 on the invasion and migration efficiency of NSCLC cells. Overexpression of STMN1 significantly increased cell invasion, which was dramatically rescued by overexpression of miR-1247 in A549 (Figure 5B) and H1299 (Figure 5B) cells. Moreover, STMN1 overexpression dramatically promoted cell wound closure in A549 (Figure 6A) and H1299 (Figure 6B) cells. MiR-1247 significantly inhibited wound closure compared with STMN1 overexpression. These results suggest that miR-1247 rescues A549 and H1299 cells from the accelerating effect of STMN1 on migration and invasion.
Figure 6.
MiR-1247 inhibited the effects of STMN1 on cell migration.
Notes: Overexpression of miR-1247 reversed the STMN1-induced acceleration of cell migration (arrows) in A549 (A) and H1299 (B) cells. Bars represent mean values ± standard deviation. **P<0.01; *P<0.05, N=3.
Abbreviations: h, hours; NC, cells transfected with mimics NC and pcDNA3.1.
Flow cytometry assay was used to evaluate whether miR-1247 regulates cell cycle through targeting of STMN1. The cell cycle was profoundly arrested at the G1/S phase after miR-1247 overexpression, an effect that was dramatically reversed by STMN1 overexpression in A549 and H1299 cells. STMN1 overexpression accelerated the cell cycle at G1/S phase (Figure 5C). These results suggested that miR-1247 rescued the STMN1-induced promotion of migration and invasion in A549 and H1299 cells by inhibiting the cell cycle by G1/S phase arrest.
Discussion
An increasing number of studies have been proved that aberrant expression of miRNAs is related to the progression of various cancers.25–28 Deng et al found that miR-1247 was downregulated in lung squamous cell carcinoma tissue samples compared with normal tissues by a chip detection assay.14 Moreover, a functional role of miR-1247 has been found in a number of cancers, including prostate cancer,12 osteoblasts from osteosarcoma,11 and pancreatic cancer.28 In addition, dysregulation of miR-1247 was associated with cancer invasion. However, miR-1247 involvement in NSCLC has not been studied. In this report, miR-1247 levels have for the first time been confirmed to be lower in human NSCLC tissues and cell lines than in normal controls. The upregulation of miR-1247 significantly promoted cell apoptosis by blocking the cell cycle at the G1/S phase. In addition, it was found that miR-1247 inhibited cell invasion and migration by targeting STMN1 in NSCLC cell lines.
The effects of miRNAs on cancer progression also depend on the function of the downstream targets that they suppress.29 Therefore, the targets of each miRNA need to be identified. A previous study had reported that STMN1 was a potential prognostic marker for patients with NSCLC, with overexpression of STMN1 shown to promote cell proliferation and invasion in NSCLC.15 Overexpression of STMN1 has been reported in patients with NSCLC resistant to paclitaxel treatment.30 In this research, STMN1 was predicted as a target gene of miR-1247 with bioinformatics methods. Moreover, the data showed that miR-1247 upregulation significantly decreased the dual luciferase activity of the 3′-UTR sequence of STMN1. Overexpression of miR-1247 was associated with a decrease in STMN1 expression in NSCLC cells. These results confirmed that STMN1 is a target gene of miR-1247.
Most importantly, the study showed that miR-1247 can rescue the effect on NSCLC cells of STMN1 on the acceleration of migration and invasion. These results indicate that miR-1247 suppressed cancer cell progression by targeting STMN1 in NSCLC cells.
DNA methylation is the main epigenetic event in human. The expression levels of miRNA and genes are frequently silenced by DNA methylation, which plays an important role in tumorigenesis.31–33 The hypermethylation of miR-1247 was first found to be associated with EMT and can be a prognostic indicator in ulcerative colitis. Is miR-1247 methylated in lung cancer? Does the level of miR-1247 methylation affect its expression? These questions are still unknown. Therefore, the expression of miR-1247 and STMN1 in normal and NSCLC cells treated with 5-aza deoxycytidine 5-Aza was tested. And it was confirmed that miR-1247 levels were significantly increased in NSCLC cells that were exposed to 5-Aza. On the contrary, STMN1 was dramatically silenced in NSCLC cells under such demethylation treatment. These results demonstrate that the expressions of miR-1247 and STMN1 are regulated by DNA methylation. Furthermore, inhibiting methylation enhanced miR-1247 expression in NSCLC cells, accounting for dramatic inhibitory effects on cell migration and invasion. Based on these findings, the methylation status of miR-1247 may serve as a potential biomarker in NSCLC. This study did not investigate the mechanism by which miR-1247 affects NSCLC cell proliferation, but the observed effects on cell motility and invasion might be influenced to some extent by changes in cell proliferation.
Conclusion
MiR-1247 was silenced by DNA methylation and negatively associated with STMN1 expression in vitro. Moreover, miR-1247 promoted cell apoptosis and restrained cell migration and invasion ability by targeting STMN1 in NSCLC cells. In addition, these effects of miR-1247 may indicate that STMN1 is a potential target gene for diagnosis and treatment of NSCLC.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Gansler T, Ganz PA, Grant M, et al. Sixty years of CA: a cancer journal for clinicians. CA Cancer J Clin. 2010;60(6):345–350. doi: 10.3322/caac.20088. [DOI] [PubMed] [Google Scholar]
- 2.Anglim PP, Alonzo TA, Laird-Offringa IA. DNA methylation-based biomarkers for early detection of non-small cell lung cancer: an update. Mol Cancer. 2008;7:81. doi: 10.1186/1476-4598-7-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verdecchia A, Francisci S, Brenner H, et al. Recent cancer survival in Europe: a 2000–02 period analysis of EUROCARE-4 data. Lancet Oncol. 2007;8(9):784–796. doi: 10.1016/S1470-2045(07)70246-2. [DOI] [PubMed] [Google Scholar]
- 4.Li S, Ma Y, Hou X, et al. MiR-185 acts as a tumor suppressor by targeting AKT1 in non-small cell lung cancer cells. Int J Clin Exp Pathol. 2015;8(9):11854–11862. [PMC free article] [PubMed] [Google Scholar]
- 5.Highlights in NSCLC from the 2014 American Society of Clinical Oncology annual meeting. Clin Adv Hematol Oncol. 2014;12(10 Suppl 18):7–16. [PubMed] [Google Scholar]
- 6.Xiao P, Liu WL. MiR-142-3p functions as a potential tumor suppressor directly targeting HMGB1 in non-small-cell lung carcinoma. Int J Clin Exp Pathol. 2015;8(9):10800–10807. [PMC free article] [PubMed] [Google Scholar]
- 7.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meltzer PS. Cancer genomics: small RNAs with big impacts. Nature. 2005;435(7043):745–746. doi: 10.1038/435745a. [DOI] [PubMed] [Google Scholar]
- 9.Fang K, Qian F, Chen YQ. MicroRNAs as regulators in normal hematopoietic and leukemia stem cells: current concepts and clinical implications. Curr Mol Med. 2012;12(5):536–546. doi: 10.2174/156652412800620002. [DOI] [PubMed] [Google Scholar]
- 10.Shi S, Lu Y, Qin Y, et al. miR-1247 is correlated with prognosis of pancreatic cancer and inhibits cell proliferation by targeting neuropilins. Curr Mol Med. 2014;14(3):316–327. doi: 10.2174/1566524014666140228120014. [DOI] [PubMed] [Google Scholar]
- 11.Zhao F, Lv J, Gan H, et al. MiRNA profile of osteosarcoma with CD117 and stro-1 expression: miR-1247 functions as an onco-miRNA by targeting MAP3K9. Int J Clin Exp Pathol. 2015;8(2):1451–1458. [PMC free article] [PubMed] [Google Scholar]
- 12.Scaravilli M, Porkka KP, Brofeldt A, et al. MiR-1247-5p is overexpressed in castration resistant prostate cancer and targets MYCBP2. Prostate. 2015;75(8):798–805. doi: 10.1002/pros.22961. [DOI] [PubMed] [Google Scholar]
- 13.Martinez-Sanchez A, Murphy CL. miR-1247 functions by targeting cartilage transcription factor SOX9. J Biol Chem. 2013;288(43):30802–30814. doi: 10.1074/jbc.M113.496729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng B, Sun Z, Jason W, Yang P. Increased BCAR1 predicts poor outcomes of non-small cell lung cancer in multiple-center patients. Ann Surg Oncol. 2013;20(Suppl 3):S701–S708. doi: 10.1245/s10434-013-3184-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nie W, Xu MD, Gan L, Huang H, Xiu Q, Li B. Overexpression of stathmin 1 is a poor prognostic biomarker in non-small cell lung cancer. Lab Invest. 2015;95(1):56–64. doi: 10.1038/labinvest.2014.124. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, Yao Y, Ming Y, et al. Downregulation of stathmin 1 in human gallbladder carcinoma inhibits tumor growth in vitro and in vivo. Sci Rep. 2016;6:28833. doi: 10.1038/srep28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang HQ, Guo X, Guo SQ, et al. STMN1 in colon cancer: expression and prognosis in Chinese patients. Eur Rev Med Pharmacol Sci. 2016;20(10):2038–2044. [PubMed] [Google Scholar]
- 18.Singer S, Malz M, Herpel E, et al. Coordinated expression of stathmin family members by far upstream sequence element-binding protein-1 increases motility in non-small cell lung cancer. Cancer Res. 2009;69(6):2234–2243. doi: 10.1158/0008-5472.CAN-08-3338. [DOI] [PubMed] [Google Scholar]
- 19.He X, Liao Y, Lu W, et al. Elevated STMN1 promotes tumor growth and invasion in endometrial carcinoma. Tumour Biol. 2016;37(7):9951–9958. doi: 10.1007/s13277-016-4869-5. [DOI] [PubMed] [Google Scholar]
- 20.Lv LV, Zhou J, Lin C, et al. DNA methylation is involved in the aberrant expression of miR-133b in colorectal cancer cells. Oncol Lett. 2015;10(2):907–912. doi: 10.3892/ol.2015.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Y, Huang JQ, Zhang X, Shen LF. MiR-129-2 functions as a tumor suppressor in glioma cells by targeting HMGB1 and is down-regulated by DNA methylation. Mol Cell Biochem. 2015;404(1–2):229–239. doi: 10.1007/s11010-015-2382-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian Y, Wei W, Li L, Yang R. Down-regulation of miR-148a promotes metastasis by DNA methylation and is associated with prognosis of skin cancer by targeting TGIF2. Med Sci Monit. 2015;21:3798–3805. doi: 10.12659/MSM.894826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li N, Zhang F, Li S, Zhou S. Epigenetic silencing of microRNA-503 regulates FANCA expression in non-small cell lung cancer cell. Biochem Biophys Res Commun. 2014;444(4):611–616. doi: 10.1016/j.bbrc.2014.01.103. [DOI] [PubMed] [Google Scholar]
- 24.Tahara T, Shibata T, Okubo M, et al. DNA methylation status of epithelial-mesenchymal transition (EMT) – related genes is associated with severe clinical phenotypes in ulcerative colitis (UC) PLoS One. 2014;9(10):e107947. doi: 10.1371/journal.pone.0107947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia Y, Zhu Y, Ma T, et al. miR-204 functions as a tumor suppressor by regulating SIX1 in NSCLC. FEBS Lett. 2014;588(20):3703–3712. doi: 10.1016/j.febslet.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 26.Jiang J, Lv X, Fan L, et al. MicroRNA-27b suppresses growth and invasion of NSCLC cells by targeting Sp1. Tumour Biol. 2014;35(10):10019–10023. doi: 10.1007/s13277-014-2294-1. [DOI] [PubMed] [Google Scholar]
- 27.Xia H, Sun S, Wang B, et al. miR-143 inhibits NSCLC cell growth and metastasis by targeting Limk1. Int J Mol Sci. 2014;15(7):11973–11983. doi: 10.3390/ijms150711973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng H, Zhang F, Lin X, et al. MicroRNA-1225-5p inhibits proliferation and metastasis of gastric carcinoma through repressing insulin receptor substrate-1 and activation of beta-catenin signaling. Oncotarget. 2016;7(4):4647–4663. doi: 10.18632/oncotarget.6615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qin J, Ke J, Xu J, et al. Downregulation of microRNA-132 by DNA hypermethylation is associated with cell invasion in colorectal cancer. Onco Targets Ther. 2015;8:3639–3648. doi: 10.2147/OTT.S91560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun R, Liu Z, Wang L, et al. Overexpression of stathmin is resistant to paclitaxel treatment in patients with non-small cell lung cancer. Tumour Biol. 2015;36(9):7195–7204. doi: 10.1007/s13277-015-3361-y. [DOI] [PubMed] [Google Scholar]
- 31.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genetics. 2002;3(6):415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 32.Tost J. DNA methylation: an introduction to the biology and the disease-associated changes of a promising biomarker. Mol Biotechnol. 2010;44(1):71–81. doi: 10.1007/s12033-009-9216-2. [DOI] [PubMed] [Google Scholar]
- 33.Lyko F, Brown R. DNA methyltransferase inhibitors and the development of epigenetic cancer therapies. J Natl Cancer Inst. 2005;97(20):1498–1506. doi: 10.1093/jnci/dji311. [DOI] [PubMed] [Google Scholar]