Abstract
Errors in the binary status of some response traits are frequent in human, animal, and plant applications. These error rates tend to differ between cases and controls because diagnostic and screening tests have different sensitivity and specificity. This increases the inaccuracies of classifying individuals into correct groups, giving rise to both false-positive and false-negative cases. The analysis of these noisy binary responses due to misclassification will undoubtedly reduce the statistical power of genome-wide association studies (GWAS). A threshold model that accommodates varying diagnostic errors between cases and controls was investigated. A simulation study was carried out where several binary data sets (case–control) were generated with varying effects for the most influential single nucleotide polymorphisms (SNPs) and different diagnostic error rate for cases and controls. Each simulated data set consisted of 2000 individuals. Ignoring misclassification resulted in biased estimates of true influential SNP effects and inflated estimates for true noninfluential markers. A substantial reduction in bias and increase in accuracy ranging from 12% to 32% was observed when the misclassification procedure was invoked. In fact, the majority of influential SNPs that were not identified using the noisy data were captured using the proposed method. Additionally, truly misclassified binary records were identified with high probability using the proposed method. The superiority of the proposed method was maintained across different simulation parameters (misclassification rates and odds ratios) attesting to its robustness.
Keywords: binary responses, misclassification, specificity, sensitivity
Introduction
It is well established that misclassification of the dependent variables adversely affects the detection power of genome-wide association studies (GWAS) and could lead to biased results.1,2 Classifying individuals into different disease classes has proven to be erroneous as binary responses are subjective measurements with no precise or quantifiable guidelines. Consequently, the outcomes from implementing GWAS using case–control studies can be misleading if the observations are inaccurate. Screening and diagnostic tests are used to identify unrecognized diseases or defects and have shown to exhibit potential for bias.3 These testing activities are used to characterize and sort individuals into two groups (eg, high/low risk) or classify them into different subclasses of the same disease or disorder. This screening process typically relies heavily on human perception; therefore, false-positive and false-negative cases are unavoidable.
In disease diagnosis, the quality of a test is often measured by its sensitivity and specificity.4 Thus, a test with low sensitivity/specificity will lead to a high false-negative/positive result. Several reviews have been published in order to assess the variation among studies and to evaluate test performances.5–7 Deeks8 pooled together estimates for sensitivity and specificity and found the average sensitivity to be 0.96. The average specificity was 0.61 exhibiting considerable variation around the mean ranging between 0.21 and 0.88. Such inaccuracy of screening tests will lead to high misdiagnostic rates in disease classification across both clinical practices and perceptual specialties.
In radiology, although false positives are of low frequency (1.5%–2%), false negatives are in excess of 25%.9 False-negative rates in cancer detection have been documented as one of the most difficult limitations.10 Published false-negative rates have ranged between 10% and 25% for breast cancer detection.11,12 Using 282 samples for breast cancer based on the sentinel lymph node biopsy, Goyal et al13 found 19 false-negative cases. Stock et al14 evaluated cervical cancer screening tests and found false-positive estimates ranging between 0.056 and 0.269. Croswell et al15 concluded that using 14 tests for cancer screening, the cumulative risk of a false positive was 60.4% and 48.8% for men and women, respectively.
False-positive and -negative rates are also prevalent in psychological disorders as it is often difficult for clinicians to distinguish between disorders due to overlapping or late development of symptoms. In the case of Alzheimer’s disease (AD), symptoms are more pronounced during later stages; therefore, diagnosis of incipient AD patients is more difficult. Two cognitive tests are generally administered for diagnosis, neurofibrillary tangles (NFTs) and the Mini-Mental State Exam. Reviews of NFT have questioned its validity as an accurate test for AD.16–18
Unfortunately, finding these errors is not simple. Even in the best-case scenario, when misclassification is suspected before analysis, retesting is often not possible and the sample must be removed, thereby reducing power of the study. Extensive research has been carried out to investigate the consequences of misclassification on the well-being of the patient19,20 as well as its effects on the accuracy of the results of studies including GWAS. GWAS aim to statistically associate genetic variants with disease status; therefore, it relies on the accuracy of both the genotypic and phenotypic data. Implementing association studies without proper data quality control measures can lead to the discovery of false associations between markers and disease. This false discovery could lead to different assessment and potentially contradictory conclusion. Using candidate gene approach, Hirschhorn et al21 concluded that out of 600 gene–disease associations reported in the literature, only 1% of these associations are likely to be true. Heterogeneity, population stratification, and noisy dependent variables were often suspected as potential explanation for the lack of replicability of GWAS results.22–25
Studies examining the effects of uncertainty found that it can lead to biased parameter estimates.26,27 A statistical approach capable of eliminating or at least attenuating the negative effects of misclassification represents an attractive solution. The Bayesian approach proposed by Rekaya et al28 made the analysis of noisy binary responses more tractable. They found, using simulated binary data with a 5.6% misclassification rate, that ignoring misclassification resulted in biased parameter estimates, with the true values falling outside the 95% high-density posterior interval. Robbins et al29 concluded that prediction power could be increased by 25% while accounting for misclassification.
Smith et al2 investigated the effects of misclassification in binary responses on GWAS results assuming the same misdiagnostic rate for cases and controls. In this study, such idea has been extended to situations where misclassification occurs with different rates for cases and controls, thus mimicking more realistic disease diagnostic scenarios. For that purpose, case–control data sets were simulated and misclassification was introduced by randomly switching the true binary status to reach the desired error rate in 5% or 7% and 0% or 3% for cases and controls, respectively. True data sets were analyzed with a standard model (M1), and noisy data sets were analyzed with threshold models either ignoring (M2) or contemplating (M3) misclassification.
Materials and methods
The methodology first presented by Rekaya et al28 and later extended and applied by Smith et al2 was adopted in this study to analyze binary data subject to misclassification where the probability of miscoding is different between cases and controls. In the presence of misclassification, the vector of observed binary responses y = (y1, y′, …yn)′ measured on n individuals (eg, clinical diagnosis for a disease) is considered a “contaminated” sample of a real unobserved responses vector r = (r1, r2, …, rn)′. The contamination could be due to several reasons including less than perfect sensitivity and specificity of a test or misdiagnosis by a clinician. Additionally, the n individuals are assumed to be genotyped for a set of single nucleotide polymorphisms (SNPs). Assessing the association between the genotyped SNPs and the trait (eg, disease status) is challenging because only the noisy data are observed. It gets even more complex when misclassification occurs with different rates for cases (false-negative rate) and controls (false-positive rate) as it is likely to be the situation with real data sets. Contrary to a common misclassification rate for both cases and controls assumed by Rekaya et al28 and Smith et al,2 specific misclassification rates for each outcome were adopted in this study, and to the best of our knowledge, this is the first time such distinction was assumed. Assuming misclassification happens with probability π1 (probability of false negatives) and π2 (probability of false positives) for cases and controls, respectively, the conditional joint distribution of the observed noisy data is:
with qi = [(1−π1)]pi + π2(1−pi)] and pi is the probability of the Bernoulli process generating the true unobserved binary response ri.
Note that when there is no misclassification (π1 = π2 = 0), then as expected, qi is equal to qi. In our case, the probability pi was assumed to be a function of the SNP effects (β). Assuming that the true unobserved data, r, is conditionally independent given β:
where pi (β) indicates that pi is a function of β (vector of SNP effects).
Let be a vector of indicator variables for the n1 case observations, where αi = 1 if is switched from case (e.g. sick) to control (e.g. healthy) and αi = 0 otherwise. Similarly, let be a vector of indicator variables for the n2 control observations, where λi = 1 if ri is switched from control to case (from zero to one) and λi = 0 otherwise. Furthermore, each αi and λi was assumed to be a Bernoulli trial with probability π1 and π2, respectively.
Given β, π1, and π2, the true data (r), α, λ and are jointly distributed as:
where n1 and n2 are the number of cases and controls, respectively. A in the previous equation, the first term in the right hand side is the likelihood of the true data. Unfortunately, the true data r is not observed. However, based on the assumed misclassification process, the relationship between y (noisy data) and r (unobserved true data) could be easily established as:
| (1) |
Notice that when ai(λi) = 0 (no misclassification), the equations in (1) reduce to ri = yi.
Using the equalities in Equation (1), the likelihood of the true data could be expressed as a function of the observed noisy data y, α, and λ. Thus, the joint distribution of the observed data (y), α, and λ is easily obtained as:
| (2) |
Finally, prior distribution was specified for all unknown parameters
| (3) |
where βmin, βmax, a1, b1, a2, and b2 are known hyper-parameters.
The joint posterior distribution of all unknown parameters is easily obtained as the product of Equations 2 and 3.
| (4) |
Following Rekaya et al28 and Smith et al,2 a data augmentation algorithm was used to implement the model in (4). A liability threshold model was used with the following relationship between the binary response and a non-observed continuous random variable, li:
with T being a subjectively specified threshold value.
At the liability scale, the model can be presented as:
| (5) |
where li is the liability for individual i, xij is the genotype for marker j, μ is an overall mean, βj is the effect of marker j and ei is a white noise. For identifiability reasons, the residual variance, var (ei), and the threshold, T, were set arbitrarily to 1 and zero, respectively.
Full conditional distributions needed for implementation using Gibbs sampler are normal for μ and β and binomial for each elements of the vectors a and λ
where a−i and λ−i are the indicator vectors for the cases and controls without the position i.
For the misclassification probabilities, their conditional distributions are proportional to:
Thus, π1 and π2 are distributed as Beta (a1 + Σai, n1 + b1 − Σai) and Beta (a1 + Σai, n1 + b1 − Σai) with Σai and Σλi being the total number of misclassified (switched) cases and control observations, respectively. It is worth mentioning that because the number of true cases and controls was unknown, n1 and n2 were set equal to the number of observed cases and controls in the first round of the iterative process and then updated to the estimated number of cases and controls thereafter.
Simulation
Typical case–control type data sets were simulated using PLINK software.30 Each data set consisted of 2000 individuals (1000 cases and 1000 controls) genotyped for 1000 common SNPs (minor allele frequency >0.05). Randomly, 15% of the SNPs were assumed to be in association with a binary response trait and the remaining 850 SNPs were considered noninfluential. The odds ratios (ORs) for the influential 150 SNPs were assigned based on the following two scenarios. A moderate scenario where 25, 35, and 90 markers of the 150 influential SNPs were assumed to have ORs of 1:4, 1:2, and 1:1.8, respectively. An extreme scenario where ORs of 1:10, 1:4, and 1:2 were specified for 25, 35, and 90 markers of the 150 influential SNPs, respectively. For each individual, a liability (quantitative phenotype) was generated as the sum of the effect of the disease SNPs and random white noise. Binary status for the simulated disease traits was assigned based on a median split of the continuous phenotype. Misclassification was artificially introduced by switching the true binary status. Randomly 5% or 7% of the cases and 0% or 3% of the controls were miscoded. To some extent, the simulated binary data mimic a clinical data generated by a test with a sensitivity of 0.95 or 0.93 and a specificity of 1 or 0.97. Furthermore, different levels of genetic complexity of the simulated response were assumed through the OR of the influential SNPs.
For two levels of miscoding for cases and controls (5% and 0% or 7% and 3%) and two OR distribution (moderate OR and extreme OR), the following data sets were simulated: 5% and 0% miscoding rates and moderate OR (D1) or extreme OR (D2); 7 and 3% miscoding rates and moderate OR (D3) or extreme OR (D4). Five replicates were simulated for each data set.
Results and discussion
To evaluate the capability of the method to identify miscoded and correctly classified observations, the posterior means (averaged over five replicates) of the true misclassification probabilities for both cases and controls were calculated. Except for scenarios where misclassification was set at 0%, misclassification probabilities were slightly underestimated but still fell within their respective 95% highest posterior density interval (Table 1). For example, when moderate ORs of the influential SNPs were used, posterior means were 0% and 4%, and 5 and 2% for D1 and D3, respectively. However, as the OR was increased for the extreme cases, these means increased to 0% and 5% (D2) and 6% and 2% (D4). Although our algorithm was designed to anticipate and account for potential misclassification, a null data set was run with no coding errors to ensure its ability to indicate no misdiagnostic errors. As expected, this analysis resulted in misclassification probabilities close to zero, with estimates of 0.001 and 0.002.
Table 1.
Summary of the posterior distribution of the misclassification probability (π) for the four simulation scenarios (averaged over five replicates)
| True | Moderate*
|
Extreme
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| PM | PSD | PM | PSD | ||||||
|
| |||||||||
| π1 | π2 | π1 | π2 | π1 | π2 | π1 | π2 | π1 | π2 |
| 5% | 0% | 0.04 | 0.002 | 0.006 | 0.0003 | 0.05 | 0.002 | 0.006 | 0.0003 |
| 7% | 3% | 0.05 | 0.02 | 0.008 | 0.004 | 0.06 | 0.02 | 0.007 | 0.004 |
Note:
Moderate effects for influential single nucleotide polymorphisms.
Abbreviations: PM, posterior mean; PSD, posterior standard deviation.
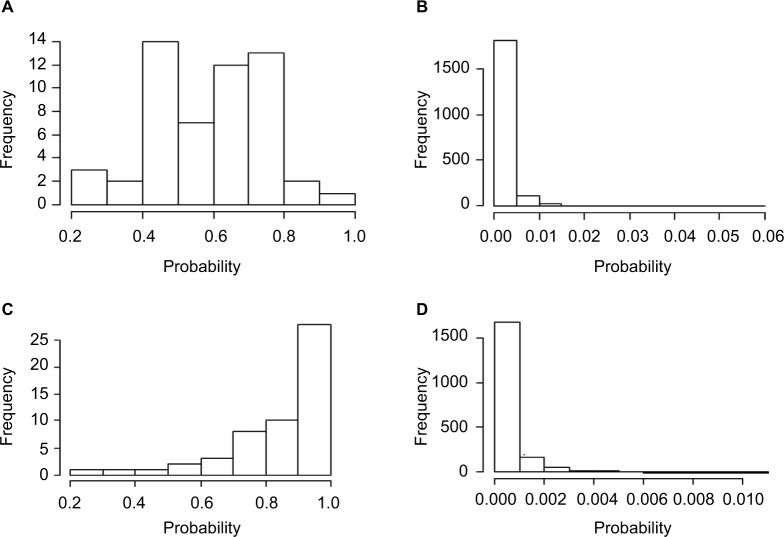
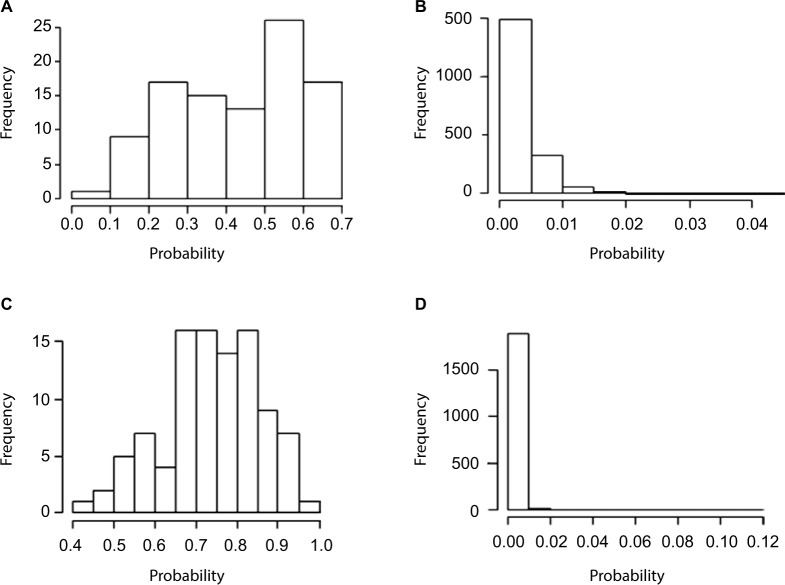
Adequate sample size is one of the major contributing factors to obtain sufficient power of GWAS. Thus, it would be beneficial to identify and correct misclassified samples rather than removing them from the study. Therefore, to continue evaluating the effectiveness of the proposed method to detect miscoded individuals, the posterior probability of an observation being misclassified was calculated (averaged over five replicates) in all four scenarios. With moderate OR and misclassification rates set to 5% for cases and 0% for controls, the 54 miscoded observations exhibited higher misclassification probability with an average of 0.58 (Figure 1A) compared to an average of 0.002 for the 1946 observations of the correctly coded group (Figure 1B). As the odds are increased for the extreme scenario (D2), the distinction became more evident. In fact, the average posterior misclassification probability of the 54 miscoded observations increased to 0.85 (Figure 1C) compared to 0.006 for the correctly coded group (Figure 1D). This is of importance as it shows our method is able to detect miscoded samples with higher probability compared to correctly coded observations. In fact, the smallest misclassification probability of the miscoded observations was 0.28 (Figure 1A) which was substantially higher than 0.06 (Figure 1B), the largest probability observed for the correctly coded group (D1). Similar estimates were obtained when misclassification rates increased to 7% for cases and 3% for controls (Figure 2). For D3 (D4), the average posterior misclassification probability was 0.43 (0.74) and 0.003 (0.002), for the miscoded (Figure 2A and C) and correctly coded (Figure 2B and D) groups, respectively.
Figure 1.
Average posterior misclassification probability for the 54 miscoded observations (A: moderate and C: extreme) and the 1946 correctly coded observations (B: moderate and D: extreme) when the misclassification rates were set to 5% and 0%.
Figure 2.
Average posterior misclassification probability for the 98 miscoded observations (A: moderate and C: extreme) and the 1902 correctly coded observations (B: moderate and D: extreme) when the misclassification rates were set to 7% and 3%.
Outside of a controlled study, there is no indication for which individuals are misdiagnosed. Thus, it is useful to evaluate the performance of the method when a subjective or heuristic criteria are used to declare misclassified samples. The results of using two cutoff values for the probability of misclassification to declare an observation as misclassified are presented in Table 2 (averaged over five replicates). Using our proposed method with a hard cutoff (p=0.5), 65 (D1) and 94% (D2) of the 54 truly miscoded samples were correctly identified. When the rate of misclassification increased to 7% for cases and 3% for controls, of the 98 miscoded observations 44 (D3; moderate OR) and 97% (D4; extreme OR) were correctly detected. Despite the rigidness of the hard cutoff approach (little variability around the designated probability), our procedure was still efficient in identifying considerable amount of misclassified observations. Once the restrictions of the cutoff probability were relaxed (cutoff value was set equal to the average of all samples misclassification probability plus two standard deviations), ~100% of the miscoded samples were identified across all scenarios except for D3 where 86% were detected. Across both cutoff probabilities for the two scenarios where the overall misclassification rate was 10%, there was a higher detection in cases than controls. This is potentially the result of higher misclassification rate in cases compared to controls; 7% versus 3%. Using real clinical data, it will be recommended to use both the classification criteria to assess the misclassification status of a sample. Additionally, other clinical information (eg, medical history) could be helpful in some cases.
Table 2.
Percent of misclassified individuals correctly identified on the basis of two cutoff probabilities across the four simulation scenarios
| Cutoff probability | D1
|
D2
|
D3
|
D4
|
||||
|---|---|---|---|---|---|---|---|---|
| Misclass | Correct | Misclass | Correct | Misclass | Correct | Misclass | Correct | |
| Hard | 0.65 | 0 | 0.94 | 0 | 0.44 | 0 | 0.97 | 0 |
| Soft | 1.00 | 0 | 0.98 | 0 | 0.86 | 0 | 1.00 | 0 |
Notes: Hard: cutoff probability was set at 0.5. Soft: cutoff probability was equal to the overall mean of the probabilities of being misclassified over the entire data set plus two standard deviations. Misclass: individuals who were misclassified. Correct: correctly coded individuals. The following data sets were simulated: 5% and 0% miscoding rates and moderate OR (D1) or extreme OR (D2); 7 and 3% miscoding rates and moderate OR (D3) or extreme OR (D4).
Abbreviation: OR, odds ratio.
In GWAS, the association between thousands of genetic variants and a phenotype is evaluated in hope of elucidating the biology of complex traits. In this instance, there is a need for unbiased and accurate identification of relevant polymorphisms. In order to assess the consequences of the presence of misclassified samples on estimating effects, the correlation between estimates of SNP effects obtained using the true (M1) and the miscoded data (M2 and M3) were calculated. For all four scenarios, the proposed approach (M3) was capable of increasing the correlation compared to the “contaminated” data (M2; Table 3). For example, for scenarios when OR of the influential SNPs were moderate, accuracies increased by 8% for D1 and 12% for D3. As the OR increased for the extreme scenarios, the same trend was observed but correlations increased by a more substantial amount. When misclassification rates were 5% and 0%, correlation increased by 0.134 and 0.217 for D2 and D4, respectively (Table 3). This indicates the ability of the method to produce consistent results and to decrease potential misclassification bias on the estimation of SNP effects. This result is important for the dissection of the genetic basis of complex traits using potentially noisy clinical data. This is the case because even without knowing the misclassification rate or the misclassified observations, the proposed method was able to enhance the signal of truly influential SNPs.
Table 3.
Correlation between true* and estimated SNP effects under four simulation scenarios using noise data analyzed with threshold models either ignoring (M2) or contemplating (M3) misclassification
| Model | 5% and 0%
|
7% and 3%
|
||
|---|---|---|---|---|
| Moderate** | Extreme | Moderate | Extreme | |
| M2 | 0.894 | 0.777 | 0.807 | 0.675 |
| M3 | 0.969 | 0.911 | 0.907 | 0.892 |
Notes:
True effects were calculated based on analysis of the true data (M1).
Moderate effects for influential SNPs. M1: true data analyzed with a standard model. M2: noisy data analyzed with threshold model ignoring misclassification. M3: noisy data analyzed with threshold model contemplating misclassification (proposed method).
Abbreviation: SNP, single nucleotide polymorphism.
The effect sizes of SNPs with true association to the phenotype should be larger in magnitude compared to non-causal SNPs. The ranking of the SNPs was observed by monitoring the most influential top 10%, and in the presence of misclassified observations (M2), the noninfluential SNPs tended to have non-zero estimates. Using scenario D4 (ignoring misclassification), eight out of the 15 most influential SNPs were not accounted for. After correction, our method (M3) was able to capture 11 out of the 15 SNPs resulting in an increase of 20% in the power of association. Even in the modest case, when misclassification rates were set at 5% for cases and 0% for controls with moderate OR of the disease associated SNPs, M2 caused a loss of 20% in power but our method reduced it to 7%. The inability to identify large portion of the most influential SNPs in the presence of misclassification will undoubtedly have negative effects on GWAS studies. In fact, it will reduce the efficiency of genomic classifiers used in diagnostics and prediction, and it will hamper the ability to identify causal genes.
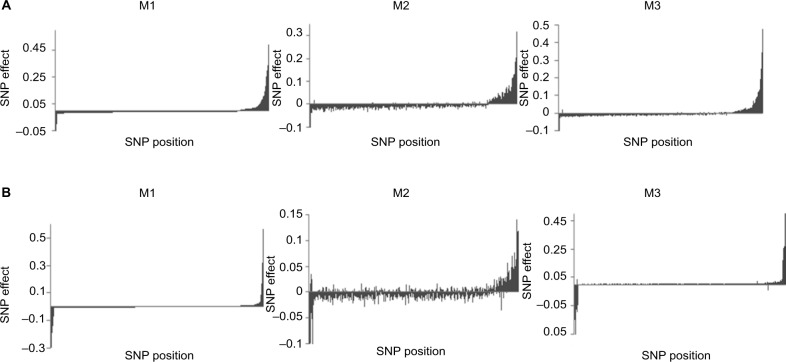
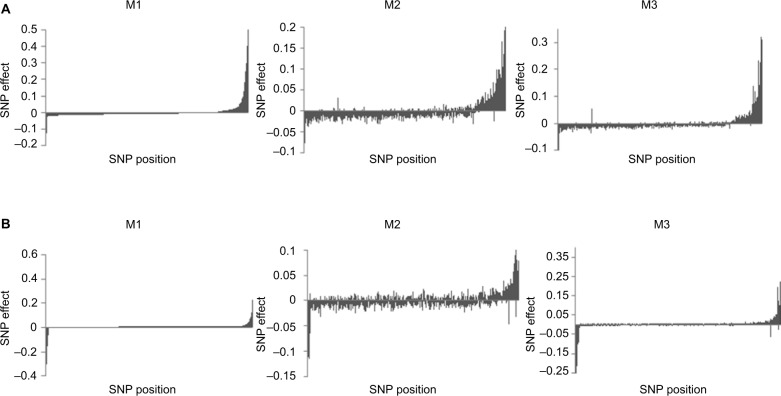
As previously mentioned, a change in rankings of the SNPs was noticed; hence, errors in estimation due to data misclassification were further investigated by examining the magnitude of the SNP effects. Based on their estimates when no misclassification was present (M1), SNP effects were ordered in decreasing order. For scenarios D1 (Figure 3A) and D2 (Figure 3B), it is evident that M2 was not able to capture the true magnitude and direction of the SNP effects when compared to our proposed method (M3). This distinction became more evident when we increased the misclassification rates to 7% for cases and 3% for controls (Figure 4). In fact, imprecise phenotyping leading to reduced estimates of effect sizes is reported as one of the limitations of GWAS.31 Accumulation of erroneous estimates from selection of nonsignificant SNPs leads to biased estimates of genetic parameters, including the variance explained by SNPs, true genetic correlations between disorders, and lower estimates of heritability.32–34 The negative effects of misclassification are expected to increase with the genetic complexity of the trait due to the increase in risk variants.35
Figure 3.
Distribution of SNP effects for 5% and 0% misclassification rates. The effects are sorted in decreasing order based on estimates using M1 when odds ratios of influential SNPs are moderate (A) and extreme (B). M1: true data analyzed with a standard model. M2: noisy data analyzed with threshold model ignoring misclassification. M3: noisy data analyzed with threshold model contemplating misclassification (proposed method).
Abbreviation: SNP, single nucleotide polymorphism.
Figure 4.
Distribution of SNP effects for 7% and 3% misclassification rates. The effects are sorted in decreasing order based on estimates using M1 when odds ratios of influential SNPs are moderate (A) and extreme (B). M1: true data analyzed with a standard model. M2: noisy data analyzed with threshold model ignoring misclassification. M3: noisy data analyzed with threshold model contemplating misclassification (proposed method).
Abbreviation: SNP, single nucleotide polymorphism.
Conclusion
High false-positive and false-negative rates of discrete responses are unavoidable for some disease traits, and correcting misclassified observations is difficult, time-consuming, and often costly to remedy. Ignoring these errors increases the uncertainty of identifying relevant associations, thus decreasing the accuracy in estimating the magnitude and direction of variant effects. This in turn will lead to an increase of false-positive results as noninfluential SNPs will tend to have inflated estimates. The proposed method was able to identify with high probability miscoded samples in both cases and controls. Cases tended to have higher probabilities than controls in part due to having a higher prevalence of being misclassified.
Our proposed method increased the accuracy of estimated SNP effects in the presence of “noisy” data which will aid in decreasing the rate of non-replicative results. Furthermore, it will reduce the false association between genetic variants and the disease of interest. It will lead to an increase in predictive power and a reduction in bias caused by classification errors. Our procedure performed well even when one of the misclassification rates was set to zero which is important when diagnostic procedures have either a high sensitivity or a high specificity. Based on the results of this simulation study, it seems reasonable to conclude that the proposed method will be effective in reducing or eliminating the negative effects of misclassification in association with the analyses of binary responses subject to outcome-specific error rates. Although the results of this studies are based on simulated OR values that are relatively high even in the moderate scenario, preliminary results from an
Acknowledgments
This study was partially supported by funding from Merial, INC. Coauthor SS was supported through an assistantship provided by the University of Georgia Graduate School.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Manchia M, Cullis J, Turecki G, Rouleau GA, Uher R, Alda M. The impact of phenotypic and genetic heterogeneity on results of genome wide association studies of complex diseases. PLoS One. 2013;8(10):e76295. doi: 10.1371/journal.pone.0076295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith S, Hay EH, Farhat N, Rekaya R. Genome wide association in the presence of misclassified binary responses. BMC Genet. 2013;14:124. doi: 10.1186/1471-2156-14-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abram E, Valesky WW. Screening and Diagnostic Tests. 2013. [Accessed December 1, 2015]. Available from: http://emedicine.medscape.com/article/773832-overview.
- 4.Bland JM, Altman DG. Diagnostic tests. 1: sensitivity and specificity. BMJ. 1994;308:1499. doi: 10.1136/bmj.308.6943.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Irwig L, Tosteson AN, Gatsonis CA, Lau J, Colditz G, Chalmers TC, Mosteller F. Guidelines for meta-analyses evaluating diagnostic tests. Ann Intern Med. 1994;120:667–676. doi: 10.7326/0003-4819-120-8-199404150-00008. [DOI] [PubMed] [Google Scholar]
- 6.Vamvakas EC. Meta-analyses of studies of diagnostic accuracy of laboratory tests: a review of concepts and methods. Arch Pathol Lab Med. 1998;122:675–686. [PubMed] [Google Scholar]
- 7.Smith-Bindman R, Kerlikowske K, Feldstein VA, et al. Endovaginal ultrasound to exclude endometrial cancer and other endometrial abnormalities. JAMA. 1998;280:1510–1517. doi: 10.1001/jama.280.17.1510. [DOI] [PubMed] [Google Scholar]
- 8.Deeks JJ. Systematic reviews of evaluations of diagnostic and screening tests. In: Egger M, Davey Smith G, Altman DG, editors. Systematic Reviews in Health Care: Meta-Analysis in Context. 2nd ed. London: BMJ Books; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Renfrew DL, Franken EA, Berbaum KS, Weigelt FH, Abu-Yousef MM. Error in radiology: classification and lessons in 182 cases presented at a problem case conference. Radiology. 1992;183:145–150. doi: 10.1148/radiology.183.1.1549661. [DOI] [PubMed] [Google Scholar]
- 10.Destounis SV, DiNitto P, Logan-Young W, Bonaccio E, Zuley ML, Willison KM. Can computer-aided detection with double reading of screening mammograms help decrease the false-negative rate? Initial experience. Radiology. 2004;232(2):578–584. doi: 10.1148/radiol.2322030034. [DOI] [PubMed] [Google Scholar]
- 11.Warren-Burhenne LJ, Wood SA, D’Orsi CJ, et al. Potential contribution of computer-aided detection to the sensitivity of screening mammography. Radiology. 2000;215:554–562. doi: 10.1148/radiology.215.2.r00ma15554. [DOI] [PubMed] [Google Scholar]
- 12.Birdwell RL, Ikeda DM, O’Shaughnessy KF, Sickles EA. Mam-mographic characteristics of 115 missed cancers later detected with screening mammography and the potential utility of computer-aided detection. Radiology. 2001;219:192–202. doi: 10.1148/radiology.219.1.r01ap16192. [DOI] [PubMed] [Google Scholar]
- 13.Goyal A, Newcombe RG, Chhabra A, Mansel RE. Factors affecting failed localisation and false-negative rates of sentinel node biopsy in breast cancer – results of the ALMANAC validation phase. Breast Cancer Res Treat. 2006;99(2):203–208. doi: 10.1007/s10549-006-9192-1. [DOI] [PubMed] [Google Scholar]
- 14.Stock EM, Stamey JD, Sankaranarayanan R, Young DM, Muwonge R, Arbyn M. Estimation of disease prevalence, true positive rate, and false positive rate of two screening tests when disease verification is applied on only screen-positives: a hierarchical model using multi-center data. Cancer Epidemiol. 2012;36(2):153–160. doi: 10.1016/j.canep.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 15.Croswell JM, Kramer BS, Kreimer AR, Prorok PC, Xu JL, Baker SG, Schoen RE. Cumulative incidence of false-positive results in repeated, multimodal cancer screening. Ann Family Med. 2009;7(3):212–222. doi: 10.1370/afm.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haroutunian V, Purohit DP, Perl DP. Neurofibrillary tangles in non-demented elderly subjects and mild Alzheimer disease. Arch Neurol. 1999;56(6):713–718. doi: 10.1001/archneur.56.6.713. [DOI] [PubMed] [Google Scholar]
- 17.Price DL, Sisodia SS. Mutant genes in familial Alzheimer’s disease and transgenic models. Ann Rev Neurosci. 1998;21:479–505. doi: 10.1146/annurev.neuro.21.1.479. [DOI] [PubMed] [Google Scholar]
- 18.Scmitt FA, Davis DG, Wekstein DR, Smith CD, Ashford JW, Markesbery WR. Preclinical AD revisited: neuropathology of cognitively normal older adults. Neurology. 2000;55:370–376. doi: 10.1212/wnl.55.3.370. [DOI] [PubMed] [Google Scholar]
- 19.Hirschfeld RM, Lewis L, Vornik LA. Perceptions and impact of bipolar disorder: how far have we really come? Results of the national depressive and manic-depressive association 2000 survey of individuals with bipolar disorder. J Clin Psych. 2003;64(2):161. [PubMed] [Google Scholar]
- 20.Bhattacharya R, Barton S, Catalan J. When good news is bad news: psychological impact of false positive diagnosis of HIV. AIDS Care. 2008;20(5):560–564. doi: 10.1080/09540120701867206. [DOI] [PubMed] [Google Scholar]
- 21.Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;2:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 22.Avery CL, Monda KL, North KE. Genetic association studies and the effect of misclassification and selection bias in putative confounders. BMC Proc. 2009;3:S48. doi: 10.1186/1753-6561-3-s7-s48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skafidas E, Testa R, Zantomio D, Chana G, Everall IP, Pantelis C. Predicting the diagnosis of autism spectrum disorder using gene pathway analysis. Mol Psychiatr. 2014;19:504–510. doi: 10.1038/mp.2012.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li A, Meyre D. Challenges in reproducibility of genetic association studies: lessons learned from the obesity field. Int J Obes (Lond) 2012;37(4):559–567. doi: 10.1038/ijo.2012.82. [DOI] [PubMed] [Google Scholar]
- 25.Wu C, DeWan A, Hoh J, Wang Z. A comparison of association methods correcting for population stratification in case-control studies. Ann Hum Genet. 2011;75(3):418–427. doi: 10.1111/j.1469-1809.2010.00639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sapp RL, Spangler Rekaya ML, Bertrand JK. A simulation study for the analysis of uncertain binary responses: application to first insemination success in beef cattle. Genet Sel Evol. 2005;37:615–634. doi: 10.1186/1297-9686-37-7-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spangler ML, Sapp RL, Rekaya R, Bertrand JK. Success at first insemination in Australian Angus cattle: analysis of uncertain binary responses. J Anim Sci. 2006;84:20–24. doi: 10.2527/2006.84120x. [DOI] [PubMed] [Google Scholar]
- 28.Rekaya R, Weigel KA, Gianola D. Threshold model for misclassified binary responses with applications to animal breeding. Biometrics. 2001;57:1123–1129. doi: 10.1111/j.0006-341x.2001.01123.x. [DOI] [PubMed] [Google Scholar]
- 29.Robbins K, Joseph S, Zhang W, Rekaya R, Bertrand JK. Classification of incipient Alzheimer patients using gene expression data: dealing with potential misdiagnosis. Online J Bioniformatics. 2006;7:22–31. [Google Scholar]
- 30.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J HumGenet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pearson TA, Manolio TA. How to interpret a genome-wide association study. J Am Med Assoc. 2008;299:1335–1344. doi: 10.1001/jama.299.11.1335. [DOI] [PubMed] [Google Scholar]
- 32.Wray NR, Lee SH, Kendler KS. Impact of diagnostic misclassification on estimation of genetic correlations using genome-wide genotypes. Eur J Human Genet. 20(6):668–674. doi: 10.1038/ejhg.2011.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet. 2011;88:294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, Nadeau JH. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010;11(6):446–450. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stringer S, Wray NR, Kahn RS, Derks EM. Underestimated effect sizes in GWAS: fundamental limitations of single SNP analysis for dichotomous phenotypes. PLoS One. 2011;6:e27964. doi: 10.1371/journal.pone.0027964. [DOI] [PMC free article] [PubMed] [Google Scholar]