Abstract
3-Hydroxy-3-methyglutaryl coenzyme A reductase (HMGCR) is a cholesterol-regulating gene with statin relevance. rs3846662 being involved in regulation of HMGCR alternative splicing, we explored its impact on HMGCR messenger RNA (mRNA) and protein levels in the brain and the associations between those levels and levels of Alzheimer’s disease pathological markers. We used brain samples derived from a cohort of 33 non-demented controls and 90 Alzheimer’s disease autopsied-confirmed cases. HMGCR mRNA levels were determined in the frontal cortex (n=114) and cerebellum (n=110) using Taqman-qPCR, and HMGCR protein levels were determined in the frontal cortex (n=117) using a commercial enzyme immunoassay. While densities of neurofibrillary tangles and senile plaques were determined in the frontal cortex (n=74), total tau, phosphorylated Tau, and beta-amyloid 1–42 levels were determined in the frontal cortex (n=94) and cerebellum (n=91) using commercial enzyme immunoassays. Despite an increase in full-length HMGCR mRNA ratio in the frontal cortex of women carrying the AA genotype, there were no associations between rs3846662 and HMGCR mRNA or protein levels. An increased Δ13 HMGCR mRNA ratio was associated with increased levels of HMGCR proteins and neurofibrillary tangles in the frontal cortex but with reduced beta-amyloid 1–42 levels in the cerebellum, suggesting a brain cell type- or a disease progression-dependent association.
Keywords: Alzheimer’s disease, HMGCR, rs3846662, Alternative splicing, Tau proteins, Beta-amyloid
Introduction
Recent evidences suggest that 3-hydroxy-3-methyglutaryl co-enzyme A reductase (HMGCR), the enzyme regulating the rate-limiting step in cholesterol biosynthesis, might act as a genetic modifier for risk, age at onset, and mild cognitive impairment (MCI) conversion to Alzheimer’s disease (AD) (Leduc et al. 2015a). More specifically, carrying a double dose of A allele at locus rs3846662 (AA or G genotype) was shown to be strongly protective against the onset and risk of AD, especially in women, and to decrease MCI conversion to AD and neurofibrillary tangle (NFT)/senile plaque (SP) accumulation in subjects also carrying the apolipoprotein ε4 risk allele (APOE4+) (Leduc et al. 2015a) (see for a review (Leduc et al. 2011a)).
The reason for the importance of this rs3846662 single nucleotide polymorphism (SNP) may lie with its involvement in regulation of HMGCR alternative splicing. Indeed, HMGCR messenger RNA (mRNA) exists in two prominent forms: full-length (FL) HMGCR and Δ13 HMGCR (Johnson et al. 2003), the latter possibly resulting in an HMGCR protein with lower or null activity (Burkhardt et al. 2008; Medina and Krauss 2009). It appears that rs3846662 is located in the binding motif of two splicing factors that are known to compete with each other to promote exon exclusion or retention (Yu et al. 2014). While the rs3846662 G allele is compatible with both motifs, the A allele disrupts the binding motif of the splicing factor promoting retention and engenders the preferential binding of heterozygous nuclear ribonucleoprotein A1 (HNRNPA1) that promotes skipping of exon 13 (Yu et al. 2014). This finding is in accordance with previous reports showing that the A allele at locus rs3846662 is associated with increased proportion of Δ13 HMGCR mRNA in vitro (Burkhardt et al. 2008; Medina et al. 2008; Simmons et al. 2011). While little is known about the effects of rs3846662 on in vivo cholesterol homeostasis in the brain, the A allele has been associated in the periphery with increased proportion of Δ13 HMGCR mRNA, in vivo, in human livers (Simmons et al. 2011); lower levels of LDL-C at baseline (Aulchenko et al. 2009; Hiura et al. 2010; Lu et al. 2010); and a lesser sensitivity and response to statin treatment, a drug inhibiting HMGCR protein (Chasman et al. 2004; Krauss et al. 2008; Medina and Krauss 2009; Chung et al. 2012). Since elevated plasma cholesterol levels (Hofman et al. 1997; Marchant et al. 2013) and treatment of hypercholesterolemia with HMGCR inhibitors (statins) in middle-aged individuals (Jick et al. 2000; Wolozin et al. 2000, 2007; Rockwood et al. 2002) appears respectively detrimental and protective against late-life development of AD, it remains unclear whether rs3846662/HMGCR affects AD risk via peripheral effects on vasculature and/or direct effects on cholesterol homeostasis in the central nervous system (CNS). However, evidence obtained in humans in recent years clearly indicates that statin utilization causes a significant reduction of phosphorylated Tau (P-Tau) proteins in the CSF, but not of total Tau (T-Tau), beta amyloid 1–40 (Aβ40), or beta-amyloid 1–42 (Aβ42) proteins (Riekse et al. 2006). Furthermore, antecedent statin use in elderly subjects with normal cognitive functions leads, over the course of several years, to a marked reduction of NFT accumulation but not of SP density, at autopsy, when compared to non-users (Li et al. 2007). Because these findings demonstrate an association between antecedent statin use and decrease P-Tau and NFT burden, we sought to examine whether lower HMGCR levels were associated with lower AD neuropathologic changes.
The current study aims to take the first steps toward exploring the impacts of rs3846662/HMGCR in the CNS affected by AD. Correlation analyses were undertaken to identify associations between HMGCR mRNA and protein levels, with brain levels of Aβ42, T-Tau, P-Tau, NFT, and SP. Since analysis of the anterior cingulate cortex using SYBR Green chemistry could not substantiate an effect of rs3846662 on HMGCR alternative splicing in the autopsied brain (Simmons et al. 2011), we used the highly specific and sensitive TaqMan chemistry to analyze the mRNA expression levels of the different HMGCR transcripts. Furthermore, as brain cell-type heterogeneity may obscure the effects of rs3846662 (Simmons et al. 2011), we determine the HMGCR mRNA levels in the frontal cortex, a region where non-neuronal cells outnumber neuronal cells by a ratio of 3.8 (Azevedo et al. 2009), and in the cerebellum, a region where neuronal cells outnumber non-neuronal cells by a ratio of 5 (Azevedo et al. 2009).
Material and Methods
Human Subjects’ Demographics
This study conformed to the Code of Ethics of the World Medical Association and was approved by the Ethics Board of the Douglas Hospital Research Centre. Every participant, or his or her legal tutor, signed an informed consent statement. Patients’ demographic characteristics are summarized in Table 1. Definite diagnosis of AD (n=90) was based on histopathological confirmation of AD according to NINCDS-ADRDA criteria (Khachaturian 1985), whereas controls (n= 33) had to be free of neurological or psychiatric diseases and of brain structural lesions (tangle and plaque indices reading <20/mm3 and <10/mm2, respectively). All brain tissues were obtained from the Douglas-Bell Canada Brain Bank, Montreal, Canada. Age at death, percent of female, percent of French Canadian descendent, and frequency of APOE4 allele are significantly different between controls and AD subjects. Except for the French Canadian status that was given for information purposes only, all other parameters were included as cofactors in the statistical analyses or accounted for by stratification (see Statistical Analysis section for more details).
Table 1.
Demographic characteristics of the studied cohort
| CTL N=33 |
AD N=90 |
|
|---|---|---|
| Age at death* | ||
| mean±SD, years | 72.6±12.0 | 78.0±8.0 |
| Age at onset | ||
| mean±SD, years | – | 69.9±9.2 |
| PMD | ||
| mean±SD, h | 30.7±21.9 | 24.1±13.0 |
| French Canadian* | ||
| No. (%) French Canadian | 23 (70) | 79 (88) |
| Sex** | ||
| No. (%) women | 12 (36) | 59 (66) |
| Apolipoprotein E4 allele*** | ||
| Frequency | 0.12 | 0.37 |
| HMGCR A allele | ||
| Frequency | 0.38 | 0.41 |
AD Alzheimer’s disease, CTL non-demented control, PMD post-mortem delay, SD standard deviation, No./N number
Statistically significant difference between the CTL and AD groups according to t test,
P≤0.05,
P≤0.01,
P≤0.001
DNA Extraction and Genotyping
DNA extraction and genotyping of HMGCR rs3846662 and APOE4 allele were performed on brain tissue samples as previously described (Leduc et al. 2015a), with DNeasy Tissue Kit (Qiagen) and pyrosequencing protocols, respectively.
RNA Extraction, Determination of HMGCR mRNA Levels, and Quality Control
RNA was extracted from cerebellum (n=119) and frontal cortex tissues (n=123) using the Maxwell® 16 Tissue LEV Total RNA Purification Kit (Promega, WI, USA) on a Maxwell® 16 LEV Instrument (Promega, WI, USA). Then, complementary DNA (cDNA) was obtained by reverse transcription on a Multigene thermocycler (Labnet International Inc.) using the High Capacity cDNA RT kit (Applied Biosystems, CA, USA) and 200 ng of total RNA. The thermocycler was set as follows: 10 min at 25 °C, 120 min at 37 °C, and 5 min at 85 °C. Finally, 20 ng of each cDNAwere applied in triplicate for real-time PCR performed on a 7500 Fast Real-Time PCR System (Applied Biosystems, CA, USA) with TaqMan® Fast Universal PCR Master Mix (Applied Biosystems, CA, USA). The following TaqMan® Gene Expression Assay Kits (Applied Biosystems, CA, USA) were used: total HMGCR, Taqman assay #Hs00168352_m1 covering exon boundary 6/7; FL HMGCR variant, custom assay covering exon boundary 13 /14 with forward primer : TGTGGCCAGCACCAATAGAG, reverse primer: CAACTGGGCCACGAGTCAT and probe : CAGAGCAATAGGTCTTGGT; Δ13 HMGCR variant, custom assay covering exon boundary 12/14 with forward primer: TCAGAACCTTCTTCTCTCCAGTACCT, reverse prime r : CACCTCCACCAAGCAAGGA , probe : CCTTACAGGGATTATAATTA; peptidylprolyl isomerase A (PPIA), Taqman assay # Hs99999904_m1; ubiquitin-conjugating enzyme E2D2 (UBE2D2), Taqman assay # Hs00366152_m1; hypoxanthine phosphoribosyltransferase 1 (HPRT1), Taqman assay # Hs99999909_m1; glyceraldehyde-3-phosphate dehydrogenase (GAPDH), Taqman assay # Hs99999905_m1; beta actin (ACTB), Taqman assay # Hs99999903_m1; and cyclin-dependent kinase inhibitor 1B (CDKN1B), Taqman assay # Hs00153277_m1. The cycling conditions were 20 s at 95 °C and 40 cycles of 3 s at 95 °C and 30 s at 60 °C. The relative gene expression of HMGCR was determined using the comparative Ct method (ΔΔCt method, Applied Biosystems User Bull. #2 Dec. 97). The efficiency of each gene expression assay kit was assessed and varied between 96 and 103 %. HEPG2 was chosen as calibrator. Selection of the most appropriate endogenous reference gene was performed as previously described (Leduc et al. 2011b) on 25 randomly selected individuals, and PPIA was selected out of five other candidates as the single most stable reference gene to use in our study. The plate-to-plate variability in relative quantities of the repeated samples was 5.2, 4.5, 6.2, and 2.3 % (coefficient of variation (CV)) for respectively total HMGCR, FL HMGCR, Δ13 HMGCR, and PPIA mRNA.
To ensure the reliability of the mRNA levels determined, several precautions were duly used. In addition to the usual quality controls, that are no reverse transcriptase controls (resulted in no amplification), no template controls (resulted in no amplification), genomic DNA controls (resulted in no amplification), confirmation of primer sets specificity on agarose gels (resulted in single band of the expected molecular weight), and determination of RNA input range linearity (linear between 16 and 1000 ng of RNA), we performed RNA purity and integrity measurements. The purity and integrity of RNA were estimated using the ratio of absorbance values at 260 and 280 nm evaluated on a Biotek Synergy H1 reader (Fisher Scientific, ON, Canada), and the RNA integrity number (RIN) determined with a Bio-Rad’s Experion instrument (Bio-Rad, CA, USA). The ratios of absorbance were all over 1.5, while RINs ranged from 2 to 8.4, with 84 % of samples over 5, the cutoff value representing good total RNA quality (Fleige and Pfaffl 2006). All Experion’s electropherograms showed no DNA contamination and visible 28S and 18S peaks, and all our qPCR products were shorter than 100 bp in length. Given that short qRT-PCR products (70–250 bp) are mostly “independent” of the RNA quality (Fleige and Pfaffl 2006), samples with a RIN≥4 were included in statistical analysis to maximize our sample size (n=114 for the frontal cortex, n=110 for the cerebellum). Results are presented as HMGCR mRNA levels and HMGCR mRNA ratios, the latter being calculated as the mRNA levels of the FL or Δ13 HMGCR variant divided by the total mRNA levels of HMGCR (any HMGCR transcripts containing exon boundary 6/7). While the FL, Δ13, or total HMGCR mRNA levels reflect the numbers of low mRNA transcripts in a given tissue, the mRNA ratios assess the effects of splicing independently of the effects on overall transcription.
Protein Extraction for HMGCR and Aβ42/Tau Protein Quantification
Frozen brain tissues were homogenized in cold phosphate-buffered saline (PBS) with protease inhibitor using a Omni beads Ruptor 24 (Omni International, GA, USA) and ceramic beads as per manufacturer’s instructions. Protein content was determined in the frontal cortex by the Pierce BCA protein assay (Promega) using bovine serum albumin as standard (Sigma) and a Synergy H1 microplate reader (BioTek, VT, USA) set at a wavelength of 562 nm. Homogenates were kept at −80 °C until use, and repeated freeze-thaw cycles were avoided.
Determination of HMGCR Protein Levels
HMGCR protein levels were measured in the frontal cortex (n=117) using a commercial sandwich enzyme immunoassay (MyBioSource, CA, USA) and a Synergy H1 microplate reader (BioTek, VT, USA). In microtiter plates pre-coated with a monoclonal antibody specific for HMGCR, samples and standards were incubated along with a standardized preparation of horseradish peroxidase (HRP)-conjugated polyclonal antibody specific for HMGCR. The protocol was conducted as per manufacturer’s instructions with the following modification: The samples and standards were incubated with the balance and conjugate solutions for 3 h at 37 °C instead of 1 h. The sensitivity of the assay is 0.1 ng/ml, and concentrations of the samples, which all fell within the standard curve range, varied between 0.2 and 1.2 ng/ml. Given the high degree of inter-lot variations (mean CV of 24.7 %), all samples were remeasured with the same lot number and the obtained intra-lot variation (mean CV of 8.8 %) was within the manufacturer’s recommendation (CV<9 %).
Determination of Aβ42, T-Tau, and P-Tau Protein Levels
Aβ42, T-Tau, and 181-P-Tau concentrations were measured in the frontal cortex (n=94) and cerebellum (n=91) using commercial enzyme immunoassays (Innotest Inc., Ghent, Belgium) and a Synergy H1 microplate reader (BioTek, VT, USA). In these assays, the wells of polystyrene microtiter plates were coated with monoclonal antibodies 21F12, AT120, or HT7 for respectively Aβ42, T-Tau, and P-Tau. The samples and standards were incubated in these wells along with biotinylated monoclonal antibodies 3D6, BT2/HT7, and AT270 for respectively Aβ42, T-Tau, and P-Tau. The protocol was conducted as per manufacturer’s instructions with the following modification: The samples were homogenates of brain tissues rather than CSF. The sensitivity of the assay was 50, 60, and 15.6 pg/ml for respectively Aβ42, T-Tau, and P-Tau. Sample concentrations, which all fell within the standard curve range, varied between the appropriate detection limit and 10,000 times the detection limit. Intra-assay and inter-assay variability measures were respectively 5.1 and 9.1 % (CV), consistent with the manufacturer’s recommendation.
Determination of NFT and SP Counts
NFT and SP densities were determined in the frontal cortex (n=74) as previously described (Etienne et al. 1986) and were consistent with the criteria used in the classification of Khachaturian (Khachaturian 1985). Briefly, paraffin-embedded sections were stained with hematoxylin and eosin, modified Bielchowsky, and alkaline Congo red preparations to visualize NFTs and SPs. Diffuse plaques were purposefully excluded from all measurements.
Statistical Analysis
All statistics were carried out using SPSS program. As expected given their high inter-individual variability, most of the data produced in this paper were non-normally distributed (assessed via Kolmogorov-Smirnov, Shapiro-Wilk tests, and visual inspection of Q-Q plots). Several transformations were attempted to obtain a normal distribution but with no success. When appropriate, parametric tests were computed, but for the most part, the non-parametric alternative had to be used. The goal of our paper was to investigate the impact of rs3846662 genotype, but most importantly of HMGCR mRNA and protein levels, on different pathological markers of AD. Given our previous findings (Leduc et al. 2015a), their effects had to be differentiated from the impacts of sex, APOE4 allele, and disease status. This raises the problem of multiple comparisons. However, the Bonferroni correction would be overly conservative, especially here, where several endpoints measure different aspects of the same phenomenon. Thus, we choose not to correct for multiple comparisons, looking instead for consistency across the results (see Discussion). Note that our study was large enough to allow, with a power of 0.80 of an alpha of 0.05, the detection of medium effect sizes.
Firstly, the impact of rs3846662 (G− vs . G+ subjects) on HMGCR mRNA and HMGCR protein brain levels were analyzed by means of Kruskal-Wallis and Mann-Whitney U tests. For all non-parametric tests, data were also split according to disease status, sex, and APOE4 allele status to examine the impacts of rs3846662 within each subgroup. Secondly, Spearman correlations were used to analyze the relationship between HMGCR mRNA or protein levels and NFT/SP accumulation and Aβ42, T-Tau, and P-Tau brain levels. These relations were analyzed with and without splitting according to disease status, sex, and APOE4 allele status. Finally, even though the data were non-normally distributed, curve estimation was used to estimate the nature of the relationship between these variables. The model with the lowest standard error of the estimate was selected as best.
Results
Frequency of rs3846662 A Allele
Previous findings indicate that homozygosity at locus rs3846662 is associated with the strongest and most significant effects (Burkhardt et al. 2008). In order to investigate the possible impact of rs3846662 on HMGCR mRNA and protein levels, a greater proportion of tissues from homozygote subjects were thus selected. Consequently, the frequency of the HMGCR A allele at locus rs3846662 (Table 1) is lower than those reported for the French Canadian (Leduc et al. 2015a) and other European populations (Medina and Krauss 2009).
Impacts of rs3846662 on HMGCR mRNA and Protein Levels
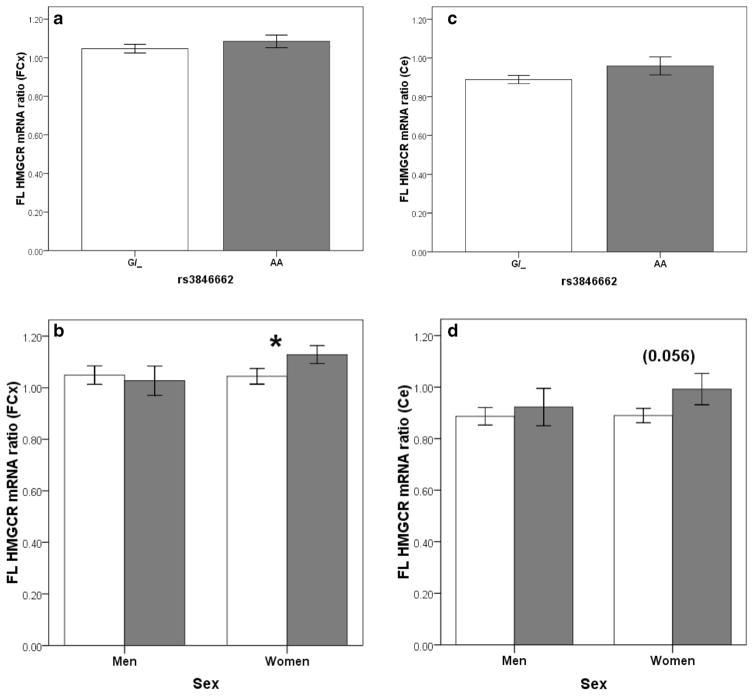
Mann-Whitney analysis of the impacts of rs3846662 on HMGCR was modestly significant at the level of the mRNA (Fig. 1a to d). Indeed, irrespective of disease status, women carrying the AA genotype (G genotype) have higher ratio of FL HMGCR mRNA ratio (i.e., mRNA prevalence of FL HMGCR over prevalence of all HMGCR transcript variants) in the frontal cortex (Fig. 1b, P=0.024) and almost significantly in the cerebellum (Fig. 1d, P=0.056). However, no associations were observed for the Δ13 HMGCR mRNA ratio (i.e., mRNA prevalence of Δ13 HMGCR over prevalence of all HMGCR transcript variants) or the FL, Δ13, and total HMGCR mRNA levels (results not shown, P>0.05). Finally, no association between rs3846662 genotype and HMGCR protein levels were observed (results not shown, P>0.05). Stratification by disease status, sex, or APOE4 allele status did not modify these findings.
Fig. 1.
FL HMGCR mRNA ratio in the frontal cortex (a, b) and the cerebellum (c, d) according to rs3846662 genotype in the whole studied cohort (a, c) and sex subgroups (b, d). Bars represent means, error bars represent SEM, and P values were obtained from Kruskal-Wallis analyses, *P≤0.05
HMGCR Biology Versus AD Pathological Markers
The states of mRNA and protein levels are set primarily by processes downstream of transcription. We therefore used Spearman correlations to investigate whether HMGCR biology was associated with AD pathological markers.
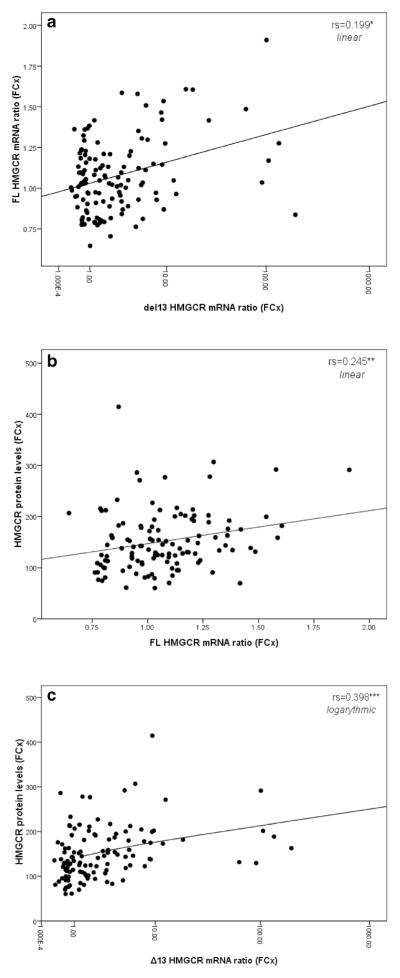
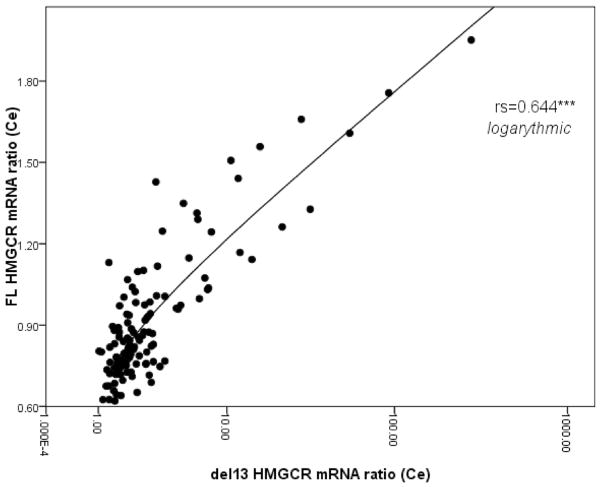
First, we observe that the FL and Δ13 HMGCR mRNA ratios do not correlate with one another in the frontal cortex (P>0.05, Fig. 2a) but do so in the cerebellum (rs 0.644 P= 6.51×10−15, Fig. 3). In the frontal cortex, HMGCR protein levels correlate with both HMGCR mRNA ratios, but most notably, with the Δ13 ratio (rs 0.209 P=0.029 and rs 0.360 P=1.13×10−4, Fig. 2b, c, respectively for FL and Δ13 ratios). The FL, Δ13, and total mRNA levels also correlated with HMGCR protein levels: Total and FL HMGCR mRNA levels are inversely correlated with the protein levels, while the Δ13 mRNA levels correlate positively with it (results not shown). Overall, these data suggest that an increase in HMGCR protein levels correlate best with an increase in both the proportion of exon 13 skipping and the numbers of Δ13 HMGCR mRNA transcripts. Except for the control group where the significance is lost, this conclusion remains valid for each subgroup tested following stratification by disease status, sex, and APOE4 allele status (results not shown).
Fig. 2.
Scatter plot between the FL and Δ13 HMGCR mRNA ratios (a) and scatter plots between FL (b) or Δ13 (c) HMGCR mRNA ratios and HMGCR protein levels. Results are for the frontal cortex. Curve estimation was used to estimate the nature of the relationship between the variables. rs represents Spearman correlation, *P≤0.05, **P≤0.01, ***P≤0.001
Fig. 3.
Scatter plot between FL and Δ13 HMGCR mRNA ratios in the cerebellum. rs represents Spearman correlation, ***P≤ 0.001
Second, in the frontal cortex, very few correlations are observed between HMGCR mRNA or protein levels and markers of AD pathology. Regarding the mRNA expression, we observed that the Δ13 HMGCR mRNA ratio correlates significantly with NFT counts (0.303, P=0.011, Table 2). Similarly, Δ13 HMGCR mRNA levels were significantly associated with NFT counts (respectively 0.265 P=0.027, Table 2). After stratification by disease status, sex, or APOE4 allele status, the significance of these correlations was lost. Furthermore, no correlations were observed between Δ13 HMGCR mRNA levels or ratio and SP accumulation, Aβ42, T-Tau, and P-Tau. Lastly, no correlations were observed between all these markers of AD pathology and FL HMGCR mRNA ratio, FL, and total HMGCR mRNA levels. Regarding the protein expression, HMGCR protein levels were correlated significantly with SP accumulation (0.252, P=0.037, Table 2) and almost significantly with NFT counts (0.230, P=0.057, Table 2). While there were no correlations with either NFT or SP counts after splitting by sex or disease status, stratification by APOE4 allele status indicated that these correlations were significant only for the APOE4-negative subgroup (0.473 P=0.011 and 0.461 P=0.013 for the NFT and SP counts, respectively, Table 2). Lastly, no correlations were observed between HMGCR protein levels and Aβ42, T-Tau, and P-Tau levels.
Table 2.
Spearman correlations between key HMGCR parameters and markers of AD pathology in the frontal cortex
| AD markers | HMGCR | Impact on significance of stratification by | HMGCR | Impact on significance of stratification by | |||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||
| Δ13 ratio | Δ13 levels | Disease | Sex | APOE4 | Protein | Disease | Sex | APOE4 | |
| NFT | 0.303* | 0.265* | Lost | Lost | Lost | 0.230 | No | No | 0.473* for E4– |
| T-Tau | No | No | No | No | No | No | No | No | No |
| P-Tau | No | No | No | No | No | No | No | No | No |
| SP | No | No | No | No | No | 0.252* | Lost | Lost | 0.461* for E4– |
| Aβ42 | No | No | No | No | No | No | No | No | No |
No no correlation
P≤0.05
In the cerebellum, we observe significant correlations between Aβ42 levels and Δ13 HMGCR mRNA ratio and Δ13 HMGCR mRNA levels (−0.351 P=0.001 and −0.359 P= 0.001 for Δ13 ratio and Δ13 mRNA levels, respectively, Table 3). Except for the control group where the significance is lost most likely due to an absence of pre-symptomatic amyloid pathology, this conclusion remains valid for each subgroup tested following stratification by disease status, sex, and APOE4 allele status (results not shown). No correlations were observed between HMGCR mRNA levels or ratios and T-Tau and P-Tau levels.
Table 3.
Spearman correlations between key HMGCR parameters and markers of AD pathology in the cerebellum
| AD markers | HMGCR | Impact on significance of stratification by | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| FL ratio | Δ13 ratio | Δ13 levels | Disease | Sex | APOE4 | |
| T-Tau | No | No | No | No | No | No |
| P-Tau | No | No | No | No | No | No |
| Aβ42 | No | −0.351*** | −0.359*** | AD only | Holds | Holds |
No no correlation
P≤0.001
Discussion
Late-onset AD is a chronic neurodegenerative disorder that represents the commonest cause of dementia and the fourth most common cause of death in the developed world (Kalaria et al. 2008). Given the absence of curative treatment and the overwhelming number of failed Alzheimer’s clinical trials witnessed over the last decade, research on AD has gradually shifted his focus toward pre-Alzheimer’s interventions and protective factors. APOE encodes the main lipid carrier protein in the CNS and is the most robustly and consistently associated gene with AD risk, with the ε4 (APOE4) and ε2 (APOE2) alleles substantially increasing and decreasing the risk level, respectively (Corder et al. 1993, 1994; Poirier et al. 1993; Saunders et al. 1993; Strittmatter et al. 1993; Benjamin et al. 1994). The recent discovery that rs3846662, which modulates the alternative splicing of HMGCR, confers a significant protection against AD risk (Leduc et al. 2015a) stressed, once again, the importance of cholesterol homeostasis for neuronal function and CNS proper functioning. However, whether the protective effects of this SNP are mediated through reduction of vascular risk factors or through the direct modulation of CNS cholesterol homeostasis remains to be clarified.
This study was designed to explore the hypothesis that the beneficial effects of HMGCR are mediated through direct modulation of CNS cholesterol homeostasis. Brain cells are believed to use HMGCR and internalization of lipoproteins via the APOE/LDLR pathway to fulfill its cholesterol needs (Poirier 2005; Yu et al. 2010). Thus, our working hypothesis is that HMGCR A allele homozygocity at locus rs3846662 ultimately results in an HMGCR with lower activity. This prompts a compensatory upregulation of the internalization of lipoproteins via the APOE/LDLR pathway, resulting in a more efficient utilization of local resources (cholesterol and ATPs) and an increase energy reserve to face unexpected neuronal insults or, neurodegenerative disorders. Even though our cross-sectional design study is not suitable to study causality, two causally related factors should cluster more frequently than is expected by chance, and therefore, a cross-sectional design should find an association between these factors (Spies et al. 2013). The purpose of this in vivo study was thus to assess if biomarkers for AD are related to local HMGCR alternative splicing. The two most predominant pathological features of AD being the extracellular SP primarily composed of Aβ peptides, and the intracellular NFT, resulting from the truncation and/or hyperphosphorylation of the microtubule-stabilizing Tau proteins (reviewed in (Adlard and Cummings 2004)); we quantified SP, Aβ42, NFT, T-Tau, and P-Tau levels in the brain. Increased levels of these specific parameters have been associated with increased brain damage and AD severity (Khatoon et al. 1992; Delacourte et al. 1999; Thal et al. 2002; Verdile et al. 2004), and lower levels are expected to correlate with the protective A allele and thus with increased Δ13 HMGCR mRNA levels/ratio.
First, let us consider the genetic portion of the study. We found that women carrying the AA genotype had slightly higher FL HMGCR mRNA ratio than women carrying at least one copy of the G allele in the frontal cortex (P<0.05, 1.13±0.03 vs 1.04± 0.03, respectively) and almost significantly in the cerebellum (P=0.056, 0.99±0.06 vs 0.89±0.03, respectively). However, no effects of the rs3846662 SNP was found on HMGCR mRNA or protein levels, indicating that the increased proportion of FL variant does not translate into higher numbers of mRNA transcripts or higher protein levels. However, it could very well translate into higher activity levels since the catalytically active HMGCR is found as a tetramer (Istvan et al. 2000). Indeed, while it is still unclear whether the Δ13 HMGCR mRNA transcript is translated in vivo into proteins, its translation into inactive monomer proteins could lead to the formation of heterotetramers with reduced enzymatic activity (Medina and Krauss 2009). Higher FL HMGCR mRNA ratio would therefore be associated with higher HMGCR activity despite unchanged HMGCR mRNA or protein levels. In any case, our finding of an increase FL HMGCR mRNA ratio in women carrying the AA genotype is unexpected. We offer two possible explanations for this finding.
Firstly, it has been suggested that the alternative splicing of HMGCR may exist primarily to correct small imbalances in cholesterol homeostasis, allowing the cell to dodge the need for stimulation of a robust transcriptional response when cholesterol disequilibrium occurs (Medina et al. 2011). Alternative splicing and rs3846662 would thus be expected to have a stronger impact on baseline lipid levels (Medina et al. 2011) and in normal conditions rather than in disease states. As such, the association between the AA genotype and increase FL HMGCR mRNA ratio observed in aging or AD subjects may not be related to its impact on HMGCR alternative splicing. In accordance with this hypothesis, a recent in vivo study of the impacts of rs3846662 on statin efficacy and lipid levels in familial hypercholesterolemia also reported increase FL HMGCR mRNA ratio/levels in women receiving statin and carrying the AA genotype (Leduc et al. 2015b). Overall, these findings suggest that the effects mediated by the AA genotype in conditions deviating from physiological conditions may be independent from its impact on HMGCR alternative splicing (Leduc et al. 2015b).
Secondly, because the significant increase in FL HMGCR mRNA ratio observed in women carrying the AA genotype would not survive Bonferroni correction, this finding might simply reflect spurious correlation arising from multiple statistical comparisons rather than true association. Simmons and colleagues also found no association between rs3846662 genotype and HMGCR mRNA prevalence in the brain, a finding they attributed to the confounding bias of cell type heterogeneity and differential expression of RNA splicing factors in the CNS (Simmons et al. 2011). However, since we quantified the HMGCR mRNA levels in brain regions with two very distinct neuronal to non-neuronal ratios, we should have been able to identify cell type-specific effects, if any. To explore whether the second assumption can explain the lack of association between the HMGCR A allele and Δ13 HMGCR mRNA levels/ratio, we performed a literature search to assess the current status of HNRNPA1 (also known as hnRNP A1) expression in the brain. An immunochemistry study performed on several mouse tissues confirmed that HNRNPA1 abundance vary between cell types, with neurons displaying much stronger staining intensity than glial cells (Kamma et al. 1995). Mouse HNRNPA1 expression in glial and neuronal cells respectively equals and surpasses that observed in hepatocytes (Kamma et al. 1995). In accordance, immunochemistry analysis of normal human tissues suggests that HNRNPA1 expression is strong in some neurons, undetectable in glial cells, and moderate in hepatocytes (Patry et al. 2003). More importantly, while there is little HNRNPA1 expression in normal human tissues (Patry et al. 2003), HNRNPA1 expression is high in proliferating and transformed cells (Biamonti et al. 1993; Patry et al. 2003) as well as most cancer (Patry et al. 2003; Pino et al. 2003; Ushigome et al. 2005; Boukakis et al. 2010; Jean-Philippe et al. 2013) and markedly reduced in senescent cells (Hubbard et al. 1995; Shimada et al. 2009) as well as in AD (Berson et al. 2013; Jean-Philippe et al. 2013). In considering these results, we speculate that rs3846662 genotype may not be significantly functional in the brain, especially in older individuals suffering of AD. Indeed, even though the A allele engenders the preferential binding of HNRNPA1 to promote exon 13 skipping (Burkhardt et al. 2008; Medina et al. 2008), its effects may be canceled by the low levels of HNRNPA1 in the brain.
Next, since rs3846662 was initially thought to alleviate AD risk via its modulation of HMGCR alternative splicing, the molecular biology portion of this study was intended to explore whether HMGCR mRNA and protein levels correlated with AD pathological markers. More specifically, we were expecting higher Δ13 HMGCR mRNA ratio/levels to correlate with lower AD pathology. While we observed only a few correlations between HMGCR mRNA ratio/levels and AD biomarkers, the disparate findings between the two brains regions raise considerable interest. In the frontal cortex, we observed an unexpected correlation between increased Δ13 HMGCR mRNA ratio/levels and increased NFT counts (Spearman correlations 0.303* and 0.265*, respectively). In contrast, in the cerebellum, we observed a significant correlation between increased Δ13 HMGCR mRNA ratio/levels and decreased Aβ42 levels. This dichotomy between the frontal cortex and cerebellum might reflect cell type-dependent effects of HMGCR or, that the beneficial effects of HMGCR are dependent upon disease severity and progression.
In line with this hypothesis, we observed that the increase in the proportion and numbers of Δ13 HMGCR mRNA transcripts in the frontal cortex correlated consistently across all subgroups (except control) with higher HMGCR protein levels. Since HMGCR protein levels are increased by 200-fold following inhibition of its activity (Goldstein and Brown 1990), whether the observed increase in protein levels is an attempt for the cell to compensate for lower HMGCR activity levels in the brain remains to be determined. Interestingly, we observed a positive correlation between increased HMGCR protein levels and increased NFT and SP but only in the APOE4-negative subgroup (Spearman correlations 0.473* and 0.416*, respectively). We have previously demonstrated that the HMGCR AA genotype do not modulate the APOE4 risk in subjects affected by full-blown AD (Leduc et al. 2015a). However, the AA genotype was shown to modulate APOE4 risk in cognitively intact and mild cognitive impairment (MCI) subjects, to delay age of onset of AD, and to decrease the lifelong accumulation of NFT and SP pathology in APOE4 carriers (Leduc et al. 2015a). The observation that HMGCR protein levels no longer modulate NFT/SP pathology in APOE4 carriers might reflect the exhaustion of the HMGCR system in end-stage AD. The modulation of NFT/SP accumulation in APOE4 carriers by the AA genotype reported previously (Leduc et al. 2015a) might occur in the early rather than later stages of the disease. Overall, this hypothesis is in line with the observation that for HMGCR inhibitors (statins) to reduce the risk of AD (Jick et al. 2000; Wolozin et al. 2000, 2007; Rockwood et al. 2002), it must be taken during a critical period, for a certain length of time and years before the expected onset of the disease (Leduc et al. 2015a); otherwise, when used in subjects with an established diagnosis, statins offer no detectable benefits (Feldman et al. 2010).
In conclusion, data obtained in vivo from a cohort of autopsied AD and non-demented control brain samples did not confirm the effects of the A allele at locus rs3846662 on HMGCR alternative splicing. The association between rs3846662 and AD may be more complicated than initially though, involving in a complex web of pleiotropic effects, epistasis, and gender interactions. While the beneficial effects of HMGCR might be brain cell type- and disease-progression dependent, our current results do not support an extensive role of local HMGCR splicing on AD pathogenesis. Given the high interindividual variability in the levels of AD biomarkers, replication of our findings in larger samples would be recommended. The use of postmortem brain tissues from subjects with different stages of AD and MCI as well as measurement of HMGCR activity should be considered in subsequent study. Finally, we remain convinced that it is still conceivable that the protective effects of rs3846662 might be mediated through vascular effects (such as arteriosclerosis). This hypothesis needs to be further investigated.
Acknowledgments
This study was supported by the Canadian Institute of Health Research (VL, JP), the Natural Sciences and Engineering Research Council of Canada (JP), the JL Levesque foundation (JP), the Canadian Stroke Network (VL), and the Leducq Fondation (RD).
Footnotes
Compliance of Ethical Standards
Conflict of Interest The authors declare that they have no conflict of interest.
References
- Adlard PA, Cummings BJ. Alzheimer’s disease—a sum greater than its parts? Neurobiol Aging. 2004;25(6):725–733. doi: 10.1016/j.neurobiolaging.2003.12.016. discussion 743–726. [DOI] [PubMed] [Google Scholar]
- Aulchenko YS, Ripatti S, Lindqvist I, et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet. 2009;41(1):47–55. doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo FA, Carvalho LR, Grinberg LT, et al. Equal numbers of neuronal and nonneuronal cells make the human brain an isometrically scaled-up primate brain. J Comp Neurol. 2009;513(5):532–541. doi: 10.1002/cne.21974. [DOI] [PubMed] [Google Scholar]
- Benjamin R, Leake A, McArthur FK, et al. Protective effect of apoE epsilon 2 in Alzheimer’s disease. Lancet. 1994;344(8920):473. doi: 10.1016/s0140-6736(94)91804-x. [DOI] [PubMed] [Google Scholar]
- Berson A, Barbash S, Shaltiel G, et al. Cholinergic-associated loss of hnRNP-A/B in Alzheimer’s disease impairs cortical splicing and cognitive function in mice. EMBO Mol Med. 2013;4(8):730–742. doi: 10.1002/emmm.201100995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biamonti G, Bassi MT, Cartegni L, et al. Human hnRNP protein A1 gene expression. Structural and functional characterization of the promoter. J Mol Biol. 1993;230(1):77–89. doi: 10.1006/jmbi.1993.1127. [DOI] [PubMed] [Google Scholar]
- Boukakis G, Patrinou-Georgoula M, Lekarakou M, Valavanis C, Guialis A. Deregulated expression of hnRNP A/B proteins in human non-small cell lung cancer: parallel assessment of protein and mRNA levels in paired tumour/non-tumour tissues. BMC Cancer. 2010;10:434. doi: 10.1186/1471-2407-10-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhardt R, Kenny EE, Lowe JK, et al. Common SNPs in HMGCR in micronesians and whites associated with LDL-cholesterol levels affect alternative splicing of exon13. Arterioscler Thromb Vasc Biol. 2008;28(11):2078–2084. doi: 10.1161/ATVBAHA.108.172288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasman DI, Posada D, Subrahmanyan L, Cook NR, Stanton VP, Jr, Ridker PM. Pharmacogenetic study of statin therapy and cholesterol reduction. JAMA. 2004;291(23):2821–2827. doi: 10.1001/jama.291.23.2821. [DOI] [PubMed] [Google Scholar]
- Chung JY, Cho SK, Oh ES, et al. Effect of HMGCR variant alleles on low-density lipoprotein cholesterol-lowering response to atorvastatin in healthy Korean subjects. J Clin Pharmacol. 2012;52(3):339–346. doi: 10.1177/0091270011398239. [DOI] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261(5123):921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Risch NJ, et al. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat Genet. 1994;7(2):180–184. doi: 10.1038/ng0694-180. [DOI] [PubMed] [Google Scholar]
- Delacourte A, David JP, Sergeant N, et al. The biochemical pathway of neurofibrillary degeneration in aging and Alzheimer’s disease. Neurology. 1999;52(6):1158–1165. doi: 10.1212/wnl.52.6.1158. [DOI] [PubMed] [Google Scholar]
- Etienne P, Robitaille Y, Wood P, Gauthier S, Nair NP, Quirion R. Nucleus basalis neuronal loss, neuritic plaques and choline acetyl-transferase activity in advanced Alzheimer’s disease. Neuroscience. 1986;19(4):1279–1291. doi: 10.1016/0306-4522(86)90142-9. [DOI] [PubMed] [Google Scholar]
- Feldman HH, Doody RS, Kivipelto M, et al. Randomized controlled trial of atorvastatin in mild to moderate Alzheimer disease. LEADe. Neurology. 2010;74:956–964. doi: 10.1212/WNL.0b013e3181d6476a. [DOI] [PubMed] [Google Scholar]
- Fleige S, Pfaffl MW. RNA integrity and the effect on the real-time qRT-PCR performance. Mol Aspects Med. 2006;27(2–3):126–139. doi: 10.1016/j.mam.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990;343(6257):425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- Hiura Y, Tabara Y, Kokubo Y, et al. Association of the functional variant in the 3-hydroxy-3-methylglutaryl-coenzyme a reductase gene with low-density lipoprotein-cholesterol in Japanese. Circ J. 2010;74(3):518–522. doi: 10.1253/circj.cj-09-0790. [DOI] [PubMed] [Google Scholar]
- Hofman A, Ott A, Breteler MM, et al. Atherosclerosis, apolipoprotein E, and prevalence of dementia and Alzheimer’s disease in the Rotterdam Study. Lancet. 1997;349(9046):151–154. doi: 10.1016/S0140-6736(96)09328-2. [DOI] [PubMed] [Google Scholar]
- Hubbard K, Dhanaraj SN, Sethi KA, et al. Alteration of DNA and RNA binding activity of human telomere binding proteins occurs during cellular senescence. Exp Cell Res. 1995;218(1):241–247. doi: 10.1006/excr.1995.1152. [DOI] [PubMed] [Google Scholar]
- Istvan ES, Palnitkar M, Buchanan SK, Deisenhofer J. Crystal structure of the catalytic portion of human HMG-CoA reductase: insights into regulation of activity and catalysis. EMBO J. 2000;19(5):819–830. doi: 10.1093/emboj/19.5.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean-Philippe J, Paz S, Caputi M. hnRNPA1: the Swiss army knife of gene expression. Int J Mol Sci. 2013;14(9):18999–19024. doi: 10.3390/ijms140918999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jick H, Zornberg GL, Jick SS, Seshadri S, Drachman DA. Statins and the risk of dementia. Lancet. 2000;356(9242):1627–1631. doi: 10.1016/s0140-6736(00)03155-x. [DOI] [PubMed] [Google Scholar]
- Johnson JM, Castle J, Garrett-Engele P, et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction micro-arrays. Science. 2003;302(5653):2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- Kalaria RN, Maestre GE, Arizaga R, et al. Alzheimer’s disease and vascular dementia in developing countries: prevalence, management, and risk factors. Lancet Neurol. 2008;7(9):812–826. doi: 10.1016/S1474-4422(08)70169-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamma H, Portman DS, Dreyfuss G. Cell type-specific expression of hnRNP proteins. Exp Cell Res. 1995;221(1):187–196. doi: 10.1006/excr.1995.1366. [DOI] [PubMed] [Google Scholar]
- Khachaturian ZS. Diagnosis of Alzheimer’s disease. Arch Neurol. 1985;42(11):1097–1105. doi: 10.1001/archneur.1985.04060100083029. [DOI] [PubMed] [Google Scholar]
- Khatoon S, Grundke-Iqbal I, Iqbal K. Brain levels of microtubule-associated protein tau are elevated in Alzheimer’s disease: a radioimmuno-slot-blot assay for nanograms of the protein. J Neurochem. 1992;59(2):750–753. doi: 10.1111/j.1471-4159.1992.tb09432.x. [DOI] [PubMed] [Google Scholar]
- Krauss RM, Mangravite LM, Smith JD, et al. Variation in the 3-hydroxyl-3-methylglutaryl coenzyme a reductase gene is associated with racial differences in low-density lipoprotein cholesterol response to simvastatin treatment. Circulation. 2008;117(12):1537–1544. doi: 10.1161/CIRCULATIONAHA.107.708388. [DOI] [PubMed] [Google Scholar]
- Leduc V, Domenger D, De Beaumont L, Lalonde D, Belanger-Jasmin S, Poirier J. Function and comorbidities of apolipoprotein e in Alzheimer’s disease. Int J Alzheimers Dis. 2011a;2011:974361. doi: 10.4061/2011/974361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leduc V, Legault V, Dea D, Poirier J. Normalization of gene expression using SYBR green qPCR: a case for paraoxonase 1 and 2 in Alzheimer’s disease brains. J Neurosci Methods. 2011b;200(1):14–19. doi: 10.1016/j.jneumeth.2011.05.026. [DOI] [PubMed] [Google Scholar]
- Leduc V, De Beaumont L, Theroux L, et al. HMGCR is a genetic modifier for risk, age of onset and MCI conversion to Alzheimer’s disease in a three cohorts study. Mol Psychiatry. 2015a;20(7):867–873. doi: 10.1038/mp.2014.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leduc V, Bourque L, Poirier J, Dufour R. Role of rs3846662 and HMGCR alternative splicing in statin efficacy and baseline lipid levels in familial hypercholesterolemia. Pharmacogenet Genomics. 2015b doi: 10.1097/FPC.0000000000000178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Larson EB, Sonnen JA, et al. Statin therapy is associated with reduced neuropathologic changes of Alzheimer disease. Neurology. 2007;69(9):878–885. doi: 10.1212/01.wnl.0000277657.95487.1c. [DOI] [PubMed] [Google Scholar]
- Lu Y, Feskens EJ, Boer JM, et al. Exploring genetic determinants of plasma total cholesterol levels and their predictive value in a longitudinal study. Atherosclerosis. 2010;213(1):200–205. doi: 10.1016/j.atherosclerosis.2010.08.053. [DOI] [PubMed] [Google Scholar]
- Marchant NL, Reed BR, Sanossian N, et al. The aging brain and cognition: contribution of vascular injury and abeta to mild cognitive dysfunction. JAMA Neurol. 2013;70(4):488–495. doi: 10.1001/2013.jamaneurol.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina MW, Krauss RM. The role of HMGCR alternative splicing in statin efficacy. Trends Cardiovasc Med. 2009;19(5):173–177. doi: 10.1016/j.tcm.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina MW, Gao F, Ruan W, Rotter JI, Krauss RM. Alternative splicing of 3-hydroxy-3-methylglutaryl coenzyme A reductase is associated with plasma low-density lipoprotein cholesterol response to simvastatin. Circulation. 2008;118(4):355–362. doi: 10.1161/CIRCULATIONAHA.108.773267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina MW, Gao F, Naidoo D, et al. Coordinately regulated alternative splicing of genes involved in cholesterol biosynthesis and uptake. PLoS One. 2011;6(4):e19420. doi: 10.1371/journal.pone.0019420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patry C, Bouchard L, Labrecque P, et al. Small interfering RNA-mediated reduction in heterogeneous nuclear ribonucleoparticule A1/A2 proteins induces apoptosis in human cancer cells but not in normal mortal cell lines. Cancer Res. 2003;63(22):7679–7688. [PubMed] [Google Scholar]
- Pino I, Pio R, Toledo G, et al. Altered patterns of expression of members of the heterogeneous nuclear ribonucleoprotein (hnRNP) family in lung cancer. Lung Cancer. 2003;41(2):131–143. doi: 10.1016/s0169-5002(03)00193-4. [DOI] [PubMed] [Google Scholar]
- Poirier J. Apolipoprotein E, cholesterol transport and synthesis in sporadic Alzheimer’s disease. Neurobiol Aging. 2005;26(3):355–361. doi: 10.1016/j.neurobiolaging.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Poirier J, Davignon J, Bouthillier D, Kogan S, Bertrand P, Gauthier S. Apolipoprotein E polymorphism and Alzheimer’s disease. Lancet. 1993;342(8873):697–699. doi: 10.1016/0140-6736(93)91705-q. [DOI] [PubMed] [Google Scholar]
- Riekse RG, Li G, Petrie EC, et al. Effect of statins on Alzheimer’s disease biomarkers in cerebrospinal fluid. J Alzheimers Dis. 2006;10(4):399–406. doi: 10.3233/jad-2006-10408. [DOI] [PubMed] [Google Scholar]
- Rockwood K, Kirkland S, Hogan DB, et al. Use of lipid-lowering agents, indication bias, and the risk of dementia in community-dwelling elderly people. Arch Neurol. 2002;59(2):223–227. doi: 10.1001/archneur.59.2.223. [DOI] [PubMed] [Google Scholar]
- Saunders AM, Strittmatter WJ, Schmechel D, et al. Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer’s disease. Neurology. 1993;43(8):1467–1472. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]
- Shimada N, Rios I, Moran H, Sayers B, Hubbard K. p38 MAP kinase-dependent regulation of the expression level and subcellular distribution of heterogeneous nuclear ribonucleoprotein A1 and its involvement in cellular senescence in normal human fibroblasts. RNA Biol. 2009;6(3):293–304. doi: 10.4161/rna.6.3.8497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons CR, Zou F, Younkin SG, Estus S. Evaluation of the global association between cholesterol-associated polymorphisms and Alzheimer’s disease suggests a role for rs3846662 and HMGCR splicing in disease risk. Mol Neurodegener. 2011;6:62. doi: 10.1186/1750-1326-6-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies PE, Verbeek MM, Sjogren MJ, de Leeuw FE, Claassen JA. Alzheimer biomarkers and clinical Alzheimer disease were not associated with increased cerebrovascular disease in a memory clinic population. Curr Alzheimer Res. 2013;11(1):40–46. doi: 10.2174/1567205010666131120101352. [DOI] [PubMed] [Google Scholar]
- Strittmatter WJ, Saunders AM, Schmechel D, et al. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90(5):1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thal DR, Rub U, Orantes M, Braak H. Phases of A beta-deposition in the human brain and its relevance for the development of AD. Neurology. 2002;58(12):1791–1800. doi: 10.1212/wnl.58.12.1791. [DOI] [PubMed] [Google Scholar]
- Ushigome M, Ubagai T, Fukuda H, et al. Up-regulation of hnRNP A1 gene in sporadic human colorectal cancers. Int J Oncol. 2005;26(3):635–640. [PubMed] [Google Scholar]
- Verdile G, Gnjec A, Miklossy J, et al. Protein markers for Alzheimer disease in the frontal cortex and cerebellum. Neurology. 2004;63(8):1385–1392. doi: 10.1212/01.wnl.0000141848.45315.a6. [DOI] [PubMed] [Google Scholar]
- Wolozin B, Kellman W, Ruosseau P, Celesia GG, Siegel G. Decreased prevalence of Alzheimer disease associated with 3-hydroxy-3-methyglutaryl coenzyme A reductase inhibitors. Arch Neurol. 2000;57(10):1439–1443. doi: 10.1001/archneur.57.10.1439. [DOI] [PubMed] [Google Scholar]
- Wolozin B, Wang SW, Li NC, Lee A, Lee TA, Kazis LE. Simvastatin is associated with a reduced incidence of dementia and Parkinson’s disease. BMC Med. 2007;5:20. doi: 10.1186/1741-7015-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Youmans KL, LaDu MJ. Proposed mechanism for lipoprotein remodelling in the brain. Biochim Biophys Acta. 2010;1801(8):819–823. doi: 10.1016/j.bbalip.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CY, Theusch E, Lo K, et al. HNRNPA1 regulates HMGCR alternative splicing and modulates cellular cholesterol metabolism. Hum Mol Genet. 2014;23(2):319–332. doi: 10.1093/hmg/ddt422. [DOI] [PMC free article] [PubMed] [Google Scholar]