Abstract
Metastasis is a multi-step process that ultimately depends on the ability of disseminating cancer cells to establish favorable communications with their microenvironment. The tumor microenvironment consists of multiple and continuously changing cellular and molecular components. One of the factors regulating the tumor microenvironment is TNF-α, a pleiotropic cytokine that plays key roles in apoptosis, angiogenesis, inflammation and immunity. TNF-α can have both pro- and anti-tumoral effects and these are transmitted via two major receptors, the 55 kDa TNFR1 and the 75 kDa TNFR2 that have distinct, as well as overlapping functions. TNFR1 is ubiquitously expressed while the expression of TNFR2 is more restricted, mainly to immune cells. While TNFR1 can transmit pro-apoptotic or pro-survival signals through a complex network of downstream mediators, the role of TNFR2 is less well understood. One of its main functions is to act as a survival factor and moderate the pro-apoptotic effects of TNFR1, particularly in immune cells. In this review, we summarize the evidence for the involvement of the TNF system in the progression of the metastatic process from its contribution to the early steps of tumor cell invasion to its role in the colonization of distant sites, particularly the liver. We show how the TNF receptors each contribute to these processes by regulating and shaping the tumor microenvironment. Current evidence and concepts on the potential use of TNF targeting agents for cancer prevention and therapy are discussed.
Keywords: TNF-α, TNFR, immune cells, metastasis, liver
1. The process of cancer metastasis - an overview
Cancer-related deaths occur mainly due to metastatic disease; understanding the molecular and cellular mechanisms underlying the process of metastasis is therefore essential to improving the survival of cancer patients. Metastasis occurs when cancer cells detach from the primary site of growth, enter the circulation and migrate to a secondary organ site where they extravasate and proliferate to give rise to secondary tumors. The ultimate site of metastases is determined by a combination of mechanical/circulatory factors [1, 2] and growth promoting interactions within the target organ microenvironment [3, 4].
The tumor microenvironment can promote metastasis in several ways. At the primary site, the microenvironment can facilitate intravasation. Inflammatory immune cells such as monocytes can induce epithelial-to-mesenchymal transition and the acquisition of a migratory phenotype in cancer cells through the production of cytokines such as tumor necrosis factor (TNF)-α and transforming growth factor (TGF)-β [5, 6]. Cells in the microenvironment can support intravasation through production of proteolytic enzymes such as metalloproteinases (MMPs) and serine proteinases such as urokinase plasminogen activator (uPA) to degrade basement membranes [7, 8]. Tumor cell extravasation can also be enhanced by host cells. Cytokines secreted by innate immune cells such as TNF-α and interleukin (IL)-6 can provide survival cues for cancer cells in the circulation [9] and also induce the expression of vascular endothelial cell adhesion molecules (CAM) that facilitate extravasation [10, 11]. Chemokines such as CXCL12 produced by endothelial cells and macrophages can mediate migration and extravasation of tumor cells that express the chemokine receptor CXCR4, thereby promoting metastasis.
1.1. The process of liver metastasis
The liver is the largest organ in vertebrates with a unique architecture suited for its diverse functions such as protein biosynthesis and detoxification of blood. Because it is the site of constant flux of antigens and bacterial products from the intestine, it is rich in inflammatory cells and mediators [12, 13]. The liver has two sources of blood supply, 80% is deoxygenated blood arriving via the portal vein and 20% is oxygen-rich blood arriving via the hepatic artery. The portal vein branches into perilobular portal veins, interlobular veins and terminal portal venules that drain into the hepatic sinusoids, as does the hepatic artery [14]. The liver sinusoids provide an extensive endothelial cell surface area for interaction with leukocytes and soluble waste macromolecules. The liver is composed of several unique cellular subpopulations.
The liver parenchyma consists mainly of hepatocytes, while the non-parenchymal compartment includes the hepatic stellate cells (HSC), liver sinusoidal endothelial cells (LSEC), and immune cells such as resident macrophages - the Kupffer cells (KC), dendritic cells and liver associated lymphocytes [14]. LSECs line the walls of hepatic sinusoids and separate the sinusoidal blood from the hepatocytes. The LSECs are morphologically [15, 16] and functionally [17, 18] distinct from endothelial cells in other organ sites. They can be activated to express CAM [19, 20] and secrete cytokines [21, 22] that can affect the course of inflammation and metastasis. The KCs are highly phagocytic and a major source of cytokines and chemokines in the liver [23, 24]. The liver-associated lymphocytes include T cells, natural killer (NK) cells and B cells that can also contribute to the local immune response to metastatic tumor cells [25].
The liver is the most common site of distant metastases from different primary tumors, followed by bone and lung [13, 26]. Despite progress in surgical techniques and targeted therapy, liver metastases remain a major cause of cancer-related death, and achieving a better understanding of the underpinning biology is therefore critical to achieving better clinical outcomes for cancer patients.
The process of liver metastasis has been divided into four major phases [27, 28]. 1. The microvascular phase, that entails arrest of circulating tumor cells in the sinusoidal vessels – an event that may result in their extravasation or death [28–30]. Inflammatory factors, nitric oxide (NO) and reactive oxygen species (ROS) released by LSEC [31] and/or sinusoidal KC [10] can either promote transendothelial migration and tumor cell survival or trigger tumor cell death at this stage; 2. The extravascular and pre-angiogenic phase, during which tumor cells extravasate into the space of Disse, and stromal cells are recruited into avascular micrometastases. During this phase, HSCs are recruited into the pre-metastatic niche and then they differentiate into myofibroblasts to promote tumor proliferation and invasion [32]; 3. The angiogenic phase, when micrometastases are vascularized through several possible interactions with the microenvironment. This phase starts when micrometastases have an average diameter greater than 300 μm and grow beyond the limits of the liver lobule. At this stage, pro-angiogenic factors such as vascular endothelial growth factor (VEGF), angiopoietins, TNF-α and IL-18 secreted by tumor-activated myofibroblasts can promote angiogenesis, while, liver-derived anti-angiogenic factors such as endostatin can inhibit the process [27, 33]; 4. The growth phase is the last stage in the process when tumor cells proliferate to establish ‘clinical’ metastases. At each of these stages, continued tumor cell growth depends on permissive interactions between metastatic tumor cells and the liver microenvironment.
2. Inflammation plays a role in cancer progression and metastasis
The host inflammatory response, orchestrated by inflammatory cells such as macrophages, neutrophils and mast cells through the release of an array of chemokines and cytokines can play opposing roles in tumor initiation, progression and metastasis. Studies in animal models have shown that NK1.1+ T cells can provide host protection against the growth of methylcholanthrene-induced fibrosarcomas through the production of the cytokine interferon (IFN)-γ. Similarly, in mouse models of prostate and breast cancer, INF-γ and perforin, when administered together, provided protection in three different tumor models controlled by innate immunity that was equivalent to protection provided by NK1.1+ T cells. In these models, direct cytotoxicity mediated by cytotoxic lymphocytes producing perforin could independently contribute antitumor effector functions that together with INF-γ controlled the initiation, growth, and spread of the tumors [34, 35]. While these examples identify host innate immunity as host protective, many studies have also highlighted the tumor-promoting effects of inflammation and the innate immune response. Epidemiological studies have linked chronic inflammation to increased risk for several cancers such as colon cancer and hepatocellular carcinoma [36, 37]. Cells transformed through oncogenes such as RET/PTC and RAS had upregulated expression of multiple pro-inflammatory cytokines such as IL-1 and IL-8 suggesting that the ability to activate an inflammatory reaction may promote the oncogenic process [38, 39]. Moreover, targeted deletions in genes encoding specific inflammatory mediators were shown to protect mice from the development of primary cancers. This was shown in nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB)-deficient mice that had a dramatic decrease in the incidence of inflammation-induced hepatocellular carcinoma [40] and in TNF-α-deficient mice that were resistant to the development of 7,12-dimetylbenz-anthracene (DMBA) and 12-O-tetradecanoyl-phorbol-13-acetate-induced skin cancer [41]. Targeting of inflammatory cells and mediators in the tumor microenvironment was also shown to reduce tumor growth [37, 42, 43]. Treatment of mice bearing orthotopically implanted pancreatic ductal adenocarcinoma cells with TNF-α enhanced tumor growth and metastasis, while inhibition of TNF-α in these mice using an anti-TNF antibody (infliximab) or the TNF-Trap Etanercept had strong antitumoral effects [36]. In a recent striking example, patients treated with antibody to colony-stimulating factor 1 (CSF-1), the major survival factor for tumor-associated macrophages, had reduced numbers of CSF-1R+CD163+ macrophages in their tumor tissues and this translated into clinically objective responses in diffuse-type giant cell tumor patients [44]. Together, these studies show that depending on the tumor type, the stage of tumor development and modalities tested, the inflammatory response may have very different effects on tumor development and progression. These opposing effects may be attributable, at least in part, to the pleotropic nature of one of the main drivers of inflammation, namely TNF-α. In this review, we focus on the complex role that TNF-α and its cell surface receptors play in tumor progression and, in particular, in the process of metastasis.
2.1. The TNF axis in inflammation and cancer
Initially identified as an endotoxin-induced cytokine that can trigger rapid hemorrhagic necrosis in mouse and human experimental tumors (hence its name) [45, 46], TNF-α has since been recognized as a major orchestrator of inflammation in non-malignant and malignant diseases [45, 47, 48]. While the major sources of this pro-inflammatory cytokine are host innate immune cells such as macrophages and neutrophils, it can also be produced by T and B lymphocytes and NK cells, and under specific conditions, by non-immune cells such as endothelial cells, mast cells, smooth and cardiac muscle cells, fibroblasts and osteoclasts [49]. Although initially thought to mediate mainly anti-tumor effects, it has become clear in recent years that TNF-α can also promote tumor progression [45, 50, 51].
2.1.1. TNF signaling
TNF-α is part of the TNF super family (TNFSF) that consists of 19 structurally related ligands that can bind to one or more of 29 members of the TNRSF [52, 53]. These receptors are characterized by the presence of 1–6 cysteine-rich domains in their extracellular region that are responsible for the binding of their respective ligands [52]. Together, these receptors regulate inflammation, immunity and cell survival [53].
TNF receptors have been classified into 2 main groups based on the presence, within their intracellular region, of a ‘death domain’ that is responsible for the ability of these receptors to cause cell death. Receptors that do not have a death domain in their intracellular region have a so-called (‘TRAF interaction motif’) ‘TIM domain’ that links them to downstream TNF receptor adaptor factor (TRAF) proteins ([54] reviewed in [55] and see diagrammatic representation in Figure 1).
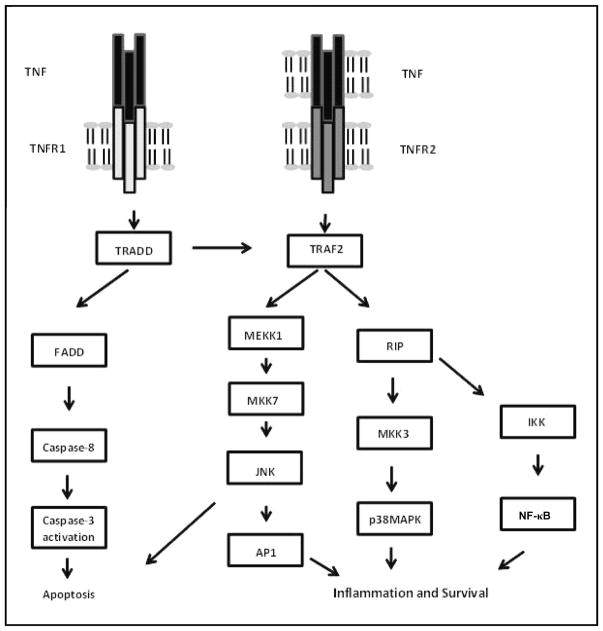
Figure 1. TNFR signaling pathways.
Following TNF stimulation, TNFR1 sequentially recruits TNFR-associated death domain (TRADD), FAS-associated death domain (FADD) and caspase-8 leading to Caspase-3 activation and apoptosis. Both receptors can also recruit TNFR-associated factor 2 (TRAF2), MAP/ERK kinase kinase 1 (MEKK1) and MAPK kinase 7 (MKK7) leading to activation of the JNK pathway and the transcription factor AP-1. Recruitment of receptor-interacting protein (RIP) via TRAF2 leads to activation of p38 MAPK via MKK3. RIP recruitment can also lead to NF-κB activation via the inhibitor of NF-κB kinase (IKK) complex. These three pathways ultimately lead to inflammation and survival. (Adapted from Aggarwal, B. B. 2003, Nat. Rev. Immunol., 3, 745–756, with permission from Macmillan Publishers Ltd.).
Receptors of the TNFR superfamily depend on the binding of adaptor proteins to activate intracellular signaling pathways, as depicted in Figure 1. Two types of adaptor proteins have been identified, those that contain a death domain including TRADD (TNF receptor associated protein with death domain) or FADD (Fas associated protein with death domain) that mediate cell death signaling [56] and a group of adaptor proteins that do not have a death domain. These include TRAF proteins that interact with the receptors either directly through the receptor’s TIM or indirectly through intermediate adaptors [54]. Depending on the adaptor complexes recruited to the receptors in response to ligand binding, different signaling pathways may be activated that lead to transcriptional activation through NF-κB or MAPK/AP-1 (Mitogen activated protein kinase/Activating protein 1) and cell survival or alternatively, to apoptosis. This diversity of signaling outcomes accounts for the diversity of biological effects exerted by TNF ligands that can vary from beneficial and host protective effects in inflammation, immunity and organogenesis, on one hand, and cellular death, on the other [55].
TNF-α is the most extensively studied of the TNF ligands. It is produced as a functional 26-kDa homotrimeric transmembrane protein that is proteolitically cleaved by the disintegrin and metalloprotease-17 (ADAM-17) (also known as TACE) to release a functional 17-kDa soluble form into the circulation [45, 49]. Both soluble and membrane-bound TNF can coexist as mono, di, or trimeric proteins. In the plasma, TNF-α appears as free or is bound to the circulating forms of one of its two cell surface receptors, namely TNF receptor 1 (TNFR1, TNFRSF1A, CD120a, p55) and TNF receptor 2 (TNFR2, TNFRSF1B, CD120b, p75). These receptors are products of two separate genes and their cellular distribution and functions are distinct. While TNFR1 can be detected in almost all cell types, TNFR2 expression is more restricted and has been documented mainly on immune cells such as subsets of T cells, as well as on myocytes, thymocytes, neuronal cells, endothelial cells and mesenchymal stem cells [57, 58]. While cell surface TNFR1 can bind to both the soluble and membrane-bound forms of TNF-α, cell surface TNFR2 binds primarily to the membrane bound form, thus, its role has been difficult to study in the laboratory and its importance, therefore, underestimated.
TNFR1 is a 434 amino acid transmembrane receptor consisting of extracellular, transmembrane and intracellular domains. The extracellular region consists of four cysteine-rich domains that are involved in ligand binding. The N-terminal cysteine-rich domain favors the preassembly of the receptor into trimeric complexes and functions to prevent spontaneous receptor autoactivation [59, 60], while the death domain (DD) characteristic of TNFR1 is in its C-terminal end. TNFR1 signaling can induce the synthesis of multiple inflammatory mediators and growth factors through NF-κB activation, or it can result in the induction of cell death through its DD via TRADD and FADD [57]. TRADD recruitment serves as a platform to assemble alternative signaling complexes that can include adaptor proteins such as TRAF2, cellular inhibitor of apoptosis (cIAP)-1, cIAP2 and receptor interacting kinase (RIP)1. This leads to activation of the nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor (Iκβ) kinase (IKK) complex, release of the inhibitor Iκβα and nuclear translocation of NF-κB and c-Jun N-terminal kinase (JNK) activation. Alternatively, the recruitment of FADD and caspase 8 results in TNF-α-induced apoptosis. NF-κB and JNK regulate the expression of a range of molecules such as transcription factors [61, 62], cytokine and chemokines such as IL-1, IFN-γ, IL-2, IL-6 and IL-8 [63–65] and cell adhesion molecules (CAM) [62, 66] that are collectively responsible for the pleiotropic effects of TNF-α.
TNFR2 is a 439 amino acid type II transmembrane protein consisting of an extracellular domain with four complementarity determining regions (CDRs), a transmembrane region and an intracellular domain. Unlike TNFR1, TNFR2 lacks a death domain and relies on TRAF2 for signaling. TRAF2 binds to a highly conserved sequence within the intracellular domain which also includes an amino acid sequence involved in TRAF2 degradation [55]. This ability to engage and/or degrade TRAF2 affects the crosstalk between TNFR2 and TNFR1 and can result in either the inhibition or the promotion of TNFR1 activities in immune or endothelial cells that express both receptors [67–70]. Thus, the ability of TNFR2 to induce TRAF2 degradation may result in the inhibition of TRAF2/cIAP1/cIAP2 complex formation and survival signaling and enhanced TNFR1-mediated cell death. The crosstalk between these receptors can therefore determine the ultimate effect of TNF-α on cell fate and whether the outcome will be cell survival, proliferation or death [55]. TNFR2 signaling can also potentiate a RIP-dependent form of cell death termed programmed necrosis induced via TNFR1, as was shown in T cells [68], or it can inhibit TNFR1-induced expression of CAM such as E-selectin and ICAM-1, as was shown in endothelial cells [70]. TNFR2 can also crosstalk with other receptor systems to regulate functions such as cell proliferation, as was shown for tumor-infiltrating T-cells stimulated by IL-2 [71] and is known to be highly expressed on an immuno-suppressive subset of tumor-infiltrating T regulatory cells (Tregs) [72] (see more details below). The role of TNFR2 in other immune and non-immune cell functions is not yet fully understood.
2.2. The TNF axis can play a dual role in tumor progression and metastasis
The critical role of TNF as a master regulator of inflammation and the immune response has been well established and the reader is referred to several excellent reviews on the subject [43, 73–78]. Here we will focus on the central role that TNF plays in orchestrating the response of the microenvironment during tumor initiation and progression with emphasis on the role of the TNF axis in metastasis. Recent evidence has made it clear that TNF signaling can play opposing roles in tumorigenesis and metastasis and may either promote or inhibit tumor progression, depending on the context, tumor type and the stage of tumor development. This dual role has resulted in a re-thinking of therapeutic strategies that target the TNF system, as discussed in greater detail below.
2.2.1. TNF-α can inhibit tumor progression
In line with the original description of TNF as an inducer of tumor cell death, several preclinical studies documented TNF-induced anti-tumorigenic effects. For example, hTNF-α was shown to cause degeneration of both syngeneic and xenografted tumors when administered to mice together with IFN-γ [78, 79]. In a system of loco-regional drug delivery by isolated limb perfusion (ILP) for soft tissue sarcoma, hTNF-α caused hemorrhagic necrosis of the tumors [77, 80] and in a rat osteosarcoma model, TNF-α caused necrosis when administered together with the chemotherapeutic drug melphalan [77]. Further studies revealed that hTNF-α can synergize with chemotherapeutic drugs by increasing the local drug concentration in the tumor tissue [76, 80]. These and other studies suggested that the anti-tumor effect of TNF-α may be due not only to direct cytotoxic activity towards tumor cells but also to the changes it can induce in the tumor microenvironment. Indeed, several studies have shown that hTNF-α triggered an infiltration of inflammatory cells such as macrophages, eosinophils, polymorphs and lymphocytes into the tumor site causing immune-mediated tumor rejection [79]. In TNF−/− mice, impaired cytotoxic activity of NK and IL-2-activated killer T cells was reported [75] and in a model of T antigen (Tag)-induced multistage carcinogenesis of pancreatic islets, CD4+ T cells were shown to home selectively into the tumor microenvironment around the islets and induce antiangiogenic chemokines that prevented tumor angiogenesis, tumor cell proliferation, and multistage carcinogenesis in a TNFR1 and INFγ dependent manner. In the absence of TNFR1 signaling, these T cells promoted angiogenesis and multistage carcinogenesis, implicating TNFR1 in anti-tumorigenic effects of T cells in this model [74]. Consistent with this conclusion, Watanabe et al. showed that administration of hTNF-α caused hemorrhaging of the capillary vessels in mouse fibrosarcoma tumors, thereby increasing leakage of chemotherapeutic drugs into the tumor tissues [81]. Taken together, these studies identified TNF-α as a tumor-inhibitory factor mediating its effects via several mechanisms including the destruction of the tumor vasculature and the induction of anti-tumor immune responses.
In clinical trials, TNF-α has not been a very successful anti-cancer agent, mainly because of its severe dose-limiting toxicity when administered systemically [82–85]. However, the addition of TNF-α to chemotherapy in the treatment of non-resectable soft tissue sarcomas of the extremities using isolated limb perfusion has had beneficial anti-tumor effects and these were attributed to enhancement of tumor-selective drug uptake and increased destruction of the tumor vasculature [86]. In the clinical setting, TNF-α may therefore have its utility in regional therapy [87], particularly when combined with chemotherapy.
2.2.2. TNF-α can also promote tumor growth
Clinical and experimental evidence implicate TNF-α in cancer initiation and progression. Strong evidence has come from TNF-α null mice. For example, when these mice were treated with the carcinogenic combination of okadaic acid and 12-O-tetradecanoylphorbol 13-acetate (TPA), a drastic reduction in the development of skin tumors relative to wild type mice was observed [41, 88], suggesting that TNF-α was important for chemical carcinogenesis in this model. TNFR1-or TNFR2-deficient mice subjected to chemical carcinogenesis also developed less skin tumors [89]. In a mouse model of inflammation-induced colon carcinogenesis, whereby administration of azoxymethane (AOM) followed by repeated dextran sulfate sodium (DSS) ingestion causes severe colonic inflammation and the mice subsequently develop multiple tumors, TNF-α was identified as a major driver of leukocyte recruitment and subsequent tumor initiation. In this model, TNFR1-deficient mice had a reduced incidence of colonic tumors as compared to wild type (WT) control mice, and the administration of a TNF-α antagonist to WT mice markedly reduced the number of colonic tumors [47]. TNF-α can promote tumor growth via several mechanisms. It can act directly on the tumor cells to enhance their growth, as was shown in a mouse model of gastric cancer [90]. In Mdr2-knockout mice that spontaneously develop cholestatic hepatitis followed by hepatocellular carcinoma (a model of inflammation-associated cancer), the inflammatory process triggers NF-κB activation in the hepatocytes through TNF-α upregulation in adjacent endothelial and inflammatory cells, increasing hepatocyte survival and proliferation and setting the stage for the onset of tumor formation. NF-κB activation was also identified as a key driver in a mouse model of colitis-associated cancer [40, 91]. Autocrine TNF-α-mediated proliferation has been documented in a wide range of cancers including leukemia [92, 93], colon, lung and pancreatic carcinoma [94, 95], glioblastoma and neuroblastoma [95–99].
TNF-α can also promote tumor expansion by upregulating MMP production in tumor cells or in the tumor microenvironment [100, 101] and (as discussed in greater detail below) plays a key role in the recruitment of tumor promoting immunosuppressive cells such as myeloid derived suppressor cells (MDSCs) and Tregs into the tumor microenvironment [102].
2.2.3. TNF-α plays a role in cancer metastasis
TNF-α has also been implicated in the process of metastasis. Increased blood levels of TNF-α have been documented in cancer patients and were found to correlate with increased incidence of metastatic diseases [103]. Increased metastasis can be mediated through different mechanisms. As mentioned above, TNF-α can enhance tumor invasiveness by acting directly on the tumor cells but it can also alter the tumor microenvironment to promote metastasis. In one example, the administration of TNF-α to mice prior to injection of B16-BL6 melanoma cells, significantly enhanced experimental pulmonary metastasis [11]. This was due to the induction of integrin α4β1 (VLA-4) - a vascular adhesion molecule (VCAM)-1 ligand on the surface of B16-BL6 cells and also induction of VCAM-1 expression on lung vascular endothelial cells, consequently enhancing tumor cell adhesion, extravasation and metastasis [104]. In several studies, animals pretreated with TNF-α were found to have increased liver metastasis. This was initially attributed to the ability of TNF-α to induce the expression of vascular endothelial CAMs such as intracellular adhesion molecule (ICAM)-1, E-selectin and VCAM-1 on LSECs, thereby enhancing tumor cell arrest and trans-endothelial migration [10, 105]. However, the role of TNF-α in the process of hepatic colonization is likely to be more complex and multilayered as multiple cells within the hepatic microenvironment express the TNF receptors [13].
2.3. The distinct roles of the TNF receptors in cancer progression
While both TNF receptors are known to play a role in tumor progression, the function of TNFR1 has been more extensively studied and is better understood. TNFR1 expression on tumor cells can promote carcinogenesis. This, for example, was shown in a mammary tumor model where TNF-α could directly induce tumor cell proliferation in vitro and in vivo and this was mediated through activation of the p42/p44 MAPK, JNK, PI3-K/Akt pathways via TNFR1 [106]. On the other hand, TNFR1 can also induce cell death. For example, TNF-α could sensitize myeloma cells to CD95L-induced but not to TRAIL-induced cell death via TNFR1-induced IKKβ-mediated upregulation of CD95 [107]. In myeloma cells co-expressing TNFR1 and TNFR2, cell death was augmented. The fate of TNFR1 expressing tumor cells reflects therefore the pleotropic effects of TNF-α and is highly context dependent [107]. As discussed above, TNFR1 plays a major role in regulating the tumor microenvironment and thereby tumor growth. TNFR1-null mice could not reject orthotopically implanted pancreatic Panc02 tumor cells and exhibited enhanced tumor progression [108] while in WT or TNFR2-deficient mice these tumors were spontaneously rejected within two weeks. Further investigation showed that loss of TNFR1 led to increased tumor infiltration by CD4+Foxp3+Treg cells and a concomitant decrease in the number of infiltrating CD8+ T cells. However, the underlying mechanism and the TNFR1-expressing cells responsible for the observed changes were not identified. In another study, Muller-Hermelink et al. found that the loss of TNFR1 expression on tumor-infiltrating CD4+ T cells altered their ability to suppress tumor growth because it blocked the production of angiogenesis inhibitors such as CXCL9 and CXCL10 [74]. This identified TNFR1 signaling as important for the tumor inhibitory activity of CD4+T cells.
The role of TNFR2 in cancer progression has only recently come to light. Rivas et al. have shown that in addition to TNFR1, TNFR2 also participated in promoting TNF-α-induced breast cancer growth [106] and Al-Lamki et al. reported that a TNF-α mutein that specifically binds TNFR2 increased TNFR2 expression and activated NF-κB and VEGFR2 signaling in renal cell carcinoma, increasing tumor cell proliferation [109]. In addition, TNFR2 can also modulate tumor growth through its effect on tumor infiltrating immune cells and appears to play mainly a tumor-promoting role by inducing an immunosuppressive microenvironment. Sasi et al. have shown that in TNFR2 knockout mice, tumor-induced angiogenesis and the incorporation of bone marrow-derived endothelial progenitor cells into a functional capillary network were decreased, resulting in decreased Lewis lung carcinoma and B16 melanoma cell growth. In the same study, the silencing of TNFR2 in the tumor cells by short-hairpin RNA increased TNF-α-induced apoptosis, implicating TNFR2 in direct and indirect effects on tumor growth [110]. More recently, the role that TNFR2 plays in regulating the tumor immune infiltrate has been revealed. A study by Chen et al. showed that TNFR2 expression defines a maximally suppressive subset of mouse CD4+CD25+FoxP3+ Treg cells. In a Lewis lung carcinoma model, these cells were shown to accumulate in the tumors and exert an immunosuppressive effect that contributed to immune evasion by tumor cells [72]. Another cell type contributing to the immunosuppressive microenvironment that allows tumors to expand are the MDSCs. TNFR2 was identified as a survival factor for these cells, mediating NF-κB-induced cellular FLICE-like inhibitory protein (c-FLIP) expression and thereby inhibiting caspase-8 and apoptosis [102]. TNFR2 is therefore emerging as a driver of the immunosuppressive microenvironment of expanding tumors, protecting them from immune attack by cytotoxic T-cells.
2.3.1. The role of the TNF receptors in metastasis
While the function of TNF and its receptors in tumor initiation and expansion at the primary site has received much attention, fewer studies have addressed the role of these receptors in the process of metastasis, although the evidence suggests that a similar dichotomy in TNF function exists at the metastatic sites. For example, Charles et al. showed that TNFR1-null bone marrow (BM) chimera mice had decreased ovarian carcinoma peritoneal metastasis in comparison to either WT or TNFR2−/− BM chimera mice and this could be attributed to a significant reduction in the production of IL-17 by CD4+ T cells and reduced recruitment of GR-1+ F4/80− cells into the ascitic fluid [111], suggesting that TNFR1 promotes peritoneal metastasis. Liver metastasis was also reportedly reduced in TNFR1-null mice and this was attributed to reduced expression of VCAM-1 on sinusoidal endothelial cells. On the other hand, we have shown that in lung and colon carcinoma cells, TNF-signaling can enhance tumor cell survival and liver metastasis by upregulating IL-6 production through crosstalk with the IGF-I receptor [112, 113].
Chopra et al. analyzed the role of TNF in lung metastasis of melanoma B16F10 cells. They found that treatment of tumor-bearing mice with rhTNF-α resulted in a significant increase in tumor burden and metastatic foci and this correlated with an increase in pulmonary infiltration by CD4+Foxp3+ T cells. Using chimera mice reconstituted with WT or TNFR2−/− BM [114], they found that loss of TNFR2 expression in immune cells resulted in reduced numbers of Treg lymphocytes in the lungs and decreased metastatic burden. Selective depletion of the Treg cells resulted in decreased metastasis, even in the presence of TNF-α treatment, suggesting that Treg-mediated immune suppression is essential to the expansion of lung metastases.
3. TNF regulates the role of different hepatic cell types in liver metastasis
3.1. Vascular sinusoidal endothelial cells
Sinusoidal endothelial cells are likely the first hepatic resident cells to be encountered by metastatic tumor cells in the liver and a barrier to tumor cell extravasation (please see a schematic representation of tumor-hepatic cell interactions in Figure 2). Specific adhesion to the sinusoidal endothelium via cell adhesion receptors induced by inflammatory factors such as TNF-α is essential for leukocyte recruitment to site of inflammation and injury and was also identified as a limiting step in the ability of tumor cells to colonize the liver [13, 27, 31, 115]. Tumor cells can induce CAM expression on the endothelium either directly through cytokine production or by triggering TNF production by inflammatory cells such as the resident KCs and circulating neutrophils. Cancer cells that express counter receptors for these CAM can utilize them to bind to the endothelium and migrate into the space of Disse. These interactions were documented in experimental liver metastasis models of melanoma, colon and lung cancer [10, 31, 116]. Inhibition of endothelial VCAM-1 and E-selectin with specific monoclonal antibodies or antisense oligonucleotides inhibited liver metastasis of melanoma, colorectal and lung carcinoma cells [31, 117, 118]. Liver sinusoidal endothelial cells express both TNFR1 and TNFR2 ([119] and Ham et al., manuscript submitted) and both have been implicated in the induction of endothelial CAM [120, 121]. However, their contributions to CAM expression, tumor cell adhesion and trans-endothelial migration in the liver remains to be fully elucidated.
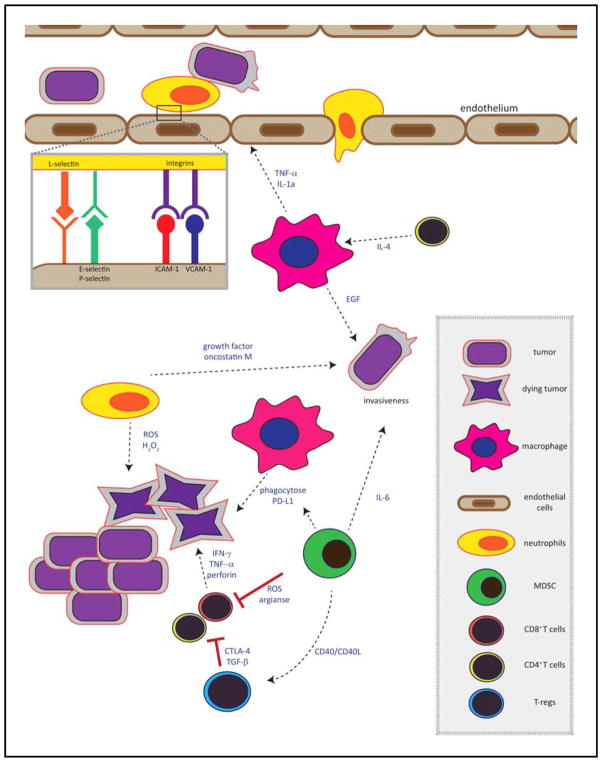
Figure 2. The immune microenvironment of metastases.
Shown is a schematic representation of the complex immune microenvironment of metastatic tumor cells in secondary sites such as the liver. The type of immune cells recruited and the cytokines produced determine tumor cell fate. The tumor cells must successfully extravasate from the blood vessels into the liver/secondary organ via interaction with vascular cell adhesion molecules or via interaction with neutrophils. At this stage tumor cells can be subject to direct cytotoxic effects mediated by cytokines such as IFN-γ, TNF-α, and perforin. Regulatory cells such as Tregs and MDSCs can block T cell-mediated cytotoxicity, allowing tumor cell growth. Growth factors such as IGF-I, PDGF and EGF and cytokines such as IL-6 within the liver microenvironment can increase tumor invasiveness and proliferation and promote tumor expansion.
Activated LSEC can also produce cytokines and chemokines that can play a role in the recruitment of immune cells and thereby contribute to the progression of metastasis. Among the inflammatory cytokines produced by LSEC are IL-1, IL-10, IL-18, TNF-α and TGF-β that are known to play a role in immune cell recruitment and activation. In addition, they can also produce chemokines such as CCL2, CCL3, CCL4, CCL5, CXCL1 and CXCL2 [119] that contribute to recruitment of immune cells. CXCL12 and CXCL9 production by LSEC was shown to mediate transendothelial migration of CD4+T cells [122].
3.2. Hepatic stellate cells
HSCs are located in the subendothelial space also known as the space of Disse between the basolateral surface of hepatocytes and the abluminal aspect of sinusoidal endothelial cells [123]. HSCs are a key player in pathological conditions of the liver such as wound repair and fibrosis and are also involved in cancer progression. Under pathophysiological conditions, the stellate cells are activated and acquire a myofibroblast-like phenotype, a process associated with increased expression of myofibroblast markers such as alpha-smooth muscle actin (α-SMA), tissue inhibitors of metalloproteinases (TIMPs) and type I collagen [123, 124]. Characteristic of activated HSCs is an excessive deposition of an altered extracellular matrix (ECM) or ‘scar’ matrix [125] rich in type I collagen, glycoproteins, proteoglycans and hyaluronan [126]. Stellate cells have emerged as central modulators of hepatic inflammation and immunity. Through the release of chemokines such as monocyte chemotactic peptide (MCP)-1, CCL21 and RANTES, HSCs can mediate the dual function of increasing fibrogenesis, as well as interacting with inflammatory cells to modify the immune response during injury [127, 128]. In an orthotopic mouse model of hepatocellular carcinoma (HCC), HSCs were shown to promote tumor growth by significantly increasing the suppressive immune cell populations of CD4+Foxp3+ Treg cells and CD11b+Gr-1+ MDSC in the tumor micro-environment [129]. Cancer and tumor-associated stromal cells can secrete paracrine factors such as TGF-β, a key regulatory molecule for ECM production [130], platelet-derived growth factor (PDGF) - the most potent proliferative cytokine for activated HSCs, fibroblast growth factor (FGF) and type 1 insulin like growth factor (IGF-1) that together activate HSC and enhance cancer progression [126, 131]. TGF-β is produced by several cell types in the liver including HSCs, KCs, hepatocytes, LSECs and platelets, but autocrine production by HSCs appears to play the major role in the upregulation of collagen synthesis [132]. Signal transduction by the type I TGF-β receptor is mediated via Smad2/3 activation and complex formation with Smad4, a transcription factor that translocates to the nucleus to regulate transcription of downstream target genes [133]. Several studies have shown that suppression of TGF-β/Smad signaling reduces collagen synthesis by HSCs and liver fibrosis [134, 135]. PDGF and PDGFR expression are also upregulated in HSCs during liver injury, resulting in activation of Ras/MEK/MAPK signaling [136, 137] and increased HSC proliferation [138–140]. In addition, HSCs can be stimulated by TNF-α to produce IL-6, thereby amplifying the acute phase response of the liver [123, 125].
HSCs play a critical role during the angiogenic (vascular) phase of liver metastasis by secreting the angiogenic factors VEGF and angiopoietin-1 or -2 that induce a pro-angiogenic microenvironment, leading to recruitment of endothelial cells that form the new microvascular network of expanding metastases [141]. TNF-α is among the numerous growth factors and cytokines implicated in HSC activation [123] but the role of the TNF axis in HSC activation in general and in the context of metastasis, in particular has not been well studied and some of the data available appear to be contradictory. For example, Houglum et al. [142] analyzed the effects of chronically elevated TNF-α serum levels on hepatic collagen metabolism in vivo using nude mice inoculated with TNF-α producing Chinese hamster ovary (CHO) cells. They found that type 1 collagen gene expression and synthesis were inhibited in the livers of these mice. Also, collagen-α1(I) gene expression was reduced in cultured stellate cells treated with TNF-α, independently of the confounding variables of stellate cell activation or proliferation. On the other hand, HSC from TNF1/TNFR2-null mice (TNFR DKO) had reduced pro-collagen-α1(I) expression, decreased proliferation and impaired PDGF-induced pro-mitogenic signaling [143]. Moreover, the authors of the latter study showed that TNFR1, but not TNFR2 had an important role in mediating HSC proliferation and the production of MMP-9 and TIMP-1, although TNF-α did not directly participate in the trans-differentiation of HSC into myofibroblasts. In another study, however, TNF-α was found to increase collagen accumulation and myofibroblast proliferation in chronic inflammation of the gastrointestinal tract, and this was reportedly mediated via TNFR2 [144]. Studies of fibrosis in other organs such as the lungs [145] and kidneys [146] also documented reduced ECM production and decreased fibrosis in TNFR-deficient mice. Thus, while TNF-α has been implicated in HSC activation and altered gene transcription, its precise role and the receptor(s) mediating its effects remain to be fully elucidated.
3.3. Kupffer cells
The resident macrophages of the liver, the KCs play an active role in liver metastasis. These cells line the hepatic sinusoids and represent 80–90% of total tissue macrophages [147]. Due to their location, KCs are the first line of defense against foreign particles absorbed from the gastrointestinal tract and the main source of cytokines and chemokines in response to liver injury or infection. They therefore play a central role in orchestrating the inflammatory microenvironment of the liver. The KCs were initially thought to mediate mainly anti-tumor effects and inhibit liver metastasis. Activation of KCs by Corynebacterium parvum and zymosan was shown to decrease liver metastasis [148] and a decrease in tumor cell adhesion to KCs correlated with increased liver metastasis [149], suggesting that KC activation was inversely correlated with metastasis formation. In an in vitro study, Gardner et al. have shown that liver macrophages co-cultured with tumor cells were activated and could phagocytose the tumor cells [150], suggesting that KCs can inhibit metastases formation by directly killing the tumor cells. However, there is evidence that KCs can also promote metastasis. Khatib et al. reported that the intrasplenic/portal injection of metastatic murine lung carcinoma H-59 and human colorectal carcinoma CX-1 cells into mice increased TNF-α and IL-1α production by KCs, triggering an inflammatory cascade that resulted in upregulated expression of sinusoidal endothelial CAM and increased trans-endothelial migration of the tumor cells [10]. Other studies have similarly documented an upregulation of CAM such as VCAM-1 on liver sinusoidal endothelial cells during the early stages of liver metastasis but identified other cytokines such as IL-18 as the drivers in this process [10, 31, 116, 151].
3.4. The neutrophils
Neutrophils play a critical role in orchestrating the innate and adaptive immune responses in various pathological conditions, including cancer [152, 153]. They are released from the BM as mature, fully differentiated Ly6G+ granulocytes, although, neutrophil precursors can be released during inflammation. In response to an inflammatory trigger, neutrophils home to the site of inflammation where they attach to the vascular endothelium and transmigrate, utilizing endothelial CAM induced by cytokines such as TNF-α [154–156]. Neutrophil mobilization towards these sites can be coordinated by the chemokines CXCL1, CXCXL2 and CXCL5 that bind to their cell surface receptor CXCR2 [157–160].
Neutrophils can play opposing roles in cancer progression in general, and the process of liver metastasis, in particular. Several clinical studies linked increased levels of tumor associated neutrophils (TANs) and the neutrophil chemoattractant IL-8 with poor outcome [161–163], suggesting that neutrophils may have a tumor-promoting effect. Indeed, neutrophils can play an important role in metastasis by promoting tumor invasiveness through cytokine production and the release of matrix degrading enzymes such as MMP-9 and elastase [164]. Neutrophils can also promote metastasis by physically interacting with, and anchoring the circulating tumor cells to the endothelium. This was demonstrated in both lung and liver metastasis models. For example, melanoma cells could attract neutrophils to the lung vascular endothelium through the release of IL-8 [159] and this in turn, induced the expression of integrin CD11b/CD18 on the neutrophils promoting their interaction with melanoma cells via the counter-receptor ICAM-1 and facilitating their trans-endothelial migration [165].
Spicer et al. using an experimental model of liver metastasis reported that neutrophil depletion by an anti-Gr-1 mAb decreased liver metastasis following the intrasplenic injection of lung carcinoma cells [166]. They showed that this was due to increased cell surface expression of CD11b on the neutrophils allowing tumor cell-neutrophil adhesion through a CD11b/ICAM-1 interaction thereby increasing tumor cell arrest on sinusoidal endothelial cells. The same group also reported that neutrophils could increase hepatic metastasis by producing extracellular traps (NETs) that entrap circulating lung carcinoma cells in the hepatic sinusoids [167]. Neutrophils can also participate in forming pre-metastatic niches in the liver. Yamamoto et al. using an orthotopic colon carcinoma model found that CXCL1 levels were significantly increased in tumor bearing mice as compared to controls and this correlated with increased neutrophil numbers in the liver prior to metastases formation [168]. Inhibition of the CXCL1 receptor CXCR2 by a neutralizing antibody abrogated liver metastasis, suggesting that neutrophil recruitment to the liver was essential for metastasis.
Tumor inhibitory functions of neutrophils have also been reported [169–171]. Fridlender et al. have recently shown that a TGFβ receptor kinase inhibitor (SM16) that blocked TGFβ signaling triggered an influx of CD11b+Ly6G+ TANs into flank and lung tumors. These neutrophils were hypersegmented, more cytotoxic to tumor cells and expressed higher levels of pro-inflammatory cytokines such as TNF-α. Depletion of these neutrophils reduced CD8+ T cell recruitment and the tumor-inhibitory effect of TGFβ signaling blockade. The authors proposed that TGFβ within the tumor microenvironment induced polarization of neutrophils from a tumor inhibitory (N1) to a tumor promoting (N2) phenotype. The contribution of this neutrophil polarization mechanism to liver metastasis remains to be elucidated.
In addition to its role in neutrophil migration, TNF-α can also regulate neutrophil survival. At high concentrations (e.g. 10 ng/ml), TNF-α was shown to induce neutrophil apoptosis, while at lower concentrations (0.1–1 ng/ml) it decreased apoptosis [172]. Apoptosis induction in neutrophils was shown to be mediated via TNFR1-activated p38 MAPK and PI3K signaling that resulted in the generation of reactive oxygen species and caspase-3 activation [173]. The specific role of TNF-α in neutrophil survival in the context of liver metastasis remains to be investigated.
3.5. Hepatocytes
Hepatocytes are the main cellular constituent of the liver parenchyma and represent up to 80% of the liver mass. While the role of the TNF axis in liver carcinogenesis and the initiation and progression of hepatocellular carcinoma has received much attention in recent years, the role of hepatocytes in general and their response to TNF-α in particular, in the context of liver metastasis is less clear. During the pre-neoplastic phase of liver carcinogenesis, TNF signaling can induce the proliferation of hepatocyte stem cells through TNFR1 and the loss of this receptor was shown to reduce tumor incidence [51]. TNF-α acted as a powerful mitogen for rat hepatocytes [174] and inhibition of this factor in adjacent endothelial and inflammatory cells resulted in apoptosis of transformed hepatocytes and failure to progress to hepatocellular carcinoma [40]. Moreover, it was shown that NF-κB activation in the liver was associated with an increased risk for hepatocarcinogenesis and was partially involved in angiogenesis, tissue invasion and metastasis [175, 176]. In an apparent contrast to the positive role that NF-κB signaling plays in heptocarcinogenesis, deletion of the upstream kinase NEMO/IKKγ in liver parenchymal cells was found to cause steatohepatitis and hepatocellular carcinoma. This was attributed to compensatory hepatocyte proliferation, inflammation, and activation of liver progenitor cells, resulting in carcinogenesis. Taken together, the results identify NF-κB activation and signaling in hepatocytes as a survival mechanism, suggesting that when impaired, TNF-α can induce cell death in these cells, leading to compensatory proliferation by other hepatocytes with increased risk of DNA mutations and tumorigenesis. In addition, NF-κB activation in non-parenchymal cells such as KCs adjacent to the hepatocytes also contributes to tumorigenesis by providing the emerging tumor cells with essential growth factors [177].
Less is known about the role of hepatocyte activation in the process of metastasis. During metastasis, hepatocytes located at the periphery of the disseminating tumor cells were shown to undergo epithelial-to-mesenchymal transition (EMT) in response to TGF-β released by tumor or non-parenchymal liver cells [13]. This could contribute to the activation of HSC and promote liver metastasis in a TNF-α-dependent manner [178].
3.6. T cells
Tumor cells can express tumor-associated antigens (TAA) that can be recognized by T cells via their T cell receptors (TCR). The two main T cells subsets, the CD4+ and CD8+ T cells, found in the tumor microenvironment play distinct and sometimes opposing roles in response to tumor growth [179]. CD8+ cytotoxic T cells can recognize antigen presented in the context of MHC class I and deliver cytotoxic hits to the target cell through the release of perforin [180–182], other secreted granzymes [183] and Fas/Fas ligand interactions [184, 185]. CD8+ T cells can also suppress tumor growth through the release of the pro-apoptotic cytokines IFN-γ [186] and TNF-α [187, 188]. T cells can also mediate tumoricidal effects through expression of other members of the TNF family such as TNF-receptor apoptosis-inducing ligand (TRAIL) that can interact with corresponding tumor cell receptors leading to cytotoxic effects [189].
The role of CD4+ T cells in tumor progression is more complex as these cells can play a dual role in malignancy, depending on their phenotype. CD4+T helper (TH)1 cells can be tumor-inhibitory through IFN-γ production and the secretion of chemokines that mediate cytotoxic CD8+ T cell expansion [180] and accumulation in the tumor site [190]. In addition, TH1 cells can recruit tumoricidal M1 macrophages to the tumor site enhancing anti-tumor cytotoxicity [191]. On the other hand, the role of TH2, TH17 and Treg cells in malignancy is less clear, and they can participate in both anti- and pro-tumor immunity.
Treg cells are characterized as CD4+CD25+FOXP3+ T cells [192] and are known to play an important role in downregulating autoimmune reactions. Treg cells can be thymus-derived natural Tregs (nTregs) or naive CD4+ T cell-induced Treg (iTregs). The tumor microenvironment, with its abundance of IL-10 [193, 194] and TGF-β [195] promotes the expansion of Treg cells. Increased Treg numbers in cancer was first noted by Woo et al. who in 2001 documented an increased presence of these cells in ovarian and non-small-cell lung cancer (NSLC) patients and showed that they secreted increased levels of TGF-β that could, in turn, affect CD8+ T cell function in vitro [196]. Since this initial report, high Treg levels have been documented in the blood and tumor sites of pancreatic, colorectal, gastric, breast, lung, ovarian and liver cancer patients and this was associated with poor prognosis [197–200]. Counter-intuitively, some studies have reported that the presence of Treg cells in certain cancers such as colon [201] and ovarian [202] carcinoma was associated with good clinical outcome. It is thus important to consider the ratio of Treg cells to effector T cells as an indicator of prognosis [203, 204].
Treg cells can downregulate tumor immuno-surveillance through several mechanisms namely, (i) through secretion of immunosuppressive cytokines such as IL-10 and TGF-β that inhibit effector T cell expansion and function [205, 206] and (ii) by initiating CD8+ T cell killing through the release of the cytolytic factors granzyme B [207], TRAIL [208] and galectin-1 ([209] and reviewed in [204]). Additionally, Treg lymphocytes express cytotoxic T lymphocyte associated protein (CTLA)-4, a cell surface protein that can contribute to a cell-to-cell-dependent suppressive mechanism by binding CD80 and CD86 molecules on antigen presenting cells (APC). This results in upregulation of indoleamine 2,3-dioxygenase (IDO) that catabolizes and depletes tryptophan necessary for effector T cell function [210].
CD8+ T cells were shown to play a role in the early stages of metastasis. For instance, in human colorectal cancer (CRC) patients, primary tumors with evidence of early metastatic invasion had lower levels of CD8+T cells and mRNA for type 1 helper effector T cell products such as INF-γ, granulysin, and granzyme B in comparison to tumors with no evidence of metastatic invasion [211], suggesting that these T cells may inhibit tumor invasion and metastasis. In contrast, CD4+ T cells have been shown to promote tumor invasion and subsequent metastasis. In CD4+ T cell-deficient mice, a significant loss of mammary tumor metastasis to the lung was seen, which was restored upon adoptive transfer of CD4+ T cells [212], indicating that CD4+ T cells could promote lung metastasis. This was attributed to IL-4 production by these cells that in turn, induced epidermal growth factor (EGF) production by tumor associated macrophages (TAM) causing increased tumor invasion. In another study, breast cancer infiltrating CD4+CD25+FOXP3+ Treg cells were found to be essential for pulmonary metastasis. Adoptive transfer of CD4+CD25+ T cells in T-cell deficient Rag1−/− mice effectively restored metastasis of mammary tumor cells to the lung [213]. This suggested that CD4+ T cells could promote cancer cell dissemination at the primary tumor site.
The role of T cells in secondary organ sites of metastasis has not been as extensively studied and is less well understood [214]. Vadrevu et al. using a breast cancer model showed that administering the complement anaphylatoxin C5a receptor could increase metastasis by suppressing effector CD8+ T cell function in the lungs [215]. This was mediated through the recruitment of immature myeloid cells in which TGF-β and IL-10 production was upregulated, resulting in Treg production and CD8+ T cell dysfunction. These results showed that inhibition of CD8+ T cells promoted lung metastasis. There is presently a paucity of information regarding the role of T cell subsets in liver metastasis. Recent studies suggest that MDSCs in the liver can accelerate liver metastases by blocking the proliferation and cytotoxicity of effector T cells, while promoting the development of Tregs [216]. However, additional studies are needed to better understand the overall contribution of T cells to liver metastasis.
The role of the TNF axis in T-Cell mediated immune modulation of metastasis has not been extensively studied. Scheurich et al., showed that resting T lymphocytes do not express TNF receptors but their expression is induced upon T cell activation. Via these receptors, TNF-α is able to exert multiple stimulatory activities such as enhancement of IL-2 receptor and HLA-DR antigen expression. TNF-α can also synergize with IL-2 to stimulate T cell proliferation and IFN-γ production [217]. Interestingly, in a study of pancreatic islet cell multistage carcinogenesis, TNFR1 and IFN-γ signaling were required for tumor-specific CD4+ T cells to produce anti-angiogenic chemokines, prevent tumor angiogenesis, tumor cell proliferation and multistage carcinogenesis. In the absence of TNFR1 or IFN-γ signaling, the T cells promoted rather than inhibited angiogenesis and multistage carcinogenesis [74].
In addition to the pro-inflammatory effects of TNF-α, there is increasing evidence of its immunosuppressive effects. TNF-α can act in concert with IL-2 to activate and expand mouse Treg cells via TNFR2. In fact, TNFR2 is expressed on 30–40% of Tregs of the peripheral activated/memory subset and TNFR2+ Treg represent the most highly suppressive Treg subset. The proportion of these cells was significantly increased in the tumor infiltrate of tumor-bearing mice [218]. In a Lewis lung carcinoma model, TNFR2 expression identified a maximally suppressive Treg subpopulation [218, 219]. In a colitis model, TNF-α signaling via TNFR2 was shown to be required for natural, but not for inducible Treg-mediated suppression [220]. In a TNF-induced experimental lung metastasis model using B16F10 melanoma cells, Chopra et al., showed that TNF treatment resulted in an increase in pulmonary Treg infiltration in a TNFR2-dependent manner and this correlated with increased tumor burden and metastatic foci. Loss of either TNF or TNFR2 in immune cells resulted in lower numbers of Treg cells within the lungs and decreased B16F10 metastasis, and a selective depletion of Treg cells attenuated metastasis even in conjunction with TNF treatment [114]. Similarly, in an experimental liver metastasis model, our laboratory found a marked reduction in the number of liver metastases in TNFR2−/− mice as compared to wild type or TNFR1−/− mice. Analysis of immune cell recruitment to the liver revealed a significant reduction in the number of Treg cells in the immune-infiltrate surrounding the sites of micrometastases in these mice in comparison to wild type mice, confirming that the accumulation of Treg cells in sites of hepatic metastases was also TNFR2-dependent (Ham et al., submitted).
3.7. Myeloid-derived suppressor cells
MDSCs represent a heterogeneous population of myeloid cells generally identified based on the expression of the cell surface markers CD11b+ and GR-1+ (in mice). This population can be further subdivided into monocytic (Mo-MDSC, CD11b+Ly6C+) and granulocytic (G-MDSC, CD11b+Ly6G+) subsets [221]. In humans, MDSCs express either one or both of the common myeloid markers CD33 and CD11b, but do not express HLA-DR [222]. Further studies have defined Mo-MDSC to include CD14+/dull [223, 224] and G-MDSC to include CD15+ [225, 226] cells. Immature myeloid cells are continuously generated in BM of healthy individuals and, normally, differentiate into mature granulocytes or monocytes able to mediate host innate immune responses [153, 227, 228]. In the tumor microenvironment, however, these cells fail to mature and can exert a tumor-promoting effect [102, 153]. Clinical studies confirmed the clinical relevance of these cells in malignant disease showing an increase in circulating monocytic MDSCs in melanoma patients and tumor-infiltrating granulocytic MDSCs in pancreatic cancer patients [221, 229, 230]. Similar findings were reported in mouse tumor models [231].
MDSCs can promote tumor growth through several mechanisms. They can exert immuno-modulatory functions and inhibit T cell proliferation. In a murine model of lung cancer, depletion of MDSCs by specific antibodies increased the number and activity of cancer-reactive NK and T effector cells, thereby decreasing tumor volume [231]. In small cell lung cancer patients, targeting of MDSC by all-trans-retinoic acid improved the immune response generated by a cancer vaccine, increasing the proportion of IFN-γ-positive CD8+ and CD4+ T cells [232]. Other studies showed that granulocytic MDSCs can suppress the activity of CD8+ T cells through production of ROS which can induce post-translational modifications in T-cell receptors and cause antigen-specific T-cell unresponsiveness [233]. Mo-MDSCs can suppress T-cell activation by producing arginase that depletes L-arginine, an essential amino acid for T cell proliferation [221]. MDSCs can also promote tumor growth by increasing the numbers of Treg cells, as was shown in B-cell lymphoma [234] and colon carcinoma [235] models. This is mediated through binding of the immune stimulatory receptor CD40 expressed on MDSCs to CD40L expressed on Treg cells to induce Treg expansion [236]. Thus, MDSCs can promote tumor growth by interfering with lymphocyte trafficking and viability, by depletion of nutrients required for T lymphocyte survival, through release of ROS causing nitration of T cell receptors, and through the activation of Tregs.
Clinical and experimental evidence suggest that MDSCs play a role in metastasis. In a clinical study, an increase in circulating MDSCs was found to correlate with cancer stage and metastatic tumor burden in patients with different solid tumors, including breast cancer [237]. Depletion of MDSCs in an animal model of metastatic breast cancer resulted in decreased spontaneous metastasis [238].
Only a few studies have investigated the mechanism(s) of metastasis promotion by MDSCs. Oh et al. showed that MDSC infiltrating a primary tumor can increase tumor cell invasiveness and increase metastasis [239]. In their study, breast cancer cell-derived factors were shown to induce IL-6 production in MDSCs and this conferred an invasive potential on the breast cancer cells through activation of STAT3 and increased production of proteinases of the ADAM family. Hence, MDSCs may contribute to metastasis by increasing tumor invasion. Liver MDSCs are phenotypically and functionally distinct from MDSCs found in lymphoid organs or the tumor sites [216]. In comparison to their splenic counterparts, they express higher Ly6C and lower Ly6G levels, indicating a monocytic lineage, express lower MHC-I and MHC-II but higher CD31, F4/80 and CD129 levels and produce higher levels of an array of pro-inflammatory cytokines and chemokines [216, 240]. Several lines of evidence suggest that hepatic MDSCs play an important role in liver metastasis. Zhao et al. showed increased levels of CD11b+GR1mid cells in the blood and liver following intrasplenic inoculation of colon and Lewis lung carcinoma cells, and depletion of CD11b+ cells by diphtheria toxin (DTR) administration in CD11b-DTR mice resulted in a marked reduction in liver metastasis [241]. Connolly et al. documented an expansion of highly immunosuppressive liver MDSCs in mice with intra-abdominal colon and pancreatic carcinomas [216]. Moreover, when co-cultured with T cells, these MDSCs had a highly suppressive effect on T cell proliferation and induced Treg expansion. Work by Ilkovitch and Lopez has shown that liver MDSCs may modulate anti-tumor immunity through interaction with KCs and the upregulation of PD-L1, a negative T-cell co-stimulatory receptor on the latter cells [240]. Further characterization of hepatic MDSCs and their tumor-promoting activities could lead to the development of novel and specific targeting strategies.
Several studies implicated TNF-α in the regulation of MDSC function. It was shown to prevent the differentiation of immature MDSCs and augment their suppressive activity [242]. Administration of a TNF-α agonist reduced the suppressive activity of MDSCs and allowed their maturation into dendritic cells and macrophages. This suppressive function appears to be mediated via TNFR2 that was recently identified as a survival factor for MDSCs. Zhao et al. using TNFR-deficient mice found that impaired peripheral accumulation of MDSCs was due to loss of TNFR2 expression on MDSCs [102]. TNFR2 signaling activated NF-κB-mediated expression of c-FLIP, an inhibitor of caspase-8, thereby promoting their survival in the tumor microenvironment. In addition, TNFR2 appears to regulate the suppressive functions of MDSCs.
Polz et al. reported that TNFR2-deficient CD11b+Ly6C+ MDSCs had decreased NO and IL-6 production and were less suppressive of T cell proliferation than their WT counterparts [243]. Hu et al. showed that transmembrane TNF (tmTNF)-α induced splenic MDSCs with enhanced suppressive activities by upregulating arginase-1 and NO synthase expression and this was mediated through activation of TNFR2 and resulted in inhibition of CD3+ T-cell proliferation [244]. Together, these findings identify the TNFR2 as a central player in the regulation of MDSC survival and functions.
4. Targeting the TNF axis for cancer therapy
On the basis of the experimental data described above, and the success of TNF antagonists in inhibiting experimental metastases in mice [36, 40, 47], a number of clinical trials of TNF-α antagonists alone, and in combination with other therapies are currently underway in cancer patients and some preliminary results are available. For example, the soluble TNF receptor Etanercept was tested in a Phase II clinical trial of 30 advanced ovarian cancer patients. A significant rise in immunoreactive TNF was seen in all patients and of the 18 patients who completed the trial, six achieved prolonged disease stabilization [245]. Adalimumab (ADA) is a fully humanized anti-TNF neutralizing IgG that was first assessed in clinical trials in 2002 [246] and there is a case report of a breast cancer patient with advanced disease and metastases to bone and lymph nodes in whom the cancer has remained stable for 3 years after initiation of ADA treatment [247]. Infliximab, another human TNF-specific neutralizing antibody showed efficacy in a phase II study of advanced renal cell cancer refractory to prior treatments with regression of hepatic metastases seen in 3 patients after 6 weeks of treatment [248]. In an excellent review, speculating on the mechanisms of action of anti-TNF treatment for cancer patients, Balkwill proposes that these could include inhibition of cytokine and chemokine production, recruitment of inflammatory cells and inhibition of angiogenesis and ECM degradation. Binding of TNF antagonists to transmembrane TNF could also induce cytotoxic pathways in immunosuppressive cells such as Treg, thereby inhibiting tumor promotion [45].
Overall, TNF-α antagonists were well-tolerated and the limited evidence available to date suggests that they may be of utility in the treatment of advanced disease. Much, however, remains to be learnt about the mechanism of action of this class of inhibitors in order to optimize their clinical use as single agents or in combination with other drugs.
5. Summary
TNF-α is a pleiotropic cytokine with diverse and opposing activities in inflammation and malignancy. TNF signaling is conveyed through 2 cell surface receptors, TNFR1 and TNFR2. There is currently a paucity of information on the role of these receptors in metastasis in general and liver metastasis, in particular. However, evidence from multiple tumor models identifies both receptors as central drivers of the response of the microenvironment to emerging and expanding tumors. While TNFR1 may convey both tumor inhibitory and tumor promoting signals through direct pro-apoptotic or pro-survival effects on tumor and immune cells, TNFR2 appears to play mainly a tumor promoting effect by regulating the survival and function of several types of immunosuppressive cells such as Treg lymphocytes and MDSCs. Increasing appreciation of the importance of the microenvironment in general and the immune microenvironment, in particular, to tumor progression has recently led to major advances in development of therapeutic strategies based on potentiation of anti-tumor immune mechanisms. Targeting of the TNF axis, TNFR2-mediated immunosuppression in particular, may offer an additional approach in the effort to alter the course of malignant disease through modulation of the immune system.
Acknowledgments
This study was made possible by grants MOP-80201 from the Canadian Institute for Health Research and PSR-SIIRI-843 from the Québec Ministère de l’Économie, de l’Innovation et des Exportations (both to PB). BH was supported by a fellowship from the McGill Integrated Cancer Research Training program, MCF was supported by the Henry R. Shibata fellowship from the Cedars Cancer Institute and by a MITACS internship and ZD by a McGill Faculty of Medicine Fellowship.
ABBREVIATIONS
- ADA
Adalimubab
- ADAM
Disintegrin and metalloproteinase
- AOM
Azoxymethane
- APC
Antigen presenting cell
- BM
Bone marrow
- CAM
Cell adhesion molecules
- CDR
Complementarity determining regions
- c-FLIP
Cellular FLICE-like inhibitory protein
- CHO
Chinese hamster ovary
- cIAP-1
Cellular inhibitor of apoptosis
- CRC
Colorectal cancer
- CSF-1
Colony-stimulating factor 1
- CTLA
Cytotoxic T lymphocyte associated protein
- DD
Death domain
- DMBA
7,12-dimetylbenz-anthracene
- DSS
Dextran sulfate sodium
- DTR
Diphtheria toxin
- ECM
Extracellular matrix
- EGF
Epidermal growth factor
- EMT
Epithelial-to-mesenchymal transition
- FADD
Fas associated protein with death domain
- FGF
Fibroblast growth factor
- G-MDSC
Granulocytic myeloid derived suppressor cell
- HCC
Hepatocellular carcinoma
- HSC
Hepatic stellate cell
- ICAM
Intracellular adhesion molecule
- IDO
Indoleamine 2,3-dioxygenase
- IFN-γ
Interferon γ
- IGF-1
Type 1 insulin like growth factor
- IKK
Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor (Ikβ) kinase
- IL
Interleukin
- ILP
Isolated limb perfusion
- iTreg
induced Treg
- JNK
c-Jun N-terminal kinase
- KC
Kupffer cell
- KO
Knockout
- LSEC
Liver sinusoidal endothelial cell
- MAPK/AP-1
Mitogen activated protein kinase/Activating protein 1
- MCP
Monocyte chemotactic peptide
- MDSC
Myeloid derived suppressor cells
- MMP
Metalloproteinase
- Mo-MDSC
Monocytic myeloid derived suppressor cell
- NF-κB
Nuclear factor kappa-light-chain enhancer of activated B cells
- NK
Natural killer
- NO
Nitric oxide
- NSLC
Non-small-cell lung cancer
- nTreg
natural Treg
- PDGF
Platelet-derived growth factor
- RIP
Receptor interacting kinase
- ROS
Reactive oxygen species
- Treg
T regulatory cell
- TAA
Tumor-associated antigen
- Tag
T antigen
- TAM
Tumor-associated macrohpage
- TAN
Tumor-associated neutrophil
- TCR
T cell receptor
- TGF-β
Transforming growth factor β
- TH
T helper
- TIM
TRAF interaction motif
- TIMP
Tissue inhibitor of metalloproteinase
- tm TNF
transmembrane TNF
- TNFR DO
TNF1/TNFR2-null mice
- TNFR1
TNF receptor 1
- TNFR2
TNF receptor 2
- TNF-α
Tumor necrosis factor α
- TNSF
TNF super family
- TPA
12-O-tetradecanoylphorbol 13-acetate
- TRADD
TNF receptor associated protein with death domain
- TRAF
TNF receptor adaptor factor
- TRAIL
TNF-receptor apoptosis-inducing ligand
- uPA
urokinase plasminogen activator
- VCAM
Vascular adhesion molecule
- VEGF
Vascular endothelial growth factor
- VLA-4
Integrin α4β1
- WT
Wild type
- α-SMA
alpha-smooth muscle actin
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors have no conflict of interest to declare.
References
- 1.Nguyen DX, Massague J. Nat Rev Genet. 2007;8:341–352. doi: 10.1038/nrg2101. [DOI] [PubMed] [Google Scholar]
- 2.Chambers AF, Groom AC, MacDonald IC. Nat Rev Cancer. 2002;2:563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 3.Weiss L, Harlos JP, Torhorst J, Gunthard B, Hartveit F, Svendsen E, Huang WL, Grundmann E, Eder M, Zwicknagl M, Cochrane HR, Stock D, Wright C, Horne CHW. J Cancer Res Clin Oncol. 1988;114:605–612. doi: 10.1007/BF00398185. [DOI] [PubMed] [Google Scholar]
- 4.Weiss L. Clin Exp Metastasis. 1992;10:191–199. doi: 10.1007/BF00132751. [DOI] [PubMed] [Google Scholar]
- 5.Fuxe J, Karlsson MC. Semin Cancer Biol. 2012;22:455–461. doi: 10.1016/j.semcancer.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 6.Ren G, Zhao X, Wang Y, Zhang X, Chen X, Xu C, Yuan ZR, Roberts AI, Zhang L, Zheng B, Wen T, Han Y, Rabson AB, Tischfield JA, Shao C, Shi Y. Cell Stem Cell. 2012;11:812–824. doi: 10.1016/j.stem.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vasiljeva O, Papazoglou A, Kruger A, Brodoefel H, Korovin M, Deussing J, Augustin N, Nielsen BS, Almholt K, Bogyo M, Peters C, Reinheckel T. Cancer Res. 2006;66:5242–5250. doi: 10.1158/0008-5472.CAN-05-4463. [DOI] [PubMed] [Google Scholar]
- 8.Mohamed MM, Cavallo-Medved D, Rudy D, Anbalagan A, Moin K, Sloane BF. Cell Physiol Biochem. 2010;25:315–324. doi: 10.1159/000276564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu TC. Cancer Res. 2007;67:6003–6006. doi: 10.1158/0008-5472.CAN-07-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khatib AM, Auguste P, Fallavollita L, Wang N, Samani A, Kontogiannea M, Meterissian S, Brodt P. Am J Pathol. 2005;167:749–759. doi: 10.1016/S0002-9440(10)62048-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okahara H, Yagita H, Miyake K, Okumura K. Cancer Res. 1994;54:3233–3236. [PubMed] [Google Scholar]
- 12.Malhi H, Gores GJ. Gastroenterology. 2008;134:1641–1654. doi: 10.1053/j.gastro.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van den Eynden GG, Majeed AW, Illemann M, Vermeulen PB, Bird NC, Hoyer-Hansen G, Eefsen RL, Reynolds AR, Brodt P. Cancer Res. 2013;73:2031–2043. doi: 10.1158/0008-5472.CAN-12-3931. [DOI] [PubMed] [Google Scholar]
- 14.Vidal-Vanaclocha F. Architectural and Functional Aspects of the Liver with Implications for Cancer Metastasis. In: Brodt P, editor. Liver Metastasis: Biology and Clinical Management. Springer Science + Business Media B. V. and Dordrecht; 2011. p. 1. online resource. [Google Scholar]
- 15.Wisse E, De Zanger RB, Charels K, van Der Smissen P, McCuskey RS. Hepatology. 1985;5:683–692. doi: 10.1002/hep.1840050427. [DOI] [PubMed] [Google Scholar]
- 16.Wisse E. J Ultrastruct Res. 1970;31:125–150. doi: 10.1016/s0022-5320(70)90150-4. [DOI] [PubMed] [Google Scholar]
- 17.Seternes T, Sorensen K, Smedsrod B. Proc Natl Acad Sci USA. 2002;99:7594–7597. doi: 10.1073/pnas.102173299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Limmer A, Ohl J, Kurts C, Ljunggren HG, Reiss Y, Groettrup M, Momburg F, Arnold B, Knolle PA. Nat Med. 2000;6:1348–1354. doi: 10.1038/82161. [DOI] [PubMed] [Google Scholar]
- 19.Scoazec JY, Feldmann G. J Hepatol. 1994;20:296–300. doi: 10.1016/s0168-8278(05)80072-8. [DOI] [PubMed] [Google Scholar]
- 20.Couvelard A, Scoazec JY, Feldmann G. Am J Pathol. 1993;143:738–752. [PMC free article] [PubMed] [Google Scholar]
- 21.Wisse E, Knook DL. Kupffer cells and other liver sinusoidal cells: proceedings of the International Kupffer Cell Symposium held in Noordwijkerhout; the Netherlands. 4–7 September, 1977; Amsterdam; New York: Elsevier/North-Holland Biomedical Press; 1977. [Google Scholar]
- 22.Karrar A, Broome U, Uzunel M, Qureshi AR, Sumitran-Holgersson S. Gut. 2007;56:243–252. doi: 10.1136/gut.2006.093906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wisse E, Braet F, Luo D, De Zanger R, Jans D, Crabbe E, Vermoesen A. Toxicol Pathol. 1996;24:100–111. doi: 10.1177/019262339602400114. [DOI] [PubMed] [Google Scholar]
- 24.Mosher B, Dean R, Harkema J, Remick D, Palma J, Crockett E. J Surg Res. 2001;99:201–210. doi: 10.1006/jsre.2001.6217. [DOI] [PubMed] [Google Scholar]
- 25.Parker GA, Picut CA. Toxicol Pathol. 2005;33:52–62. doi: 10.1080/01926230590522365. [DOI] [PubMed] [Google Scholar]
- 26.Alagusundaramoorthy SS, Gedaly R. World J Gastroenterol. 2014;20:14348–14358. doi: 10.3748/wjg.v20.i39.14348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vidal-Vanaclocha F. The Tumor Microenvironment at Different Stages of Hepatic Metastasis. In: Brodt P, editor. Liver Metastasis: Biology and Clinical Management. Springer Science + Business Media B. V. and Dordrecht; 2011. p. 1. online resource. [Google Scholar]
- 28.Weiss L. Cancer Metastasis Rev. 1992;11:227–235. doi: 10.1007/BF01307179. [DOI] [PubMed] [Google Scholar]
- 29.Barbera-Guillem E, Smith I, Weiss L. Int J Cancer. 1993;53:298–301. doi: 10.1002/ijc.2910530221. [DOI] [PubMed] [Google Scholar]
- 30.Roos E, Dingemans KP, van de Pavert IV, van den Bergh-Weerman MA. Br J Cancer. 1978;38:88–99. doi: 10.1038/bjc.1978.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vidal-Vanaclocha F, Fantuzzi G, Mendoza L, Fuentes AM, Anasagasti MJ, Martin J, Carrascal T, Walsh P, Reznikov LL, Kim SH, Novick D, Rubinstein M, Dinarello CA. Proc Natl Acad Sci USA. 2000;97:734–739. doi: 10.1073/pnas.97.2.734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olaso E, Santisteban A, Bidaurrazaga J, Gressner AM, Rosenbaum J, Vidal-Vanaclocha F. Hepatology. 1997;26:634–642. doi: 10.1002/hep.510260315. [DOI] [PubMed] [Google Scholar]
- 33.Solaun MS, Mendoza L, De Luca M, Gutierrez V, Lopez MP, Olaso E, Lee Sim BK, Vidal-Vanaclocha F. Hepatology. 2002;35:1104–1116. doi: 10.1053/jhep.2002.32528. [DOI] [PubMed] [Google Scholar]
- 34.Kaplan DH, Shankaran V, Dighe AS, Stockert E, Aguet M, Old LJ, Schreiber RD. Proc Natl Acad Sci USA. 1998;95:7556–7561. doi: 10.1073/pnas.95.13.7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Street SE, Cretney E, Smyth MJ. Blood. 2001;97:192–197. doi: 10.1182/blood.v97.1.192. [DOI] [PubMed] [Google Scholar]
- 36.Egberts JH, Cloosters V, Noack A, Schniewind B, Thon L, Klose S, Kettler B, von Forstner C, Kneitz C, Tepel J, Adam D, Wajant H, Kalthoff H, Trauzold A. Cancer Res. 2008;68:1443–1450. doi: 10.1158/0008-5472.CAN-07-5704. [DOI] [PubMed] [Google Scholar]
- 37.Thun MJ, Henley SJ, Gansler T. Novartis Found Symp. 2004;256:6–21. discussion 22–28, 49–52, 266–269. [PubMed] [Google Scholar]
- 38.Borrello MG, Alberti L, Fischer A, Degl’innocenti D, Ferrario C, Gariboldi M, Marchesi F, Allavena P, Greco A, Collini P, Pilotti S, Cassinelli G, Bressan P, Fugazzola L, Mantovani A, Pierotti MA. Proc Natl Acad Sci USA. 2005;102:14825–14830. doi: 10.1073/pnas.0503039102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J, Yang G, Thompson-Lanza JA, Glassman A, Hayes K, Patterson A, Marquez RT, Auersperg N, Yu Y, Hahn WC, Mills GB, Bast RC., Jr Cancer Res. 2004;64:1655–1663. doi: 10.1158/0008-5472.can-03-3380. [DOI] [PubMed] [Google Scholar]
- 40.Pikarsky E, Porat RM, Stein I, Abramovitch R, Amit S, Kasem S, Gutkovich-Pyest E, Urieli-Shoval S, Galun E, Ben-Neriah Y. Nature. 2004;431:461–466. doi: 10.1038/nature02924. [DOI] [PubMed] [Google Scholar]
- 41.Moore RJ, Owens DM, Stamp G, Arnott C, Burke F, East N, Holdsworth H, Turner L, Rollins B, Pasparakis M, Kollias G, Balkwill F. Nat Med. 1999;5:828–831. doi: 10.1038/10552. [DOI] [PubMed] [Google Scholar]
- 42.Balkwill FR, Naylor MS, Malik S. Eur J Cancer. 1990;26:641–644. doi: 10.1016/0277-5379(90)90097-d. [DOI] [PubMed] [Google Scholar]
- 43.Balkwill F. Nat Rev Cancer. 2004;4:540–550. doi: 10.1038/nrc1388. [DOI] [PubMed] [Google Scholar]
- 44.Ries CH, Cannarile MA, Hoves S, Benz J, Wartha K, Runza V, Rey-Giraud F, Pradel LP, Feuerhake F, Klaman I, Jones T, Jucknischke U, Scheiblich S, Kaluza K, Gorr IH, Walz A, Abiraj K, Cassier PA, Sica A, Gomez-Roca C, de Visser KE, Italiano A, Le Tourneau C, Delord JP, Levitsky H, Blay JY, Ruttinger D. Cancer Cell. 2014;25:846–859. doi: 10.1016/j.ccr.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 45.Balkwill F. Nat Rev Cancer. 2009;9:361–371. doi: 10.1038/nrc2628. [DOI] [PubMed] [Google Scholar]
- 46.Carswell EA, Old LJ, Kassel RL, Green S, Fiore N, Williamson B. Proc Natl Acad Sci USA. 1975;72:3666–3670. doi: 10.1073/pnas.72.9.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Popivanova BK, Kitamura K, Wu Y, Kondo T, Kagaya T, Kaneko S, Oshima M, Fujii C, Mukaida N. J Clin Invest. 2008;118:560–570. doi: 10.1172/JCI32453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang H, Bocchetta M, Kroczynska B, Elmishad AG, Chen Y, Liu Z, Bubici C, Mossman BT, Pass HI, Testa JR, Franzoso G, Carbone M. Proc Natl Acad Sci USA. 2006;103:10397–10402. doi: 10.1073/pnas.0604008103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bradley JR. J Pathol. 2008;214:149–160. doi: 10.1002/path.2287. [DOI] [PubMed] [Google Scholar]
- 50.Komori A, Yatsunami J, Suganuma M, Okabe S, Abe S, Sakai A, Sasaki K, Fujiki H. Cancer Res. 1993;53:1982–1985. [PubMed] [Google Scholar]
- 51.Knight B, Yeoh GC, Husk KL, Ly T, Abraham LJ, Yu C, Rhim JA, Fausto N. J Exp Med. 2000;192:1809–1818. doi: 10.1084/jem.192.12.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bodmer JL, Schneider P, Tschopp J. Trends Biochem Sci. 2002;27:19–26. doi: 10.1016/s0968-0004(01)01995-8. [DOI] [PubMed] [Google Scholar]
- 53.Locksley RM, Killeen N, Lenardo MJ. Cell. 2001;104:487–501. doi: 10.1016/s0092-8674(01)00237-9. [DOI] [PubMed] [Google Scholar]
- 54.Dempsey PW, Doyle SE, He JQ, Cheng G. Cytokine Growth Factor Rev. 2003;14:193–209. doi: 10.1016/s1359-6101(03)00021-2. [DOI] [PubMed] [Google Scholar]
- 55.Cabal-Hierro L, Lazo PS. Cell Signal. 2012;24:1297–1305. doi: 10.1016/j.cellsig.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 56.Journal of Nutrition, Growth and Cancer. Food & Nutrition Press; Westport, Conn: 1983. p. v. [Google Scholar]
- 57.Faustman D, Davis M. Nat Rev Drug Discov. 2010;9:482–493. doi: 10.1038/nrd3030. [DOI] [PubMed] [Google Scholar]
- 58.Speeckaert MM, Speeckaert R, Laute M, Vanholder R, Delanghe JR. Am J Nephrol. 2012;36:261–270. doi: 10.1159/000342333. [DOI] [PubMed] [Google Scholar]
- 59.Tuma R, Russell M, Rosendahl M, Thomas GJ., Jr Biochemistry. 1995;34:15150–15156. doi: 10.1021/bi00046a022. [DOI] [PubMed] [Google Scholar]
- 60.Chan FK, Chun HJ, Zheng L, Siegel RM, Bui KL, Lenardo MJ. Science. 2000;288:2351–2354. doi: 10.1126/science.288.5475.2351. [DOI] [PubMed] [Google Scholar]
- 61.Gupta S, Campbell D, Derijard B, Davis RJ. Science. 1995;267:389–393. doi: 10.1126/science.7824938. [DOI] [PubMed] [Google Scholar]
- 62.Read MA, Whitley MZ, Gupta S, Pierce JW, Best J, Davis RJ, Collins T. J Biol Chem. 1997;272:2753–2761. doi: 10.1074/jbc.272.5.2753. [DOI] [PubMed] [Google Scholar]
- 63.Vila-del Sol V, Punzon C, Fresno M. J Immunol. 2008;181:4461–4470. doi: 10.4049/jimmunol.181.7.4461. [DOI] [PubMed] [Google Scholar]
- 64.Tak PP, Firestein GS. J Clin Invest. 2001;107:7–11. doi: 10.1172/JCI11830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Neurath MF, Becker C, Barbulescu K. Gut. 1998;43:856–860. doi: 10.1136/gut.43.6.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Min W, Pober JS. J Immunol. 1997;159:3508–3518. [PubMed] [Google Scholar]
- 67.Weiss T, Grell M, Hessabi B, Bourteele S, Muller G, Scheurich P, Wajant H. J Immunol. 1997;158:2398–2404. [PubMed] [Google Scholar]
- 68.Chan FK, Shisler J, Bixby JG, Felices M, Zheng L, Appel M, Orenstein J, Moss B, Lenardo MJ. J Biol Chem. 2003;278:51613–51621. doi: 10.1074/jbc.M305633200. [DOI] [PubMed] [Google Scholar]
- 69.Moynagh PN. J Cell Sci. 2005;118:4589–4592. doi: 10.1242/jcs.02579. [DOI] [PubMed] [Google Scholar]
- 70.Leeuwenberg JF, van Tits LJ, Jeunhomme TM, Buurman WA. Cytokine. 1995;7:457–462. doi: 10.1006/cyto.1995.0062. [DOI] [PubMed] [Google Scholar]
- 71.Trentin L, Zambello R, Bulian P, Cerutti A, Enthammer C, Cassatella M, Nitti D, Lise M, Agostini C, Semenzato G. Br J Cancer. 1995;71:240–245. doi: 10.1038/bjc.1995.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen X, Subleski J, Kopf H, Howard O, Männel D, Oppenheim J. J Immunol (Baltimore, Md: 1950) 2008;180:6467–6471. doi: 10.4049/jimmunol.180.10.6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Watanabe N, Niitsu Y, Umeno H, Sone H, Neda H, Yamauchi N, Maeda M, Urushizaki I. Cancer Res. 1988;48:650–653. [PubMed] [Google Scholar]
- 74.Muller-Hermelink N, Braumuller H, Pichler B, Wieder T, Mailhammer R, Schaak K, Ghoreschi K, Yazdi A, Haubner R, Sander CA, Mocikat R, Schwaiger M, Forster I, Huss R, Weber WA, Kneilling M, Rocken M. Cancer Cell. 2008;13:507–518. doi: 10.1016/j.ccr.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 75.Baxevanis CN, Voutsas IF, Tsitsilonis OE, Tsiatas ML, Gritzapis AD, Papamichail M. Eur J Immunol. 2000;30:1957–1966. doi: 10.1002/1521-4141(200007)30:7<1957::AID-IMMU1957>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 76.de Wilt JH, ten Hagen TL, de Boeck G, van Tiel ST, de Bruijn EA, Eggermont AM. Br J Cancer. 2000;82:1000–1003. doi: 10.1054/bjoc.1999.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Manusama ER, Stavast J, Durante NM, Marquet RL, Eggermont AM. Eur J Surg Oncol. 1996;22:152–157. doi: 10.1016/s0748-7983(96)90671-x. [DOI] [PubMed] [Google Scholar]
- 78.Brouckaert PG, Leroux-Roels GG, Guisez Y, Tavernier J, Fiers W. Int J Cancer. 1986;38:763–769. doi: 10.1002/ijc.2910380521. [DOI] [PubMed] [Google Scholar]
- 79.Balkwill FR, Lee A, Aldam G, Moodie E, Thomas JA, Tavernier J, Fiers W. Cancer Res. 1986;46:3990–3993. [PubMed] [Google Scholar]
- 80.van der Veen AH, de Wilt JH, Eggermont AM, van Tiel ST, Seynhaeve AL, ten Hagen TL. Br J Cancer. 2000;82:973–980. doi: 10.1054/bjoc.1999.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Watanabe N, Niitsu Y, Umeno H, Kuriyama H, Neda H, Yamauchi N, Maeda M, Urushizaki I. Cancer Res. 1988;48:2179–2183. [PubMed] [Google Scholar]
- 82.Cai W, Kerner ZJ, Hong H, Sun J. Biochem Insights. 2008;2008:15–21. [PMC free article] [PubMed] [Google Scholar]
- 83.Beutler B, Milsark IW, Cerami AC. J Immunol. 2008;181:7–9. [PubMed] [Google Scholar]
- 84.Kettelhut IC, Fiers W, Goldberg AL. Proc Natl Acad Sci USA. 1987;84:4273–4277. doi: 10.1073/pnas.84.12.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tracey KJ, Beutler B, Lowry SF, Merryweather J, Wolpe S, Milsark IW, Hariri RJ, Fahey TJ, 3rd, Zentella A, Albert JD, Shires TC, Cerami A. Science. 1986;234:470–474. doi: 10.1126/science.3764421. [DOI] [PubMed] [Google Scholar]
- 86.Verhoef C, de Wilt JH, Grunhagen DJ, van Geel AN, ten Hagen TL, Eggermont AM. Curr Treat Options Oncol. 2007;8:417–427. doi: 10.1007/s11864-007-0044-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.van Horssen R, Ten Hagen TL, Eggermont AM. Oncologist. 2006;11:397–408. doi: 10.1634/theoncologist.11-4-397. [DOI] [PubMed] [Google Scholar]
- 88.Suganuma M, Okabe S, Marino MW, Sakai A, Sueoka E, Fujiki H. Cancer Res. 1999;59:4516–4518. [PubMed] [Google Scholar]
- 89.Arnott CH, Scott KA, Moore RJ, Robinson SC, Thompson RG, Balkwill FR. Oncogene. 2004;23:1902–1910. doi: 10.1038/sj.onc.1207317. [DOI] [PubMed] [Google Scholar]
- 90.Oshima H, Ishikawa T, Yoshida GJ, Naoi K, Maeda Y, Naka K, Ju X, Yamada Y, Minamoto T, Mukaida N, Saya H, Oshima M. Oncogene. 2014;33:3820–3829. doi: 10.1038/onc.2013.356. [DOI] [PubMed] [Google Scholar]
- 91.Greten FR, Eckmann L, Greten TF, Park JM, Li ZW, Egan LJ, Kagnoff MF, Karin M. Cell. 2004;118:285–296. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 92.Duncombe AS, Heslop HE, Turner M, Meager A, Priest R, Exley T, Brenner MK. J Immunol. 1989;143:3828–3834. [PubMed] [Google Scholar]
- 93.Elbaz O, Mahmoud LA. Leuk Lymphoma. 1994;12:191–195. doi: 10.3109/10428199409059589. [DOI] [PubMed] [Google Scholar]
- 94.Kalthoff H, Roeder C, Gieseking J, Humburg I, Schmiegel W. Proc Natl Acad Sci USA. 1993;90:8972–8976. doi: 10.1073/pnas.90.19.8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schmiegel W, Roeder C, Schmielau J, Rodeck U, Kalthoff H. Proc Natl Acad Sci USA. 1993;90:863–867. doi: 10.1073/pnas.90.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Digel W, Stefanic M, Schoniger W, Buck C, Raghavachar A, Frickhofen N, Heimpel H, Porzsolt F. Blood. 1989;73:1242–1246. [PubMed] [Google Scholar]
- 97.Giri DK, Aggarwal BB. J Biol Chem. 1998;273:14008–14014. doi: 10.1074/jbc.273.22.14008. [DOI] [PubMed] [Google Scholar]
- 98.Liu RY, Fan C, Mitchell S, Chen Q, Wu J, Zuckerman KS. Cancer Res. 1998;58:2217–2223. [PubMed] [Google Scholar]
- 99.Wu S, Boyer CM, Whitaker RS, Berchuck A, Wiener JR, Weinberg JB, Bast RC., Jr Cancer Res. 1993;53:1939–1944. [PubMed] [Google Scholar]
- 100.Hagemann T, Robinson SC, Schulz M, Trumper L, Balkwill FR, Binder C. Carcinogenesis. 2004;25:1543–1549. doi: 10.1093/carcin/bgh146. [DOI] [PubMed] [Google Scholar]
- 101.Stuelten CH, DaCosta Byfield S, Arany PR, Karpova TS, Stetler-Stevenson WG, Roberts AB. J Cell Sci. 2005;118:2143–2153. doi: 10.1242/jcs.02334. [DOI] [PubMed] [Google Scholar]
- 102.Zhao X, Rong L, Li X, Liu X, Deng J, Wu H, Xu X, Erben U, Wu P, Syrbe U, Sieper J, Qin Z. J Clin Invest. 2012;122:4094–4104. doi: 10.1172/JCI64115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ardizzoia A, Lissoni P, Brivio F, Tisi E, Perego MS, Grassi MG, Pittalis S, Crispino S, Barni S, Tancini G. J Biol Regul Homeost Agents. 1992;6:103–107. [PubMed] [Google Scholar]
- 104.Luo JL, Maeda S, Hsu LC, Yagita H, Karin M. Cancer Cell. 2004;6:297–305. doi: 10.1016/j.ccr.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 105.Sturm JW, Magdeburg R, Berger K, Petruch B, Samel S, Bonninghoff R, Keese M, Hafner M, Post S. Int J Cancer. 2003;107:11–21. doi: 10.1002/ijc.11320. [DOI] [PubMed] [Google Scholar]
- 106.Rivas MA, Carnevale RP, Proietti CJ, Rosemblit C, Beguelin W, Salatino M, Charreau EH, Frahm I, Sapia S, Brouckaert P, Elizalde PV, Schillaci R. Exp Cell Res. 2008;314:509–529. doi: 10.1016/j.yexcr.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 107.Rauert H, Stuhmer T, Bargou R, Wajant H, Siegmund D. Cell Death Dis. 2011;2:e194. doi: 10.1038/cddis.2011.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chopra M, Lang I, Salzmann S, Pachel C, Kraus S, Bauerlein CA, Brede C, Garrote AL, Mattenheimer K, Ritz M, Schwinn S, Graf C, Schafer V, Frantz S, Einsele H, Wajant H, Beilhack A. PLoS One. 2013;8:e75737. doi: 10.1371/journal.pone.0075737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Al-Lamki RS, Sadler TJ, Wang J, Reid MJ, Warren AY, Movassagh M, Lu W, Mills IG, Neal DE, Burge J, Vandenebeele P, Pober JS, Bradley JR. Am J Pathol. 2010;177:943–954. doi: 10.2353/ajpath.2010.091218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sasi SP, Yan X, Enderling H, Park D, Gilbert HY, Curry C, Coleman C, Hlatky L, Qin G, Kishore R, Goukassian DA. Oncogene. 2012;31:4117–4127. doi: 10.1038/onc.2011.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Charles KA, Kulbe H, Soper R, Escorcio-Correia M, Lawrence T, Schultheis A, Chakravarty P, Thompson RG, Kollias G, Smyth JF, Balkwill FR, Hagemann T. J Clin Invest. 2009;119:3011–3023. doi: 10.1172/JCI39065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Brodt P. Oncoimmunology. 2012;1:1601–1603. doi: 10.4161/onci.21424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li S, Wang N, Brodt P. Cancer Res. 2012;72:865–875. doi: 10.1158/0008-5472.CAN-11-1357. [DOI] [PubMed] [Google Scholar]
- 114.Chopra M, Riedel SS, Biehl M, Krieger S, von Krosigk V, Bauerlein CA, Brede C, Jordan Garrote AL, Kraus S, Schafer V, Ritz M, Mattenheimer K, Degla A, Mottok A, Einsele H, Wajant H, Beilhack A. Carcinogenesis. 2013;34:1296–1303. doi: 10.1093/carcin/bgt038. [DOI] [PubMed] [Google Scholar]
- 115.Elices MJ, Osborn L, Takada Y, Crouse C, Luhowskyj S, Hemler ME, Lobb RR. Cell. 1990;60:577–584. doi: 10.1016/0092-8674(90)90661-w. [DOI] [PubMed] [Google Scholar]
- 116.Mendoza L, Carrascal T, De Luca M, Fuentes AM, Salado C, Blanco J, Vidal-Vanaclocha F. Hepatology. 2001;34:298–310. doi: 10.1053/jhep.2001.26629. [DOI] [PubMed] [Google Scholar]
- 117.Brodt P, Fallavollita L, Bresalier RS, Meterissian S, Norton CR, Wolitzky BA. Int J Cancer. 1997;71:612–619. doi: 10.1002/(sici)1097-0215(19970516)71:4<612::aid-ijc17>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 118.Khatib AM, Fallavollita L, Wancewicz EV, Monia BP, Brodt P. Cancer Res. 2002;62:5393–5398. [PubMed] [Google Scholar]
- 119.Connolly MK, Bedrosian AS, Malhotra A, Henning JR, Ibrahim J, Vera V, Cieza-Rubio NE, Hassan BU, Pachter HL, Cohen S, Frey AB, Miller G. J Immunol. 2010;185:2200–2208. doi: 10.4049/jimmunol.1000332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kitakata H, Nemoto-Sasaki Y, Takahashi Y, Kondo T, Mai M, Mukaida N. Cancer Res. 2002;62:6682–6687. [PubMed] [Google Scholar]
- 121.Chandrasekharan UM, Siemionow M, Unsal M, Yang L, Poptic E, Bohn J, Ozer K, Zhou Z, Howe PH, Penn M, DiCorleto PE. Blood. 2007;109:1938–1944. doi: 10.1182/blood-2006-05-020875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Schrage A, Wechsung K, Neumann K, Schumann M, Schulzke JD, Engelhardt B, Zeitz M, Hamann A, Klugewitz K. Hepatology. 2008;48:1262–1272. doi: 10.1002/hep.22443. [DOI] [PubMed] [Google Scholar]
- 123.Friedman SL. Physiol Rev. 2008;88:125–172. doi: 10.1152/physrev.00013.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kocabayoglu P, Friedman SL. Front Biosci (Schol Ed) 2013;5:217–230. doi: 10.2741/s368. [DOI] [PubMed] [Google Scholar]
- 125.Sato M, Suzuki S, Senoo H. Cell Struct Funct. 2003;28:105–112. doi: 10.1247/csf.28.105. [DOI] [PubMed] [Google Scholar]
- 126.Gressner AM, Bachem MG. Digestion. 1995;56:335–346. doi: 10.1159/000201257. [DOI] [PubMed] [Google Scholar]
- 127.Friedman SL. Gastroenterol. 2008;134:1655–1669. doi: 10.1053/j.gastro.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Benyon RC, Arthur MJ. Semin Liver Dis. 2001;21:373–384. doi: 10.1055/s-2001-17552. [DOI] [PubMed] [Google Scholar]
- 129.Zhao W, Zhang L, Xu Y, Zhang Z, Ren G, Tang K, Kuang P, Zhao B, Yin Z, Wang X. Lab Invest. 2014;94:182–191. doi: 10.1038/labinvest.2013.139. [DOI] [PubMed] [Google Scholar]
- 130.Senoo H, Kojima N, Sato M. Vitam Horm. 2007;75:131–159. doi: 10.1016/S0083-6729(06)75006-3. [DOI] [PubMed] [Google Scholar]
- 131.Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SF, Csiszar K, Giaccia A, Weninger W, Yamauchi M, Gasser DL, Weaver VM. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gressner AM. J Hepatol. 1995;22:28–36. [PubMed] [Google Scholar]
- 133.Massague J. Nat Rev Mol Cell Biol. 2000;1:169–178. doi: 10.1038/35043051. [DOI] [PubMed] [Google Scholar]
- 134.Kinoshita K, Iimuro Y, Otogawa K, Saika S, Inagaki Y, Nakajima Y, Kawada N, Fujimoto J, Friedman SL, Ikeda K. Gut. 2007;56:706–714. doi: 10.1136/gut.2006.092460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tu X, Zheng X, Li H, Cao Z, Chang H, Luan S, Zhu J, Chen J, Zang Y, Zhang J. Toxicol Sci. 2015;146:157–169. doi: 10.1093/toxsci/kfv081. [DOI] [PubMed] [Google Scholar]
- 136.Pinzani M, Marra F, Carloni V. Liver. 1998;18:2–13. doi: 10.1111/j.1600-0676.1998.tb00120.x. [DOI] [PubMed] [Google Scholar]
- 137.Pinzani M. Front Biosci. 2002;7:d1720–1726. doi: 10.2741/A875. [DOI] [PubMed] [Google Scholar]
- 138.Tsai TH, Shih SC, Ho TC, Ma HI, Liu MY, Chen SL, Tsao YP. PLoS One. 2014;9:e95443. doi: 10.1371/journal.pone.0095443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bartneck M, Warzecha KT, Tacke F. Hepatobiliary Surg Nutr. 2014;3:364–376. doi: 10.3978/j.issn.2304-3881.2014.11.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kwiecinski M, Elfimova N, Noetel A, Tox U, Steffen HM, Hacker U, Nischt R, Dienes HP, Odenthal M. Lab Invest. 2012;92:978–987. doi: 10.1038/labinvest.2012.70. [DOI] [PubMed] [Google Scholar]
- 141.Smedsrod B, Le Couteur D, Ikejima K, Jaeschke H, Kawada N, Naito M, Knolle P, Nagy L, Senoo H, Vidal-Vanaclocha F, Yamaguchi N. Liver Int. 2009;29:490–501. doi: 10.1111/j.1478-3231.2009.01979.x. [DOI] [PubMed] [Google Scholar]
- 142.Houglum K, Buck M, Kim DJ, Chojkier M. Am J Physiol. 1998;274:G840–847. doi: 10.1152/ajpgi.1998.274.5.G840. [DOI] [PubMed] [Google Scholar]
- 143.Tarrats N, Moles A, Morales A, Garcia-Ruiz C, Fernandez-Checa JC, Mari M. Hepatol. 2011;54:319–327. doi: 10.1002/hep.24388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Theiss AL, Simmons JG, Jobin C, Lund PK. J Biol Chem. 2005;280:36099–36109. doi: 10.1074/jbc.M505291200. [DOI] [PubMed] [Google Scholar]
- 145.Ortiz LA, Lasky J, Gozal E, Ruiz V, Lungarella G, Cavarra E, Brody AR, Friedman M, Pardo A, Selman M. Am J Respir Crit Care Med. 2001;163:244–252. doi: 10.1164/ajrccm.163.1.2002123. [DOI] [PubMed] [Google Scholar]
- 146.Guo G, Morrissey J, McCracken R, Tolley T, Liapis H, Klahr S. Am J Physiol Renal Physiol. 2001;280:F777–785. doi: 10.1152/ajprenal.2001.280.5.F777. [DOI] [PubMed] [Google Scholar]
- 147.Kolios G, Valatas V, Kouroumalis E. World J Gastroenterol. 2006;12:7413–7420. doi: 10.3748/wjg.v12.i46.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Pearson HJ, Anderson J, Chamberlain J, Bell PR. Cancer Immunol Immunother. 1986;23:214–216. doi: 10.1007/BF00205652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Meterissian S, Steele GD, Jr, Thomas P. Clin Exp Metastasis. 1993;11:175–182. doi: 10.1007/BF00114975. [DOI] [PubMed] [Google Scholar]
- 150.Gardner CR, Wasserman AJ, Laskin DL. Hepatol. 1991;14:318–324. [PubMed] [Google Scholar]
- 151.Auguste P, Fallavollita L, Wang N, Burnier J, Bikfalvi A, Brodt P. Am J Pathol. 2007;170:1781–1792. doi: 10.2353/ajpath.2007.060886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Pham CT. Nat Rev Immunol. 2006;6:541–550. doi: 10.1038/nri1841. [DOI] [PubMed] [Google Scholar]
- 153.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Nat Rev Immunol. 2012;12:253–268. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Monson KM, Dowlatshahi S, Crockett ET. J Inflamm (Lond) 2007;4:11. doi: 10.1186/1476-9255-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kakkar AK, Lefer DJ. Curr Opin Pharmacol. 2004;4:154–158. doi: 10.1016/j.coph.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 156.Radi ZA, Kehrli ME, Jr, Ackermann MR. J Vet Intern Med. 2001;15:516–529. doi: 10.1892/0891-6640(2001)015<0516:camlta>2.3.co;2. [DOI] [PubMed] [Google Scholar]
- 157.Zhang XW, Liu Q, Wang Y, Thorlacius H. Br J Pharmacol. 2001;133:413–421. doi: 10.1038/sj.bjp.0704087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Burdon PC, Martin C, Rankin SM. Blood. 2005;105:2543–2548. doi: 10.1182/blood-2004-08-3193. [DOI] [PubMed] [Google Scholar]
- 159.Cacalano G, Lee J, Kikly K, Ryan AM, Pitts-Meek S, Hultgren B, Wood WI, Moore MW. Science. 1994;265:682–684. doi: 10.1126/science.8036519. [DOI] [PubMed] [Google Scholar]
- 160.Mei J, Liu Y, Dai N, Favara M, Greene T, Jeyaseelan S, Poncz M, Lee JS, Worthen GS. Immunity. 2010;33:106–117. doi: 10.1016/j.immuni.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Jensen HK, Donskov F, Marcussen N, Nordsmark M, Lundbeck F, von der Maase H. J Clin Oncol. 2009;27:4709–4717. doi: 10.1200/JCO.2008.18.9498. [DOI] [PubMed] [Google Scholar]
- 162.Wislez M, Rabbe N, Marchal J, Milleron B, Crestani B, Mayaud C, Antoine M, Soler P, Cadranel J. Cancer Res. 2003;63:1405–1412. [PubMed] [Google Scholar]
- 163.Bellocq A, Antoine M, Flahault A, Philippe C, Crestani B, Bernaudin JF, Mayaud C, Milleron B, Baud L, Cadranel J. Am J Pathol. 1998;152:83–92. [PMC free article] [PubMed] [Google Scholar]
- 164.Kessenbrock K, Plaks V, Werb Z. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Huh SJ, Liang S, Sharma A, Dong C, Robertson GP. Cancer Res. 2010;70:6071–6082. doi: 10.1158/0008-5472.CAN-09-4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Spicer JD, McDonald B, Cools-Lartigue JJ, Chow SC, Giannias B, Kubes P, Ferri LE. Cancer Res. 2012;72:3919–3927. doi: 10.1158/0008-5472.CAN-11-2393. [DOI] [PubMed] [Google Scholar]
- 167.Cools-Lartigue J, Spicer J, McDonald B, Gowing S, Chow S, Giannias B, Bourdeau F, Kubes P, Ferri L. J Clin Invest. 2013;123:3446–3458. doi: 10.1172/JCI67484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Yamamoto M, Kikuchi H, Ohta M, Kawabata T, Hiramatsu Y, Kondo K, Baba M, Kamiya K, Tanaka T, Kitagawa M, Konno H. Cancer Res. 2008;68:9754–9762. doi: 10.1158/0008-5472.CAN-08-1748. [DOI] [PubMed] [Google Scholar]
- 169.Dissemond J, Weimann TK, Schneider LA, Schneeberger A, Scharffetter-Kochanek K, Goos M, Wagner SN. J Invest Dermatol. 2003;121:936–938. doi: 10.1046/j.1523-1747.2003.12475.x. [DOI] [PubMed] [Google Scholar]
- 170.Lichtenstein A. Blood. 1986;67:657–665. [PubMed] [Google Scholar]
- 171.Fridlender ZG, Sun J, Kim S, Kapoor V, Cheng G, Ling L, Worthen GS, Albelda SM. Cancer Cell. 2009;16:183–194. doi: 10.1016/j.ccr.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.van den Berg JM, Weyer S, Weening JJ, Roos D, Kuijpers TW. Journal of Leukocyte Biology. 2001;69:467–473. [PubMed] [Google Scholar]
- 173.Geering B, Gurzeler U, Federzoni E, Kaufmann T, Simon HU. Blood. 2011;117:5953–5962. doi: 10.1182/blood-2010-11-322206. [DOI] [PubMed] [Google Scholar]
- 174.Iocca HA, Isom HC. Am J Pathol. 2003;163:465–476. doi: 10.1016/s0002-9440(10)63676-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Bond M, Fabunmi RP, Baker AH, Newby AC. FEBS Lett. 1998;435:29–34. doi: 10.1016/s0014-5793(98)01034-5. [DOI] [PubMed] [Google Scholar]
- 176.Koch AE, Polverini PJ, Kunkel SL, Harlow LA, DiPietro LA, Elner VM, Elner SG, Strieter RM. Science. 1992;258:1798–1801. doi: 10.1126/science.1281554. [DOI] [PubMed] [Google Scholar]
- 177.Wullaert A, van Loo G, Heyninck K, Beyaert R. Endocr Rev. 2007;28:365–386. doi: 10.1210/er.2006-0031. [DOI] [PubMed] [Google Scholar]
- 178.Meindl-Beinker NM, Dooley S. J Gastroenterol Hepatol. 2008;23(Suppl 1):S122–127. doi: 10.1111/j.1440-1746.2007.05297.x. [DOI] [PubMed] [Google Scholar]
- 179.Kershaw MH, Westwood JA, Darcy PK. Nat Rev Cancer. 2013;13:525–541. doi: 10.1038/nrc3565. [DOI] [PubMed] [Google Scholar]
- 180.Hadrup S, Donia M, Thor Straten P. Cancer Microenviron. 2013;6:123–133. doi: 10.1007/s12307-012-0127-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Henkart PA. Immunity. 1994;1:343–346. doi: 10.1016/1074-7613(94)90063-9. [DOI] [PubMed] [Google Scholar]
- 182.Kagi D, Ledermann B, Burki K, Seiler P, Odermatt B, Olsen KJ, Podack ER, Zinkernagel RM, Hengartner H. Nature. 1994;369:31–37. doi: 10.1038/369031a0. [DOI] [PubMed] [Google Scholar]
- 183.Trapani JA, Davis J, Sutton VR, Smyth MJ. Curr Opin Immunol. 2000;12:323–329. doi: 10.1016/s0952-7915(00)00094-7. [DOI] [PubMed] [Google Scholar]
- 184.Seki N, Brooks AD, Carter CR, Back TC, Parsoneault EM, Smyth MJ, Wiltrout RH, Sayers TJ. J Immunol. 2002;168:3484–3492. doi: 10.4049/jimmunol.168.7.3484. [DOI] [PubMed] [Google Scholar]
- 185.Caldwell SA, Ryan MH, McDuffie E, Abrams SI. J Immunol. 2003;171:2402–2412. doi: 10.4049/jimmunol.171.5.2402. [DOI] [PubMed] [Google Scholar]
- 186.Detjen KM, Farwig K, Welzel M, Wiedenmann B, Rosewicz S. Gut. 2001;49:251–262. doi: 10.1136/gut.49.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Ratner A, Clark WR. J Immunol. 1993;150:4303–4314. [PubMed] [Google Scholar]
- 188.Barth RJ, Jr, Mule JJ, Spiess PJ, Rosenberg SA. J Exp Med. 1991;173:647–658. doi: 10.1084/jem.173.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Wiley SR, Schooley K, Smolak PJ, Din WS, Huang CP, Nicholl JK, Sutherland GR, Smith TD, Rauch C, Smith CA, Goodwin RG. Immunity. 1995;3:673–682. doi: 10.1016/1074-7613(95)90057-8. [DOI] [PubMed] [Google Scholar]
- 190.Marzo AL, Kinnear BF, Lake RA, Frelinger JJ, Collins EJ, Robinson BW, Scott B. J Immunol. 2000;165:6047–6055. doi: 10.4049/jimmunol.165.11.6047. [DOI] [PubMed] [Google Scholar]
- 191.Haabeth OA, Bogen B, Corthay A. Oncoimmunol. 2012;1:1146–1155. doi: 10.4161/onci.21542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Mougiakakos D, Choudhury A, Lladser A, Kiessling R, Johansson CC. Adv Cancer Res. 2010;107:57–117. doi: 10.1016/S0065-230X(10)07003-X. [DOI] [PubMed] [Google Scholar]
- 193.Seo N, Hayakawa S, Takigawa M, Tokura Y. Immunol. 2001;103:449–457. doi: 10.1046/j.1365-2567.2001.01279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Hara M, Kingsley CI, Niimi M, Read S, Turvey SE, Bushell AR, Morris PJ, Powrie F, Wood KJ. J Immunol. 2001;166:3789–3796. doi: 10.4049/jimmunol.166.6.3789. [DOI] [PubMed] [Google Scholar]
- 195.Knutson KL, Disis ML, Salazar LG. Cancer Immunol Immunother. 2007;56:271–285. doi: 10.1007/s00262-006-0194-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Woo EY, Chu CS, Goletz TJ, Schlienger K, Yeh H, Coukos G, Rubin SC, Kaiser LR, June CH. Cancer Res. 2001;61:4766–4772. [PubMed] [Google Scholar]
- 197.Wolf AM, Wolf D, Steurer M, Gastl G, Gunsilius E, Grubeck-Loebenstein B. Clin Cancer Res. 2003;9:606–612. [PubMed] [Google Scholar]
- 198.Liyanage UK, Moore TT, Joo HG, Tanaka Y, Herrmann V, Doherty G, Drebin JA, Strasberg SM, Eberlein TJ, Goedegebuure PS, Linehan DC. J Immunol. 2002;169:2756–2761. doi: 10.4049/jimmunol.169.5.2756. [DOI] [PubMed] [Google Scholar]
- 199.Ormandy LA, Hillemann T, Wedemeyer H, Manns MP, Greten TF, Korangy F. Cancer Res. 2005;65:2457–2464. doi: 10.1158/0008-5472.CAN-04-3232. [DOI] [PubMed] [Google Scholar]
- 200.Ichihara F, Kono K, Takahashi A, Kawaida H, Sugai H, Fujii H. Clin Cancer Res. 2003;9:4404–4408. [PubMed] [Google Scholar]
- 201.Correale P, Rotundo MS, Del Vecchio MT, Remondo C, Migali C, Ginanneschi C, Tsang KY, Licchetta A, Mannucci S, Loiacono L, Tassone P, Francini G, Tagliaferri P. J Immunother. 2010;33:435–441. doi: 10.1097/CJI.0b013e3181d32f01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Leffers N, Gooden MJ, de Jong RA, Hoogeboom BN, ten Hoor KA, Hollema H, Boezen HM, van der Zee AG, Daemen T, Nijman HW. Cancer Immunol Immunother. 2009;58:449–459. doi: 10.1007/s00262-008-0583-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Sato E, Olson SH, Ahn J, Bundy B, Nishikawa H, Qian F, Jungbluth AA, Frosina D, Gnjatic S, Ambrosone C, Kepner J, Odunsi T, Ritter G, Lele S, Chen YT, Ohtani H, Old LJ, Odunsi K. Proc Natl Acad Sci USA. 2005;102:18538–18543. doi: 10.1073/pnas.0509182102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Facciabene A, Motz GT, Coukos G. Cancer Res. 2012;72:2162–2171. doi: 10.1158/0008-5472.CAN-11-3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Loser K, Apelt J, Voskort M, Mohaupt M, Balkow S, Schwarz T, Grabbe S, Beissert S. J Immunol. 2007;179:365–371. doi: 10.4049/jimmunol.179.1.365. [DOI] [PubMed] [Google Scholar]
- 206.Letterio JJ. Oncogene. 2005;24:5701–5712. doi: 10.1038/sj.onc.1208922. [DOI] [PubMed] [Google Scholar]
- 207.Grossman WJ, Verbsky JW, Tollefsen BL, Kemper C, Atkinson JP, Ley TJ. Blood. 2004;104:2840–2848. doi: 10.1182/blood-2004-03-0859. [DOI] [PubMed] [Google Scholar]
- 208.Akinnusi ME, El Solh AA. Inflamm Allergy Drug Targets. 2007;6:201–209. doi: 10.2174/187152807783334346. [DOI] [PubMed] [Google Scholar]
- 209.Garin MI, Chu CC, Golshayan D, Cernuda-Morollon E, Wait R, Lechler RI. Blood. 2007;109:2058–2065. doi: 10.1182/blood-2006-04-016451. [DOI] [PubMed] [Google Scholar]
- 210.Cederbom L, Hall H, Ivars F. Eur J Immunol. 2000;30:1538–1543. doi: 10.1002/1521-4141(200006)30:6<1538::AID-IMMU1538>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 211.Pages F, Berger A, Camus M, Sanchez-Cabo F, Costes A, Molidor R, Mlecnik B, Kirilovsky A, Nilsson M, Damotte D, Meatchi T, Bruneval P, Cugnenc PH, Trajanoski Z, Fridman WH, Galon J. N Engl J Med. 2005;353:2654–2666. doi: 10.1056/NEJMoa051424. [DOI] [PubMed] [Google Scholar]
- 212.DeNardo DG, Barreto JB, Andreu P, Vasquez L, Tawfik D, Kolhatkar N, Coussens LM. Cancer Cell. 2009;16:91–102. doi: 10.1016/j.ccr.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Tan W, Zhang W, Strasner A, Grivennikov S, Cheng JQ, Hoffman RM, Karin M. Nature. 2011;470:548–553. doi: 10.1038/nature09707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Sceneay J, Smyth MJ, Moller A. Cancer Metastasis Rev. 2013;32:449–464. doi: 10.1007/s10555-013-9420-1. [DOI] [PubMed] [Google Scholar]
- 215.Vadrevu SK, Chintala NK, Sharma SK, Sharma P, Cleveland C, Riediger L, Manne S, Fairlie DP, Gorczyca W, Almanza O, Karbowniczek M, Markiewski MM. Cancer Res. 2014;74:3454–3465. doi: 10.1158/0008-5472.CAN-14-0157. [DOI] [PubMed] [Google Scholar]
- 216.Connolly MK, Mallen-St Clair J, Bedrosian AS, Malhotra A, Vera V, Ibrahim J, Henning J, Pachter HL, Bar-Sagi D, Frey AB, Miller G. Journal of Leukocyte Biology. 2010;87:713–725. doi: 10.1189/jlb.0909607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Scheurich P, Thoma B, Ucer U, Pfizenmaier K. J Immunol. 1987;138:1786–1790. [PubMed] [Google Scholar]
- 218.Chen X, Subleski JJ, Kopf H, Howard OM, Mannel DN, Oppenheim JJ. J Immunol. 2008;180:6467–6471. doi: 10.4049/jimmunol.180.10.6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Chen X, Subleski JJ, Hamano R, Howard OM, Wiltrout RH, Oppenheim JJ. Eur J Immunol. 2010;40:1099–1106. doi: 10.1002/eji.200940022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Housley WJ, Adams CO, Nichols FC, Puddington L, Lingenheld EG, Zhu L, Rajan TV, Clark RB. J Immunol. 2011;186:6779–6787. doi: 10.4049/jimmunol.1003868. [DOI] [PubMed] [Google Scholar]
- 221.Keskinov AA, Shurin MR. Immunobiology. 2015;220:236–242. doi: 10.1016/j.imbio.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 222.Almand B, Clark JI, Nikitina E, van Beynen J, English NR, Knight SC, Carbone DP, Gabrilovich DI. J Immunol. 2001;166:678–689. doi: 10.4049/jimmunol.166.1.678. [DOI] [PubMed] [Google Scholar]
- 223.Poschke I, Mougiakakos D, Hansson J, Masucci GV, Kiessling R. Cancer Res. 2010;70:4335–4345. doi: 10.1158/0008-5472.CAN-09-3767. [DOI] [PubMed] [Google Scholar]
- 224.Filipazzi P, Valenti R, Huber V, Pilla L, Canese P, Iero M, Castelli C, Mariani L, Parmiani G, Rivoltini L. J Clin Oncol. 2007;25:2546–2553. doi: 10.1200/JCO.2006.08.5829. [DOI] [PubMed] [Google Scholar]
- 225.Ko JS, Zea AH, Rini BI, Ireland JL, Elson P, Cohen P, Golshayan A, Rayman PA, Wood L, Garcia J, Dreicer R, Bukowski R, Finke JH. Clin Cancer Res. 2009;15:2148–2157. doi: 10.1158/1078-0432.CCR-08-1332. [DOI] [PubMed] [Google Scholar]
- 226.Rodriguez PC, Ernstoff MS, Hernandez C, Atkins M, Zabaleta J, Sierra R, Ochoa AC. Cancer Res. 2009;69:1553–1560. doi: 10.1158/0008-5472.CAN-08-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.van Ginderachter JA, Beschin A, De Baetselier P, Raes G. Eur J Immunol. 2010;40:2976–2985. doi: 10.1002/eji.201040911. [DOI] [PubMed] [Google Scholar]
- 228.Nicholson LB, Raveney BJ, Munder M. Curr Mol Med. 2009;9:23–29. doi: 10.2174/156652409787314499. [DOI] [PubMed] [Google Scholar]
- 229.Khaled YS, Ammori BJ, Elkord E. J Immunol Res. 2014;2014:879897. doi: 10.1155/2014/879897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Meyer C, Cagnon L, Costa-Nunes CM, Baumgaertner P, Montandon N, Leyvraz L, Michielin O, Romano E, Speiser DE. Cancer Immunol Immunother. 2014;63:247–257. doi: 10.1007/s00262-013-1508-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Srivastava MK, Zhu L, Harris-White M, Kar UK, Huang M, Johnson MF, Lee JM, Elashoff D, Strieter R, Dubinett S, Sharma S. PLoS One. 2012;7:e40677. doi: 10.1371/journal.pone.0040677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Iclozan C, Antonia S, Chiappori A, Chen DT, Gabrilovich D. Cancer Immunol Immunother. 2013;62:909–918. doi: 10.1007/s00262-013-1396-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Kusmartsev S, Nefedova Y, Yoder D, Gabrilovich DI. J Immunol. 2004;172:989–999. doi: 10.4049/jimmunol.172.2.989. [DOI] [PubMed] [Google Scholar]
- 234.Serafini P, Mgebroff S, Noonan K, Borrello I. Cancer Res. 2008;68:5439–5449. doi: 10.1158/0008-5472.CAN-07-6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Huang B, Pan PY, Li Q, Sato AI, Levy DE, Bromberg J, Divino CM, Chen SH. Cancer Res. 2006;66:1123–1131. doi: 10.1158/0008-5472.CAN-05-1299. [DOI] [PubMed] [Google Scholar]
- 236.Pan PY, Ma G, Weber KJ, Ozao-Choy J, Wang G, Yin B, Divino CM, Chen SH. Cancer Res. 2010;70:99–108. doi: 10.1158/0008-5472.CAN-09-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Diaz-Montero CM, Salem ML, Nishimura MI, Garrett-Mayer E, Cole DJ, Montero AJ. Cancer Immunol Immunother. 2009;58:49–59. doi: 10.1007/s00262-008-0523-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Simpson KD, Templeton DJ, Cross JV. J Immunol. 2012;189:5533–5540. doi: 10.4049/jimmunol.1201161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Oh K, Lee OY, Shon SY, Nam O, Ryu PM, Seo MW, Lee DS. Breast Cancer Res. 2013;15:R79. doi: 10.1186/bcr3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Ilkovitch D, Lopez DM. Cancer Res. 2009;69:5514–5521. doi: 10.1158/0008-5472.CAN-08-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Zhao L, Lim SY, Gordon-Weeks AN, Tapmeier TT, Im JH, Cao Y, Beech J, Allen D, Smart S, Muschel RJ. Hepatol. 2013;57:829–839. doi: 10.1002/hep.26094. [DOI] [PubMed] [Google Scholar]
- 242.Sade-Feldman M, Kanterman J, Ish-Shalom E, Elnekave M, Horwitz E, Baniyash M. Immunity. 2013;38:541–554. doi: 10.1016/j.immuni.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 243.Polz J, Remke A, Weber S, Schmidt D, Weber-Steffens D, Pietryga-Krieger A, Muller N, Ritter U, Mostbock S, Mannel DN. Immun Inflamm Dis. 2014;2:121–130. doi: 10.1002/iid3.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Hu X, Li B, Li X, Zhao X, Wan L, Lin G, Yu M, Wang J, Jiang X, Feng W, Qin Z, Yin B, Li Z. J Immunol. 2014;192:1320–1331. doi: 10.4049/jimmunol.1203195. [DOI] [PubMed] [Google Scholar]
- 245.Madhusudan S, Muthuramalingam SR, Braybrooke JP, Wilner S, Kaur K, Han C, Hoare S, Balkwill F, Ganesan TS. J Clin Oncol. 2005;23:5950–5959. doi: 10.1200/JCO.2005.04.127. [DOI] [PubMed] [Google Scholar]
- 246.Sedger LM, McDermott MF. Cytokine Growth Factor Rev. 2014;25:453–472. doi: 10.1016/j.cytogfr.2014.07.016. [DOI] [PubMed] [Google Scholar]
- 247.Ben Musa R, Usha L, Hibbeln J, Mutlu EA. World J Gastroenterol. 2014;20:5912–5917. doi: 10.3748/wjg.v20.i19.5912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Maisey N. Cancer Invest. 2007;25:589–593. doi: 10.1080/07357900701359700. [DOI] [PubMed] [Google Scholar]