Abstract
Elongation Factor-2 Kinase (eEF2K) in an unusual mammalian enzyme that has one known substrate, elongation factor-2. It belongs to a class of kinases, called alpha kinases, that has little sequence identity to the > 500 conventional protein kinases, but performs the same reaction and has similar catalytic residues. The phosphorylation of eEF2 blocks translation elongation, which is thought to be critical to regulating cellular energy usage. Here we report a system for discovering new substrates of alpha kinases and identify the first new substrates of eEF2K including AMPK and alpha4, and determine a sequence motif for the kinase that shows a requirement for threonine residues as the target of phosphorylation. These new substrates suggest that eEF2K has a more diverse role in regulating cellular energy usage that involves multiple pathways and regulatory feedback.
Keywords: eEF2K, eEF2, translation elongation, energy homeostasis, alpha4, AMPK
1. Introduction
Regulation of cellular energy homeostasis is an essential feature of mammalian cells and involves nearly every major biological pathway. While several central energy-regulating pathways have been well characterized, others have remained poorly understood, including the elongation factor-2 kinase (eEF2K). Regulation of translation elongation is an important place to manage cellular energy needs, because translation is one of the most energy intensive processes in the cell, and elongation is the step in translation that requires the most energy, yet little is known about regulation at this level(1–3). The major known mechanism to regulate translation elongation is through the phosphorylation of elongation factor-2 (eEF2), the only known substrate of eEF2K(4,5). The phosphorylation of eEF2 is thought to regulate energy usage by decreasing its affinity for the ribosome and blocking translation elongation(5,6). There is increasing appreciation for the importance of eEF2K in maintaining cellular energy levels, in particular for survival of cancer cells, for example in early stages of tumor development, suggesting a broader role in signaling pathways. eEF2K is tightly regulated through phosphorylation by several major signaling pathways including AMPK, mTOR, ERK, and PKA(7–12). Since no other substrates have been identified for eEF2K besides autophosphorylation, it has been thought that eEF2K represents a terminal effector of energy homeostasis. As eEF2K is an alpha kinase, standard kinase tools have not been applicable to its investigation(13,14). Recent evidence has shown a conserved role for eEF2K in the cellular response to nutrient deprivation and a role in tumor survival(15,16). In certain malignancies, including neuroblastoma and medulloblastoma, eEF2K expression correlates with poor patient survival(15,17). Despite the fundamental importance in responding to nutrient levels, a clear understanding of how the activity of this kinase helps the tumor survive remains elusive. Here, we report a system for finding substrates of alpha kinases like eEF2K and the discovery of several novel substrates of eEF2K, including alpha4, the essential regulatory subunit of protein phosphatase 2A (PP2A) involved in glutamine homeostasis(18,19); and AMPK, the key regulator of cellular energy homeostasis(20,21). With the identification and confirmation of new substrates of eEF2K we provide a sequence motif, which will guide discovery of further eEF2K substrates. Together, these substrates suggest that it is not only through eEF2 that eEF2K regulates energy usage, but rather multiple substrates involved in feedback of energy metabolism.
2. Experimental Procedures
2.1. Plasmids and reagents
Plasmids containing the cDNA for eEF2K as well as the substrates were obtained from DNASU. AMPK kinase dead plasmid was a kind gift from David Carling. MBP was obtained from Millipore.
2.2. Protein Purification
Full-length eEF2K was cloned from a plasmid containing eEF2K cDNA (DNASU) into a modified pET 47b vector (EMD) with an N-terminal Sumo fusion tag instead of the HRV3C cleavage site. The plasmid was transformed into BL21 Rosetta 2 Cells (EMD). An overnight culture was used to inoculate (1:500) 1 L of LB medium containing 50 µg/ml kanacmycin, 30 µg/ml chloramphenicol, and 500 µM zinc sulfate and grown at 37 °C until the cells reached an OD600 of 1.0 to 1.3, at which point they were transferred to 16 °C and induced overnight with 0.2 mM IPTG. The next day, cells were harvested and resuspended in TBS (pH 8.0, 250 mM NaCl) and lysed with a microfluidizer. After pelleting the cell debris, the lysate was loaded onto a column containing Ni-NTA (Qiagen) for IMAC purification. The column was equilibrated with TBS supplemented with 40 mM imidazole pH 8.0, washed with TBS with 50 mM imidazole, and eluted with 4 ml of TBS with 250 mM imidazole. The eluate was then supplemented with 0.5 mM THP (EMD) and concentrated with Amicon centrifugal filters (Millipore). The tag was then cleaved with Sumo protease (Life Sensors) overnight at 4 °C. The protein was then purified with ion exchange chromatography, on a 1 ml HiTrap Q column (GE Lifesciences) using a gradient from Tris pH 8.0 with 40 mM NaCl to Tris pH 8.0 with 500 mM NaCl. The correct fractions were then collected and concentrated and purified further on a Superdex200 gel filtration column (GE Lifescience). Finally, the protein was concentrated to 10 mg/ml. Mutants for the analog sensitive kinase were generated using standard Quichchange mutagenesis and purified in the same way as the wild-type protein. AMPK kinase dead and kinase dead T482A mutants were expressed in BL21 cells as previously reported(22).
2.3. Labeling of cell lysate
HeLa cell lysate was generated from 1L frozen HeLa pellets lysed with a sonicator in HEPES pH 7.5 buffer. Lysates were labeled for 1 hr at room temperature in reactions containing lysate, 3 µM eEF2K kinase (or zero kinase control or EGTA control), and 250 µM N6-Furfuryl ATP γ−thiophosphate (Axxora), 250 µM ATP, 50 mM HEPES pH 7.5, 10 mM MnCl2, 5 µM calmodulin, 150 µM CaCl2, and 3 mM GTP to block nonspecific labeling. The reaction was then quenched with 100 mM EDTA, and a small aliquot was removed for western analysis by incubating it with 0.4 mg/ml paranitrobenzomesylate (PNBM) for 45 minutes at room temperature. The PNBM-labeled lysate was then boiled with SDS sample buffer, and the samples were run on a Western blot using the thiophosphate ester primary antibody (Epitomics) at a dilution of 1:10,000 overnight at 4 °C, followed by anti-rabbit secondary antibody with IR-Dye 800 (Li-Cor) and analyzed on a Li-Cor Odyssey.
2.4. Mass spectrometry analysis of substrates
The remaining thiophoshate-labeled sample was enriched and analyzed for mass spectrometry analysis as previously reported(23,24). In short, the samples were denatured with urea and cleaved with mass spectrometry grade trypsin (Promega) overnight at 37 °C. The peptides were then purified on a Sep-Pak column, and enriched by binding to Sulfolink iodacetyl resin (pierce) overnight at room temperature in the dark. Finally, samples were washed and eluted with oxone and purified by Zip-tip (Millipore). Desalted peptides were resuspended into 10 µL of 0.1% Formic Acid. Peptides were then loaded on to a nanoACQUITY (Waters) UPLC instrument for reversed-phase chromatography with a C18 column (BEH130, 1.7-µm bead size, 100 µm × 100 mm) in front of an LTQ Orbitrap Velos. The LC was operated at a 600nL/min flow rate and peptides were separated over an 80 minute gradient from 2–50% Buffer B (Buffer A: water and 0.1% formic acid, Buffer B: acetonitrile and 0.1% formic acid). Survey scans were recorded over a 350 – 1800 m/z range and MS/MS fragmentation was performed using ETD on the top 8 peaks. Peaklists were generated with the UCSF program PAVA and searched against the SwissProt Homo Sapiens database (downloaded June 27, 2013, 20,264 entries) using Protein Prospector (version 5.10.10). Data was searched with a 20 ppm tolerance for parent and fragment ions, allowing for standard variable modifications and S/T/Y phosphorylation.
2.5. Validation of substrates
Substrates were cloned into a Tag-2 vector (Agilent) containing an N-terminal Flag tag. After mini-prepping the substrates, they were transfected into HEK293T cells using Lipofectamine LTX (Life Technologies) for 48 hours. After transfection, cells were washed with PBS and lysed with TBS containing 1% Triton-X (Sigma) and protease and phosphatase inhibitors (Roche) for 30 minutes on ice. Cells were then pelleted and the clarified lysate was loaded onto Flag magnetic beads (Sigma) pre-equilibrated with lysis buffer. The substrates were incubated with the beads overnight at 4 °C and then washed with lysis buffer and eluted with 3X Flag peptide. The eluted substrates were then incubated with kinase in a reaction containing 10 mM MgCl2, 100 nM purified eEF2K, 50 mM HEPES pH 7.5, 5 µM calmodulin, 150 µM CaCl2 and ATP containing 150 µM cold ATP and 0.25 µCi/µl 32P γ-ATP (Perkin Elmer) for 30 minutes at room temperature. The reaction was then quenched by boiling the samples with SDS sample buffer and analyzed by PAGE. The gel was dried and imaged onto a phosphor screen and analyzed on a Typhoon. For AMPK, validation was performed using the thiophosphate ester antibody and a Dylight 680 anti-His antibody (Pierce) after performing an enzymatic reaction with 0.5 mM ATP-γ-S, 40 nM eEF2K, and 10 µM AMPK holoenzyme. The amount of phosphorylation decrease was quantified and normalized using densitometry analysis of the anti-thiophosphate and anti-His blot respectively, using LI-COR Image Studio Light software. The AMPK motif was searched using ScanProsite(25) and compared to the PhosphoSite database(26).
3. Results
3.1. Development of an analog-sensitive eEF2K
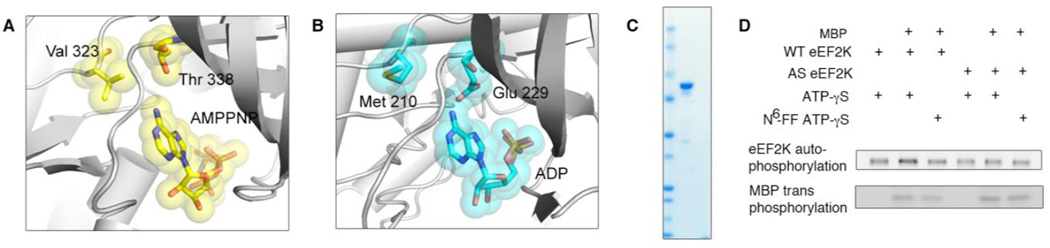
We speculated that eEF2K could have additional substrates along with eEF2, and the phosphorylation of these substrates with eEF2 would help explain the kinase’s importance and mechanism. Therefore, we employed an analog-sensitive(AS)/covalent capture approach to identify additional substrates. This approach relies on engineering a mutant version of the kinase that can accept a bulky analog of ATP that cannot be efficiently used by any other kinase(27,28). This ATP analog can then be used to transfer thiophosphate rather than phosphate to its substrates, which serves as a chemical handle for detection or enrichment and mass spectrometry analysis(23,29). eEF2K is a calcium/calmodulin dependent kinase that belongs to the alpha kinase family, which has negligible sequence identity to the conventional protein kinase family. Therefore, we had to explore novel “gatekeeper” residues that could be mutated to accept bulkier ATP analogs. While there is no structure of eEF2K, there are crystal structures of other alpha kinase family members, so we were able to generate a homology model of eEF2K using the structure of Myosin Heavy Chain Kinase(30) (PDB code 3LMH). The residue that most closely overlays with conventional gatekeeper residues (Fig. 1A) is glutamate, which makes likely hydrogen bonds to the adenine in the active site and has been shown to be essential for activity in the alpha kinase family (Fig. 1B). Therefore, we chose to mutate Met 210, which is not conserved and occupies space in the pocket close to the N6 position of the adenine base. We first developed a recombinant bacterial expression system for the full-length kinase (Fig. 1C), which allowed us to easily produce mutants of the kinase in large quantities. After testing multiple mutants of recombinantly expressed full-length protein, we discovered that M210C showed substantial activity with the bulky analog of ATP, N6-Furfuryl ATP γ-thiophosphate (FF-ATPγS). While conventional protein kinases show poor activity with bulky ATP analogs, the apparently larger atypical active site of wild-type eEF2K still showed activity with sterically enlarged ATP. However, the AS mutant of eEF2K showed some improved tolerance for FF-ATPγS, so we employed this mutant for mass spectrometry identification of novel substrates.
Figure 1. Development of an analog sensitive kinase system for eEF2K.
(A) Active site of Src, indicating gatekeeper residues. Val 323 was initially considered, but Thr 338 became the successful gatekeeper residue. (B) Homology model of eEF2K active site, based on the structure of MHCK. (C) Purification of recombinant full-length eEF2K protein in bacteria, showing the coomassie-stained gel of the final protein. (D) Analysis of eEF2K wild type and mutant usage of ATP analogs for autophosphorylation and transphosphorylation, using MBP as a substrate. The reactions were performed with ATPγS and blotted with MAb 51–8.
3.2. Discovery of new substrates of eEF2K
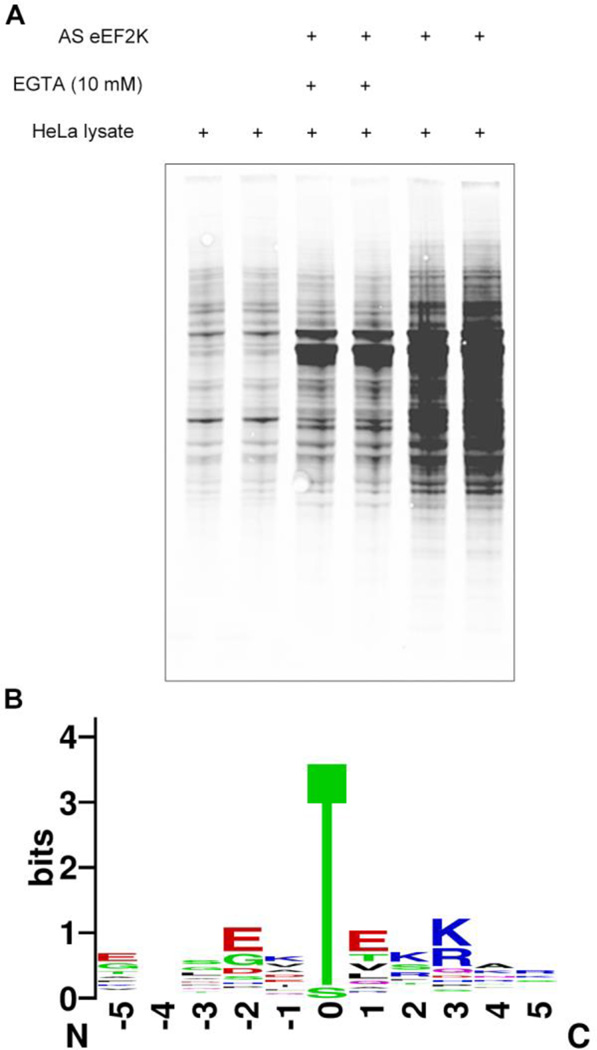
To identify substrates we incubated HeLa cell lysate with purified AS-eEF2K. For background controls we also used lysate alone or AS-eEF2K with EGTA to remove phosphorylation by eEF2K that requires calcium, as eEF2K is calcium dependent kinase. After phosphorylation we monitored the levels of lysate labeling using MAb 51–8 that recognizes a chemically modified thiophosphate(29). As seen in Fig. 2A, substantial phosphorylation specific to eEF2K is seen in the lysate suggesting a range of substrates for eEF2K. Therefore, we analyzed the labeled lysate by iodacetyl-enrichment/oxidative release of substrate peptides and mass spectrometry identification of peptides. We analyzed the peptides with ETD mass spectrometry (see Supplementary Table 1 for all of the peptides found). After filtering against the control samples, the resulting set of peptides showed several phosphopeptides specific to the AS-eEF2K sample (Table 1). In addition to autophosphorylation of eEF2K and phosphorylation of eEF2 at the known Thr57 site, several new targets were identified. The substrates showed a strong sequence motif consisting of acidic residues in the −2 position and basic residues in the +3 position and almost exclusively threonine as the phosphorylated residue (Fig. 2B). The strong threonine preference was consistent with earlier peptide library analysis of eEF2K(31), further supporting the validity of the substrates. The selectivity for threonine is unusual, but not unprecedented in the kinome. A few other kinases have been reported to be threonine specific, including MST1 and MST4. It was recently reported that the position after the DFG motif, termed the “DFG+1 residue”, is responsible for the selectivity for serine vs. threonine, with β-branched amino acids in the DFG+1 position being responsible for the threonine selectivity(32). Although no structural information is known for eEF2K, it does contain a DFG motif that could have the same function in conventional protein kinases. For eEF2K, the DFG+1 residue is an aspartate, which occurs only one other time in the entire kinome, in Trio kinase.
Figure 2. Discovery of substrates of eEF2K.
(A) Phosphorylation of substrates in HeLa lysate was detected using the thiophosphate adduct antibody. Each reaction was performed in duplicate. A small amount of each reaction was removed for this Western blot analysis. The rest was captured on resin for mass spectrometry analysis. (B) Sequence motif for substrates detected in mass spectrometry experiments selectively with eEF2K and calcium. Position 0 shows a strong preference for threonine. Motif was generated using WebLogo(47)
Table 1. Selected phosphorylated hits identified from eEF2K mass spectrometry analysis.
Example peptides discovered with the phosphosite shown in bold. We note that for several of proteins multiple sites were identified. For the full list, see Supplementary Table 1. These substrates are ones that were identified as phosphopeptides and were enriched in the AS kinase samples relative to the lysate only controls.
| Symbol | Name | Phosphopeptide |
|---|---|---|
| EEF2K | Eukaryotic elongation factor 2 kinase | DSGYPSEKRGE |
| EEF2 | Elongation factor 2 | GETRFTDTRKD |
| HSPD1 | Heat shock protein 60 | TKDGVTVAKSI |
| PTMA | Prothymosin alpha | TSSEITTKDLK |
| HNRNPA1 | Heterogeneous nuclear ribonucleoprotein A1 | VSREDSQRPGA |
| SRC8 | Src substrate cortactin | EYQGKTEKHAS |
| NDRG1 | N-myc downstream-regulated gene 1 protein | TSLDGTRSRSH |
| MAP4 | Microtubule-associated protein 4 | TEAAATTRKPE |
| HN1L | Hematological and neurological expressed 1- like protein |
FGSPVTATSRL |
| IGBP1 (alpha4) |
Immunoglobulin-binding protein 1 | EDDEQTLHRAR |
| SARNP | SAP domain-containing ribonucleoprotein | GTTEDTEAKKR |
| SAFB2 | Scaffold attachment factor B2 | VISVKTTSRSK |
| PCNP | PEST proteolytic signal-containing nuclear protein |
AIGSQTTKKAS |
| PRP31 | U4/U6 small nuclear ribonucleoprotein Prp31 | ERLGLTEIRKQ |
3.3. Validation of novel substrates
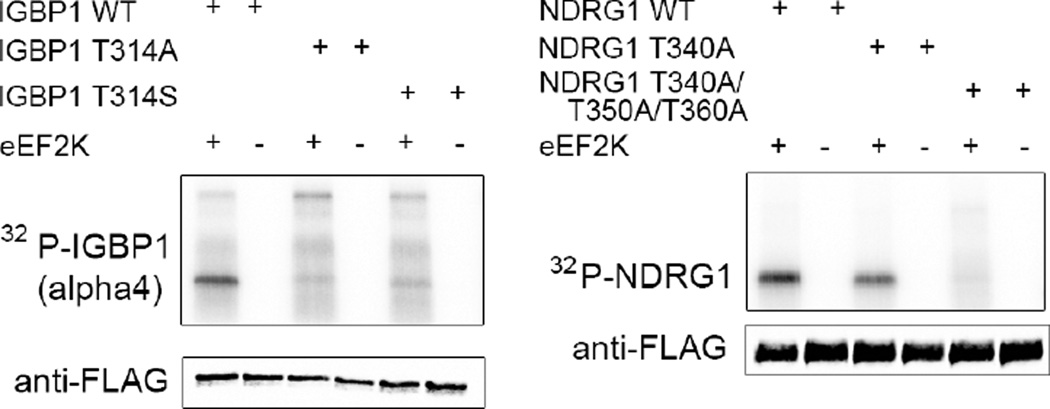
We then validated substrates using an orthogonal in vitro phosphorylation assay with wild-type eEF2K enzyme and either wild-type or phosphosite mutants of the substrates to establish that they were indeed phosphorylated by eEF2K and that the sites we identified were correct. Therefore, we expressed and purified twelve of the top substrates as Flag-tagged full-length proteins in HEK293 cells. We phosphorylated the immunoprecipitated substrates using eEF2K and γ-32P-ATP. Of the twelve new substrates, two in particular showed strong in vitro phosphorylation in vitro and total loss of signal upon phosphosite mutation: alpha4 (also known as IGBP1) and NDRG1 (Fig. 3). Mutation of the alpha4 site to alanine or serine abrogated phosphorylation validating the site and reinforcing the remarkably strong preference for threonine. NDRG1 contains a set of three repeats containing the phosphorylation site so all 3 sites were mutated resulting in complete loss of phosphorylation. The discovery of alpha4 was particularly interesting as a substrate of eEF2K because it has been linked to nutrient response, as an essential protein required for glutamine deprivation, a key nutrient for cancer cells. Furthermore, alpha4 is a regulatory subunit of PP2A(33), which is the phosphatase that acts on phospho-eEF2(34). Alpha4 regulates PP2a by binding to and stabilizing the protein to protect it from degradation(35). Therefore, it is possible that alpha4 acts in a feedback loop for the eEF2K-eEF2 axis. As further evidence of its role in energy regulation, it acts on mTOR signaling by regulating dephosphorylation of eIF4E and S6 Kinase(36). The function of NDRG1 has been debated but it is well known as a substrate of AKT, is completely repressed by Myc(37), and is thought to have a role in the cell’s response to stress from several sources including hypoxia and heavy metals(38–41).
Figure 3. Discovery and validation of two new substrates of eEF2K: IGBP1 (alpha 4) and NDRG1.
Wild type and mutant IGBP1 and NDRG1 proteins were purified from HEK293T cells and phosphorylated using 32P-γ-ATP. The samples were analyzed by SDS-PAGE followed by autoradiography. Samples were also analyzed by western blot for loading prior to addition of the 32P-γ-ATP.
3.4. Bioinformatics discovery of additional substrates and confirmation of substrate motif
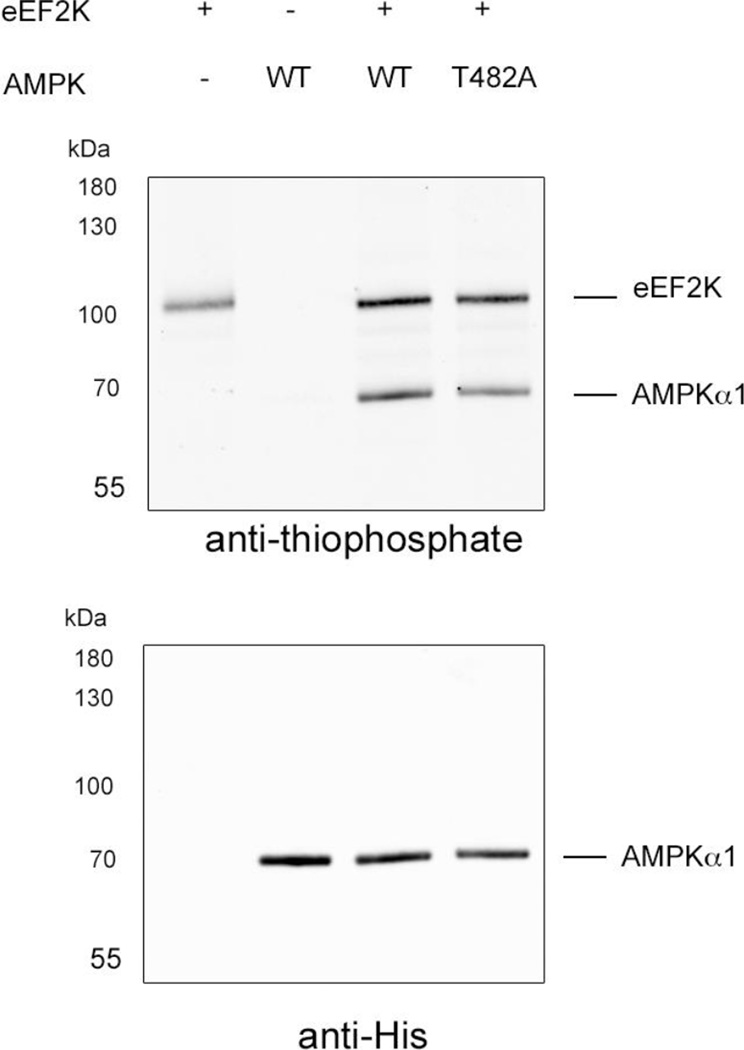
To find additional substrates, validate our sequence motif, and overcome false negatives from the substrate capture method (see discussion below), we scanned the proteome for the motif (that have a glutamate or aspartate in the −2 position and a lysine or arginine in the +3 position and a threonine at the 0 position) and filtering for known phosphorylation sites led to the identification of the alpha subunit of AMPK at Thr 482. While eEF2K has been long known to be a substrate of AMPK(7), the reverse has never been found. The phosphorylation site on AMPK was previously found in a proteomics screen, but it was not known which kinase is responsible. Moreover, the phosphorylation occurs on a regulatory tail of AMPK, proximal to other regulatory phosphorylation sites by GSK3, suggesting a potential negative feedback mechanism(42). We expressed the AMPK holo enzyme with a recombinant multicistronic bacterial expression system and looked for phosphorylation using wild-type eEF2K and ATPγS (Fig. 4). We then saw strong phosphorylation of the alpha subunit that was decreased by 18% by a T482A mutation, validating the phosphorylation by eEF2K of AMPK. Because only a partial reduction of phosphorylation is observed in the mutant, likely additional sites of phosphorylation exist on AMPK by eEF2K, further enhancing the connection between these two major energy-regulating enzymes.
Figure 4. eEF2K phosphorylates AMPK on T482.
Recombinant AMPK kinase dead was expressed recombinantly in bacteria as a tricistronic holoenzyme comprising a,b, and g subunits. The protein was then phosphorylated using ATPγS and analyzed by western blot. The phosphorylation is seen on the α subunit, which is also the one His-tagged for purification.
4. Discussion
The analog sensitive technique is a powerful approach for identifying novel substrates of protein kinase. Here we have extended it to an unusual family of kinases that bears no sequence identity to the large family of conventional eukaryotic protein kinases. We have noticed that the substrate capture and release approach possesses can lead to false negative identifications due to the following circumstances: 1) low protein abundance, 2) the presence of a cysteine in a tryptic peptide containing a phosphorothioate will not be released in following the capture step, 3) spacing of lysine or arginine residues in the phosphorylation motif can lead to missed cleavages or very short peptides which are missed by the LC/MS. In order to mitigate these drawbacks we have found that the kinase motif discovered by the thiophosphate capture is a useful tool that can enable a bioinformatics based method to search the proteome(43). We have used these two approaches, the chemical genetic and bioinformatics methods, as a way to find new substrates of protein kinase, both conventional and atypical.
The alpha-kinase eEF2K was initially discovered based on the calcium stimulated phosphorylation of a 100kD protein in cell lysates incubated with 32P-ATP(44). It was always a mystery that eEF2K would have only one substrate, given the huge number of substrates known for most kinases and given major role of eEF2K in integrating energy signals in the cell. Here, we have found additional substrates for the kinase that add to the complex picture of energy-regulating signaling networks. These substrates suggest that eEF2K has a broader role in regulating cellular energy than just eEF2 regulation, through phosphorylation of an array of substrates involved in these processes. We note that eEF2 is one of the most abundant proteins in cells(45,46). Therefore, it is possible that additional substrates could only be phosphorylated either based on localization or after eEF2 is fully phosphorylated. We speculate that additional substrates could be a feedback mechanism for enabling translation to start again when nutrients are available. Lastly, we report that we have extended the analog-sensitive approach towards alpha kinases, which should facilitate discovery of new substrates of this challenging family of kinases. Future structural work on eEF2K should shed insight onto the mechanisms of substrate recognition by eEF2K and other alpha kinases.
Supplementary Material
Research highlights.
eEF2K has multiple substrates beyond eEF2, including AMPK and alpha4.
Atypical kinases can be made analog sensitive for discovering new substrates.
eEF2K has an unusual substrate motif that requires threonine.
Acknowledgments
This work was supported by the Howard Hughes Medical Institute and NIH U19AI109622 and R01AI099245 to K.M.S. Mass spectrometry was provided by the Bio-Organic Biomedical Mass Spectrometry Resource at UCSF (A.L. Burlingame, Director), supported by the Biomedical Technology Research Centers program of the NIH National Institute of General Medical Sciences, NIH NIGMS 8P41GM103481 (the Thermo Scientifi LTQ-Orbitrap Velos is specifically supported by P41GM103481 and Howard Hughes Medical Institute). M.B.L. is a Merck fellow of the Helen Hay Whitney Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Browne GJ, Proud CG. Regulation of peptide-chain elongation in mammalian cells. Eur J Biochem. 2002;269:5360–5368. doi: 10.1046/j.1432-1033.2002.03290.x. [DOI] [PubMed] [Google Scholar]
- 2.Buttgereit F, Brand MD. A hierarchy of ATP-consuming processes in mammalian cells. The Biochemical journal. 1995;312(Pt 1):163–167. doi: 10.1042/bj3120163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.White-Gilbertson S, Kurtz DT, Voelkel-Johnson C. The role of protein synthesis in cell cycling and cancer. Mol Oncol. 2009;3:402–408. doi: 10.1016/j.molonc.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitsui K, Brady M, Palfrey HC, Nairn AC. Purification and characterization of calmodulin-dependent protein kinase III from rabbit reticulocytes and rat pancreas. J Biol Chem. 1993;268:13422–13433. [PubMed] [Google Scholar]
- 5.Ryazanov AG, Shestakova EA, Natapov PG. Phosphorylation of elongation factor 2 by EF-2 kinase affects rate of translation. Nature. 1988;334:170–173. doi: 10.1038/334170a0. [DOI] [PubMed] [Google Scholar]
- 6.Carlberg U, Nilsson A, Nygard O. Functional properties of phosphorylated elongation factor 2. Eur J Biochem. 1990;191:639–645. doi: 10.1111/j.1432-1033.1990.tb19169.x. [DOI] [PubMed] [Google Scholar]
- 7.Browne GJ, Finn SG, Proud CG. Stimulation of the AMP-activated protein kinase leads to activation of eukaryotic elongation factor 2 kinase and to its phosphorylation at a novel site, serine 398. J Biol Chem. 2004;279:12220–12231. doi: 10.1074/jbc.M309773200. [DOI] [PubMed] [Google Scholar]
- 8.Browne GJ, Proud CG. A novel mTOR-regulated phosphorylation site in elongation factor 2 kinase modulates the activity of the kinase and its binding to calmodulin. Mol Cell Biol. 2004;24:2986–2997. doi: 10.1128/MCB.24.7.2986-2997.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pavur KS, Petrov AN, Ryazanov AG. Mapping the Functional Domains of Elongation Factor-2 Kinase†. Biochemistry. 2000;39:12216–12224. doi: 10.1021/bi0007270. [DOI] [PubMed] [Google Scholar]
- 10.Fabrini R, De Luca A, Stella L, Mei G, Orioni B, Ciccone S, Federici G, Lo Bello M, Ricci G. Monomer-dimer equilibrium in glutathione transferases: a critical re-examination. Biochemistry. 2009;48:10473–10482. doi: 10.1021/bi901238t. [DOI] [PubMed] [Google Scholar]
- 11.Tavares CD, O'Brien JP, Abramczyk O, Devkota AK, Shores KS, Ferguson SB, Kaoud TS, Warthaka M, Marshall KD, Keller KM, Zhang Y, Brodbelt JS, Ozpolat B, Dalby KN. Calcium/calmodulin stimulates the autophosphorylation of elongation factor 2 kinase on Thr-348 and Ser-500 to regulate its activity and calcium dependence. Biochemistry. 2012;51:2232–2245. doi: 10.1021/bi201788e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Li W, Williams M, Terada N, Alessi DR, Proud CG. Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO J. 2001;20:4370–4379. doi: 10.1093/emboj/20.16.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drennan D, Ryazanov AG. Alpha-kinases: analysis of the family and comparison with conventional protein kinases. Prog Biophys Mol Biol. 2004;85:1–32. doi: 10.1016/S0079-6107(03)00060-9. [DOI] [PubMed] [Google Scholar]
- 14.Middelbeek J, Clark K, Venselaar H, Huynen MA, van Leeuwen FN. The alpha-kinase family: an exceptional branch on the protein kinase tree. Cell Mol Life Sci. 2010;67:875–890. doi: 10.1007/s00018-009-0215-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leprivier G, Remke M, Rotblat B, Dubuc A, Mateo AR, Kool M, Agnihotri S, El-Naggar A, Yu B, Somasekharan SP, Faubert B, Bridon G, Tognon CE, Mathers J, Thomas R, Li A, Barokas A, Kwok B, Bowden M, Smith S, Wu X, Korshunov A, Hielscher T, Northcott PA, Galpin JD, Ahern CA, Wang Y, McCabe MG, Collins VP, Jones RG, Pollak M, Delattre O, Gleave ME, Jan E, Pfister SM, Proud CG, Derry WB, Taylor MD, Sorensen PH. The eEF2 kinase confers resistance to nutrient deprivation by blocking translation elongation. Cell. 2013;153:1064–1079. doi: 10.1016/j.cell.2013.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tekedereli I, Alpay SN, Tavares CD, Cobanoglu ZE, Kaoud TS, Sahin I, Sood AK, Lopez-Berestein G, Dalby KN, Ozpolat B. Targeted silencing of elongation factor 2 kinase suppresses growth and sensitizes tumors to doxorubicin in an orthotopic model of breast cancer. PLoS ONE. 2012;7:e41171. doi: 10.1371/journal.pone.0041171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Molenaar JJ, Koster J, Zwijnenburg DA, van Sluis P, Valentijn LJ, van der Ploeg I, Hamdi M, van Nes J, Westerman BA, van Arkel J, Ebus ME, Haneveld F, Lakeman A, Schild L, Molenaar P, Stroeken P, van Noesel MM, Ora I, Santo EE, Caron HN, Westerhout EM, Versteeg R. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–593. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- 18.Kong M, Ditsworth D, Lindsten T, Thompson CB. Alpha4 is an essential regulator of PP2A phosphatase activity. Mol Cell. 2009;36:51–60. doi: 10.1016/j.molcel.2009.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reid MA, Wang WI, Rosales KR, Welliver MX, Pan M, Kong M. The B55alpha subunit of PP2A drives a p53-dependent metabolic adaptation to glutamine deprivation. Mol Cell. 2013;50:200–211. doi: 10.1016/j.molcel.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 20.Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol. 2011;13:1016–1023. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hardie DG, Schaffer BE, Brunet A. AMPK: An Energy-Sensing Pathway with Multiple Inputs and Outputs. Trends Cell Biol. 2015 doi: 10.1016/j.tcb.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neumann D, Woods A, Carling D, Wallimann T, Schlattner U. Mammalian AMP-activated protein kinase: functional, heterotrimeric complexes by co-expression of subunits in Escherichia coli. Protein Expr Purif. 2003;30:230–237. doi: 10.1016/s1046-5928(03)00126-8. [DOI] [PubMed] [Google Scholar]
- 23.Hertz NT, Wang BT, Allen JJ, Zhang C, Dar AC, Burlingame AL, Shokat KM. Chemical genetic approach for kinase-substrate mapping by covalent capture of thiophosphopeptides and analysis by mass spectrometry. Curr Protoc Chem Biol. 2010;2:15–36. doi: 10.1002/9780470559277.ch090201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ultanir SK, Hertz NT, Li G, Ge WP, Burlingame AL, Pleasure SJ, Shokat KM, Jan LY, Jan YN. Chemical genetic identification of NDR1/2 kinase substrates AAK1 and Rabin8 Uncovers their roles in dendrite arborization and spine development. Neuron. 2012;73:1127–1142. doi: 10.1016/j.neuron.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Castro E, Sigrist CJ, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, Bairoch A, Hulo N. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006;34:W362–W365. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hornbeck PV, Chabra I, Kornhauser JM, Skrzypek E, Zhang B. PhosphoSite: A bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics. 2004;4:1551–1561. doi: 10.1002/pmic.200300772. [DOI] [PubMed] [Google Scholar]
- 27.Shah K, Liu Y, Deirmengian C, Shokat KM. Engineering unnatural nucleotide specificity for Rous sarcoma virus tyrosine kinase to uniquely label its direct substrates. Proc Natl Acad Sci U S A. 1997;94:3565–3570. doi: 10.1073/pnas.94.8.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Specht KM, Shokat KM. The emerging power of chemical genetics. Curr Opin Cell Biol. 2002;14:155–159. doi: 10.1016/s0955-0674(02)00317-4. [DOI] [PubMed] [Google Scholar]
- 29.Allen JJ, Li M, Brinkworth CS, Paulson JL, Wang D, Hubner A, Chou WH, Davis RJ, Burlingame AL, Messing RO, Katayama CD, Hedrick SM, Shokat KM. A semisynthetic epitope for kinase substrates. Nat Methods. 2007;4:511–516. doi: 10.1038/nmeth1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye Q, Crawley SW, Yang Y, Cote GP, Jia Z. Crystal structure of the alpha-kinase domain of Dictyostelium myosin heavy chain kinase A. Sci Signal. 2010;3:ra17. doi: 10.1126/scisignal.2000525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crawley SW, Cote GP. Determinants for substrate phosphorylation by Dictyostelium myosin II heavy chain kinases A and B and eukaryotic elongation factor-2 kinase. Biochim Biophys Acta. 2008;1784:908–915. doi: 10.1016/j.bbapap.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 32.Chen C, Ha BH, Thevenin AF, Lou HJ, Zhang R, Yip KY, Peterson JR, Gerstein M, Kim PM, Filippakopoulos P, Knapp S, Boggon TJ, Turk BE. Identification of a major determinant for serine-threonine kinase phosphoacceptor specificity. Mol Cell. 2014;53:140–147. doi: 10.1016/j.molcel.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McConnell JL, Watkins GR, Soss SE, Franz HS, McCorvey LR, Spiller BW, Chazin WJ, Wadzinski BE. Alpha4 is a ubiquitin-binding protein that regulates protein serine/threonine phosphatase 2A ubiquitination. Biochemistry. 2010;49:1713–1718. doi: 10.1021/bi901837h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Redpath NT, Proud CG. Activity of protein phosphatases against initiation factor-2 and elongation factor-2. Biochem J. 1990;272:175–180. doi: 10.1042/bj2720175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.LeNoue-Newton M, Watkins GR, Zou P, Germane KL, McCorvey LR, Wadzinski BE, Spiller BW. The E3 ubiquitin ligase- and protein phosphatase 2A (PP2A)-binding domains of the Alpha4 protein are both required for Alpha4 to inhibit PP2A degradation. J Biol Chem. 2011;286:17665–17671. doi: 10.1074/jbc.M111.222414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu E, Knutzen CA, Krauss S, Schweiger S, Chiang GG. Control of mTORC1 signaling by the Opitz syndrome protein MID1. Proc Natl Acad Sci U S A. 2011;108:8680–8685. doi: 10.1073/pnas.1100131108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J, Chen S, Zhang W, Zhang J, Liu X, Shi H, Che H, Wang W, Li F, Yao L. Human differentiation-related gene NDRG1 is a Myc downstream-regulated gene that is repressed by Myc on the core promoter region. Gene. 2008;417:5–12. doi: 10.1016/j.gene.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 38.Agarwala KL, Kokame K, Kato H, Miyata T. Phosphorylation of RTP an ER stress-responsive cytoplasmic protein. Biochem Biophys Res Commun. 2000;272:641–647. doi: 10.1006/bbrc.2000.2833. [DOI] [PubMed] [Google Scholar]
- 39.Lachat P, Shaw P, Gebhard S, van Belzen N, Chaubert P, Bosman FT. Expression of NDRG1, a differentiation-related gene, in human tissues. Histochem Cell Biol. 2002;118:399–408. doi: 10.1007/s00418-002-0460-9. [DOI] [PubMed] [Google Scholar]
- 40.Zhou D, Salnikow K, Costa M. Cap43, a novel gene specifically induced by Ni2+ compounds. Cancer Res. 1998;58:2182–2189. [PubMed] [Google Scholar]
- 41.Ellen TP, Ke Q, Zhang P, Costa M. NDRG1, a growth and cancer related gene: regulation of gene expression and function in normal and disease states. Carcinogenesis. 2008;29:2–8. doi: 10.1093/carcin/bgm200. [DOI] [PubMed] [Google Scholar]
- 42.Suzuki T, Bridges D, Nakada D, Skiniotis G, Morrison SJ, Lin JD, Saltiel AR, Inoki K. Inhibition of AMPK catabolic action by GSK3. Mol Cell. 2013;50:407–419. doi: 10.1016/j.molcel.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lipp JJ, Marvin MC, Shokat KM, Guthrie C. SR protein kinases promote splicing of nonconsensus introns. Nat Struct Mol Biol. 2015;22:611–617. doi: 10.1038/nsmb.3057. [DOI] [PubMed] [Google Scholar]
- 44.Ryazanov AG. Ca2+/calmodulin-dependent phosphorylation of elongation factor 2. FEBS Lett. 1987;214:331–334. doi: 10.1016/0014-5793(87)80081-9. [DOI] [PubMed] [Google Scholar]
- 45.Geiger T, Wehner A, Schaab C, Cox J, Mann M. Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.014050. M111 014050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nagaraj N, Wisniewski JR, Geiger T, Cox J, Kircher M, Kelso J, Paabo S, Mann M. Deep proteome and transcriptome mapping of a human cancer cell line. Mol Syst Biol. 2011;7:548. doi: 10.1038/msb.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.