Abstract
The simultaneous measurement of T cell function with recovery of individual T cells would greatly facilitate characterizing antigen-specific responses both in vivo and in model systems. We have developed a microraft array methodology that automatically measures the ability of individual T cells to kill a population of target cells and viably sorts specific cells into a 96-well plate for expansion.
A human T cell culture was generated against the influenza M1p antigen. Individual microrafts on a 70 × 70 array were loaded with on average 1 CD8+ cell from the culture and a population of M1p presenting target cells. Target cell killing, measured by fluorescence microscopy, was quantified in each microraft. The rates of target cell death among the individual CD8+ T cells varied greatly; however, individual T cells maintained their rates of cytotoxicity throughout the time course of the experiment enabling rapid identification of highly cytotoxic CD8+ T cells. Microrafts with highly active CD8+ T cells were individually transferred to wells of a 96-well plate, using a needle-release device coupled to the microscope. Three sorted T cells clonally expanded. All of these expressed high-avidity T cell receptors for M1p/HLA*02:01 tetramers, and 2 of the 3 receptors were sequenced.
While this study investigated single T cell cytotoxicity rates against simple targets with subsequent cell sorting, future studies will involve measuring T cell mediated cytotoxicity in more complex cellular environments, enlarging the arrays to identify very rare antigen specific T cells, and measuring single cell CD4+ and CD8+ T cell proliferation.
Keywords: Single-cell analysis, T cell cloning, Microraft array
Introduction
Antigen-specific, cytotoxic CD8+ T cell responses are an essential component in the adaptive immune system’s ability to control both viral infections and cancer. Multiple types of experiments designed to quantify the number of antigen-specific CD8+ T cells and measure the cytotoxic activity of a T cell population are routinely used to characterize CD8+ responses; however, many standard immune monitoring assays have limitations with respect to sample size (or cell number) requirements, sample processing, assay time or cost that make detailed characterization of immune responses problematic.
To measure the fraction of antigen-specific CD8+ T cells in a T cell population, which is important in measuring immune responses and immunodominance in vivo as well as in the expansion of antigen-specific T cells in culture, peptide/MHC (major histocompatibility complex) tetramers are often used.1, 2 Tetramer-based enumeration is expensive however, and requires a unique tetramer per antigen to be tested, which is problematic particularly in the field of personalized cancer neo-antigen discovery.3, 4 Furthermore, this assay, by itself, does not provide a measurement of T cell effector function or activity. For functional assessment of individual T cells in a bulk population, intracellular cytokine flow cytometry,5 which can be coupled with tetramer enumeration,6-8 or ELISPOT analysis can be employed.9, 10 These methods report cytokine secretion in response to a stimulus, such as T cell receptor (TCR) binding to its cognate peptide/MHC. Compared to cytokine secretion, a more direct measure of cytotoxicity is the measurement of cell surface expression of CD107, which is contained in T cell cytotoxic granules and whose surface expression is associated with cytotoxicity.11 While all of these methods give information on T cell function, none of them directly measure individual T cell mediated killing, the most significant property of CD8+ T cells, nor do they provide any data regarding the time dependence as the measurement is only made at the conclusion of the experiment.
Beyond the quantification of antigen-specific T cells and measurements of their function, there is often a need for clonal expansion of individual T cells to allow further characterization or to produce clonal populations that can be used as probes for monitoring antigen presentation under different situations and in different cell types. Clonal T cell populations can be expanded using limiting dilution methods and by fluorescence activated cell sorting (FACS); however, both methods have drawbacks. Limiting dilution has no selection method so, while it is often a more efficient method for clonal T cell expansion compared to FACS, many expanded clonal populations do not have the antigen-specific properties of interest.12-14 FACS can provide specific cell sorting based upon the expression of cell surface markers, including TCRs specific to peptide/MHC tetramers.15, 16 FACS negatively affects cell viability however because of the temperature and pressures applied to the T cells in the cell sorter.17 Although cell viability can be furthermore reduced by the enhanced activation of Src kinases through ligation of the TCR with multimeric peptide/MHC tetramers; although, some of the deficiencies with peptide/MHC induced cell death can be mitigated using reversible peptide/MHC tetramers.18 Surface CD107 expression on T cells after antigenic stimulation has also been used to isolate individual CD8+ T cells.19 However, this method does not directly measure cell killing, and the measurement is only made at the time of sorting, whereas CD107 expression varies with time.
In this project we have developed a methodology to identify, isolate and clonally expand antigen specific CD8+ T cells based upon the longitudinal measurement of their killing of antigen expressing target cells. The methodology was developed using microraft arrays with each microraft designed to contain a population of fluorescently labeled antigen-presenting target cells and 1 CD8+ T cell. A cytotoxicity dye in the media allowed for the measurement of target cell death over time. Over the 6-h time course of the experiments, there was tremendous heterogeneity in the rate of target cell killing by individual CD8+ T cells; however, the rates of killing remained relatively constant for single cells, allowing for the identification of highly cytotoxic CD8+ T cells in as little as 2 h. Microrafts that contained a CD8+ T cell and showed a high rate of target cell death were transferred from the array to a well in a 96-well plate using a previously designed needle-release device, and the sorted T cells were clonally expanded for further characterization including measurement of TCR affinity to a peptide/MHC tetramer and sequencing of the TCRα and TCRβ CDR3 regions.
Experimental
Detailed descriptions of the materials and methods used in these experiments are provided in the supplementary methods section. Brief descriptions of the experimental methods are provided below.
Fabrication of microraft arrays
Microraft arrays, consisting of PDMS microwells containing releasable, magnetic elements were created using previously described methods.20-22 The arrays for this series of experiments contained 4900 microwells (70 × 70 array) constructed on SU-8 pillars (200 × 200 × 200 μm, spaced 100 μm apart) coated with poly(styrene-co-acrylic acid) (PS-AA) co-mixed with γFe2O3 nanoparticles.21
Microscopy Setup
An MVX10 MacroView upright microscope (Olympus, Center Valley, PA) equipped with an ORCA-Flash 4.0 CMOS camera (Hamamatsu, Bridgewater, NJ) was used to acquire bright field and fluorescence micrographs. The MVX10 MacroView was fitted with a PS3H122 Motorized Focus Drive and a H138A motorized XY translational stage manipulated by a ProScan H31 Controller and a PJ2J100 joystick (Prior Scientific Inc., Rockland, MA). A Lambda 10-3 optical filter changer was utilized to control an emission filter wheel (LB10-NWE), an excitation filter wheel with SmartShutter (LB10-NWIQ) and a stand-alone SmartShutter shutter (IQ25-SA) (Sutter Instrument, Novato, CA). A sedat filter set (89000 – ET – Sedat Quad; Chroma Technology Corp, Bellows Falls, VT) containing 5 excitation filters (350 ± 50 nm, 402 ± 15 nm, 490 ± 20 nm, 555 ± 25 nm, 645 ± 30 nm), 4 emission filters (455 ± 50 nm, 525 ± 36 nm, 605 ± 52 nm, 705 ± 72 nm) and a multiband dichroic enabled measurement of fluorescence in the blue, green, red and far red wavelengths. A lumen 200 arc lamp (Prior Scientific Inc., Rockland, MA) provided light for fluorescence excitation. All microscopy equipment was controlled by custom software written in MATLAB (MathWorks, Natick, MA) and using a Micro-Manager (Open Imaging, San Francisco, CA) core.23, 24 A custom-made incubator was made to fit around the microscopy set-up to regulate temperature, humidity and CO2 concentration during time-lapse microscopy experiments.
Preparation of microraft arrays for the cytotoxicity assays
Prior to experiments on the microraft array, the array was treated for 5 min in a plasma cleaner (Harrick Plasma, Ithaca, NY). The array was then washed with 100% ethanol and subsequently washed ×5 with PBS. The microraft array was coated in bovine gelatin in PBS and incubated at. The gelatin solution was aspirated and the array was washed with PBS before plating cells.
Generation of dendritic cells (DCs) from CD34+ progenitors
Dendritic cells were differentiated from CD34+ cells using a modification of a previously described method.25 CD34+ cells were isolated from cryopreserved leukapheresis products obtained from the Hematopoietic Progenitor Cell Laboratory at UNC Hospitals using the CD34 Microbead Kit UltraPure (Miltenyi Biotec). CD34+ cells were incubated for 12 d in CTS AIM V media with 10% human AB serum (complete media, CM), supplemented with GM-CSF, Flt3-ligand, SCF and IL-4 as described in the supplemental methods to yield immature DCs that were cryopreserved for future use. Immature DCs were differentiated into mature DCs by incubation with GM-CSF, IL-4 and TNF-α for 2 d and GM-CSF, IL-4, TNF-α, IFN-α and IL-6 for 2 d. Matured DCs were co-incubated with peptides (M1p or PR1) for at least 18 h.
Generation of cytotoxic T lymphocytes (CTLs)
PBMCs were isolated from a leukapheresis product from the same donor using Ficoll-Paque PLUS, and monocytes were removed by plastic adherence. Non-adherent cells (NADs) were collected, and CD8+ cells from the NAD fraction were isolated by magnetic separation using the CD8+ T cell isolation kit (Miltenyi Biotec).
The cytotoxic T lymphocyte (CTL) culture was initiated by incubating the CD8+ T cells with M1p pulsed DCs in CM supplemented with IL-21.26 After a 3 d incubation, the cells were supplemented with CM plus IL-7 and IL-15 every 2 d. CTLs were restimulated with M1p-pulsed DCs 11 d after culture initiation in CM containing IL-21, IL-7 and IL-15. IL-2 was added 19 d after initiation of the culture.26 The CTLs were restimulated 21 and 34 d after culture initiation and were cryopreserved in aliquots 41 d after initiation of the culture.
Tetramer analysis
Ten μL of APC-labeled M1p/HLA-A*02:01 tetramer or negative tetramer was added to each sample with PE-Cy7-anti-CD8 antibody and a FITC labeled lineage mix of CD4, CD14, CD16 and CD19 antibodies. The cells were washed and resuspended. DAPI live/dead dye was added. Sample acquisition was performed on a FACSCanto flow cytometer (BD Biosciences) and data was analyzed using FlowJo version 7.6.5 (FlowJo LLC, Ashland, OR).
Bulk culture cytotoxicity assay
Cyropreserved CTLs were restimulated with M1p pulsed DCs in CM supplemented with IL-2, IL-7 and IL-15 4 d prior to the cytotoxicity assay. Differentiated DCs (targets, T) were stained with DDAO-SE in DPBS, incubated overnight with M1p or PR1 peptide, washed and resuspended. CTLs (effectors, E) were stained with Cell Tracker Green CMFDA dye and resuspended in CM. CTLs were co-incubated with DCs at 1:1 E:T ratio or a 2:1 E:T ratio. After 6 h, cells were resuspended, treated with LIVE/DEAD Fixable Violet Dead Cell Stain and fixed in 2% formaldehyde. Cells were analyzed on a Miltenyi MACSQuant flow cytometer (Miltenyi Biotec) with data analysis using FlowJo version 7.6.5.
Restimulation of CTLs and isolation of CD8+ cells prior to plating on the microraft arrays
Cryopreserved CTLs were thawed and restimulated with M1p pulsed DCs. After 3 d, the CTLs were isolated using the CD8+ T cell isolation kit and maintained in CM supplemented with IL-7, IL-15 and IL-2. CD8+ T cells were plated on microraft arrays 2 to 3 d later.
Single T cell cytotoxicity assay
The single cell cytotoxicity assay was constructed in a similar method to that reported by Varadarajan et al.27 DCs were pulsed with either M1p or PR1 peptides. Both sets of DCs were stained with Hoechst 33342 and Sytox Green prior to plating. Cells were deposited on microraft arrays in phenol-red free RPMI 1640 supplemented with human AB serum, penicillin, streptomycin and HEPES. For each cytotoxicity assay, two microraft arrays were prepared: one with M1p pulsed DCs added and the other with PR1 pulsed DCs. DCs were added at cell:microraft ratios of 30:1. Arrays were centrifuged to pull the DCs into the microrafts, and the media was aspirated. CD8+ T cells from the M1p culture were stained with CellTracker Deep Red and Sytox Green and added to both arrays at a cell:microraft ratio of 1:1. The arrays were again centrifuged and the media was aspirated. Culture media containing Sytox Green was placed over each array, and each array was covered with a cover slip. Arrays were imaged every 30 min for 6 h while in the incubator housing the microscope.
Image acquisition
A customized MATLAB program controlled the microscope to acquire bright field and fluorescence images of the microraft array at designated time points. A graphical user interface (GUI) was designed to permit input of user-selected parameters such as fluorescence channel selection, camera exposure, microraft array geometry, microraft array numbers and time-lapse conditions.28, 29 A 5% overlap between imaged fields of view was used in all experiments.
Image processing and analysis
In parallel with the image acquisition, the MATLAB program processed and analyzed the acquired images. The bright field images were used to identify individual microraft locations. Flat-field correction was performed on each bright field image to correct for uneven illumination intensity.30 Otsu’s method was then used to calculate a threshold for each image, and the image was converted to binary.31 The binary images were processed to fill the interior of each microraft border and remove any resulting objects that were larger than 1.5× or smaller than 0.5× the microraft size eliminating debris. The positions of all microraft were identified at each time point.
Fluorescence images were processed in a similar fashion. A top hat filter was applied with a disk structuring element having a radius twice the nominal diameter of a cell to remove background fluorescence.32, 33 Otsu’s method was used to determine a threshold for each image, and the threshold was applied to convert each image to binary.31 The pixel locations and intensity values of each fluorescence image were determined for each individual microraft. A watershed algorithm was applied to each image in the far red channel (corresponding to cells stained with CellTracker Deep Red) to count the number of individual cells on each microraft.34-36
Selection of cells for release
The locations of individual microrafts were determined using the image processing and analysis software. Microrafts containing a single CD8+ cell were using CellTracker Deep Red fluorescence. These microrafts were sorted based on the increase in Sytox Green fluorescence intensity. Automatically identified microrafts were re-screened after gelatin encapsulation to ensure that a single CellTracker Deep Red-positive cell remained on the microraft.
Microraft/cell release and transfer to a 96-well plate
Upon completion of the cytotoxicity assay, the arrays were overlaid with a thin layer of gelatin as described previously.20 The incubator surrounding the microscope was cooled to 24°C just prior to gelatin overlay of the array. The culture media above the array was replaced with 5 wt% bovine gelatin in PBS and the array was centrifuged. The array was then incubated for 10 min at 37°C, washed and then incubated at 4°C for 5 min to solidify the gelatin within the microrafts. Cold (4°C) culture media was overlaid onto the arrays. Previous experiments using the microraft array configuration and gelatin encapsulation have shown that 97.8% of cells remain in their initial microwells throughout the entire gelatin encapsulation and release process.20
Microraft isolation was performed by actuating a small needle (10 μm tip, 100 μm base, 5 mm long) to puncture the PDMS of the microraft array and eject individual microrafts as described previously.20-22 Released rafts were then captured by a magnetic wand mounted on a computer controlled 3-axis motor and deposited into a 96-microwell plate as described previously.20
Preparation of feeder cells for T cell expansion
PBMCs were isolated from 3 different buffy coats using Ficoll-Paque PLUS or Lymphoprep and irradiated at 35 Gy. Equal numbers of PBMCs from each buffy coat were combined in CM with IL-7, IL-15 and IL-2 and plated at a concentration of 1 × 105/well in 96 well round bottom plates.
Limiting dilution
CD8+ CTLs were serially diluted in CM to 5 cells/mL. One hundred μL was added to each well of a 96 well plate containing feeder cells, for an expected concentration of 0.5 CTLs/well.
T cell expansion
Microrafts containing selected CTLs were isolated as described above in to 96-well plates containing feeder cells. Cells released from the microrafts and cells plated by limiting dilution were expanded in parallel using minor modifications of the protocol described by Perna el al.37 OKT3 antibody was added 2 to 3 d after plating the cells, and the cells were fed weekly by adding CM supplemented with IL-7, IL-15, IL-2 and OKT3 antibody. Expanding single cell clones were transferred to 24 well plates between d 11 and 19 after plating. Tetramer staining was performed 2 d later. Tetramer positive clones were further expanded with a mix of irradiated PBMCs and an EBV-immortalized lymphoblastoid cell line (EBV-LCL) as feeder cells in CM supplemented with IL-2, IL-7, IL-15 and OKT3 antibody.37 CD8+ cells were isolated using Miltenyi’s CD8+ T cell isolation kit, and cell pellets were frozen at −80°C for TCR α and β chain CDR3 sequencing.
Analysis of TCR α- and β-chain CDR3 sequences
RNA was isolated from the B8, D10 and F9 CD8+ T cell clones using Qiagen’s RNeasy Micro Kit and stored in aliquots at −80°C. Analysis of TCR α- and β-chain rearrangements was performed using multiplex RT-PCR as described by Kim et al. and Seitz et al.38, 39 The initial RT-PCR reaction was performed using the OneStep RT-PCR kit.38 TCR β-chains were amplified from 1 μL of the RT-PCR product in a run-off reaction, followed by semi-nested PCR using the Fast Start High Fidelity PCR System. Reactions that produced PCR products visible by agarose gel electrophoresis were purified using the Qiagen PCR Purification Kit and sequenced.38 TCR α-chains were amplified by nested touchdown PCR using 1 μL of the RT-PCR reaction using the Fast Start High Fidelity PCR System.38 PCR products were analyzed by agarose gel electrophoresis, purified using the Qiagen PCR Purification Kit, and sequenced using the Cα-in primer.38 Sequencing was performed at the UNC Genome Analysis Facility (University of North Carolina, Chapel Hill, NC), and sequences were analyzed to determine the V and J alleles as well as the CDR3 sequence using IMGT/V-Quest (http://www.imgt.org/IMGT_vquest/vquest).40, 41 One clone (F9) did not produce readable sequences using the protocol described above, possibly due to poor quality RNA. The TCRβ-chain rearrangement of this clone was analyzed using a modification of another multiplexed RT-PCR protocol for single cell TCRβ analysis.42
Statistics
Cytotoxicity results on the microraft array were compared using the nonparametric Mann-Whitney U-test. All statistics were performed in MATLAB. The k-medoids algorithm was used to cluster single CD8+ T cell responses.43 The Kruskal-Wallis test was used to compare the clustered groups above. This test is a nonparametric version of a one-way ANOVA test, and an extension of the Mann-Whitney U test to more than 2 groups. A multiple comparison test was used to identify which groups had a statistically significant difference.44
Results and discussion
Microraft arrays for selection of nonadherent cells
Our group and others have developed microarrays for the functional analysis of rare cells.27, 45, 46 In this project, we aimed to develop several technological advances to make an automated device that could analyze the interactions and changes in multiple cell populations simultaneously over time and then viably sort cells of interest for further characterization. One of the most important functional characteristics of CD8+ T cells is antigen specific cytotoxicity, so the measurement of single-cell cytotoxicity with subsequent cell sorting was chosen to be the first immunologic assay developed on the microraft arrays. To develop this assay, methods to analyze 2 cell types and 3 fluorescence channels were developed, with this initial assay being able to measure the cytotoxic ability of >1,000 individual T cells against target cells.
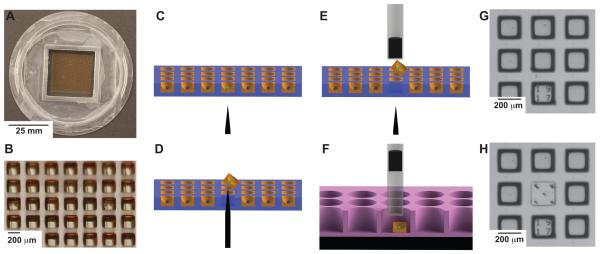
The microraft arrays in this study are comprised of a regular pattern (70 × 70) of transparent microcarriers or microrafts on which cells are cultured and assayed over time by microscopy (Figure 1A, B). The microrafts fabricated from a magnetic polystyrene possess a concave surface of 120 μm depth enhancing retention of cells on their surfaces during assay setup and performance. After the identification of cells of interest based upon a fluorescent read-out over time, five percent gelatin in PBS is poured on the array and gelled encapsulating the cells within the concave surface of their microraft. To release a selected microraft with cells from the array, a microneedle situated under the microscope stage pushes through the PDMS membrane underlying the microraft and releases the selected microraft from the array (Figure 1C, D). The microraft with gel-encapsulated cell(s) is then readily captured by an overlaying magnetic wand dipped into the media above the array (Figure 1E). The wand with the captured microraft is then placed into the well of a 96-well plate. The microraft is deposited by the dissipation of surface tension forces created by the residual fluid droplet on the wand tip and by repulsion of the wand magnet by a block magnet underlying the 96-well plate. The microraft is then pulled down to the bottom of the well by the block magnet (Figure 1F). Microrafts can be visualized by bright field microscopy, and the release of microrafts can also be confirmed by their presence in the 96-well plate and absence on the array (Figure 1G, H).
Figure 1. Microraft based selection of cells.
The microraft array in these studies is a 2.5 cm × 2.5 cm array containing 70 × 70 microrafts. Each microraft is 200 μm × 200 μm × 200 μm in outer dimensions and separated from adjacent microrafts by a 100 μm gap. The cassette walls surrounding the array are 1 cm high allowing for excess media to be applied over the microrafts (Figure 1A, B). Fluorescence from each microraft can be measured. In this example a “green cell” is identified (Figure 1C). The array is encapsulated in gelatin, and the needle release mechanism pushes through the PDMS to release the identified microraft (Figure 1D). The magnetic wand attached to the microscope objective captures the released microraft (Figure 1E). The wand is moved to a well of a 96-well plate where a block magnet pulls the microraft to the bottom of the well (Figure 1F). A 3 × 3 section of the array is shown using bright field microscopy (Figure 1G). The needle release ejected the microraft in the middle. In this example 5 needle penetrations were used to demonstrate the accuracy of the device and to show that the elastomeric PDMS readily reseals without loss of media despite repeated puncturing (Figure 1H).
Design of T cell mediated cytotoxicity assay
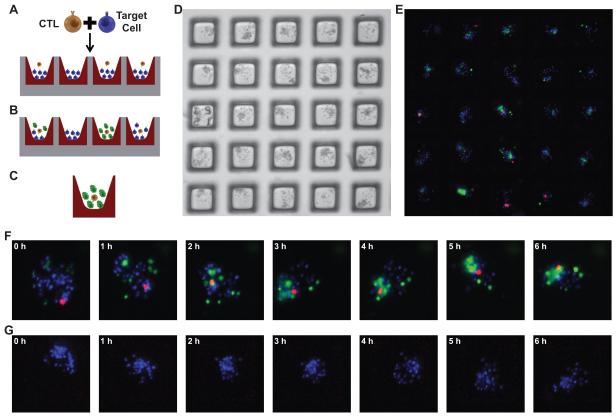
While the microraft array format can be employed for multiple types of experiments and cells, this series of experiments developed an assay for the selection and capture of T cells based upon their cytotoxicity.27 The microraft arrays were seeded with autologous DCs (“target” cells) that had been pulsed with either M1p or PR1 peptides and labeled with Hoechst dye. Targets were applied to the array at a ratio of 30 cells per microraft (147,000 cells per array). CD8+ T cells were labeled with CellTracker Deep Red and placed onto the array at a cell:microraft ratio of 1:1 in order to maximize the number of microrafts that contained a single T cell. Cells settled onto the array in a stochastic manner so that the number in each well across the array followed the Poisson distribution.22, 47 By the Poisson distribution, roughly 1/3 of the wells (36.8% or 1803 wells) are predicted to possess a single CD8+ T cell, 26.4% (1294) are predicted to contain >1 T cell and 36.8% (1803) are expected to have 0 T cells. The media overlaying the array contained Sytox Green, a DNA binding dye that is membrane impermeable. The assay was intended to identify microrafts that contained a single T cell and showed a high rate of cytotoxicity as evidenced by increasing green fluorescence over a 6 h time course (Figure 2A-C). Because both the target cells and T cells were applied to the media over the microraft array, individual microrafts had different numbers and ratios of target and T cells. (Figure 2D, E). Fluorescence images were obtained every 30 min for 6 h and the development of green fluorescence over time was measured for each microraft. (Figure 2F, G).
Figure 2. Design of T cell mediated cytotoxicity assay.
The microraft array was designed to identify the best M1p specific CTLs. The arrays were loaded with Hoechst labeled “target” cells (peptide pulsed DCs) at a density of 30 cells per microraft and co-incubated with CTLs from an M1p culture at a mean density of 1 cell per microraft. (Figure 2A). Over time target cells die and take up Sytox Green that is present in the media (Figure 2B). Microrafts with the greatest increase in green fluorescence are identified as likely having the most cytotoxic T cells (Figure 2C). Both the targets and T cells distribute into the microrafts according to the Poisson distribution. A 5 × 5 microraft array shows multiple different ratios of cells throughout the array (Figure 2D, E). Fluorescence images for 2 of the 4900 microrafts are shown at 1 h time points (images were collected every 30 min). A microraft with 1 CTL (red cell) shows increased killing (green fluorescence) over time (Figure 2F), and another microraft without a CTL shows no killing over time (Figure 2G).
M1p Specific T cell culture
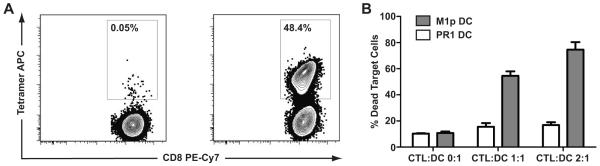
To evaluate the microraft cytotoxicity assay, we tested a T cell culture that was generated against the influenza M1p antigen. M1p/HLA-A*02:01 tetramer enumeration showed that 48.4% of CD8+ T cells were specific for M1p (Figure 3A). The bulk culture displayed antigen specific cytotoxicity against M1p pulsed autologous DCs compared to autologous DCs pulsed with the leukemia associated antigen PR1 (Figure 3B).48
Figure 3. M1p specific T cell culture.
A T cell culture was generated against the influenza M1p antigen, and the percentage of M1p-specific CD8+ T cells was enumerated using M1p/HLA-A*02:01 tetramers and a negative control HLA-A*02:01 tetramer, along with an anti-CD8 antibody and a lineage marker mix of CD4, CD14, CD16 and CD19 antibodies. Tetramer staining was analyzed by gating on live cells (DAPI negative), lymphocytes (FSC vs SSC), and CD8+Lineage− cells. Only the CD8+Lineage− cells are shown on the plots. Over 99% of the cells that fell in the live lymphocyte gates were CD8+Lineage−. Non-specific binding to the negative control tetramer was low (0.05% of CD8+ T-cells), while 48.4% of CD8+ T-cells had high avidity interactions with the M1p/HLA-A*02:01 tetramer (Figure 3A). The culture also demonstrated antigen specific cytotoxicity against M1p pulsed autologous DCs compared to PR1 pulsed autologous DCs (Figure 3B). For the cytotoxicity assay, the FSC vs SSC plot was gated to remove debris in the lower left hand corner, and DDAO-SE positive target cells (DCs) were selected on a DDAO-SE vs CMFDA plot. The percentage of dead target cells shown is the percentage of DDAO-SE positive DCs that also stained positive with the Fixable Violet Dead Cell Stain.
Automated analysis of cytotoxicity on microraft arrays
The analyses performed on the microraft arrays compared development of green fluorescence over time in microrafts that contained either 0 or 1 CD8+ T cell. Microrafts with 0 T cells were considered the control and reflected the rate of spontaneous target cell death over the time of the investigation. A total magnification of 4× was used for scanning microrafts, resulting in a pixel size of 1.62 μm/pixel. This magnification was chosen because it allowed for easy identification of single cells while maintaining a large field of view to minimize image acquisition time. Each 70 × 70 microraft array required 49 images per channel at this magnification with a 5% overlap of images. Image acquisition of a single array using bright field (100 ms camera exposure) plus 3 fluorescence channels (200 ms camera exposure each) required 216 ± 4 s to complete (n = 10). An autofocus algorithm was used to maintain focus for each image and required 99 ± 2 s to complete. All of the automatically acquired focal planes were accurate to within the microscope objective’s depth of field at 4× magnification (±21.8 μm) compared to the manually selected focal planes (n = 50). The resulting total microraft array scan time (autofocus and image acquisition) was 315 ± 5 s.
In parallel with the image acquisition, the MATLAB program processed and analyzed the acquired images. The MATLAB GUI was used to select the desired combination of bright field and fluorescence channels for imaging. The processing time of each set of images was faster than the microscope stage movement and image acquisition, so no additional time was added to the total scan time. The bright field images were used to identify individual microraft locations. Due to the elastomeric nature of PDMS, image analysis was needed to accurately locate the exact positions of microrafts on the array. The microraft segmentation method consisted of background estimation, flat-field correction, thresholding and morphological filtering. The vast majority of microrafts (99.8 ± 0.8%) were correctly identified without false positives (n = 100 images, 100-121 microrafts per image). The microrafts that were not correctly identified were missed due to debris obscuring the microraft.
The fluorescence images were analyzed to determine the intensity, area, position and number of cells displaying each fluorophore (Hoechst, Sytox Green and CellTracker Deep Red). Top hat filtering and Otsu’s thresholding was applied to each image to produce a binary mask of the cells on the microraft array. Within the mask created by each fluorophore, the intensity, location and number of pixels was recorded for each microraft. The number of T cells on each microraft was counted from the far-red fluorescence after implementation of the watershed algorithm. Of 401 microrafts manually examined, the segmentation algorithm resulted in a false positive rate of 4.5% and a false negative rate of 7.5% corresponding to a sensitivity and specificity of 93.0% (95% CI, 88.5% - 96.1%) and 96.5% (95% CI, 93.0% - 98.6%), respectively. False negatives were often due to two or more cells in very close proximity that were not successfully separated by the algorithm or due to cells with very low-intensity far-red fluorescence. False positives were due to light from a very bright cell refracted off the edge of the microraft. The watershed segmentation method ultimately resulted in a sensitivity of 96 ± 16% in identifying cells with CellTracker Deep Red fluorescence and a false cell identification rate of 1.5 ± 9.0% (n = 401 microrafts). The watershed segmentation was most accurate when low numbers of cells were located on a microraft. For this reason, the data obtained from the watershed segmentation was only used to count T cells.
Cytotoxicity or cell killing was measured by summing the Sytox Green pixel intensity within the Hoechst fluorescence area on each microraft. Using the Hoechst positive regions on each microraft as a mask for the Sytox Green fluorescence greatly reduced spurious measurements due to debris and disintegrating cells and their fragments. The effector cell count and cytotoxicity information was recorded for each microraft at each time point to generate temporal traces of cytotoxicity corresponding to specific numbers of effector cells present.
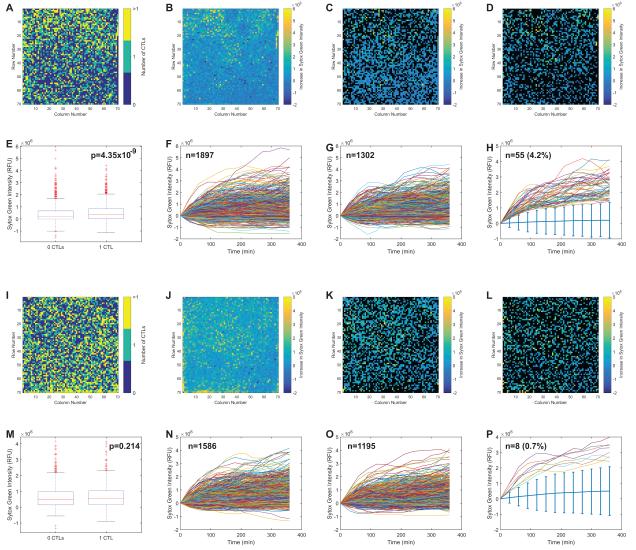
Figure 4 shows the results of 2 microraft arrays analyzed in parallel. The first array (Figure 4A-H) contained M1p-pulsed DCs as targets and the second array (Figure 4I-P) contained PR1-pulsed DCs. Because the assay was intended to identify individual highly cytotoxic T cells, the arrays were first analyzed to identify microrafts with 0, 1 or >1 CD8+ T cell (Figure 4A, I), and the increase in green fluorescence at each microraft was measured (Figure 4B, J). These general analyses showed that the T cells distributed evenly over the array, and there did not appear to be any particular regions on the arrays with abnormally strong increases or decreases in green fluorescence. Several different methods were considered to analyze the inter-microraft cytotoxicity. In the initial analysis, the microrafts were segregated into those with 0 T cells and those with 1 T cell (microrafts with >1 T cell were excluded). The difference in green fluorescence at t = 6 h and t = 0 h was measured for each microraft. The differences in fluorescence across the arrays are shown (Figure 4C, D, K, L). This representation proved to be difficult to visualize and interpret. However, when the fluorescence differences observed in the 0 T cell microrafts were compared to the differences observed in the 1 T cell microraft, there was a statistically significant difference observed in the M1p array but not the PR1 array (Figure 4E, M). Since the assay monitors cellular processes over time, more information on a per T cell basis is available than the simple differences over 2 time points. The change in green fluorescence over time for each microraft was measured for microrafts with 0 T cells (Figure 4F, N) and 1 T cell (Figure 4G, O). While many microrafts exhibited similar rates of increase in green fluorescence, there were some microrafts that exhibited high rates of apparent cytotoxicity. To identify these microrafts, the median rate of green fluorescence ± 4 median absolute deviations for the 0 T cell microrafts in each array was calculated. The rate of green fluorescence increase for all microrafts with 1 T cell was obtained, and only microrafts with green fluorescence > 4 median absolute deviations from the median at all time points were selected as “best killers”. A total of 55 microrafts met this criterion in the M1p array and 8 met the criterion on the PR1 array (Figure 4H, P).
Figure 4. Overall analysis of cytotoxicity on microraft arrays.
Two arrays are compared: the first with M1p-pulsed DCs as targets (Figure 4A-H) and the second PR1-pulsed DCs as targets (Figure 4I-P). The fluorescence measurements for each microraft are shown in processed 70 × 70 array images (Figure 4A-D, 4I-L), where each box represents the appropriate fluorescent measurement for 1 microraft. T cells distribute evenly across the arrays with some microrafts having 0, some having 1 and some having >1 CTL present, measured by enumerating the numbers of segmented far-red labeled cells per microraft (Figure 4A, I). After 6 h, both arrays have a modest increase in green fluorescence (cell death) throughout the entire array (Figure 4B, J). Comparisons were made between the green fluorescence produced in microrafts with 0 CTLs and those with 1 CTL. In these representations only microrafts with the appropriate numbers of CTLs (i.e. 0 or 1) are colored. Quantified green fluorescence at t = 6 h (minus green fluorescence at t = 0 h) for microrafts with 0 CTLs (Figure 4C, K) and 1 CTL (Figure 4D, L) is shown with the other microrafts represented in black. The differences in mean fluorescence change between microrafts with 0 and 1 CTL were small, but statistically significant for the M1p array (Figure 4E) but not the PR1 array (Figure 4M). The array can measure the change in green fluorescence per microraft during the time course of the experiment. The fluorescence over time traces for all of the microrafts with 0 CTLs (Figure 4F and N) and 1 CTL (Figure 4G, O) are shown. The microrafts with the “best killers” were identified as having 1 CTL with green fluorescence > 4 median absolute deviations above the median (denoted by the light blue bars) at all time points. There were 55 of these microrafts on the M1p array and 8 on the PR1 array (Figure 4H, P).
Single CD8+ T cell cytotoxicity rates remain stable over 6 hours
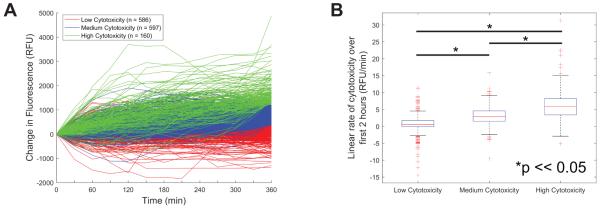
In addition to identifying antigen-specific cytotoxic T cells for further characterization (Figure 4) the rate of target cell death throughout the array was analyzed with normalization to the number of target cells per microraft. Data from microrafts containing a single CD8+ T cell were clustered using the k-medioids algorithm. The population of M1p specific CD8+ T cells was clustered into 3 groups based upon rate of cytotoxicity (Figure 5A). Cytotoxicity was measured as the development of green fluorescence normalized to the blue fluorescence per microraft. Over the 6 h time course of the experiments, the rate of cytotoxicity for each cell remained relatively constant meaning that cells with high rates of cytotoxicity as measured over a 6 h time course could be identified within 2 h of the experiment (Figure 5B).
Figure 5. Single CD8+ T cell cytotoxicity rates.
Data from microrafts containing a single CTL were clustered using the K-medoids algorithm. The data were clustered into three groups based on the total change in green fluorescence (Sytox Green) from 0 to 6 h normalized to the area of target cells (Hoechst+ fluorescence). These clusters were labeled as “Low Cytotoxicity” (n = 580), “Medium Cytotoxicity” (n = 603), and “High Cytotoxicity” (n = 160) (Figure 5A). A linear fit was performed on the first 2 h of the fluorescence/cytotoxicity data (first 5 time points). There was a statistically significant difference in the slope of the fits for all identified clusters. This indicates that it may be possible to separate Low, Medium and High cytotoxicity after the first 2 h of a cytotoxicity assay (Figure 5B).
Isolation and expansion of T cells based upon cytotoxicity
Individual CD8+ T cell expansion was performed on the original T cell culture by limiting dilution. Cells were diluted to an expected density of 1 cell per 2 wells into a 96-well plate and incubated for 3 weeks. The 96 most active T cells from the M1p array were sorted using the microraft release device into a 96-well plate and again allowed to clonally expand for 3 weeks. The limiting dilution and microraft release experiments were performed twice. Of the 192 microraft release procedures, 122 resulted in the single selected microraft being deposited into a well. From the two 96-well plates that received cells by limiting dilution, 4 wells demonstrated T cell expansion; however, none of these clones demonstrated specificity for M1p by tetramer analysis (data not shown). Of the 122 wells with single microrafts, 3 clonal populations were expanded. All 3 of these CD8+ T cell populations demonstrated high avidity binding to M1p/tetramer (Figure 6A-L). While the expansion efficiency of sorted cells was not high, it is comparable to that of peptide/MHC based FACS, which is the technique usually employed to isolate and clonally expand antigen-specific T cells. Initial studies with tetramer based FACS for clonal expansion reported expansion rates of 2 to 13% of sorted cells.49 Limiting dilution, the gentlest clonal T cell expansion strategy employed, yielded a similar number of clones competent to expand as that yielded by our gelatin encapsulation and microraft release method. This similarity suggests that our antigen-specific sorting method is not substantially harsher on T cells than limiting dilution, and that the overall low clonal expansion efficiency is more likely related to the health of the initial T cell culture. We do note that the tested T cell culture for these experiments had been stimulated and then cryopreserved for 8 months prior to use in the cytotoxicity and expansion experiments described in this paper. This prolonged cryopreservation and the resulting thawing of the T cell culture could have substantially influenced the surviving T cells’ ability to expand robustly. It is unlikley that the CellTracker Deep Red dye used to identify CTLs on the array impacted their long-term viability in culture since this dye, and the related compound CFDA SA (commonly referred to as CFSE) have been used extensively to measure primary T cell expansion for both in vitro and in vivo T cell functional experiments.50, 51 Using the device, we were able to isolate 1 CD8+ T cell every 20 s based upon the cytotoxicity measurements. This method of single cell isolation is roughly 3× faster than micromanipulation, which has been used on microwell platforms previously, and it minimizes the risk of cross contamination by not repetitively using microcapillaries for cell transfer.46, 52
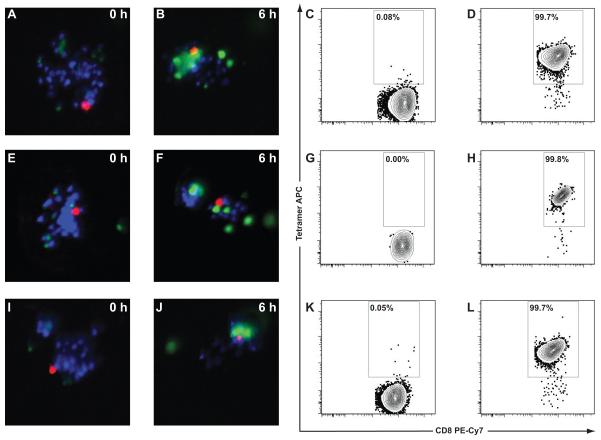
Figure 6. Isolation and expansion of T cells based upon cytotoxicity.
The CTLs from the M1p array with the greatest measured cytotoxicity were sorted using the microraft release device into a 96-well plate and allowed to clonally expand for 2 weeks. Three clones expanded: CTL3-MR-B8 (Figure 6A-D), CTL3-MR-D10 (Figure 6E-H), and CTL-MR-F9 (Figure 6I-L). The t = 0 and t = 6 h fluorescence for each of their respective microrafts are shown (Figure 6A-B, E-F, I-J). After clonal expansion TCR avidity for M1p/HLA-A*02:01 tetramer was measured. Negative tetramers were again used to define the tetramer gate (Figure 6C, G, K), and all 3 clonal populations had high avidity interactions with M1p/HLA-A*02:01 tetramer (Figure 6D, H, L). Tetramer staining was analyzed by gating on live cells (DAPI negative), lymphocytes (FSC vs SSC), and CD8+Lineage− cells. Only the CD8+Lineage− cells are shown on the plots.
While the primary focus of this study was to develop the microraft array platform to identify antigen specific CTLs and expand clonal CTL populations (Figure 6), a secondary aim was to expand on the method to determine the sequence of the clonal CTL’s TCR because these data could be used for the development of immunotherapeutics. TCRs exist as heterodimeric protein complexes consisting of a TCRα chain paired with a TCRβ chain, which are expressed on the T cell’s surface. The paired chains bind with high affinity to the target peptide/MHC, which were in this instance the M1p/HLA-A*02:01 complex. The portions of the TCRα and TCRβ chains that impart the specificity of the interaction with the target peptide/MHC are known as the CDR3 regions. If the CDR3 regions and flanking V and J segments of the TCRα and TCRβ chains are known, full-length transgenic TCR constructs can be produced and transfected into primary human T cells to change their specificity towards the target peptide/MHC. This approach could have a broad application in cancer immunotherapy where a cancer patient’s T cells could be transduced with a transgenic TCR, containing TCRα and TCRβ chain sequences, that targets a cancer-specific peptide/MHC complex.53
We performed TCRα and TCRβ CDR3 sequencing on the sorted and expanded T-cell clones obtained in our cytotoxicity assay. RNA was extracted from small aliquots of the clonal populations, and the CDR3 regions of the TCRα and TCRβ chains were amplified for sequencing using a multi-primer (to enable amplification of all potential flanking V and J segments) RT-PCR protocol modified from single-cell TCR sequencing methods. The CDR3 regions, and their associated flanking V and J segments, for TCRβ chains was determined for all 3 M1p/HLA-A*02:01 specific clonal populations, and the TCRα V, CDR3 and J sequences were determined for 2 of the clones (Table 1). With these data, it would be possible to produce viral vectors containing full-length TCRα and TCRβ chains that could be transduced into primary T cells for targeting influenza infected cells expressing the M1p/HLA-A*02:01 complex.
Table 1.
Seven T-cell clones were expanded using either microraft selection (denoted -MR-) or limiting dilution (denoted -LD-). Three of 3 MR selected clones expressed high avidity TCRs towards M1p/HLA-A*02:01 tetramer. The V segment, CDR3 sequence, and J segment for both the TCRα and TCRβ chain were sequenced on the first 2 clones, but only the TCRβ sequence could be determined for the third clone. None of the clones identified by LD expressed high avidity TCRs. Therefore, their TCRs were not sequenced.
| Clone | M1p Tetramer | TCRα | TCRβ |
|---|---|---|---|
| CTL3-MR-B8 | Positive | V19-CALSEAGTGGSYIPTF-J6 | V19-CASSMFVGQPQHF-J1-5 |
| CTL3-MR-D10 | Positive | V41-CAVSVEETSGSRLTF-J58 | V19-CASSFFHNNEQFF-J2-1 |
| CTL3-MR-F9 | Positive | ND | V19-CASSIRSSYEQYF-J2-7 |
| CTL3-LD-B2 | Negative | ND | ND |
| CTL3-LD-C4 | Negative | ND | ND |
| CTL3-LD-C8 | Negative | ND | ND |
| CTL3-LD-G3 | Negative | ND | ND |
Conclusions
In conclusion, we have developed an automated microraft array that can measure the cytotoxic ability of individual CD8+ T cells and isolate selected cells for clonal expansion and further analysis - in this case tetramer based avidity measurements and TCRα and TCRβ CDR3 sequencing. This study provided us with several insights into CD8+ T cell mediated cytotoxicity.
When examined at a single T cell level there is tremendous heterogeneity of cytotoxicity. Even though 48.4% of the tested culture was M1p specific (Figure 3A), single cell analysis showed no clear demarcation between “antigen-specific” and non-specific CD8+ T-cells. Rather there was a broad range in cytotoxicity (Figure 5A), which emphasizes the differences between static measurements of TCR avidity (via peptide/HLA tetramer) and functional analysis. While the rates of cell mediated cytotoxicity varied greatly among the T cell population, individual CD8+ T cells maintained a relatively consistent rate of target cell death throughout the 6 h time frame of the assay. This consistent rate of cell killing enabled the identification of the most highly cytotoxic individual CD8+ T cells within 2 h (Figure 5). The ability to determine the activity of small numbers of effector T cells in very short time scales could have a significant impact on immune monitoring as T cell therapies are developed for clinical use. Such assays could be used to screen T cells cultures for antigen specificity against multiple antigens to reduce the risk of off-target cytotoxicity that has been observed in early T cell therapy studies.54-56
This automated platform can be modified to address other issues in immunology. While the pilot study was intended to rapidly identify “best killers” in an enriched population, the array geometry can be easily modified to identify rare cytotoxic T cells. Microraft arrays having individual microrafts measuring 50 μm × 50 μm with a 20 μm gap have been fabricated on a 2.5 cm × 2.5 cm array yielding >125,000 microrafts on a single array, and the array size can also be scaled to further increase the number of elements, which could enable the identification and subsequent cloning of rare tumor antigen-specific CD8+ T cells.
While modifying the array geometry and assay time scale could allow for discovery of rare antigen specific T cells, the array can also be used to interrogate the effects of multiple cellular populations interacting at the same time. While this study only interrogated 2 cell populations, further cells types, such as myeloid derived suppressor cells (MDSC)57 and regulatory T cells (Treg)58 could also be added to the arrays to evaluate their relative abilities to inhibit cytotoxic T cell function over a broad spectrum of conditions. Because the assay also measures target cell death, as opposed to a surrogate marker like T cell CD107 expression, studies investigating target cell resistance to T cell mediated cytotoxicity could also be designed.
In addition to measuring cytotoxicity, experiments on the microraft arrays could also be designed to measure individual T cell proliferation over a period of a few days. Proliferation would also be expected to be an antigen-specific response; however, the measurement of T cell division would provide a general read out for both CD8+ (cytotoxic) and CD4+ (helper) responses with the subsequent possibility of sorting clonal populations of antigen specific CD8+ and CD4+ T cells.
Supplementary Material
Acknowledgments
Funding: This work was supported by a University Cancer Research Fund Innovation Award and by NIH (CA177993).
Footnotes
N.L.A. & C.E.S. disclose a financial interest in Cell Microsystems, a licensee of the microraft technology used in this manuscript.
References
- 1.Altman JD, Moss PA, Goulder PJ, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ, Davis MM. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 2.Ogg GS, Jin X, Bonhoeffer S, Dunbar PR, Nowak MA, Monard S, Segal JP, Cao Y, Rowland-Jones SL, Cerundolo V, Hurley A, Markowitz M, Ho DD, Nixon DF, McMichael AJ. Science. 1998;279:2103–2106. doi: 10.1126/science.279.5359.2103. [DOI] [PubMed] [Google Scholar]
- 3.Brown SD, Warren RL, Gibb EA, Martin SD, Spinelli JJ, Nelson BH, Holt RA. Genome Res. 2014;24:743–750. doi: 10.1101/gr.165985.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tran E, Ahmadzadeh M, Lu YC, Gros A, Turcotte S, Robbins PF, Gartner JJ, Zheng Z, Li YF, Ray S, Wunderlich JR, Somerville RP, Rosenberg SA. Science. 2015;350:1387–1390. doi: 10.1126/science.aad1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pala P, Hussell T, Openshaw PJ. J Immunol Methods. 2000;243:107–124. doi: 10.1016/s0022-1759(00)00230-1. [DOI] [PubMed] [Google Scholar]
- 6.Appay V, Nixon DF, Donahoe SM, Gillespie GM, Dong T, King A, Ogg GS, Spiegel HM, Conlon C, Spina CA, Havlir DV, Richman DD, Waters A, Easterbrook P, McMichael AJ, Rowland-Jones SL. J Exp Med. 2000;192:63–75. doi: 10.1084/jem.192.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Appay V, Rowland-Jones SL. J Immunol Methods. 2002;268:9–19. doi: 10.1016/s0022-1759(02)00195-3. [DOI] [PubMed] [Google Scholar]
- 8.Gea-Banacloche JC, Migueles SA, Martino L, Shupert WL, McNeil AC, Sabbaghian MS, Ehler L, Prussin C, Stevens R, Lambert L, Altman J, Hallahan CW, de Quiros JC, Connors M. J Immunol. 2000;165:1082–1092. doi: 10.4049/jimmunol.165.2.1082. [DOI] [PubMed] [Google Scholar]
- 9.Czerkinsky CC, Nilsson LA, Nygren H, Ouchterlony O, Tarkowski A. J Immunol Methods. 1983;65:109–121. doi: 10.1016/0022-1759(83)90308-3. [DOI] [PubMed] [Google Scholar]
- 10.Hill PC, Brookes RH, Fox A, Jackson-Sillah D, Jeffries DJ, Lugos MD, Donkor SA, Adetifa IM, de Jong BC, Aiken AM, Adegbola RA, McAdam KP. PLoS Med. 2007;4:e192. doi: 10.1371/journal.pmed.0040192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Betts MR, Brenchley JM, Price DA, De Rosa SC, Douek DC, Roederer M, Koup RA. J Immunol Methods. 2003;281:65–78. doi: 10.1016/s0022-1759(03)00265-5. [DOI] [PubMed] [Google Scholar]
- 12.Chapuis AG, Ragnarsson GB, Nguyen HN, Chaney CN, Pufnock JS, Schmitt TM, Duerkopp N, Roberts IM, Pogosov GL, Ho WY, Ochsenreither S, Wolfl M, Bar M, Radich JP, Yee C, Greenberg PD. Sci Transl Med. 2013;5:174ra127. doi: 10.1126/scitranslmed.3004916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho WY, Nguyen HN, Wolfl M, Kuball J, Greenberg PD. J Immunol Methods. 2006;310:40–52. doi: 10.1016/j.jim.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 14.Riddell SR, Greenberg PD. J Immunol Methods. 1990;128:189–201. doi: 10.1016/0022-1759(90)90210-m. [DOI] [PubMed] [Google Scholar]
- 15.Dunbar PR, Ogg GS, Chen J, Rust N, van der Bruggen P, Cerundolo V. Curr Biol. 1998;8:413–416. doi: 10.1016/s0960-9822(98)70161-7. [DOI] [PubMed] [Google Scholar]
- 16.Yee C, Savage PA, Lee PP, Davis MM, Greenberg PD. Journal of immunology. 1999;162:2227–2234. [PubMed] [Google Scholar]
- 17.Mollet M, Godoy-Silva R, Berdugo C, Chalmers JJ. Biotechnol Bioeng. 2008;100:260–272. doi: 10.1002/bit.21762. [DOI] [PubMed] [Google Scholar]
- 18.Guillaume P, Baumgaertner P, Angelov GS, Speiser D, Luescher IF. J Immunol. 2006;177:3903–3912. doi: 10.4049/jimmunol.177.6.3903. [DOI] [PubMed] [Google Scholar]
- 19.Rubio V, Stuge TB, Singh N, Betts MR, Weber JS, Roederer M, Lee PP. Nat Med. 2003;9:1377–1382. doi: 10.1038/nm942. [DOI] [PubMed] [Google Scholar]
- 20.Attayek PJ, Hunsucker SA, Wang Y, Sims CE, Armistead PM, Allbritton NL. Anal Chem. 2015;87:12281–12289. doi: 10.1021/acs.analchem.5b03579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gach PC, Wang Y, Phillips C, Sims CE, Allbritton NL. Biomicrofluidics. 2011;5:32002–3200212. doi: 10.1063/1.3608133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Phillips C, Xu W, Pai JH, Dhopeshwarkar R, Sims CE, Allbritton N. Lab Chip. 2010;10:2917–2924. doi: 10.1039/c0lc00186d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Curr Protoc Mol Biol. 2010 doi: 10.1002/0471142727.mb1420s92. Chapter 14, Unit14 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edelstein AD, Tsuchida MA, Amodaj N, Pinkard H, Vale RD, Stuurman N. J Biol Methods. 2014;1 doi: 10.14440/jbm.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dees EC, McKinnon KP, Kuhns JJ, Chwastiak KA, Sparks S, Myers M, Collins EJ, Frelinger JA, Van Deventer H, Collichio F, Carey LA, Brecher ME, Graham M, Earp HS, Serody JS. Cancer Immunol Immunother. 2004;53:777–785. doi: 10.1007/s00262-004-0520-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolfl M, Greenberg PD. Nat Protoc. 2014;9:950–966. doi: 10.1038/nprot.2014.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Varadarajan N, Julg B, Yamanaka YJ, Chen H, Ogunniyi AO, McAndrew E, Porter LC, Piechocka-Trocha A, Hill BJ, Douek DC, Pereyra F, Walker BD, Love JC. J Clin Invest. 2011;121:4322–4331. doi: 10.1172/JCI58653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brent RP. Algorithms for minimiziation without derivatives. Prentice-Hall; Englewood Cliffs, NJ: 1973. [Google Scholar]
- 29.Brent RP. Algorithms for minimization without derivatives. Courier Corporation; 2013. [Google Scholar]
- 30.Leong FJ, Brady M, McGee JO. J Clin Pathol. 2003;56:619–621. doi: 10.1136/jcp.56.8.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Otsu N. Automatica. 1975;11:23–27. [Google Scholar]
- 32.Serra J. Vol. 1. Academic Press; 1982. [Google Scholar]
- 33.Soille P. Springer Science and Business Media. 2013.
- 34.Malpica N, de Solorzano CO, Vaquero JJ, Santos A, Vallcorba I, Garcia-Sagredo JM, del Pozo F. Cytometry. 1997;28:289–297. doi: 10.1002/(sici)1097-0320(19970801)28:4<289::aid-cyto3>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 35.Meyer F. Signal processing. 1994;31:113–125. [Google Scholar]
- 36.Ng HP, Ong SH, Foong KW, Goh PS, Nowinski WL. Comput Biol Med. 2008;38:171–184. doi: 10.1016/j.compbiomed.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 37.Perna SK, De Angelis B, Pagliara D, Hasan ST, Zhang L, Mahendravada A, Heslop HE, Brenner MK, Rooney CM, Dotti G, Savoldo B. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:106–117. doi: 10.1158/1078-0432.CCR-12-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim SM, Bhonsle L, Besgen P, Nickel J, Backes A, Held K, Vollmer S, Dornmair K, Prinz JC. PloS one. 2012;7:e37338. doi: 10.1371/journal.pone.0037338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seitz S, Schneider CK, Malotka J, Nong X, Engel AG, Wekerle H, Hohlfeld R, Dornmair K. Proc Natl Acad Sci U S A. 2006;103:12057–12062. doi: 10.1073/pnas.0604247103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brochet X, Lefranc MP, Giudicelli V. Nucleic Acids Res. 2008;36:W503–508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guidicelli V, Brochet X, Lefranc MP. Cold Spring Harbor Protocols. 2011;6:695–714. doi: 10.1101/pdb.prot5633. [DOI] [PubMed] [Google Scholar]
- 42.Hunsucker SA, McGary CS, Vincent BG, Enyenihi AA, Waugh JP, McKinnon KP, Bixby LM, Ropp PA, Coghill JM, Wood WA, Gabriel DA, Sarantopoulos S, Shea TC, Serody JS, Alatrash G, Rodriguez-Cruz T, Lizee G, Buntzman AS, Frelinger JA, Glish GL, Armistead PM. Cancer immunology research. 2015;3:228–235. doi: 10.1158/2326-6066.CIR-14-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park H-S, Jun C-H. Expert Systems with Applications. 2009;36:3336–3341. doi: 10.1016/j.eswa.2007.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hochberg Y, Tamhane AC. Multiple Comparisons Procedures. Wiley; 1987. [Google Scholar]
- 45.Liadi I, Singh H, Romain G, Rey-Villamizar N, Merouane A, Adolacion JR, Kebriaei P, Huls H, Qiu P, Roysam B, Cooper LJ, Varadarajan N. Cancer immunology research. 2015;3:473–482. doi: 10.1158/2326-6066.CIR-14-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Varadarajan N, Kwon DS, Law KM, Ogunniyi AO, Anahtar MN, Richter JM, Walker BD, Love JC. Proc Natl Acad Sci U S A. 2012;109:3885–3890. doi: 10.1073/pnas.1111205109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taswell C. J Immunol Methods. 1984;72:29–40. doi: 10.1016/0022-1759(84)90430-7. [DOI] [PubMed] [Google Scholar]
- 48.Molldrem J, Dermime S, Parker K, Jiang YZ, Mavroudis D, Hensel N, Fukushima P, Barrett AJ. Blood. 1996;88:2450–2457. [PubMed] [Google Scholar]
- 49.Dunbar PR, Chen JL, Chao D, Rust N, Teisserenc H, Ogg GS, Romero P, Weynants P, Cerundolo V. Journal of immunology. 1999;162:6959–6962. [PubMed] [Google Scholar]
- 50.Lyons AB. Immunol Cell Biol. 1999;77:509–515. doi: 10.1046/j.1440-1711.1999.00864.x. [DOI] [PubMed] [Google Scholar]
- 51.Zhou W, Kang HC, O'Grady M, Chambers KM, Dubbels B, Melquist P, Gee KR. Journal of Biological Methods. 2016;3:7. [Google Scholar]
- 52.Choi JH, Ogunniyi AO, Du M, Du M, Kretschmann M, Eberhardt J, Love JC. Biotechnol Prog. 2010;26:888–895. doi: 10.1002/btpr.374. [DOI] [PubMed] [Google Scholar]
- 53.Arber C, Feng X, Abhyankar H, Romero E, Wu MF, Heslop HE, Barth P, Dotti G, Savoldo B. J Clin Invest. 2015;125:157–168. doi: 10.1172/JCI75876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cameron BJ, Gerry AB, Dukes J, Harper JV, Kannan V, Bianchi FC, Grand F, Brewer JE, Gupta M, Plesa G, Bossi G, Vuidepot A, Powlesland AS, Legg A, Adams KJ, Bennett AD, Pumphrey NJ, Williams DD, Binder-Scholl G, Kulikovskaya I, Levine BL, Riley JL, Varela-Rohena A, Stadtmauer EA, Rapoport AP, Linette GP, June CH, Hassan NJ, Kalos M, Jakobsen BK. Sci Transl Med. 2013;5:197ra103. doi: 10.1126/scitranslmed.3006034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Linette GP, Stadtmauer EA, Maus MV, Rapoport AP, Levine BL, Emery L, Litzky L, Bagg A, Carreno BM, Cimino PJ, Binder-Scholl GK, Smethurst DP, Gerry AB, Pumphrey NJ, Bennett AD, Brewer JE, Dukes J, Harper J, Tayton-Martin HK, Jakobsen BK, Hassan NJ, Kalos M, June CH. Blood. 2013;122:863–871. doi: 10.1182/blood-2013-03-490565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Warren EH, Fujii N, Akatsuka Y, Chaney CN, Mito JK, Loeb KR, Gooley TA, Brown ML, Koo KK, Rosinski KV, Ogawa S, Matsubara A, Appelbaum FR, Riddell SR. Blood. 2010;115:3869–3878. doi: 10.1182/blood-2009-10-248997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gabrilovich DI, Nagaraj S. Nat Rev Immunol. 2009;9:162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mills KH. Nat Rev Immunol. 2004;4:841–855. doi: 10.1038/nri1485. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.