Abstract
The Drosophila dopaminergic (DA) system consists of a relatively small number of neurons clustered throughout the brain and ventral nerve cord. Previous work shows that clusters of DA neurons innervate different brain compartments, which in part accounts for functional diversity of the DA system. In this paper, we analyzed the association between DA neuron clusters and specific brain lineages, developmental and structural units of the Drosophila brain which provide a framework of connections that can be followed throughout development. The hatching larval brain contains six groups of primary DA neurons (born in the embryo), which we assign to six distinct lineages. We can show that all larval DA clusters persist into the adult brain. Some clusters increase in cell number during late larval stages while others do not become DA-positive until early pupa. Ablating neuroblasts with hydroxyurea (HU) prior to onset of larval proliferation (generates secondary neurons) confirms these added DA clusters are primary neurons born in the embryo, rather than secondary neurons. A single cluster that becomes DA-positive in the late pupa, PAM1/lineage DALcm1/2, forms part of a secondary lineage which can be ablated by larval HU application. By supplying lineage information for each DA cluster, our analysis promotes further developmental and functional analyses of this important system of neurons.
Keywords: Drosophila, brain, development, dopamine neuron, lineage, RRID: AB_528404, RRID: AB_528402, RRID:AB_2314331, RRID:AB_572268, RRID:BDSC_8848, RRID:BDSC_5130
Introduction
The dopaminergic system of the vertebrate brain consists of relatively small populations of brain stem neurons with widespread projections throughout most regions of the central nervous system. Brain centers involved in complex “high order” functions such as the selection and execution of motor behavior, salience attribution, or motivation rely on dopaminergic neuronal input and function. Dopamine receptors are the site of action of many drugs applied in the treatment of mood disorders and psychosis (Winterer and Weinberger, 2004; Rolls et al., 2008). Dopamine-containing (DA) neurons with anatomical and functional attributes that are similar to those established for the vertebrate brain exist in all animals with a nervous system. In Drosophila, the dopaminergic system plays a part in many functions, including addictive behavior, arousal and sleep, learning and memory, locomotor activity, aggression, and metabolic signaling (Wolf and Heberlein, 2003; Birman, 2005; Foltenyi et al., 2007; Waddell, 2010; Riemensperger et al., 2013; Alekseyenko et al., 2013; Bjordal et al., 2014). Recent studies have identified specific subsets of DA neurons to which discrete roles can be attributed (Liu et al., 2012; Musso et al., 2015; Yamagata et al., 2015). Particular interest has been directed to the Drosophila DA neurons in studies of genes whose homologs are involved in Parkinson's disease in humans (Guo, 2010; Riemensperger et al., 2013). To gain additional insights into the mechanisms by which dopamine controls behavior, structural analyses of DA circuits are of central importance.
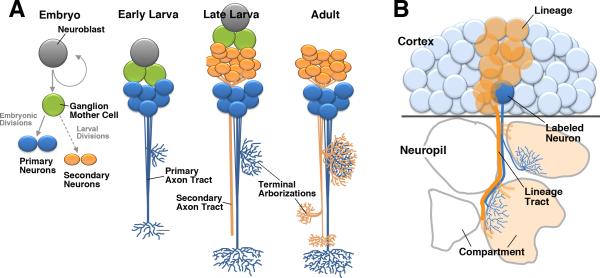
The anatomy of Drosophila DA neurons has been characterized for the larval and adult stage, using antisera against dopamine (DA), tyrosine hydroxylase (TH), and reporter constructs for the TH gene (Budnik and White, 1988; Nässel and Elekes, 1992; Friggi-Grelin et al., 2003; Blanco et al., 2011). In the current paper we establish the identity between larval and adult DA neurons by analyzing the pattern of these cells at successive pupal stages. Furthermore, we assign individual clusters of DA neurons and their projections to specific brain lineages, which represent a topological framework to which all neurons in the Drosophila brain can be mapped. The central brain (supraesophageal ganglion minus optic lobes) is formed by a stereotyped set of approximately 100 paired lineages (Younossi-Hartenstein et al., 1996; Dumstrei et al., 2003; Urbach and Technau, 2003; Pereanu and Hartenstein, 2006; Cardona et al., 2010a; Ito et al., 2013; Lovick et al., 2013; Wong et al., 2013; Yu et al., 2013). Each lineage is derived from one neuroblast that proliferates during an early, embryonic phase and a later, larval phase (Hartenstein et al., 2008; Larsen et al., 2009). Embryonically, each neuroblast generates a small cluster of 10-20 neurons that differentiate and assemble into the larval brain (Larsen et al., 2009; Fig.1A). Neuroblasts then undergo a second, larval phase of proliferation adding more neurons (100-150) to each lineage (Bello et al., 2008; Hartenstein et al., 2008). Neurons of one lineage form a compact cell cluster in the outer layer (cortex) of the brain and project their axons in one cohesive bundle. First, embryonically born (primary) neurons form the primary axon tracts (PATs); axons of larvally born (secondary) neurons follow the PATs and add to them the secondary axon tracts (SATs; Larsen et al., 2009; Lovick et al., 2015; Fig.1A, B). The axonal and dendritic branches of a given lineage form arborizations that typically spread within discrete, invariant compartments of the brain (Hartenstein et al., 2008; Kumar et al., 2009; Spindler and Hartenstein, 2010, 2011; Ito et al., 2013; Wong et al., 2013; Yu et al., 2013). The axon tract of a lineage, together with the compartments filled by its arborizations, forms a structural entity that can be followed from the larva into the adult (Lovick et al., 2013). Lineages form the basis of the neuropil compartments and long axon tracts of the brain.
Fig.1.
Lineages as the ordering principle of Drosophila neuroanatomy. Neuroblasts born in the embryo give rise to primary and secondary neurons through an invariant sequence of asymmetric divisions (A). Primary neurons differentiate in the late embryo and form form tracts (PATs) and terminal arborizations in the larval brain neuropile (B). During later larval stages (C) neuroblasts produce secondary neurons which form secondary axon tracts (SATs) along their primary neuronal siblings. Secondary neurons, along with the majority of primary neurons, differentiate during metamorphosis to form the adult brain (D).
In the present paper, we assign the neurons of the Drosophila DA (DA) system to specific lineages and follow the development of the projection of these neurons from the early larva to adult. Previous work had shown that the clusters of DA neurons are associated with different compartments, including the mushroom body, central complex, and central visual centers (Zhang et al., 2007; Lebestky et al., 2009; Mao and Davis, 2009; Selcho et al., 2009; Aso et al., 2010; Kong et al., 2010; Waddell, 2010). This anatomical diversity, at least in part, accounts for the diversity of functions of the DA system. To unravel the function of the DA system, it will be necessary to manipulate dopamine release or reception in specific groups of neurons. Knowing the parent lineages of the clusters of DA neurons will make it easier to identify Gal4-driver lines that can be used to mark or manipulate these neuron clusters. Our developmental analysis paves the way for more detailed questions into the rewiring of the brain during metamorphosis, and into the control of TH regulation, which will be of particular importance when using Drosophila as a model to study fundamental aspects of neurodegenerative diseases such as Parkinson's.
Material and Methods
Immunohistochemistry
The following primary antibodies were used (Table 1): mouse anti-Neurotactin (Nrt, BP106; RRID:AB_528404), mouse anti-Neuroglian (Nrg, BP104; RRID:AB_528402), and rat anti-DNcadherin (DN-Ex #8; RRID:AB_2314331) antibodies from Developmental Studies Hybridoma Bank (DSHB, University of Iowa, Iowa City, Iowa; each diluted 1:10) and mouse anti-TH (Immunostar, #22941 used at 1:300; RRID:AB_572268). For antibody labeling standard procedures were followed (e.g., Ashburner, 1989). For fluorescent staining, the following secondary antibodies were used: Alexa Fluor 546 goat anti-Mouse IgG (H+L) (#A11030; Invitrogen, Carlsbad, CA; used at 1:500) and Cy5 goat anti-Rat IgG (H+L) (112-175-143; Jackson Immunoresearch, West Grove, PA; used at 1:400).
Table 1.
Primary antibodies used for immunohistochemistry
| Antibody | Immunogen | Source/RRID | Dilution | Specificity |
|---|---|---|---|---|
| Rat anti-DN-cadherin | Exon 8 (a.a. residues 1210-1272) of Drosophila CadN gene | Developmental Studies Hybridoma Bank (DSHB), Iowa City, Iowa, DN-Ex #8; RRID:AB_2314331 | 1:10 | Detected by Western Blot of DN-cadherin transfected S2 cells and immunolabeling of Drosophila embryos (Iwai et al., 1997) |
| Mouse anti-Neuroglian | 18 N-terminal a.a. residues of the Drosophila Neuroglian gene | DSHB, BP104; RRID:AB_528402 | 1:10 | Protein purification by immunoaffinity chromatography using the neuroglian antibody and microsequencing (Bieber et al., 1989) |
| Mouse anti-Neurotactin | 1-280 aminoterminal a.a. residues of Drosophila Neurotactin gene | DSHB, BP106; RRID:AB_528404 | 1:10 | Immunolabeling in embryos matches pattern fusion protein made from Neurotactin cDNA (Hortsch et al., 1990) and an additional monoclonal Neurotactin antibody (Piovant and Léna, 1988) |
| Mouse anti-Tyrosine hydroxylase | Full length tyrosine hydroxylase (TH) from rat PC12 cells; recognizes 34 kDa catalytic core | Immunostar, 22941; RRID:AB_572268 | 1:300 | Immunolabeling of rat catecholamine neurons (TH required for synthesis catecholamine) |
All larvae and adults were grown at 25C on standard food media. Adults were aged 3 to 5 days post-eclosion before dissection. For antibody labeling, standard procedures were followed (Ashburner, 1989). Briefly, dissected brains were fixed with phosphate buffered saline (PBS), pH 7.4, containing 4% paraformaldehyde for 25 – 30 min's. They were then washed with 1x PBS, pH 7.4, containing 0.1 % Triton X-100 for 3 × 10 min's. Samples were then incubated in blocking buffer (2% bovine serum albumin (BSA) in 1X PBS, pH 7.4, containing 0.1 % Triton X-100) for 1 hour at RT. They were then incubated with primary antibody diluted in blocking buffer overnight at 4C. They were then washed 3 × 15 min in 1X PBS, pH 7.4, containing 0.1 % Triton X-100 @ RT. The samples were washed in blocking buffer 1 × 20 min's. They were then incubated with secondary antibody diluted in blocking buffer overnight at 4C. Samples were washed in 1X PBS, pH 7.4, containing 0.1 % Triton X-100 for 3 × 15 min's and mounted in Vectashield (Vector Laboratories).
Brains were viewed in the confocal microscope (20X or 40X objective; MRC 1024ES microscope with Radiance 2000 using Laser sharp 2000, version 5.2 build 824 software; Bio-Rad, Hercules, CA). Confocal sections were taken at 2-μm intervals for all preparations. We used Imagej1.41d for image analysis and to generate merged confocal sections.
Markers
An overview of the key features of the primary antibodies used are shown in Table 1. Details of their production and specificity are summarized below.
The DN-cadherin antibody (DSHB DN-EX #8), a marker for neuropil, is a mouse monoclonal antibody raised against a peptide encoded by exon 8, amino acid residues 1210-1272 of the Drosophila CadN gene. The antibody detected two major bands, 300 kDa and 200 kDa molecular weights on Western blot of S2 cells only after transfection with a cDNA encoding the DN-cadherin protein (Iwai et al., 1997). In addition, the specificity of this antibody was tested with immunostaining of Drosophila embryos. Signal was hardly detectable in homozygous mutant, l(2)36DaM19 with nonsense mutation causes premature termination of protein translation. In contrast, this antibody gave a signal in mutant embryos with N-cadherin transgene.
The Neurotactin antibody (DSHB BP106) labels secondary neurons and their axons. It is a mouse monoclonal antibody raised against the first 280 aminoterminal amino acid residues (Hortsch et al., 1990) of the Drosophila Nrt gene. The monoclonal antibody detected the same Drosophila embryonic pattern to that of a polyclonal antisera raised against a fusion protein using part of the Neurotactin cDNA (Hortsch et al., 1990). In addition, another monoclonal antibody, MAb E1C, against Neurotactin gave a similar expression pattern in Drosophila embryos to that of BP106 (Piovant and Léna, 1988).
The Neuroglian antibody (DSHB BP104) labels secondary neurons and axons in the adult brain. It is a mouse monoclonal antibody from a library generated against isolated Drosophila embryonic nerve cords (Bieber et al., 1989). The Nrg antibody was used to purify protein from whole embryo extracts by immunoaffinity chromatography. Protein microsequencing of the purified protein was performed to determine that the 18 N-terminal amino acids that is identical to the sequence determined for the N-terminus of the protein based on a full-length cDNA clone (Bieber et al., 1989).
The Tyrosine hydroxylase antibody (Immunostar, #22941) labels DA neurons. It is a mouse monoclonal antibody raised against full length tyrosine hydroxylase (TH) purified to homogeneity from rat PC12 cells. Partial digestion with the protease chymotrypsin indicates that the antibody recognizes the 34 kDa catalytic core of TH. The antiserum shows significant labeling of rat catecholamine neuron systems employing indirect immunofluorescent and biotin/avidin-HRP techniques.
For marking DA neurons and inducing single cell clones among this neuronal population we used the following lines: Hs-flp (Basler and Struhl, 1994); TH-Gal4 (Friggi-Grelin et al., 2003; #8848, RRID:BDSC_8848, Bloomington Drosophila Stock Center, Bloomington, IN); UAS-mCD8::GFP (Lee and Luo, 1999; #5130, RRID:BDSC_5130, BDSC);. UAS < Cd2, y+ > CD8::GFP (Wong et al., 2002).
Generation of clones
Flies homozygous for hsflp ; THgal4; UAS > cd2, y+ > CD8::GFP were allowed to lay eggs for 2 days at 25C. Early (first or second instar) larvae were heat shocked at 37C for 5-10 minutes to induce the flip-out in random TH-Gal4-positive neurons, generating clones. Animals were allowed to grow at 25C until eclosion and dissected at day 3 to 5 days post-eclosion.
Generation of Three-Dimensional Models
Staged Drosophila larval and adult brains labeled with suitable markers were viewed as whole-mounts by confocal microscopy [Biorad MRC 1024ES microscope using Biorad Lasersharp version 3.2 software; lenses: 40x oil (NA 1.0; WD 0.17)]. Complete series of optical sections were taken at 2-μm intervals. Digitized images of confocal sections were imported into the Amira (www.amiravis.com) program. All models were generated using the Amira software package. Surface rendered digital models were created by manually labeling each lineage and neuropile compartment within a series of confocal images. The Amira program also allows one to register confocal stacks with each other, and to adjust virtual lighting, camera angle, transparency, reflection and other parameters in a straightforward manner.
Hydroxyurea (HU) Ablation Experiments
Hydroxyurea (HU, Sigma-Aldrich, #H8627-5G) acts as a DNA-synthesis inhibitor which blocks the normal function of nucleotide reductase and is lethal to S-phase cells (Furst and Mahowald, 1985). HU has been used in Drosophila to ablate adult muscle precursors (Broadie and Bate, 1991) as well as central brain neuroblasts (de Belle and Heisenberg, 1994). The procedure for preparation of HU was adapted from Broadie and Bate (1991). HU was administered to fly larvae through the diet. Briefly, HU was dissolved in MilliQ ddH20 at a concentration of 50mg/ml. The dissolved HU was then added to partially melted fly medium to achieve a final concentration of 5mg/ml. After thorough mixing, the HU medium was poured onto 60 × 15mm petri dishes to cool. Food plates were made fresh for each experiment. To ablate neuroblasts, precisely staged larvae were allowed to grow on standard medium at 25°C in petri dishes until time of ablation. Larvae were quickly transferred via blunted forceps to food plates containing 5mg/ml of HU for four hours. This is sufficient time for the HU to accumulate to doses high enough to kill actively dividing neuroblasts (Broadie and Bate, 1991). After four hours, larvae were transferred to petri dishes containing standard medium and grown until dissected as either wandering L3 or adults. Fly stocks and larvae for experiments were grown at 25°C.
Results
Lineage identity of the DA neurons of the larval brain
DA neurons of the early and late larval brain were labeled by anti-TH and TH-Gal4 driving UAS-mCD8::GFP. Both markers, used in the same brain specimen, labeled identical clusters of neurons, confirming the findings of Friggi-Grelin et al. (2002). Nässel and Elekes (1992) concluded that the expression of TH and DA “probably label the same neurons.” Neurons labeled by anti-DA or anti-TH/TH-Gal4-positive neurons can be expected to contain DA, even though that in itself is admittedly no proof that they are dopaminergic, in the sense that they utilize dopamine as a transmitter. That being said, we will in the following refer to these neurons as “DA neurons”, following the convention established in the recent literature (e.g., Waddell, 2010; Yamagata et al., 2015). We compared the pattern of DA clusters in the early larva (L1) and late larva (L3). The DA population comprises two dorsomedial clusters (DM1/2) and two lateral clusters (DL1/2). DM1 and DL2 are split into two smaller units each (DM1a/b and DL2a/b, respectively) (Fig.2A). Since most larval DA neurons appear as differentiated cells already in L1 (Figs.2B, C; 3A), they constitute embryonically born primary neurons, a supposition confirmed by the analysis of Blanco et al (2011) who could assign the seven clusters to seven separate neuroblast clones induced in the embryo. One cluster, DM2, is not yet represented at L1 (Fig.2B); cells of this cluster gradually express TH-Gal4 during the L3 stage (Fig.3A). In mid-L3 larvae, DM2 is typically represented by a single TH-Gal4-positive neuron (not shown); by late L3 the number has increased to about 6 (Figs.3A; 4C). We conclude that, even though all DA neurons are born in the embryo, they initiated TH expression at different stages in the larva. As shown below, several groups of primary neurons remain TH-Gal4-negative throughout the larva and initiate expression during metamorphosis.
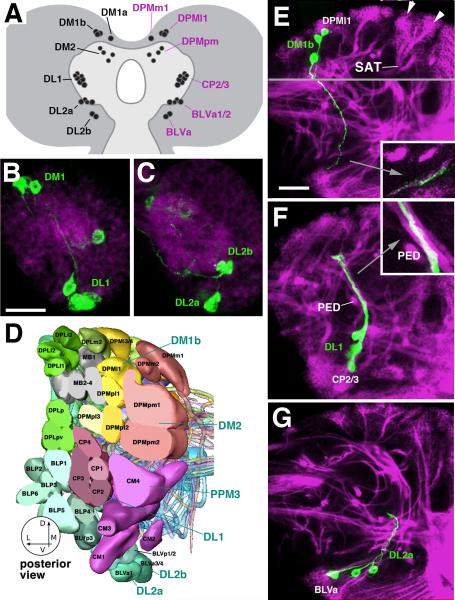
Fig.2.
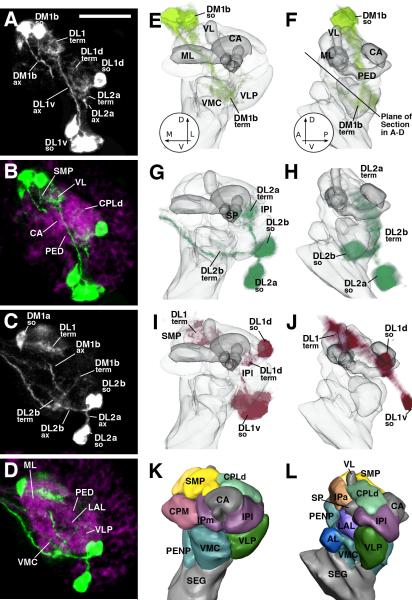
Lineage identity of larval DA neurons. A: Schematic drawing of late larval brain introducing the nomenclature of DA neuron clusters (left side). Lineage identity of these neurons, established in this paper, is shown on right side. B, C: Z-projections generated from five consecutive frontal confocal sections of L1 brain hemisphere (B: posterior level, C: central level. Dorsal is up, medial to the left. DA neurons are labeled by TH-Gal4 > UAS-mCD8::GFP (green); brain neuropile is labeled by anti-DN-cadherin (magenta). E-G: Z-projections of frontal confocal sections of third instar larval brain hemisphere in which DA neurons were labeled with TH-Gal4 > UAS-mCD8::GFP (green). Secondary lineages are marked by anti-Neurotactin (magenta). Dorsal is up, medial to the left. Each lineage forms a cluster of neuronal cell bodies at the periphery of the brain (examples shown by arrowheads in E) from which a tract of axons (secondary axon tract, SAT) projects radially into the neuropile. D: 3D digital model of secondary lineages of late larval brain. Posterior view, only lineages of posterior half of brain are shown. Different colors identify lineage groups, as defined in Pereanu and Hartenstein, (2006). DA neuronal clusters associated with lineages are highlighted by blue lettering. E: DM1b neurons have axons that form part of the tract of lineage DPMl1; inset shows close association of (primary) DM1b neurons and (secondary) DPMl1 axon tract. F: DL1 neurons are associated with the lineage pair CP2/3, which forms a conspicuous tract that crosses over the peduncle (PED; shown at high magnification in inset). G: DL2a cluster is associated with the BLVa1/2 lineage group. Scale bar: 20μm (B, C); 25μm (E-G)
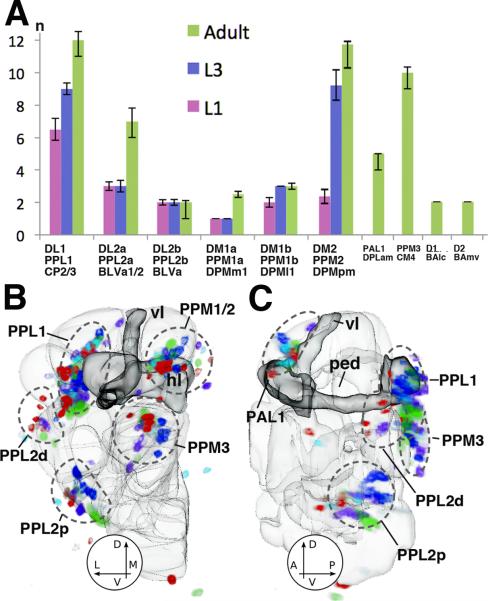
Fig.3.
Cell numbers and variability of Drosophila DA neurons. (A) Plot of cell numbers per one brain hemisphere for different clusters of DA neurons at early larval stage (L1), late larval stage (L3), and adult (n=5). (B, C) Variability in number and position of DA neurons in the adult brain. Both panels show 3D digital models of left brain hemisphere in posterior view (A) and lateral view (B). Outlines of compartments (light gray) and mushroom body (dark gray) are shown as reference. Labeled cell bodies of DA neurons of five registered brain hemispheres are shown in different colors. One color (e.g., red) represents all DA neurons of one brain hemisphere. The overlay of five brains still allows to recognize the main clusters of DA neurons (PPL1, PPL2d/p, PPM1-3); however, there is considerable scatter of somata within clusters. Abbreviations of compartments: ML medial lobes of the mushroom body; PED peduncle; VL vertical lobes of the mushroom body.
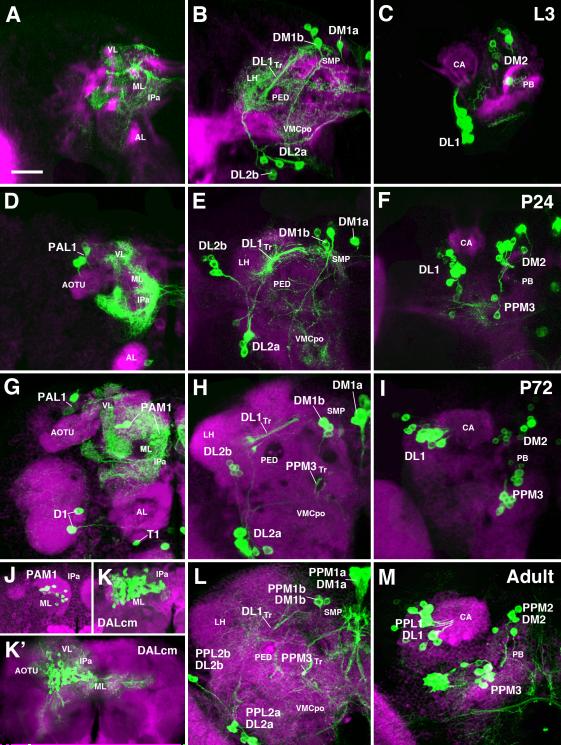
Fig.4.
Development of pattern of DA neurons from larva to adult. All panels present z-projections of frontal confocal sections of brain hemisphere at late third instar larval stage (A-C), 24h pupa (D-F), 72h pupa (G-I), and adult (J-M). Dorsal up, medial to the right. DA neurons are labeled with TH-Gal4 > UAS-mCD8::GFP (green). Secondary lineages are labeled by anti-Neurotactin (red in A-C) or anti-Neuroglian (red in D-I, L, M). Panels of the left column (A D, G, J-K’) represent anterior slice of the brain at the level of the mushroom body lobes (ML medial lobe; VL vertical lobe). Middle column (B, E, H, L) shows posterior slice at level right behind central complex; right column (C, F, I, M) represents z-projections far posterior, at level of calyx (CA) and protocerebral bridge (PB). Clusters of DA neurons, located in the anterior brain (left column) or posterior brain (middle and right column) are annotated. Individual clusters can be followed throughout metamorphosis based on location and axonal trajectory. For adult (bottom row), both larval and adult annotation is provided (e.g. PPL2b/DL2b). Note DA clusters that become TH-positive during metamorphosis: PPM3 and PAL1, corresponding to the lineages CM4 and DPLam, respectively, are recognizable from 24h onward (D, F); PAM1 first differentiates in the 72h pupa (G, J-K’). Also note strong reduction of terminal arborizations of DA neurons during pupa (E, H), compared to larva (B) or adult (L). Other abbreviations: AL antennal lobe; AOTU anterior optic tubercle; CA calyx; IPa anterior domain of inferior protocerebrum; LH lateral horn; ML medial lobe; PED peduncle; PB protocerebral bridge; SMP superior medial protocerebrum; VL vertical lobe; VLP ventrolateral protocerebrum; VMCpo posterior domain of ventromedial cerebrum. Bar: 25μm
To determine the specific lineage identity of the DA clusters we mapped the trajectory of their primary axon tracts relative to the secondary tracts in the late larval brain, for which a complete lineage atlas has been established (Pereanu and Hartenstein, 2006; Cardona et al., 2010a; Fig.2D). The underlying assumption is, as explained in the introduction and shown in Fig.1, that the axon tracts of secondary lineages fasciculate with, or are at least are very close to, the axon tracts formed by primary neurons. Such a spatial relationship has been shown for many lineages for which both primary and secondary components were labeled together (Larsen et al., 2009; Das et al., 2013; Lovick et al., 2015). To visualize the SATs, brains were labeled with anti-Neurotactin (Nrt; magenta in Fig.2E-G). It is apparent that most of the GFP-positive axons of DA neurons fasciculate with specific SATs, which we then took as the criterion to assign a given DA neuron to the corresponding lineage. A summary of all DA clusters including the lineages they associate with and the neuropil compartments they innervate is presented in Table 2.
Table 2.
DA neuron clusters (first column: larval clusters; second column: adult clusters), neuron type (primary or secondary; third column), associated lineages (fourth column; upper term: nomenclature after Pereanu and Hartenstein, 2006; lower term in green: Nomenclature after), and main projections in neuropil (fifth column)
| Larval Cluster | Adult Cluster | Neuron Type | Lineage | Innervation |
|---|---|---|---|---|
| DL1 | PPL1 | 1° | CP2/3 DL1/2 |
Vertical Lobe of Mushroom Body (VL) Superior Medial Protocerebrum (SMP) Anterior Inferior Protocerebrum (IPa) Superior Lateral Protocerebrum and Lateral Horn (SLP, LH; CPLd in Larva) |
| DL2a | PPL2a | 1° | BLVa1/2 LHa2/4 |
Calyx of Mushroom Body (CA) Superior Lateral Protocerebrum (SLP) Calyx of Mushroom Body (CA) |
| DL2b | PPL2b | 1° | BLVa LHa |
Lobula Complex (LO) Superior Lateral Protocerebrum (SLP) Ventromedial Cerebrum (VMC) |
| DM1a | PPM1a | 1° | DPMm1 DM1 |
Subesophageal Zone (SEZ) Ventromedial Cerebrum (VMC) Subesophageal Zone (SEZ) Lateral Accessory Lobe (LAL) |
| DM1b | PPM1b | 1° | DPMl1 PSp3 |
Ventromedial Cerebrum (VMC) Subesophageal Zone (SEZ) |
| DM2 | PPM2 | 1° | DPMpm1/2 | * |
| PPM3 | 1° | DM2/3 CM4 DM4 |
Central Complex (CC) Lateral Accessory Lobe (LAL) Bulb (BU) Ventromedial Cerebrum (VMC) Superior Medial Protocerebrum (SMP) |
|
| PAL1 | 1° | DPLam | Ventromedial Cerebrum (VMC) | |
| PAM1 | 2° | VLPd2 DALcm1/2 CREa2 |
Medial Lobe of Mushroom Body (ML) Superior Medial Protocerebrum (SMP) Anterior Inferior Protocerebrum (IPa) |
|
| D1 | 1° | BAlc ALl1 |
Antennal Lobe (AL) | |
| T1 | 1° | BAmv1/2 LALv1 or VESa2 |
Ventromedial Cerebrum (VMC) |
DM1 and DM2
DM1, located more anteriorly than DM2, contains 1-2 medial neurons (here termed DM1a) with descending axons. This trajectory corresponds to that of lineage DPMm1. A second, slightly more laterally located component of DM1 (here termed DM1b; 2-3 cells; here and in the following we refer to cell number refer per brain hemisphere) fasciculates with the SAT of lineage DPMl1 (Fig.2E). DM2, located further posteriorly, behind the protocerebral bridge (Nässel and Ekeles, 1992) is formed by a larger group of neurons (4-6 somata). They are typically stained more faintly, and their axons cannot be followed for a long distance. However, axons enter the neuropil alongside the SATs of the two DPMpm lineages, which suggests that DM2 neurons form part of one or both of these lineages (not shown).
DL1
The DL1 cluster comprises 6-8 cells that follow the tract formed by the centro-posterior lineages CP2/3 (Fig.2F). Blanco et al (2011) concluded that two neuroblast lineages contributed to the DL1 cluster, suggesting that both CP2 and CP3 (which share a common tract: Pereanu and Hartenstein, 2006; Cardona et al., 2010a) produce DA neurons. Further supporting this idea is the observation that in the early larval brain the DL1 cluster is typically split into a large, ventral group and a small, dorsal group of cells (Fig.2B); these two groups might correspond to the CP2 and CP3 lineages, respectively.
DL2
The second cluster, DL2, is located further ventrally and anteriorly than DL1 (Budnik and White, 1988; Nässel and Elekes, 1992). Cell bodies fall within two subgroups, termed here DL2a and b, and are located closest to the BLVa group of lineages, formed by four lineages with tracts projecting dorsally. The tract of the pair BLVa1/2 reaches up into the dorsal protocerebrum (see below), whereas the tract of the BLVa3/4 pair is shorter. Axons of the DL2 clusters of DA neurons do not fasciculate directly with either of these SATs. DL2a axons are close to the tract (and terminal arborization) defined by the BLVa1/2 lineage pair (Fig.2G). Cell bodies of the small, ventral group of DA neurons, DL2b, are located closest to the BLVa3/4 pair of lineages (not shown).
Developmental increase of primary DA neurons during the larval period
Several DA clusters, notably DL1 and DM2, increase in size from early larva to adult. In the case of DM2, L1 larvae had between 0 and 4 neurons (Fig.2D), whereas in late larvae and pupae the number went up to more than 10 (Figs.3A; 4C, F, I, M). In DL1, the increase is less dramatic, but still significant, from 6-8 cells in the early larva to 12-14 cells in the adult (Fig.3A). The additional DA-positive neurons could be either primary neurons, born in the embryo and expressing TH-Gal4 later than other DA neurons, or secondary neurons, born during the second phase of neuroblast proliferation that takes place in the larva. To distinguish between these possibilities we ablated neuroblasts prior to onset of larval proliferation by feeding the drug hydroxyurea (HU), which blocks cells in mitosis, and ultimately leads to the demise of neuroblasts (de Belle and Heisenberg, 1994). HU application does not prevent the addition of DA neurons belonging to the DM2 and DL1 clusters during later larval stages (Fig.5A, B), supporting the notion that these neurons are primary neurons born in the embryo, prior to the HU pulse.
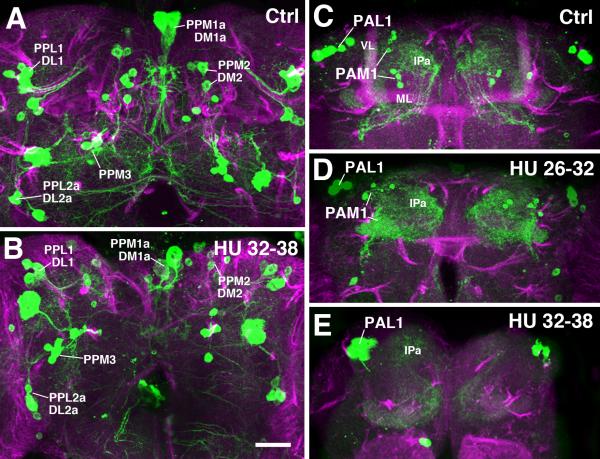
Fig.5.
Pattern of DA neurons in adult brain following HU ablation of neuroblasts at early larval stage. Panels present z- projections of frontal confocal sections of adult brain. Dorsal up, medial at center of panels. DA neurons are labeled with TH-Gal4 > UAS-mCD8::GFP (green). Secondary lineages are labeled by anti-Neuroglian (magenta). A, B: Posterior brain. All DA clusters seen in the control brain (A; see also Fig.3L, M) are visible after a HU pulse from 32h to 38h (B), which ablates most secondary lineages. C-E: Anterior brain; control (C), HU pulse 26-32h (D), HU pulse 32-38h (E). Whereas neither HU pulse interferes with the development of the PAL1 cluster, the late pulse (E) eliminates PAM1, indicating that this cluster comprises secondary neurons.
Other abbreviations: IPa anterior domain of inferior protocerebrum; ML medial lobe; VL vertical lobe. Bar: 25μm
Primary DA clusters of the larva persist into the adult brain
We analyzed the pattern of DA neurons expressing TH-Gal4>UAS-mCD8::GFP during three pupal stages (P12, P24, P72) and in the adult brain. As for the larval brain, we compared the expression of TH-Gal4 driving UAS-mCD8::GFP to the pattern of neurons labled by anti-TH. In the adult brain, a sizeable fraction of the PAM1 neurons is not labeled by TH-Gal4, confirming the intitial analysis of Friggi-Grelin et al. (2003). In addition, a small subset of neurons forming part of the PPM2 and PPM3 clusters is anti-TH-positive, but does not express TH-Gal4.
Our data demonstrate that all clusters of primary DA neurons described for the larva persist in the adult. Whereas terminal arborizations of DA neurons are pruned during metamorphosis and may grow back in an adult-specific pattern (see below), the cell bodies and principal trajectories of axons are recognizeable at all pupal stages (Fig.4). As a result, larval groups of DA neurons could be unequivocally assigned to adult groups defined in previous papers (Nässel and Elekes, 1992). DM1 and DM2 correspond to PPM1 and PPM2, respectively; DL1 and DL2 become PPL1 and PPL2.
To the primary neurons listed above, several DA clusters are added during metamorphosis. One can subdivide these into two groups: (1) primary neurons that start expressing TH in the pupa; (2) secondary neurons born during the larval period.
PPM3 and PAL1: primary neurons of lineages CM4 and DPLam that show delayed TH expression during metamorphosis
The PPM3 cluster appears during early pupal stages (Fig.4F, I). Based on its location and trajectory, this group forms part of the CM4 lineage. It appears that the PPM3 neurons are primary neurons: already in the early pupa, when they first become TH-Gal4-positive, their cell bodies are large in comparison to the immature secondary neurons and they are scattered, rather than forming a tight cluster. Furthermore, application of HU at early larval stages did not prevent the appearance of PPM3 in the pupa and adult (Fig.5B). This implies that the PPM3 neurons already existed in the larva and do not begin expressing TH-Gal4 until during metamorphosis. In the 24h pupa, 2-3 PPM3 cell bodies are faintly visible (Fig.4F), and by P72 expression has spread to 9-10 cells (Fig.4I).
The second group of primary neurons that become TH-Gal4-positive in the early pupa is the PAL1 cluster, located in the anterior cortex; its axons project posteriorly and ventrally, fasciculating with the SAT of the DPLam lineage (Fig.4D, G). As stated above for PPM3, the PAL1 neuronal cluster persists after applying pulses at stages when DPLam proliferates, confirming the primary nature of this cluster (Fig.5D, E)
Two small groups of DA neurons which have not received much attention in the previous literature, but where reported in the careful anatomical study by Nassel and Ekeles (1992), become visible in the anterior brain at 72h after puparium formation (apf; Fig.4G). The D1 cluster, projecting along the tract of the BAlc lineage which contains local interneurons and projection neurons of the antennal lobe (Das et al., 2013), has 1-2 cells; T1, projecting along the tract of lineages BAmv1/2 into the ventromedial cerebrum and periesophageal neuropil, contains a single cell
PAM1: Secondary neurons of the DALcm1/2 lineage expressing TH
The PAM1 cluster starts expressing TH-Gal4 at around 48h apf (Fig.4G, J). The cluster contains in the order of 30-40 (approximately 100 neurons when labeling with anti-TH) small, closely packed somata located in the anterior cortex. Axons form a short tight bundle that fasciculates with the SATs formed by DALcm1/2 and the dorsal BAmd1 hemilineage (Fig.4K, K’). In accordance with the projection pattern described for these lineages (Wong et al., 2013), PAM1 neurons form a dense network of short terminal arbors in the medial lobe of the mushroom body (see below). PAM1 represents the only group of secondary neurons among the DA system, since HU application at 32-38h after hatching was able to eliminate this cluster in the adult (Fig.5E). By contrast, HU pulses between 20 and 32h left the cluster intact (Fig.5D), indicating that PAM1 neurons belong to DALcm1/2, which is born after 32h (Lovick and Hartenstein, 2015).
Attempts have been made in the past to demonstrate defects in the Drosophila DA neurons through analysis of mutations of genes affecting the DA system in vertebrates (Pesah et al., 2005; Sang et al., 2007). The results suggest that defects, if existing at all, are minor. A parameter that can cause debate is variability of neuronal number and projection among wild type animals. Sykes et al. (2004) found a considerable variability in the number of DA neurons in the adult ventral nerve cord. To address this issue we counted cells identified as DA neurons by anti-TH and/or TH-Gal4>UAS-mCD8::GFP for early larva, late larva, and adult (Fig.3A). In addition, the set of DA neurons of five registered adult brains were superimposed to address the question of spatial variability (Fig.3B, C). Cell counts show that variability in number (standard deviation) ranges around 10% for most groups (omitting the PAM1 cluster, which contains a large number of small, secondary neurons, most of which are negative for TH-Gal4>UAS-mCD8::GFP). Thus, the number of DA neurons of a given group rarely varies more than ± 1 among different specimens (Fig.3A). The same applies for DA neurons at larval stages (data not shown). Note that the (minor) variability could be accounted for by slight differences in labeling intensity. The spatial scatter of cell body location, on the other hand, is considerable for all groups (Fig.3B, C).
The groups of DA neurons can be assigned to discrete neuropil compartments
During the first larval instar, the arborization of the DA neurons is relatively sparse. This makes it possible to analyze in preparations in which all DA neurons are labeled using TH-Gal4 how the individual neuron clusters distribute their neurites to different neuropil compartments (Fig.6). There are only few neuropil compartments in the late larval or adult brain that do not receive input from at least one DA neuron. It is generally assumed that compartments, some delineated more easily than others, represent structural and functional units of the insect brain. Compartment boundaries are formed by a higher density of glial processes and systems of fiber bundles (Younossi-Hartenstein et al., 2003; Pereanu et al., 2010) and can be visualized by markers for glia or neurites, such as anti-DNcadherin employed in this work. Compartments defined for the larva persist in the adult brain; in addition, several compartments, notably those of the central complex, are formed de novo during metamorphosis. We will employ the nomenclature introduced for the adult brain (Pereanu et al., 2010; Ito et al., 2014) when referring to compartments throughout development.
Fig.6.
Terminal arborization of DA neurons in early larval brain. A-D: Z-projections of confocal sections of L1 brain hemisphere. DA neurons are labeled by TH-Gal4 > UAS-mCD8::GFP (white in A, C; green in B, D). In B and D, neuropil compartments are visualized by anti-DN-cadherin (magenta). Plane of confocal sections is tilted at a 45deg angle relative to the horizontal plane, as shown by the line in panel F. E-L: 3D digital models of first instar larval brain hemisphere. Models of left column (E, G, I, K) show anterior view (dorsal up, medial to the left); right column presents lateral view (dorsal up, anterior to the left). In E-J, neuropil compartments are shown semitransparent and in gray color; compartments are rendered in different colors in panels K and L. E-J present volume renderings of the three most conspicuous DA clusters of the early larva in different colors (E, F: DM1 light green; G, H: DL2 dark green; I, J: DL1 magenta). In all panels, DA clusters are annotated in a manner that points out the location of somata (so), axon tract (ax), and terminal arborization (term). Abbreviations of compartments: AL antennal lobe; CA calyx of mushroom body; VL vertical lobe of mushroom body; IPa anterior domain of inferior protocerebrum; IPl lateral domain of inferior protocerebrum; IPm medial domain of inferior protocerebrum; CPLd primordium of superior lateral protocerebrum and lateral horn; CPM primordium of central complex and posterior inferior protocerebrum; LAL lateral accessory lobe; ML medial lobe of mushroom body; PED peduncle; PENP periesophageal neuropil; SEG subesophageal ganglion; SMP superior medial protocerebrum; SP spur; VLP ventrolateral protocerebrum; VMC ventromedial cerebrum; Bar: 20μm
In the following, we will summarize the major projections of the DA neuronal clusters that is visible in the early larval brain. The innervation of the mushroom body and central complex has been described in detail for the late third instar larva (Selcho et al., 2009) and adult (Mao and Davis, 2009; Waddell, 2013; Alekseyenko et al., 2013). We will only briefly mention these projections and refer to the cited papers for detail.
DL1/PPL1 neurons innervate the superior protocerebrum and the mushroom body
The DL1 group of larval DA neurons, forming part of the CP2/3 lineage, projects along the oblique posterior fascicle (obP) anteriorly and medially. Terminal branches at the distal tips of the axons are relatively focused around the vertical lobe (VL) of the mushroom body and in the medially adjacent superior medial protocerebrum (SMP) and anterior inferior protocerebrum (IPa; Fig.6A, B, I, J; Table 2). Several DL1 neurons have also proximal branches in the CPLd compartment, a thin layer of dorsal neuropil that will grow into the superior lateral protocerebrum and lateral horn of the adult brain (Fig.6A, B, I; for position and nomenclature of compartments see Fig.6K, L). In the adult, PPL1 cells have a similar projection pattern (Mao and Davis, 2009).
DL2/PPL2 neurons innervate the superior lateral protocerebrum and calyx
DL2a neurons of the larva (BLVa1/2 lineage) project long unbranched neurites dorsally; distal terminal branches are confined to the calyx and centro-posterior lateral (CPLd) compartment, where they intermingle with proximal branches of DL1 (Fig.6A, B, G, H; Table 2). In the adult, PPL2a neurons also have distal branches in the calyx and deep layers of the superior lateral protocerebrum (SLP), descendant of the larval CPLd (Waddell, 2010).
The second subgroup of DL2/PPL2, DL2b/PPL2b, has neurites extending posteriorly and forming terminal arborizations in the posterior domain of the ventromedial cerebrum (VMC; Fig.6C, D, G, H; Table 2). Some or all axons continue across the midline. In L3, this projection is also visible; in addition, neurites of DL2b neurons now also project vertically, joining the bundle formed by DL2a neurons towards the calyx and CPLd. In the pupa and adult, DA expression in PPL2b is faint or absent (not shown).
DM1/PPM1 and PAL1 neurons descend to the basal brain
Larval DM1 neurons have a similar overall shape as the DL1 and DL2 neurons, extending long, unbranched neurites that form mostly distal terminal arbors. In this case, neurites follow a ventral course. DM1b extend along the DPPT (dorso-posterior protocerebral tract), the tract associated with the DPMl1 lineage which DM1 forms part of, and terminate in a wide domain covering much of the ventromedial cerebrum (VMC) and anteriorly adjacent lateral accessory lobe (LAL; Fig.6A, C, E, F; Table 2). PPM1b neurons have a similar morphology in the adult (not shown). DM1a neurons project ventrally at a more medial location, following the medial cervical connective (not shown). Their terminations are in the tritocerebrum and subesophageal ganglion (the subesophageal zone or SEZ; Ito et al., 2014), where they overlap with arbors of subesophageal DA neurons (not considered here). DM2 neurons express TH in late larvae. Located in the posterior cortex, neurites extend forward in the longitudinal superior medial fascicle (loSM), following the trajectory of the DPMpm1/2 lineages (not shown). Due to the wide overlap of terminal arbors of DA neurons in the late larva it is not possible to delineate the arborization of DM2 (delineated by an asterisk in Table 2).
The PAL1 group of neurons starts expressing TH around 24h after puparium formation. These cells, presumably primary neurons of the DPLam lineage, project a long, unbranched tract posteriorly and medially, across the peduncle (Fig.4D, G; Table 2), and form widespread, bilateral terminal branches in the ventromedial cerebrum (VMC), a compartment surrounding the great commissure.
PPM3 neurons innervate the central complex
The PPM3 group of neurons, members of the CM4 lineage, comprises 5-8 cells with long unbranched neurites projecting anteriorly along the medial equatorial fascicle (MEF; Fig.4F). PPM3 neurons have widespread distal terminal branches in the central complex and neighboring compartments, including the lateral accessory lobe and bulbs (LAL, BU; both closely connected with the central complex; Hanesch et al., 1989), the superior medial protocerebrum (SMP), the anterior domain of the inferior protocerebrum (IPa), and the ventromedial cerebrum (VMC; Fig.4L; Table 2). It is evident that individual members of the PPM3 cluster have distinctly different structural phenotypes, as described in a recent study by Alekseyenko et al. (2013).
PAM1/DALcm1/2 neurons innervate the medial lobes of the MB and adjacent protocerebral domains
The last group of DA neurons, PAM1, stands out by the fact that its neurons are quite clearly secondary neurons. As described in detail by previous authors (e.g., Aso et al., 2014), PAM1 neurons have relatively short, bifurcated neurites with terminal arbors in small regions of the medial lobes of the mushroom body (ML) and surrounding inferior and superior protocerebrum (IPa, SIP, SMP; Table 2).
Discussion
Lineage-based subdivision of the Drosophila DA system
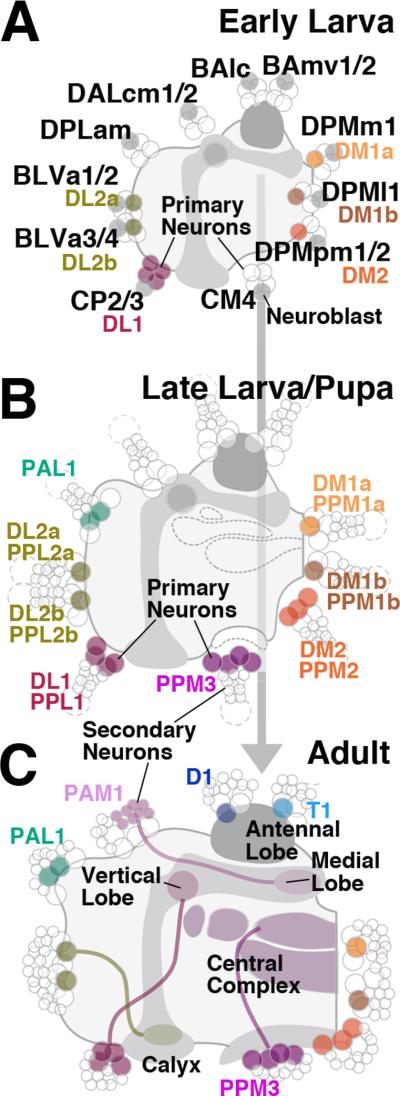
The first objective of this paper was to assign the brain DA neurons to neural stem cell lineages and follow them from their time of appearance to the adult stage. At least six different lineages contain primary neurons that express TH during the larval stage. Primary neurons of four additional lineages initiate TH expression past the onset of metamorphosis (summarized in Fig.7A, B). Our second goal was to establish whether primary DA neurons visible in the larva are maintained in the adult, or are replaced by secondary neurons. The former is the case, as shown by the continued presence of labeled (antibody or TH-Gal4> UAS-mCD8::GFP) clusters of neurons at corresponding positions at closely spaced pupal stages (Fig.4; summarized in Fig.7B, C), and the fact that application of HU during larval stages does not eliminate DA neurons in the adult (Fig.5). Only one group of DA neurons, PAM1, consists of secondary neurons of lineage DALcm1/2, born in the larva, and differentiating during late pupal stages (Fig.7C). Interestingly, a recent study (Rohwedder et al., 2015) in which a different antibody directed against TH was used, discovered four additional primary neurons (pPAM) that express DA, and that, based on their branching pattern (around the medial lobe of the mushroom body) belong to the lineage DALcm1/2.
Fig.7.
Schematic representation of the system DA neurons at early larval stages (A), late larval/early pupal stages (B), and the adult (C). Nomenclature of DA neurons is shown in association with the names of the corresponding lineages. Primary neurons are indicated by large circles, secondary neurons by small circles. The innervation of the different compartments of the mushroom body and central complex of the adult brain by clusters PPL1, PPL2, PAM1, and PPM3 is indicated.
Projections of the different groups/lineages of DA neurons can be assigned to different compartments of the brain. Notably, PPL1 (CP2/3 lineage) innervates the lobes of the mushroom body and the adjacent IPa compartment in the anterior protocerebrum; PPM3 (CM4 lineage) branches widely in the different compartments of the central complex and adjacent LAL, and PPL2 (BLVa1/2) projects to the posterior SLP. Within a given group, DA neurons differ further in regard to their exact branching pattern, as shown here by marking individual neurons of the PPM3/CM4 group, and in previous reports (Mao and Davis, 2009; Waddell et al., 2010; Alekseyenko et al., 2013) that focused on the mushroom body-related PPL1/CP2/3 neurons. PPL1 neurons fall into five different types innervating different domains within the vertical lobe (Mao and Davis, 2009). PPM3 neurons project to different compartments of the central complex; for example, one subset innervates the upper layers of the fan shaped body (FB) and the ellipsoid body (EB); another one is restricted to the lower FB and noduli (NO; see also Kong et al., 2010; Alekseynko et al., 2013). These findings suggest that primary neurons of the CP2/3 lineage have a unique identity, which coincides with numerous previous studies of primary neurons belonging to other lineages (Bossing et al., 1996; Schmidt et al., 1997; Schmid et al., 1999).
At present, not much information is available about the primary lineages that include the clusters of DA neurons of the larval brain. Primary neurons are produced in the embryo by the proliferatory activity of approximately 100 pairs of neuroblasts. These cells undergo 5-8 rounds of division, producing lineages of 10-16 cells each (Larsen et al., 2009). Approximately 30% of primary neurons undergo programmed cell death at the embryo-larva transition, reducing the average size per larval primary lineage to about 10 (Larsen et al., 2009). Still, this number exceeds the number of DA neurons in any of the clusters in the larval brain, implying that not all cells of a given lineage express the DA phenotype. Future studies, employing lineage-specific markers, are required to establish these additional details about the DA neuronal genealogy.
Location and projection pattern of DA neurons have been mapped for numerous insect species, among them locusts (Homberg, 2002) and bees (Schäfer and Rehder, 1989; Schürmann et al., 1989; Kirchhof et al., 1999). In all species, DA neurons are distributed in discrete clusters, scattered over much of the brain surface, suggesting that as in Drosophila, these neurons form part of multiple lineages. In some cases, it is evident that lineages producing DA neurons are homologous. For example, the C1/C2 clusters of DA neurons in the bee brain (Schäfer and Rehder, 1989) quite clearly correspond in cell body location and projection (to the lobes of the mushroom body) to PAM1/DALcm1/2. Likewise, C3 of the bee may correspond to a combination of PPL1/CP and PPM3/CM4. C3 emits two separate fiber bundles, one of which projects and ramifies towards the central complex; this could represent the PPM3/CM4 group in fly which projects via the MEF fascicle to the central complex. The second contingent of bee C3 DA neurons projects towards the lobes of the mushroom body in the anterior neuropil; this group may correspond to PPL1/CP in Drosophila. Aside from the above mentioned similarities, one very obvious difference exists in the antennal lobe with regards to in the distribution of DA neurons and their terminal arbors. In Drosophila, only two neurons which start expressing TH during metamorphosis are associated with this compartment, whereas in other insects (like bee), the antennal lobe receives dense projections from DA neurons located adjacent to it (Kirchhof et al., 1999). It will be an interesting and rewarding task to establish, lineage by lineage, the similarities and differences in the anatomy of the DA system (as well as other groups of neurons sharing similar neurotransmitter expression patterns) among different insect taxa, and to correlate these patterns with physiological and behavioral parameters.
Functional neuroanatomy of the DA system
DA neurons play a role in many aspects of insect brain function, which is not surprising since most parts of the brain are innervated by these neurons. Predictably, DA neurons projecting towards the mushroom body will be involved in learning and memory (Aso et al., 2010; 2014; Waddell, 2010; 2013), whereas DA neurons associated with other compartments will have other functions. It is of course possible that intricate, not yet resolved interactions take place between the different DA neurons. One fundamental role of DA neurons appears to be to modulate overall motor activity (endogenous arousal; sleep vs wakefulness), as well as the responsiveness of brain neurons to specific sensory stimuli (exogenous arousal; Lebestky et al., 2009).
Three clusters of DA neurons of the adult brain, PPL1, PPL2, and PAM1, innervate the vertical lobe/spur (also called “heel”), the calyx, and the medial lobe of the mushroom body (MB), respectively. Experimental studies link each of these clusters to memory formation. PPL1 neurons were identified as part of the circuit required for aversive odor memory (Claridge-Chang et al., 2009), but (in part) also play a role in reward learning. Thus, several PPL1 neurons carry information reflecting energy build up during the formation of long term memory (Musso et al., 2015). In the larva, the DL1 (=PPL1 in adult), but also other DA groups (DM1, DM2) sense amino acid imbalances and control food intake (Bjordal et al., 2014). PAM1 neurons, innervating different domains of the medial lobes of the MB, also form part of the pathway that carries both aversive (electric shock) and reward stimuli (e.g., sweet taste; water) towards the MB (Aso et al., 2012; Lin et al., 2014; reviewed in Waddell, 2013). Different subsets of PAM1 neurons appear to be involved in the formation of short term and long term memories (Yamagata et al., 2015). Aggressive behavior is modulated by DA neurons of the PPM3 group whose neurons widely innervate the central complex (Alekseyenko et al., 2013). Members of the PPM3 cluster of DA neurons also control arousal and ethanol-induced locomotion (Lebestky et al., 2009; Kong et al., 2010).
An important step forward in our understanding of DA neuronal function would be to decipher how DA synapses, as well as different DA receptors, are integrated into the microcircuitry of the brain. Progress towards this has been made in the mammalian prefrontal cortex, where DA axons innervate both projection neurons (glutamatergic, excitatory pyramidal cells) as well as local interneurons (inhibitory GABAergic cells). Pyramidal neurons form direct, reciprocal excitatory connections; in addition, local different types of interneurons form recurrent inhibitory and disinhibitory loops (Goldman-Rakic, 1999; Goldman-Rakic et al., 2000; Druga, 2009). The electrical activity of this structured network depends on the balance between excitatory and inhibitory elements; for example, elevated excitatory synapse activity, or depressed inhibitory synapse activity, would result in an abnormal spread of activation throughout the prefrontal cortex. Dopamine plays a key role in modulating this balance (Winterer and Weinberger, 2004; Rolls et al., 2008; Glausier et al., 2009). The exact pattern of dopamine receptor expression in pyramidal neurons and inhibitory interneurons, and of the distribution of DA neurons within the microcircuitry of the prefrontal cortex, specifies the role of the DA system.
The relationship between DA terminals, DA receptors, projection neurons, and local interneurons has not been worked out in the Drosophila brain. Thus, it is not clear for the mushroom body, central complex, or any other compartment innervated by DA neurons what type of neuron is the direct target of the DA input. Attempts to reconstruct neuronal connectivity, in the larval brain, from serial section electron microscopy (EM; Cardona et al., 2009, 2010b; Fushiki et al., 2016) will hopefully provide answers to these questions. Thus, given their characteristic projection, branching pattern, and possibly ultrastructural characteristics (such as size and shape of transmitter vesicles), it should be possible to identify DA neurons in the serial EM stacks, and reconstruct their exact position within the neuronal circuits of different brain compartments. The association of DA neurons with specific lineages plays an important role within this undertaking. Thus, trajectories of primary lineages, including those containing the DA neurons, have been mapped for the early larval brain (Hartenstein et al., 2015). As a result, we can recognize DA lineages and establish their connectivity with the help of the serial EM stack that is currently being reconstructed by Cardona and colleagues.
Acknowledgements
We thank the members of the Hartenstein and Guo laboratories for critical discussions during the preparation of this manuscript. We are grateful to the Bloomington Stock Center, R. Axel, and G. Struhl for fly strains and the Developmental Studies Hybridoma Bank and Immunostar for antibodies.
Manuscript supported by: This work was supported by NIH grant R01 NS054814 to V.H., NIH grant RO1 NS48396 to M.G, and USPHS National Research Service Award (No. GM07104) to J.K.L.
Footnotes
We declare no conflict of interest.
References
- Alekseyenko OV, Chan YB, Li R, Kravitz EA. Single dopaminergic neurons that modulate aggression in Drosophila. Proc Natl Acad Sci U S A. 2013;110:6151–6156. doi: 10.1073/pnas.1303446110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M. Drosophila. A laboratory manual. Cold Spring Harbor laboratory Press; Cold Spring Harbor, NY: 1989. pp. 214–217. [Google Scholar]
- Aso Y, Siwanowicz I, Bräcker L, Ito K, Kitamoto T, Tanimoto H. Specific dopaminergic neurons for the formation of labile aversive memory. Curr Biol. 2010;20(16):1445–1451. doi: 10.1016/j.cub.2010.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso Y, Herb A, Ogueta M, Siwanowicz I, Templier T, Friedrich AB, Ito K, Scholz H, Tanimoto H. Three dopamine pathways induce aversive odor memories with different stability. PLoS Genet. 2012;8(7):e1002768. doi: 10.1371/journal.pgen.1002768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso Y, Sitaraman D, Ichinose T, Kaun KR, Vogt K, Belliart-Guérin G, Plaçais PY, Robie AA, Yamagata N, Schnaitmann C, Rowell WJ, Johnston RM, Ngo TT, Chen N, Korff W, Nitabach MN, Heberlein U, Preat T, Branson KM, Tanimoto H, Rubin GM. Mushroom body output neurons encode valence and guide memory-based action selection in Drosophia. Elife. 2014;3:e04580. doi: 10.7554/eLife.04580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler K, Struhl G. Compartment boundaries and the control of Drosophila limb pattern by hedgehog protein. Nature. 1994;368:208–214. doi: 10.1038/368208a0. [DOI] [PubMed] [Google Scholar]
- Bello BC, Izergina N, Caussinus E, Reichert H. Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Dev. 2008;3:5. doi: 10.1186/1749-8104-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieber AJ, Snow PM, Hortsch M, Patel NH, Jacobs JR, Traquina ZR, Schilling J, Goodman CS. Drosophila neuroglian: a member of the immunoglobulin superfamily with extensive homology to the vertebrate neural adhesion molecule L1. Cell. 1989;59:447–460. doi: 10.1016/0092-8674(89)90029-9. [DOI] [PubMed] [Google Scholar]
- Birman S. Arousal mechanisms: speedy flies don't sleep at night. Curr Biol. 2005;15:R511–513. doi: 10.1016/j.cub.2005.06.032. [DOI] [PubMed] [Google Scholar]
- Bjordal M, Arquier N, Kniazeff J, Pin JP, Léopold P. Sensing of amino acids in a dopaminergic circuitry promotes rejection of an incomplete diet in Drosophila. Cell. 2014;156:510–521. doi: 10.1016/j.cell.2013.12.024. [DOI] [PubMed] [Google Scholar]
- Blanco J, Pandey R, Wasser M, Udolph G. Orthodenticle is necessary for survival of a cluster of clonally related dopaminergic neurons in the Drosophila larval and adult brain. Neural Dev. 2011;6:34. doi: 10.1186/1749-8104-6-34. doi: 10.1186/1749-8104-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossing T, Udolph G, Doe CQ, Technau GM. The embryonic central nervous system lineages of Drosophila melanogaster. I. Neuroblast lineages derived from the ventral half of the neuroectoderm. Dev Biol. 1996;179:41–64. doi: 10.1006/dbio.1996.0240. [DOI] [PubMed] [Google Scholar]
- Broadie KS, Bate M. The development of adult muscles in Drosophila: ablation of identified muscle precursor cells. Development. 1991;113:103–118. doi: 10.1242/dev.113.1.103. [DOI] [PubMed] [Google Scholar]
- Budnik V, White K. Catecholamine-containing neurons in Drosophila melanogaster: distribution and development. J Comp Neurol. 1988;268:400–413. doi: 10.1002/cne.902680309. [DOI] [PubMed] [Google Scholar]
- Cardona A, Saalfeld S, Tomancak P, Hartenstein V. Drosophila brain development: closing the gap between a macroarchitectural and microarchitectural approach. Cold Spring Harb Symp Quant Biol. 2009;74:235–248. doi: 10.1101/sqb.2009.74.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona A, Saalfeld S, Arganda I, Pereanu W, Schindelin J, Hartenstein V. Identifying neuronal lineages of Drosophila by sequence analysis of axon tracts. J Neurosci. 2010a;30:7538–7553. doi: 10.1523/JNEUROSCI.0186-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona A, Saalfeld S, Preibisch S, Schmid B, Cheng A, Pulokas J, Tomancak P, Hartenstein V. An integrated micro- and macroarchitectural analysis of the Drosophila brain by computer-assisted serial section electron microscopy. PLoS Biol. 2010b;8(10):e1000502. doi: 10.1371/journal.pbio.1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claridge-Chang A, Roorda RD, Vrontou E, Sjulson L, Li H, Hirsh J, Miesenböck G. Writing memories with light-addressable reinforcement circuitry. Cell. 2009;139:405–415. doi: 10.1016/j.cell.2009.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Gupta T, Davla S, Prieto-Godino LL, Diegelmann S, Reddy OV, Raghavan KV, Reichert H, Lovick J, Hartenstein V. Neuroblast lineage-specific origin of the neurons of the Drosophila larval olfactory system. Dev Biol. 2013;373:322–337. doi: 10.1016/j.ydbio.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Belle JS, Heisenberg M. Associative odor learning in Drosophila abolished by chemical ablation of mushroom bodies. Science. 1994;263:692–695. doi: 10.1126/science.8303280. [DOI] [PubMed] [Google Scholar]
- Druga R. Neocortical inhibitory system. Folia Biol (Praha) 2009;55:201–217. [PubMed] [Google Scholar]
- Dumstrei K, Wang F, Nassif C, Hartenstein V. Early development of the Drosophila brain: V. Pattern of postembryonic neuronal lineages expressing DE-cadherin. J Comp Neurol. 2003;455:451–462. doi: 10.1002/cne.10484. [DOI] [PubMed] [Google Scholar]
- Foltenyi K, Andretic R, Newport JW, Greenspan RJ. Neurohormonal and neuromodulatory control of sleep in Drosophila. Cold Spring Harb Symp Quant Biol. 2007;72:565–571. doi: 10.1101/sqb.2007.72.045. [DOI] [PubMed] [Google Scholar]
- Friggi-Grelin F, Coulom H, Meller M, Gomez D, Hirsh J, Birman S. Targeted gene expression in Drosophila dopaminergic cells using regulatory sequences from tyrosine hydroxylase. J Neurobiol. 2003;54:618–627. doi: 10.1002/neu.10185. [DOI] [PubMed] [Google Scholar]
- Furst A, Mahowald AP. Cell division cycle of cultured neural precursor cells from Drosophila. Dev Biol. 1985;112:467–476. doi: 10.1016/0012-1606(85)90419-1. [DOI] [PubMed] [Google Scholar]
- Fushiki A, Zwart MF, Kohsaka H, Fetter RD, Cardona A, Nose A. A circuit mechanism for the propagation of waves of muscle contraction in Drosophila. Elife. 2016;5:e13253. doi: 10.7554/eLife.13253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glausier JR, Khan ZU, Muly EC. Dopamine D1 and D5 receptors are localized to discrete populations of interneurons in primate prefrontal cortex. Cereb Cortex. 2009;19:1820–1834. doi: 10.1093/cercor/bhn212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman-Rakic PS. The physiological approach: functional architecture of working memory and disordered cognition in schizophrenia. Biol Psychiatry. 1999;46:650–661. doi: 10.1016/s0006-3223(99)00130-4. [DOI] [PubMed] [Google Scholar]
- Goldman-Rakic PS, Muly EC, 3rd, Williams GV. D(1) receptors in prefrontal cells and circuits. Brain Res Brain Res Rev. 2000;31:295–301. doi: 10.1016/s0165-0173(99)00045-4. [DOI] [PubMed] [Google Scholar]
- Guo M. What have we learned from Drosophila models of Parkinson's disease? Prog Brain Res. 2010;184:3–16. doi: 10.1016/S0079-6123(10)84001-4. [DOI] [PubMed] [Google Scholar]
- Hanesch U, Fischbach KF, Heisenberg M. Neuronal architecture of the central complex in Drosophila melanogaster. Cell Tiss Res. 1989;257:343–366. [Google Scholar]
- Hartenstein V, Spindler S, Pereanu W, Fung S. The development of the Drosophila larval brain. Adv Exp Med Biol. 2008;628:1–31. doi: 10.1007/978-0-387-78261-4_1. [DOI] [PubMed] [Google Scholar]
- Hartenstein V, Younossi-Hartenstein A, Lovick J, Kong A, Omoto J, Ngo K, Viktorin G. Lineage-associated tracts defining the anatomy of the Drosophila first instar larval brain. Dev Biol. 2015 doi: 10.1016/j.ydbio.2015.06.021. doi: 10.1016/j.ydbio.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homberg U. Neurotransmitters and neuropeptides in the brain of the locust. Microsc Res Tech. 2002;56:189–209. doi: 10.1002/jemt.10024. [DOI] [PubMed] [Google Scholar]
- Hortsch M, Patel NH, Bieber AJ, Traguina ZR, Goodman CS. Drosophila neurotactin, a surface glycoprotein with homology to serine esterases, is dynamically expressed during embryogenesis. Development. 1990;110:1327–1340. doi: 10.1242/dev.110.4.1327. [DOI] [PubMed] [Google Scholar]
- Ito M, Masuda N, Shinomiya K, Endo K, Ito K. Systematic analysis of neural projections reveals clonal composition of the Drosophila brain. Curr Biol. 2013;23:644–655. doi: 10.1016/j.cub.2013.03.015. [DOI] [PubMed] [Google Scholar]
- Ito K, Shinomiya K, Ito M, Armstrong JD, Boyan G, Hartenstein V, Harzsch S, Heisenberg M, Homberg U, Jenett A, Keshishian H, Restifo LL, Rössler W, Simpson JH, Strausfeld NJ, Strauss R, Vosshall LB, Insect Brain Name Working Group A systematic nomenclature for the insect brain. Neuron. 2014;81:755–65. doi: 10.1016/j.neuron.2013.12.017. [DOI] [PubMed] [Google Scholar]
- Iwai Y, Usui T, Hirano S, Steward R, Takeichi M, Uemura T. Axon patterning requires DN-cadherin, a novel neuronal adhesion receptor, in the Drosophila embryonic CNS. Neuron. 1997;19:77–89. doi: 10.1016/s0896-6273(00)80349-9. [DOI] [PubMed] [Google Scholar]
- Kirchhof BS, Homberg U, Mercer AR. Development of dopamineimmunoreactive neurons associated with the antennal lobes of the honey bee, Apis mellifera. J Comp Neurol. 1999;411:643–653. [PubMed] [Google Scholar]
- Kong EC, Woo K, Li H, Lebestky T, Mayer N, Sniffen MR, Heberlein U, Bainton RJ, Hirsh J, Wolf FW. A pair of dopamine neurons target the D1-like dopamine receptor DopR in the central complex to promote ethanol-stimulated locomotion in Drosophila. PLoS One. 2010;5(4):e9954. doi: 10.1371/journal.pone.0009954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Fung S, Lichtneckert R, Reichert H, Hartenstein V. Arborization pattern of engrailed-positive neural lineages reveal neuromere boundaries in the Drosophila brain neuropil. J Comp Neurol. 2009;517:87–104. doi: 10.1002/cne.22112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen C, Shy D, Spindler SR, Fung S, Pereanu W, Younossi-Hartenstein A, Hartenstein V. Patterns of growth, axonal extension and axonal arborization of neuronal lineages in the developing Drosophila brain. Dev Biol. 2009;335:289–304. doi: 10.1016/j.ydbio.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebestky T, Chang JS, Dankert H, Zelnik L, Kim YC, Han KA, Wolf FW, Perona P, Anderson DJ. Two different forms of arousal in Drosophila are oppositely regulated by the dopamine D1 receptor ortholog DopR via distinct neural circuits. Neuron. 2009;64:522–536. doi: 10.1016/j.neuron.2009.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Owald D, Chandra V, Talbot C, Huetteroth W, Waddell S. Neural correlates of water reward in thirsty Drosophila. Nat Neurosci. 2014;17:1536–1542. doi: 10.1038/nn.3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Plaçais PY, Yamagata N, Pfeiffer BD, Aso Y, Friedrich AB, Siwanowicz I, Rubin GM, Preat T, Tanimoto H. A subset of dopamine neurons signals reward for odour memory in Drosophila. Nature. 2012;488:512–516. doi: 10.1038/nature11304. [DOI] [PubMed] [Google Scholar]
- Lovick JK, Ngo KT, Omoto JJ, Wong DC, Nguyen JD, Hartenstein V. Postembryonic lineages of the Drosophila brain: I. Development of the lineage-associated fiber tracts. Dev Biol. 2013;384:228–257. doi: 10.1016/j.ydbio.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovick JK, Hartenstein V. Hydroxyurea-mediated neuroblast ablation establishes birth dates of secondary lineages and addresses neuronal interactions in the developing Drosophila brain. Dev Biol. 2015;402:32–47. doi: 10.1016/j.ydbio.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovick JK, Kong A, Omoto JJ, Ngo KT, Younossi-Hartenstein A, Hartenstein V. Patterns of growth and tract formation during the early development of secondary lineages in the Drosophila larval brain. Dev Neurobiol. 2015 doi: 10.1002/dneu.22325. doi: 10.1002/dneu.22325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z, Davis RL. Eight different types of dopaminergic neurons innervate the Drosophila mushroom body neuropil: anatomical and physiological heterogeneity. Front Neural Circuits. 2009;3:5. doi: 10.3389/neuro.04.005.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musso PY, Tchenio P, Preat T. Delayed dopamine signaling of energy level builds appetitive long-term memory in Drosophila. Cell Rep. 2015;10:1023–1031. doi: 10.1016/j.celrep.2015.01.036. [DOI] [PubMed] [Google Scholar]
- Nässel DR, Elekes K. Aminergic neurons in the brain of blowflies and Drosophila: dopamine- and tyrosine hydroxylase-immunoreactive neurons and their relationship with putative histaminergic neurons. Cell Tissue Res. 1992;267:147–167. doi: 10.1007/BF00318701. [DOI] [PubMed] [Google Scholar]
- Pereanu W, Hartenstein V. Neural lineages of the Drosophila brain: a three-dimensional digital atlas of the pattern of lineage location and projection at the late larval stage. J Neurosci. 2006;26:5534–5553. doi: 10.1523/JNEUROSCI.4708-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereanu W, Kumar A, Jennett A, Reichert H, Hartenstein V. Development-based compartmentalization of the Drosophila central brain. J Comp Neurol. 2010;518:2996–3023. doi: 10.1002/cne.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesah Y, Burgess H, Middlebrooks B, Ronningen K, Prosser J, Tirunagaru V, Zysk J, Mardon G. Whole-mount analysis reveals normal numbers of dopaminergic neurons following misexpression of alpha-Synuclein in Drosophila. Genesis. 2005;41:154–159. doi: 10.1002/gene.20106. [DOI] [PubMed] [Google Scholar]
- Piovant M, Léna P. Membrane glycoproteins immunologically related to the human insulin receptor are associated with presumptive neuronal territories and developing neurones in Drosophila melanogaster. Development. 1988;103:145–156. doi: 10.1242/dev.103.1.145. [DOI] [PubMed] [Google Scholar]
- Wang Y, Pu Y, Shen P. Neuropeptide-gated perception of appetitive olfactory inputs in Drosophila larvae. Cell Rep. 2013;3:820–830. doi: 10.1016/j.celrep.2013.02.003. [DOI] [PubMed] [Google Scholar]
- Riemensperger T, Issa AR, Pech U, Coulom H, Nguyễn MV, Cassar M, Jacquet M, Fiala A, Birman S. A single dopamine pathway underlies progressive locomotor deficits in a Drosophila model of Parkinson disease. Cell Rep. 2013;5:952–960. doi: 10.1016/j.celrep.2013.10.032. [DOI] [PubMed] [Google Scholar]
- Rohwedder A, Wenz NL, Stehle B, Huser A, Yamagata N, Zlatic M, Truman JW, Tanimoto H, Saumweber T, Gerber B, Thum AS. Four individually identified paired dopamine neurons signal reward in larval Drosophila. Curr Biol. 2015 doi: 10.1016/j.cub.2016.01.012. in press. [DOI] [PubMed] [Google Scholar]
- Rolls ET, Loh M, Deco G, Winterer G. Computational models of schizophrenia and dopamine modulation in the prefrontal cortex. Nat Rev Neurosci. 2008;9:696–709. doi: 10.1038/nrn2462. [DOI] [PubMed] [Google Scholar]
- Sang TK, Chang HY, Lawless GM, Ratnaparkhi A, Mee L, Ackerson LC, Maidment NT, Krantz DE, Jackson GR. A Drosophila model of mutant human parkin-induced toxicity demonstrates selective loss of dopaminergic neurons and dependence on cellular dopamine. J Neurosci. 2007;27:981–992. doi: 10.1523/JNEUROSCI.4810-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer S, Rehder V. Dopamine-like immunoreactivity in the brain and suboesophageal ganglion of the honeybee. J Comp Neurol. 1989;280:43–58. doi: 10.1002/cne.902800105. [DOI] [PubMed] [Google Scholar]
- Schmid A, Chiba A, Doe CQ. Clonal analysis of Drosophila embryonic neuroblasts: neural cell types, axon projections and muscle targets. Development. 1999;126:4653–4689. doi: 10.1242/dev.126.21.4653. [DOI] [PubMed] [Google Scholar]
- Schmidt H, Rickert C, Bossing T, Vef O, Urban J, Technau GM. The embryonic central nervous system lineages of Drosophila melanogaster. II. Neuroblast lineages derived from the dorsal part of the neuroectoderm. Dev Biol. 1997;189:186–204. doi: 10.1006/dbio.1997.8660. [DOI] [PubMed] [Google Scholar]
- Schürmann FW, Elekes K, Geffard M. Dopamine-like immunoreactivity in the bee brain. Cell Tiss Res. 1989;256:399–410. [Google Scholar]
- Selcho M, Pauls D, Han KA, Stocker RF, Thum AS. The role of dopamine in Drosophila larval classical olfactory conditioning. PLoS One. 2009;4(6):e5897. doi: 10.1371/journal.pone.0005897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spindler SR, Hartenstein V. The Drosophila neural lineages: a model system to study brain development and circuitry. Dev Genes Evol. 2010;220:1–10. doi: 10.1007/s00427-010-0323-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spindler SR, Hartenstein V. Bazooka mediates secondary axon morphology in Drosophila brain lineages. Neural Dev. 2011;6:16. doi: 10.1186/1749-8104-6-16. doi: 10.1186/1749-8104-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes PA, Norman HS, Condron BG. Variation in serotonergic and dopaminergic neuronal survival in the central nervous system of adult Drosophila. Cell Tissue Res. 2004;317:327–331. doi: 10.1007/s00441-004-0940-4. [DOI] [PubMed] [Google Scholar]
- Urbach R, Technau GM. Molecular markers for identified neuroblasts in the developing brain of Drosophila. Development. 2003;130:3621–3637. doi: 10.1242/dev.00533. [DOI] [PubMed] [Google Scholar]
- Van Swinderen B, Andretic R. Dopamine in Drosophila: setting arousal thresholds in a miniature brain. Proc Biol Sci. 2011;278:906–913. doi: 10.1098/rspb.2010.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell S. Dopamine reveals neural circuit mechanisms of fly memory. Trends Neurosci. 2010;33:457–464. doi: 10.1016/j.tins.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell S. Reinforcement signalling in Drosophila; dopamine does it all after all. Curr Opin Neurobiol. 2013;23:324–9. doi: 10.1016/j.conb.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterer G, Weinberger DR. Genes, dopamine and cortical signal-to-noise ratio in schizophrenia. Trends Neurosci. 2004;27:683–690. doi: 10.1016/j.tins.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Wolf FW, Heberlein U. Invertebrate models of drug abuse. J Neurobiol. 2003;54:161–178. doi: 10.1002/neu.10166. [DOI] [PubMed] [Google Scholar]
- Wong AM, Wang JW, Axel R. Spatial representation of the glomerular map in the Drosophila protocerebrum. Cell. 2002;109:229–241. doi: 10.1016/s0092-8674(02)00707-9. [DOI] [PubMed] [Google Scholar]
- Wong DC, Lovick JK, Ngo KT, Borisuthirattana W, Omoto JJ, Hartenstein V. Postembryonic lineages of the Drosophila brain: II. Identification of lineage projection patterns based on MARCM clones. Dev Biol. 2013;384:258–289. doi: 10.1016/j.ydbio.2013.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata N, Ichinose T, Aso Y, Plaçais PY, Friedrich AB, Sima RJ, Preat T, Rubin GM, Tanimoto H. Distinct dopamine neurons mediate reward signals for short- and long-term memories. Proc Natl Acad Sci U S A. 2015;112:578–583. doi: 10.1073/pnas.1421930112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Awasaki T, Yu HH, He Y, Ding P, Kao JC, Lee T. Diverse neuronal lineages make stereotyped contributions to the Drosophila locomotor control center, the central complex. J Comp Neurol. 2013;521(12):Spc1. doi: 10.1002/cne.23339. doi: 10.1002/cne.23366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Nassif C, Green P, Hartenstein V. Early neurogenesis of the Drosophila brain. J Comp Neurol. 1996;370:313–329. doi: 10.1002/(SICI)1096-9861(19960701)370:3<313::AID-CNE3>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Salvaterra P, Hartenstein V. Early development of the Drosophila brain IV. Larval neuropile compartments defined by glial septa. J Comp Neurol. 2003;455:435–450. doi: 10.1002/cne.10483. [DOI] [PubMed] [Google Scholar]
- Yu HH, Awasaki T, Schroeder MD, Long F, Yang JS, He Y, Ding P, Kao JC, Wu GY, Peng H, Myers G, Lee T. Clonal development and organization of the adult Drosophila central brain. Curr Biol. 2013;23:633–643. doi: 10.1016/j.cub.2013.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Guo JZ, Peng Y, Xi W, Guo A. Dopamine-mushroom body circuit regulates saliency-based decision-making in Drosophila. Science. 2007;316:1901–1904. doi: 10.1126/science.1137357. [DOI] [PubMed] [Google Scholar]