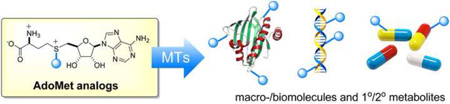
Abstract
S-Adenosyl-l-methionine (AdoMet) is an essential enzyme cosubstrate in fundamental biology with an expanding range of biocatalytic and therapeutic applications. In recent years, technologies enabling the synthesis and utilization of novel functional AdoMet surrogates have rapidly advanced. Developments highlighted within this brief review include improved syntheses of AdoMet analogs, unique S-adenosyl-l-methionine isosteres with enhanced stability, and corresponding applications in epigenetics, proteomics and natural product/small molecule diversification (‘alkylrandomization‘).
Keywords: Methyltransferase, methione adenosyltransferase, S-adenosylmethionine, SAM, halogenase
Graphical abstract
Introduction
Methyltransferase (MT)-catalyzed S-adenosyl-l-methionine (AdoMet, SAM, or SAMe)-dependent methylation is a key enzymatic reaction that enables the functional modulation of a vast array of biomolecules ranging from small metabolites to macromolecules (Fig. 1a; Fig. 2a) [1–5]. Consistent with this, alterations in methylation are associated with a wide range of human pathologies and variability in drug response [2–4]. Despite great advances in methylation-dependent bioinformatics and disease-associated biomarkers, the study of intracellular MT spatial/temporal resolution, specificity and/or function remains a challenge [3,4]. Within this context, the early proof of concept studies revealing synthetic non-native AdoMet analogs to function as efficient cosubstrates for DNA [6] or natural product (NP) [7] MTs inspired a range of subsequent conceptually similar strategies to study NP [8–11], protein [12–18], and nucleic acid [19–23] methylation. Subsequent development of permissive enzyme-based strategies for the synthesis of differentially S-alkylated AdoMet analogs has further simplified access to these unique cosubstrates [11,14,19,24] and also facilitated emerging cell-based applications [14]. Within this context, this brief review attempts to highlight recent advances in the generation and application of differentially S/Se-alkylated AdoMet analogs and what are perceived to be key remaining challenges in further advancing the impact of these unique reagents. While a wide array of AdoMet adenosyl and/or l-methionine (l-Met) chain modified analogs have been pursued within the context of inhibitor design, it is important to note that these fall outside the scope of this review [25–27].
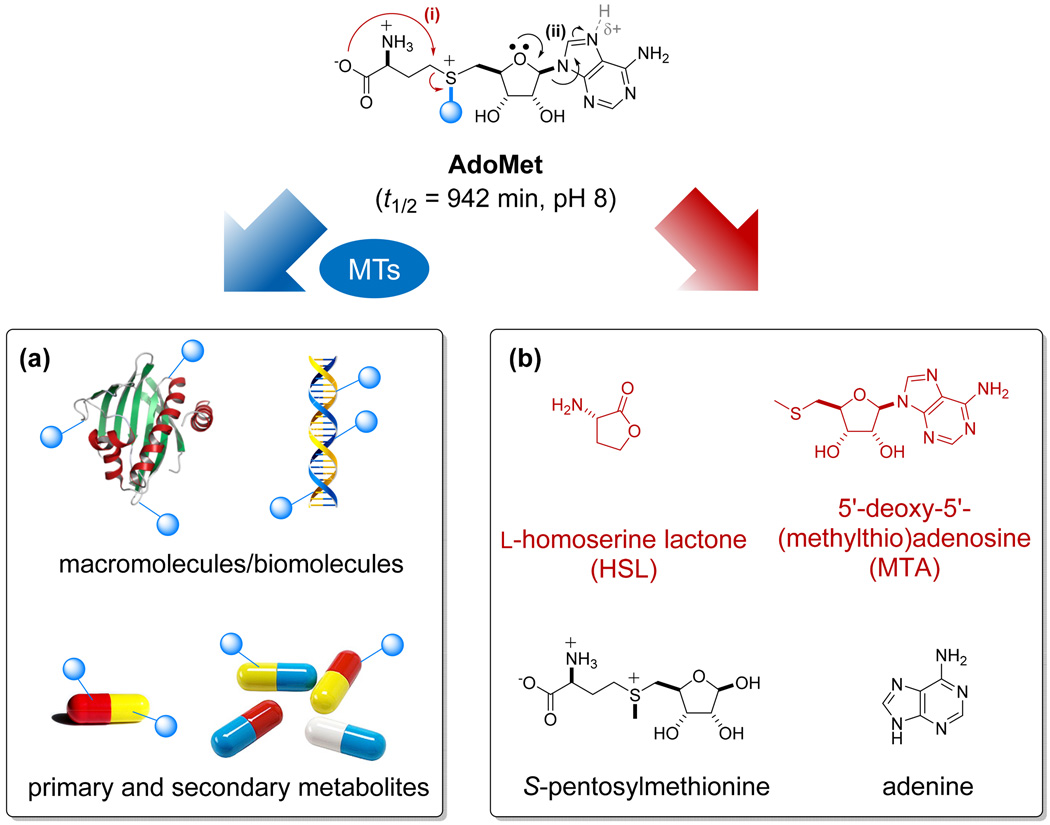
Figure 1. Representative AdoMet utilization and degradation pathways.
(a) AdoMet serves as a critical alkyl donor in most MT-catalyzed reactions within the context of modifying nucleic acids, proteins and small molecule-based metabolites (blue sphere signifies methyl in native systems). (b) AdoMet chemically degrades via intramolecular cyclization (pathway i) and depurination (pathway ii).
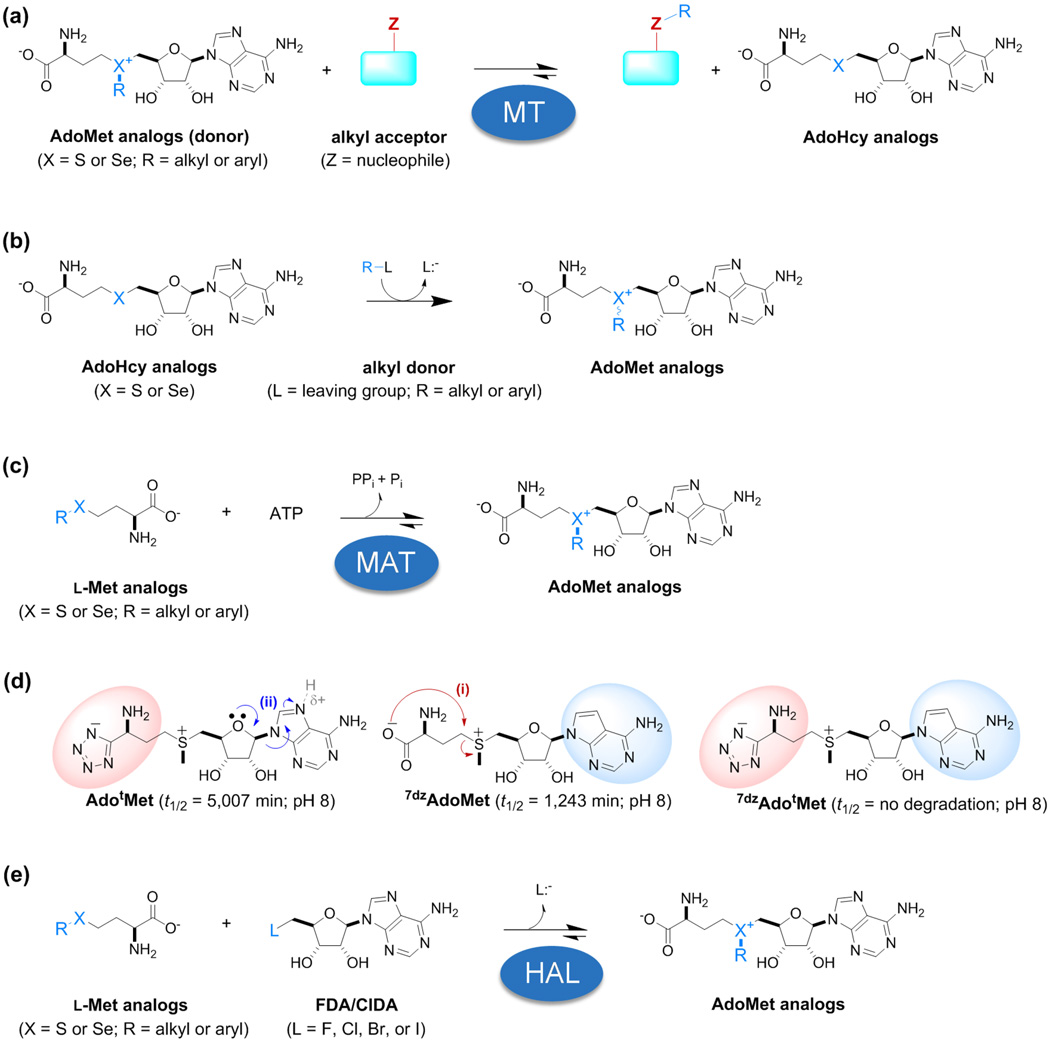
Figure 2. Key reactions and reagents.
(a) General MT-catalyzed reaction scheme (AdoHcy, S-adenosyl-l-homocysteine; also known as SAH). MTs can catalyze C-, O-, N- or S-methylation. (b) Typical synthetic strategy for AdoMet analog chemical synthesis where common leaving groups include halides, triflates, mesylates and tosylates. (c) General methionine adenosyl transferase (MAT; also known as S-adenosylmethionine synthetase/synthase, SAMS)-catalyzed reaction scheme. (d) Stabilized functional AdoMet surrogates afforded via MAT-catalyzed turnover of (S)-3-(methylthio)-1-(1H-tetrazol-5-yl)propan-1-amine (tetrazole-l-methionine, l-tMet) and ATP or l-tMet and 7-deaza-ATP (7dzATP) to give AdotMet and 7dzAdotMet, respectively. (e) General halogenase-catalyzed reaction scheme (HAL: adenosyl-chloride synthase, SalL, or adenosyl-fluoride synthase, FDAS).
Chemical synthesis of AdoMet analogs
Differentially S/Se-alkylated AdoMet surrogates have been constructed via both chemical and enzyme-catalyzed synthesis, the former of which is briefly summarized within this section with an emphasis on analogs demonstrated as functional cosubstrates for downstream AdoMet-utilizing enzymes. Alkylation of S-adenosyl-l-homocysteine (AdoHcy) with alkyl halides in HCOOH/AcOH in the presence or absence of Lewis acid (AgClO4 or AgOTf) as the predominate synthetic strategy of choice, has enabled the synthesis of >20 chemically diverse S/Se-alkylated AdoMets (Fig. 2b). Table 1 highlights functionally active analogs synthesized to date, where subtle variations from the conventional synthetic strategy are noted. While synthetic strategies opened the door to the interrogation of methyltransferases [6,7,21,28,29], typical synthetic yields range from 3% to 90% of (S/R)-sulfonium diastereomeric mixtures where residual starting materials (AdoHcy, a potent product inhibitor of AdoMet-utilizing enzymes) are commonly detrimental to the target enzymes to be studied [30,31]. Thus, purification via reverse-phase chromatography [32–34] or cation-exchange HPLC [35] is typically required, the nature of which often restricts practical scale. AdoMet chemical lability can also be disadvantageous to lengthy synthetic manipulations and/or purification schemes where intramolecular cyclization, depurination and sulfonium epimerization contribute to AdoMet t1/2 (Fig. 1b) [19,20,35,36].
Table 1.
Summary of AdoMet Analog Syntheses and Applications.
| Entry | Thio/Seleno-Alkyl Substitution | Heteroatom | Method for Analog Synthesis | Analog Application | ||||
|---|---|---|---|---|---|---|---|---|
| Chemical | MAT | Halogenase | NA | P | SM | |||
| 1 | S | [37,68] | [68] | |||||
| 2 | S | [69–72] | [69–72] | |||||
| 3 | S | [53] | [73] | |||||
| 4 | S | [83] | ||||||
| 5 | S, Se | [6] | [8,11,24,37,83] | [19] | [6] | [19] | [8,11] | |
| 6 | S, Se | [9,12,13,33,54,63] | [11,24] | [52] | [12,13,33] | [9–11] | ||
| 7 | S, Se | [11,24] | [11] | |||||
| 8 | S, Se | [6,9,32,59,62,64] | [8,11,24,83] | [19] | [6,32] | [17–19,59,62,64–66] | [8,9,11] | |
| 9 | S, Se | [6,59] | [11,24,37,84] | [19] | [6] | [84] | ||
| 10 |  |
S, Se | [11,24] | |||||
| 11 | S | [6,9,20,32,54] | [6,20,32,54] | [18] | [9] | |||
| 12 | S, Se | [63] | [11,24] | [63] | ||||
| 13 | S | [9] | [11,24] | [17] | [9] | |||
| 14 | S, Se | [11,24] | ||||||
| 15 | 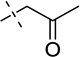 |
S | [77] | [77] | ||||
| 16 | S, Se | [11,24,37,84] | [19] | |||||
| 17 | 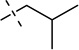 |
S | [11,24] | |||||
| 18 | S, Se | [13,50,59,60,62,63] | [14] | [50,53] | [13,14,17,18,59,60,63,65] | |||
| 19 | S, Se | [59,62,63] | [63] | |||||
| 20 | 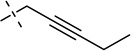 |
S | [20] | [20] | ||||
| 21 | S | [11,24] | ||||||
| 22 | 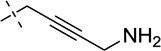 |
S | [20] | [20] | ||||
| 23 | S | [17] | [17] | |||||
| 24 | S, Se | [59,63] | [14] | [14,17,59,63,65] | ||||
| 25 | S | [11,24] | ||||||
| 26 | 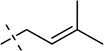 |
S, Se | [11,24] | |||||
| 27 | Se | [11,24] | ||||||
| 28 | S | [11,24] | ||||||
| 29 | S | [11,24] | ||||||
| 30 | 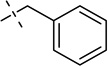 |
S | [9] | [19] | [19] | [9] | ||
| 31 | S | [17] | [17] | |||||
| 32 | S, Se | [59] | [59] | |||||
| 33 | S, Se | [15,16] | [11,24] | [15,16] | ||||
| 34 | 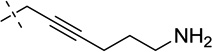 |
S | [20] | [20] | ||||
| 35 | 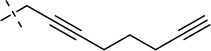 |
S | [20] | [20] | ||||
| 36 | S | [62] | [18,62] | |||||
| 37 | 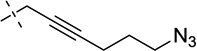 |
S | [20] | [20] | ||||
| 38 | 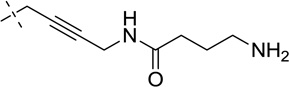 |
S | [20,22] | [22] | ||||
| 39 | 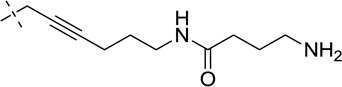 |
S | [20,52,54] | [20,52] | ||||
Chemoenzymatic synthesis of AdoMet analogs
The complement to conventional AdoMet cosubstrate synthesis is enzyme-catalyzed production. Two distinct enzymes have been employed (methionine adenosyltransferases and halogenases, Table 1), a main advantage of which is the potential to directly couple AdoMet analog production to downstream utilization reactions and thereby circumvent the fundamental AdoMet stability issues and/or the need for tedious purification procedures noted in previous section.
Methionine adenosyltransferases (MATs, EC 2.5.1.6)
MATs (also known as S-adenosylmethionine synthetase/synthase, SAMS) catalyze the formation of AdoMet from adenosine triphosphate (ATP) and (l-Met) as a predominate strategy for AdoMet production in nature (Fig. 2c). Within this context, Singh et al. surveyed the capabilities of a representative set of wild-type bacterial, archaeal and mammalian MATs with 44 structurally diverse differentially S/Se-alkylated l-Met analogs. This cumulative effort highlighted human MAT II catalytic alpha subunit (hMAT2A) and the archael thermophilic Methanocaldococcus jannaschii MAT (mMAT) as notably permissive [11]. Using the same suite of putative substrates, Wang and Singh et al. reported similar promiscuity for the archael Sulfolobus solfataricus MAT (sMAT) and, notably, the corresponding first structural elucidation for a thermostable MAT (sMAT, PDB ID 4HPV) and corresponding non-native ligand-bound complex (S-adenosylethionine, AdoEth; PDB ID 4L2Z) [24]. The Luo group also reported the successful hMAT2A-catalyzed synthesis of two differentially S-alkylated AdoMet analogs carrying bulky chemoselective handles and the design of key hMAT2A mutants to improve activity toward targeted non-native l-Met analogs [14]. In a similar fashion, a wild-type MAT from Bacillus subtilis was recently reported to accept four of 11 differentially S-alkylated methionine analogs tested along with key mutants that displayed improved proficiency, permissivity and an apparent reduction in product (AdoMet) inhibition [37]. In addition to l-Met analogs bearing alternative S-alkyl groups, six different carboxyl- and/or amino-modified l-Met analogs were also recently assessed for their viability as alternative cosubstrates of pathogenic bacterial MATs [38]. Cumulatively, well over 50 l-Met analogs have been assessed as putative substrates for a wide array of wild-type and mutant MATs within the last 5 years toward enabling non-native AdoMet production and, in many cases, subsequent utilization in coupled systems [8,11,14,24,37–39]. In addition, tetrazole-based surrogates AdotMet and 7dzAdotMet (Fig. 2d) recently generated via hMAT2A-catalyzed synthesis were demonstrated to serve as functional cosubstrates for the prototypical class I MT DnrK involved in daunorubicin biosynthesis [40]. This latter study notably highlighted a dramatic improvement in the corresponding 7dzAdotMet isostere stability where structure elucidation of DnrK ligand-bound structures also revealed AdotMet to occupy the AdoMet site with a slight shift toward the DnrK-bound acceptor coinciding with a slight improvement in kcat.
Halogenases (SalL, EC 2.5.1.94; FDAS, EC 2.5.1.63)
The innovative application of two wild-type microbial halogenases (5′-chloro-5′-deoxyadenosine synthase or adenosyl-chloride synthase, SalL; 5′-fluoro-5′-deoxyadenosine synthase or adenosyl-fluoride synthase, FDAS) have recently been reported for differentially S-alkylated AdoMet production. SalL and FDAS catalyze the reversible formation of l-Met and 5′-chloro or 5’-fluoro-5′-deoxyadenosine (ClDA or FDA, respectively) from AdoMet and chloride or fluoride, respectively, where the equilibrium typically favors the reactants (Fig. 2e) [41–45]. In this pioneering work [19], SalL and FDAS were found to catalyze the production of six differentially S-alkylated AdoMet analogs from their respective l-Met analogs and commercially available ClDA or FDA. Structure-based rational design of SalL (PDB ID 2Q6I and 2Q6L) and FDAS (PDB ID 1RQR) mutants also led to catalytic improvements with targeted non-native substrates [19]. In addition, SalL-catalyzed AdoMet analog production has been successfully coupled to the model MTs arginine methyltransferase 1 (PRMT1), DNA MT HhaI and the natural product MT MtfA [19,46]. Reminiscent of the 7dzAdotMet isosteres described in the prior section, SalL-catalyzed synthesis of the thieno[3,4-d]pyrimidine-based thAdoMet was also recently reported [47]. While thAdoMet stability was not assessed, thAdoMet served as a functional cosubstrate for the model DNA MT M.TaqI. Like MATs, halogenases importantly enable coupling to downstream AdoMet-utilizing processes in vitro. Whether ClDA/FDA uptake (compared to readily available cellular ATP for MAT) impacts cell-based applications remains to be determined.
AdoMet analog applications
The pioneering applications of non-native AdoMet cosubstrates in MT-catalyzed reactions to afford non-native alkylation of DNA [6] and the indolocarbazole rebeccamycin [7] reported in 2006 by Weinhold group and the Rajski/Thorson collaborative team, respectively, served as the key proof of concept for an array of subsequent innovative advances and applications (Table 1). This section briefly summarizes recent representative examples in the context of modifying nucleic acids, proteins and complex natural products.
Nucleic acids
DNA/RNA methylation plays a key role in epigenetic regulation of gene expression where the vast temporal and spatial complexity presents a notable technological challenge to molecular and mechanistic study, further complicated by the high structural conservation among nucleic acid MTs (NAMTs) [48,49]. While cytosine methylation is a highly conserved modification across many species and among the best understood nucleic acid modifications, many other nucleic acid methylation events also contribute to epigenetic regulation [49]. AdoMet analogs present a valuable new tool to study these essential processes via MT-catalyzed installation of isotopic or chemoselective handles as a framework for epigenetic mapping [6,20–23,50–52]. This concept has been further extended to track RNA modification [53–55]
Proteins
Protein methylation is a key post-translational protein function modulator as exemplified by the role of histone and transcription factor methylation in cellular differentiation and proliferation [56–58]. Here again, the structural conservation among protein MTs (PMTs), vast array of protein targets, and corresponding temporal and spatial occurrence present significant experimental barriers [3]. As with the nucleic acid strategies highlighted in the previous section, AdoMet analogs also enable selective installation of novel chemoselective handles to track and identify methylation events catalyzed by PMTs [12–18,57–66], the proof of concept of which was first demonstrated by Weinhold and coworkers using the wild-type PMT Dim-5 and AdoMet analog 18 (Table 1) [60]. Interestingly, while 18 is also a validated substrate of other wild-type PMTs and NAMTs [50,53], Luo and collaborators more recently reported the need for engineered PMTs to accommodate this AdoMet analog in the pursuit of putative bioorthogonal reagents to study PMT-catalyzed methylation events in vitro and living cells [14,17,18,59,63,65,67]. Isotopic tags have also been installed using corresponding PMTs and suitably-labeled AdoMet analogs (Table 1, entries 1 – 3) [68–73].
Natural products
Natural product (NP) methylation is a highly prevalent biosynthetic reaction where natural product methyltransferase (NPMT)-catalyzed regio/stereospecific O-, N-, S- and/or C-methylation of the fundamental NP core can contribute to bioactivity modulation [5,74]. Within this context, AdoMet analogs in conjunction with both NPMT domains of large multi-functional modular enzyme complexes and standalone late-stage tailoring NPMTs have enabled NP ‘alkylrandomization’ (i.e., differential alkylation, the terminology reminiscent of NP ‘glycorandomization’ [75,76]) to afford novel coumarins [9], fungal polyketides [10], indolocarbazoles [7,11], macrolides [8], nonribosomal peptides [46] and related small molecules [9,40,77]. Importantly, these technologies present a clear complement to conventional synthesis to extend NP structure-activity relationships (SAR) via selective NPMT-catalyzed installation of non-native alkyl groups, protecting groups and/or uniquely functionalized handles for subsequent downstream chemoselective diversification where C-MTs also offer new avenues to potentially access synthetically difficult C-C bond-forming operations [9].
Conclusions
As exemplified by the platform development and innovative applications summarized within this brief review, S-alkylated AdoMet analogs serve as useful chemical biology tools, where practical access has paved the way for a rapidly expanding array of opportunities in fundamental discovery and targeted synthesis. A perceived area for considerable growth in this regard are cell-based applications, the key for which will be the development of universal bioorthogonal AdoMet surrogate/catalyst pairings with high catalytic turnover and exquisite selectivity. ‘Bump-and-hole’ technologies [78], such as those pioneered by Shokat and colleagues [79], serve as the basis for similar AdoMet adenine-modified strategies to achieve MT bioorthogonality as exemplified by the early work of Schultz and Gray [80] and a more recent example by the Zhou group [81]. Alternatively, Luo and collaborators have pursued putative bioorthogonality via targeting specific AdoMet S-alkyl modifications [17,67]. This growing precedent suggests a vibrant future for cell-based, and possibly even whole animal, applications where the fundamental key to achieving true bioorthogonality will depend on the development of AdoMet surrogate/catalyst pairings that display suitable selectivity for the targeted process/reaction over native biochemical processes/enzymes [82].
Highlights.
AdoMet is one of the most essential cosubstrates in nature.
Practical access to AdoMet analogs enables new tools, technologies, leads and discoveries.
Both synthetic and chemoenzymatic strategies for AdoMet production have been advanced.
Chemoenzymatic strategies set the stage for cell-based or whole animal applications.
Bioorthogonal catalyst/AdoMet pairings are anticipated to have a dramatic impact.
Acknowledgments
Past and present work in the Thorson lab and the University of Kentucky Center for Pharmaceutical Research and Innovation is supported in part by NIH R37 AI52188, NIH R01 CA203257, NIH R24 OD21479, the National Center for Advancing Translational Sciences (UL1TR000117), the University of Kentucky College of Pharmacy and the University of Kentucky Markey Cancer Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Struck A-W, Thompson ML, Wong LS, Micklefield J. S-Adenosyl-methionine-dependent methyltransferases: Highly versatile enzymes in biocatalysis, biosynthesis and other biotechnological applications. [Internet] Chembiochem. 2012;13:2642–2655. doi: 10.1002/cbic.201200556. [DOI] [PubMed] [Google Scholar]
- 2.Bottiglieri T. S-Adenosyl-l-methionine (SAMe): From the bench to the bedside—molecular basis of a pleiotrophic molecule. Am. J. Clin. Nutr. 2002;76:1151S–1157S. doi: 10.1093/ajcn/76/5.1151S. [DOI] [PubMed] [Google Scholar]
- 3.Cacabelos R. Epigenomic networking in drug development: From pathogenic mechanisms to pharmacogenomics [Internet] Drug Dev. Res. 2014;75:348–365. doi: 10.1002/ddr.21219. [DOI] [PubMed] [Google Scholar]
- 4.Lu SC, Mato JM. S-Adenosylmethionine in liver health, injury, and cancer. Physiol. Rev. 2012;92:1515–1542. doi: 10.1152/physrev.00047.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liscombe DK, Louie GV, Noel JP. Architectures, mechanisms and molecular evolution of natural product methyltransferases [Internet] Nat. Prod. Rep. 2012;29:1238–1250. doi: 10.1039/c2np20029e. [DOI] [PubMed] [Google Scholar]
- 6.Dalhoff C, Lukinavičius G, Klimašauskas S, Weinhold E. Direct transfer of extended groups from synthetic cofactors by DNA methyltransferases [Internet] Nat. Chem. Biol. 2006;2:31–32. doi: 10.1038/nchembio754. [DOI] [PubMed] [Google Scholar]
- 7.Zhang C, Weller RL, Thorson JS, Rajski SR. Natural product diversification using a non-natural cofactor analogue of S-adenosyl-l-methionine [Internet] J. Am. Chem. Soc. 2006;128:2760–2761. doi: 10.1021/ja056231t. [DOI] [PubMed] [Google Scholar]
- 8. Law BJC, Struck A-W, Bennett MR, Wilkinson B, Micklefield J. Site-specific bioalkylation of rapamycin by the RapM 16-O-methyltransferase [Internet] Chem. Sci. 2015;6:2885–2892. doi: 10.1039/c5sc00164a. This study highlights a chemoenzymatic strategy for rapalog generation. Specifically, the authors reported a coupled system comprised of mutant hMAT2A (I322V) and RapM to enable the generation of rapamycin derivatives with alternative 16-O-akyl modifications.
- 9.Stecher H, Tengg M, Ueberbacher BJ, Remler P, Schwab H, Griengl H, Gruber-Khadjawi M. Biocatalytic Friedel-Crafts alkylation using non-natural cofactors [Internet] Angew. Chemie Int. Ed. 2009;48:9546–9548. doi: 10.1002/anie.200905095. [DOI] [PubMed] [Google Scholar]
- 10.Winter JM, Chiou G, Bothwell IR, Xu W, Garg NK, Luo M, Tang Y. Expanding the structural diversity of polyketides by exploring the cofactor tolerance of an inline methyltransferase domain. [Internet] Org. Lett. 2013;15:2011–2014. doi: 10.1021/ol401723h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Singh S, Zhang J, Huber TD, Sunkara M, Hurley K, Goff RD, Wang G, Zhang W, Liu C, Rohr J, et al. Facile chemoenzymatic strategies for the synthesis and utilization of S-adenosyl-l-methionine analogues [Internet] Angew. Chemie Int. Ed. 2014;53:3965–3969. doi: 10.1002/anie.201308272. This study highlights the broadest survey to date of MATs in the context of AdoMet analog production. Specifically, 5 diverse MATs were assessed with 44 structurally diverse differentially S/Se-alkylated l-Met analogs and the corresponding application of a MAT/RebM-coupled system to afford a small set of novel differentially alkylated indolocarbazoles was also reported.
- 12.Binda O, Boyce M, Rush JS, Palaniappan KK, Bertozzi CR, Gozani O. A chemical method for labeling lysine methyltransferase substrates [Internet] ChemBioChem. 2011;12:330–334. doi: 10.1002/cbic.201000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Willnow S, Martin M, Lüscher B, Weinhold E. A selenium-based click AdoMet analogue for versatile substrate labeling with wild-type protein methyltransferases [Internet] Chem Bio Chem. 2012;13:1167–1173. doi: 10.1002/cbic.201100781. [DOI] [PubMed] [Google Scholar]
- 14.Wang R, Islam K, Liu Y, Zheng W, Tang H, Lailler N, Blum G, Deng H, Luo M. Profiling genome-wide chromatin methylation with engineered posttranslation apparatus within living cells [Internet] J. Am. Chem. Soc. 2013;135:1048–1056. doi: 10.1021/ja309412s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Islam K, Bothwell I, Chen Y, Sengelaub C, Wang R, Deng H, Luo M. Bioorthogonal profiling of protein methylation using azido derivative of S-adenosyl-l-methionine. J. Am. Chem. Soc. 2012;134:5909–5915. doi: 10.1021/ja2118333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blum G, Islam K, Luo M. Current Protocols in Chemical Biology. John Wiley & Sons, Inc.; 2013. Bioorthogonal profiling of protein methylation (BPPM) using an azido analog of S-adenosyl-l-methionine [Internet] pp. 45–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Islam K, Chen Y, Wu H, Bothwell IR, Blum GJ, Zeng H, Dong A, Zheng W, Min J, Deng H, et al. Defining efficient enzyme-cofactor pairs for bioorthogonal profiling of protein methylation [Internet] Proc. Natl. Acad. Sci. 2013;110:16778–16783. doi: 10.1073/pnas.1216365110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guo H, Wang R, Zheng W, Chen Y, Blum G, Deng H, Luo M. Profiling substrates of protein arginine N-methyltransferase 3 with S-adenosyl-l-methionine analogues. ACS Chem. Biol. 2014;9:476–484. doi: 10.1021/cb4008259. This study highlights an application of the concept termed Bioorthogonal Profiling of Protein Methylation (BPPM). Specifically, the authors reported the mutant PRMT3 (M233G) as uniquely permissive and to afford a putative cellular bioorthogonal platform from which more than 80 putative novel PRMT3 protein substrates were indentified.
- 19.Thomsen M, Vogensen SB, Buchardt J, Burkart MD, Clausen RP. Chemoenzymatic synthesis and in situ application of S-adenosyl-l-methionine analogs. [Internet] Org. Biomol. Chem. 2013;11:7606–7610. doi: 10.1039/c3ob41702f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lukinavičius G, Tomkuvienė M, Masevičius V, Klimašauskas S. Enhanced chemical stability of AdoMet analogues for improved methyltransferase-directed labeling of DNA [Internet] ACS Chem. Biol. 2013;8:1134–1139. doi: 10.1021/cb300669x. [DOI] [PubMed] [Google Scholar]
- 21.Pljevaljcić G, Schmidt F, Weinhold E. Sequence-specific methyltransferase-induced labeling of DNA (SMILing DNA). [Internet] Chem Bio Chem. 2004;5:265–269. doi: 10.1002/cbic.200300739. [DOI] [PubMed] [Google Scholar]
- 22.Lukinavičius G, Lapienė V, Staševskij Z, Dalhoff C, Weinhold E, Klimašauskas S. Targeted labeling of DNA by methyltransferase-directed transfer of activated groups (mTAG) [Internet] J. Am. Chem. Soc. 2007;129:2758–2759. doi: 10.1021/ja0691876. [DOI] [PubMed] [Google Scholar]
- 23.Gottfried A, Weinhold E. Sequence-specific covalent labelling of DNA. [Internet] Biochem. Soc. Trans. 2011;39:623–628. doi: 10.1042/BST0390623. [DOI] [PubMed] [Google Scholar]
- 24. Wang F, Singh S, Zhang J, Huber TD, Helmich KE, Sunkara M, Hurley KA, Goff RD, Bingman CA, Morris AJ, et al. Understanding molecular recognition of promiscuity of thermophilic methionine adenosyltransferase sMAT from Sulfolobus solfataricus [Internet] FEBS J. 2014;281:4224–4239. doi: 10.1111/febs.12784. This study highlights the first determined X-ray crystal structure of a thermophilic MAT and the first X-ray crystal structure of a MAT bound to a non-native functional AdoMet surrogate (S-adenosylethionine, AdoEth).
- 25.Cai X-C, Kapilashrami K, Luo M. Methods in Enzymology. Academic Press; 2016. Synthesis and assays of inhibitors of methyltransferases [Internet] [DOI] [PubMed] [Google Scholar]
- 26.Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: A new frontier for drug discovery [Internet] Nat. Rev. Drug Discov. 2012;11:384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- 27.Zhang J, Zheng YG. SAM/SAH analogs as versatile tools for SAM-dependent methyltransferases [Internet] ACS Chem. Biol. 2015;11:583–597. doi: 10.1021/acschembio.5b00812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pljevaljcic G, Pignot M, Weinhold E. Design of a new fluorescent cofactor for DNA methyltransferases and sequence-specific labeling of DNA. [Internet] J. Am. Chem. Soc. 2003;125:3486–3492. doi: 10.1021/ja021106s. [DOI] [PubMed] [Google Scholar]
- 29.Comstock LR, Rajski SR. Efficient synthesis of azide-bearing cofactor mimics [Internet] J. Org. Chem. 2004;69:1425–1428. doi: 10.1021/jo035485z. [DOI] [PubMed] [Google Scholar]
- 30.Khani-Oskouee S, Jones JP, Woodard RW. Stereochemical course of the biosynthesis of 1-aminocyclopropane-1-carboxylic acid. I. Role of the asymmetric sulfonium pole and the α-amino acid center [Internet] Biochem. Biophys. Res. Commun. 1984;121:181–187. doi: 10.1016/0006-291x(84)90704-6. [DOI] [PubMed] [Google Scholar]
- 31.Borchardt RT, Wu YS. Potential inhibitors of S-adenosylmethionine-dependent methyltransferases. 5. Role of the asymmetric sulfonium pole in the enzymatic binding of S-adenosyl-l-methionine. [Internet] J. Med. Chem. 1976;19:1099–1103. doi: 10.1021/jm00231a004. [DOI] [PubMed] [Google Scholar]
- 32.Dalhoff C, Lukinavičius G, Klimašauskas S, Weinhold E. Synthesis of S-adenosyl-l-methionine analogs and their use for sequence-specific transalkylation of DNA by methyltransferases [Internet] Nat. Protoc. 2006;1:1879–1886. doi: 10.1038/nprot.2006.253. [DOI] [PubMed] [Google Scholar]
- 33.Bothwell IR, Islam K, Chen Y, Zheng W, Blum G, Deng H, Luo M. Se-Adenosyl-l-selenomethionine cofactor analogue as a reporter of protein methylation [Internet] J. Am. Chem. Soc. 2012;134:14905–14912. doi: 10.1021/ja304782r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang J, Klinman JP. High-performance liquid chromatography separation of the (S,S)- and (R,S)-forms of S-adenosyl-l-methionine [Internet] Anal. Biochem. 2015;476:81–83. doi: 10.1016/j.ab.2015.02.004. This study reports an advance in the chromatographic resolution of (S,S)- and (R,S)-AdoMet.
- 35.Hoffman JL. Chromatographic analysis of the chiral and covalent instability of S-adenosyl-l-methionine [Internet] Biochemistry. 1986;25:4444–4449. doi: 10.1021/bi00363a041. [DOI] [PubMed] [Google Scholar]
- 36.Iwig DF, Booker SJ. Insight into the polar reactivity of the onium chalcogen analogues of S-adenosyl-l-methionine. [Internet] Biochemistry. 2004;43:13496–13509. doi: 10.1021/bi048693+. [DOI] [PubMed] [Google Scholar]
- 37. Dippe M, Brandt W, Rost H, Porzel A, Schmidt J, Wessjohann LA. Rationally engineered variants of S-adenosylmethionine (SAM) synthase: Reduced product inhibition and synthesis of artificial cofactor homologues [Internet] Chem. Commun. 2015;51:3637–3640. doi: 10.1039/c4cc08478k. This study highlights the study and engineering of B. subtilis MAT. Specifically, the authors report notable production of the desired catalysts in an E. coli heterologous host where the rationally designed I317V mutant afforded enhanced turnover with targeted S-alkyl substition and decreased product inhibition.
- 38.Zano SP, Bhansali P, Luniwal A, Viola RE. Alternative substrates selective for S-adenosylmethionine synthetases from pathogenic bacteria [Internet] Arch. Biochem. Biophys. 2013;536:64–71. doi: 10.1016/j.abb.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wijayasinghe YS, Blumenthal RM, Viola RE. Producing proficient methyl donors from alternative substrates of S-adenosylmethionine synthetase [Internet] Biochemistry. 2014;53:1521–1526. doi: 10.1021/bi401556p. This study further highlights the potential for MATs from pathogenic bacteria to utilize L-Met surrogates and the corresponding AdoMet analogs produced to serve as substrates of model DNA MTs and mammalian catechol MT. In particular, this paper and other recently reported work by the authors expose novel functional modifications of the L-Met carboxylate/amine.
- 40. Huber TD, Wang F, Singh S, Johnson BR, Zhang J, Sunkara M, Van Lanen SG, Morris AJ, Phillips GN, Jr, Thorson JS. Functional AdoMet isosteres resistant to classical AdoMet degradation pathways. ACS Chem. Biol. 2016 doi: 10.1021/acschembio.6b00348. This study highlights the synthesis and study of AdoMet analogs designed for improved stability. Specifically, the authors report the MAT-catalyzed synthesis of tetrazole-based and 7-deaza-adenosine-based surrogates AdotMet and 7dzAdotMet that display notably improved stability over AdoMet. Biochemical and structural studies in the context of the prototypical class I MT DnrK were also reported.
- 41.Eustáquio AS, Pojer F, Noel JP, Moore BS. Discovery and characterization of a marine bacterial SAM-dependent chlorinase [Internet] Nat. Chem. Biol. 2008;4:69–74. doi: 10.1038/nchembio.2007.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deng H, O’Hagan D. The fluorinase, the chlorinase and the Duf-62 enzymes. Curr. Opin. Chem. Biol. 2008;12:582–592. doi: 10.1016/j.cbpa.2008.06.036. [DOI] [PubMed] [Google Scholar]
- 43.Zhu X, Robinson DA, McEwan AR, O’Hagan D, Naismith JH. Mechanism of enzymatic fluorination in Streptomyces cattleya. J. Am. Chem. Soc. 2007;129:14597–14604. doi: 10.1021/ja0731569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dong C, Huang F, Deng H, Schaffrath C, Spencer JB, O’Hagan D, Naismith JH. Crystal structure and mechanism of a bacterial fluorinating enzyme [Internet] Nature. 2004;427:561–565. doi: 10.1038/nature02280. [DOI] [PubMed] [Google Scholar]
- 45.Deng H, Cobb SL, McEwan AR, McGlinchey RP, Naismith JH, O’Hagan D, Robinson DA, Spencer JB. The fluorinase from Streptomyces cattleya is also a chlorinase [Internet] Angew. Chemie Int. Ed. 2006;45:759–762. doi: 10.1002/anie.200503582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lipson JM, Thomsen M, Moore BS, Clausen RP, La Clair JJ, Burkart MD. A tandem chemoenzymatic methylation by S-adenosyl-l-methionine. [Internet] Chem Bio Chem. 2013;14:950–953. doi: 10.1002/cbic.201300221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vranken C, Fin A, Tufar P, Hofkens J, Burkart MD, Tor Y. Chemoenzymatic synthesis and utilization of a SAM analog with an isomorphic nucleobase. [Internet] Org. Biomol. Chem. 2016;14:6189–6192. doi: 10.1039/c6ob00844e. The study extends the prior pioneering work by the same team on the study and application of SalL. Specifically, the authors report the ability of SalL to accept 5'-chloro-5'-deoxythienoadenosine and the corresponding thAdoMet isostere to serve as a functional cosubstrate in M.TaqI-catalyzed DNA alkylation.
- 48.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE-a database for DNA restriction and modification: Enzymes, genes and genomes. Nucleic Acids Res. 2015;43:D298–D299. doi: 10.1093/nar/gku1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith ZD, Meissner A. DNA methylation: Roles in mammalian development. [Internet] Nat. Rev. Genet. Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 50. Vranken C, Deen J, Dirix L, Stakenborg T, Dehaen W, Leen V, Hofkens J, Neely RK. Super-resolution optical DNA mapping via DNA methyltransferase-directed click chemistry [Internet] Nucleic Acids Res. 2014;42:e50–e50. doi: 10.1093/nar/gkt1406. This study highlights the application of AdoMet analogs in high resolution genome optical mapping. The authors demonstrated 3 of 11 wild-type MTs in the presence of AdoMet analog 18 (Table 1) to enable efficient chemoselective Cu(I)-mediated Huisgen 1,3-dipolar cycloaddition for selective fluorophore labeling of DNA and the subsequent application in optical mapping of the T7 phage genome.
- 51.Levy-Sakin M, Grunwald A, Kim S, Gassman NR, Gottfried A, Antelman J, Kim Y, Ho SO, Samuel R, Michalet X, et al. Toward single-molecule optical mapping of the epigenome. ACS Nano. 2014;8:14–26. doi: 10.1021/nn4050694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Neely RK, Dedecker P, Hotta J, Urbanavičiūtė G, Klimašauskas S, Hofkens J. DNA fluorocode: A single molecule, optical map of DNA with nanometre resolution [Internet] Chem. Sci. 2010;1:453–460. [Google Scholar]
- 53.Motorin Y, Burhenne J, Teimer R, Koynov K, Willnow S, Weinhold E, Helm M. Expanding the chemical scope of RNA:methyltransferases to site-specific alkynylation of RNA for click labeling [Internet] Nucleic Acids Res. 2011;39:1943–1952. doi: 10.1093/nar/gkq825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tomkuviene M, Clouet-d’Orval B, Černiauskas I, Weinhold E, Klimašauskas S. Programmable sequence-specific click-labeling of RNA using archaeal box C/D RNP methyltransferases [Internet] Nucleic Acids Res. 2012;40:6765–6773. doi: 10.1093/nar/gks381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Plotnikova A, Osipenko A, Masevičius V, Vilkaitis G, Klimašauskas S. Selective covalent labeling of miRNA and siRNA duplexes using HEN1 methyltransferase [Internet] J. Am. Chem. Soc. 2014;136:13550–13553. doi: 10.1021/ja507390s. This study highlights a recent extension of non-native AdoMet-based RNA modification to small RNA duplexes representative of miRNAs and siRNAs. Specifically, the authors report a methyltransferase-directed Transfer of Activated Groups (mTAG) approach to afford efficient modification and chemoselective labeling of small RNAs using Arabidopsis thaliana HEN1 as the catalyst.
- 56.Greer EL, Shi Y. Histone methylation: A dynamic mark in health, disease and inheritance. [Internet] Nat. Rev. Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luo M. Current chemical biology approaches to interrogate protein methyltransferases. [Internet] ACS Chem. Biol. 2012;7:443–463. doi: 10.1021/cb200519y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dillon SC, Zhang X, Trievel RC, Cheng X. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 2005;6:227.1–227.10. doi: 10.1186/gb-2005-6-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bothwell IR, Luo M. Large-scale, protection-free synthesis of Se-adenosyl-l-selenomethionine analogues and their application as cofactor surrogates of methyltransferases. Org. Lett. 2014;16:3056–3059. doi: 10.1021/ol501169y. This study highlights a practical advance in the synthesis of differentially Se-alkylated AdoMet reagents based on the simplification of the synthetic strategy to afford Se-AdoHcy on gram scale.
- 60.Peters W, Willnow S, Duisken M, Kleine H, Macherey T, Duncan KE, Litchfield DW, Lüscher B, Weinhold E. Enzymatic site-specific functionalization of protein methyltransferase substrates with alkynes for click labeling [Internet] Angew. Chemie Int. Ed. 2010;49:5170–5173. doi: 10.1002/anie.201001240. [DOI] [PubMed] [Google Scholar]
- 61.Osborne T, Roska RLW, Rajski SR, Thompson PR. In situ generation of a bisubstrate analogue for protein arginine methyltransferase 1. J. Am. Chem. Soc. 2008;130:4574–4575. doi: 10.1021/ja077104v. [DOI] [PubMed] [Google Scholar]
- 62.Wang R, Zheng W, Yu H, Deng H, Luo M. Labeling substrates of protein arginine methyltransferase with engineered enzymes and matched S-adenosyl-l-methionine analogues [Internet] J. Am. Chem. Soc. 2011;133:7648–7651. doi: 10.1021/ja2006719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Islam K, Zheng W, Yu H, Deng H, Luo M. Expanding cofactor repertoire of protein lysine methyltransferase for substrate labeling. [Internet] ACS Chem. Biol. 2011;6:679–684. doi: 10.1021/cb2000567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Y, Pan Y, Yang W, Liu W, Zou H, Zhao ZK. Protein arginine allylation and subsequent fluorophore targeting [Internet] Chem Bio Chem. 2013;14:1438–1443. doi: 10.1002/cbic.201300176. [DOI] [PubMed] [Google Scholar]
- 65.Wang R, Ibáñez G, Islam K, Zheng W, Blum G, Sengelaub C, Luo M. Formulating a fluorogenic assay to evaluate S-adenosyl-l-methionine analogues as protein methyltransferase cofactors. [Internet] Mol. Biosyst. 2011;7:2970–2981. doi: 10.1039/c1mb05230f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhang Y, Pan Y, Liu W, Zhou YJ, Wang K, Wang L, Sohail M, Ye M, Zou H, Zhao ZK. In vivo protein allylation to capture protein methylation candidates [Internet] Chem. Commun. 2016;52:6689–6692. doi: 10.1039/c6cc02386j. This study highlights a recent application of AdoMet analogs as an enabling tool in PMT substrate identification. Specifically, the authors reported efficient uptake and use of S-allyl-AdoMet in yeast where a subsequent chemoselective palladium(II)-catalyzed oxidative Heck reaction presented the basis for isotopic labelling/biotinylation and the ultimate identification of >150 putative PMT substrate hits.
- 67.Wang R, Luo M. A journey toward bioorthogonal profiling of protein methylation inside living cells. [Internet] Curr. Opin. Chem. Biol. 2013;17:729–737. doi: 10.1016/j.cbpa.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang K, Zhou YJ, Liu H, Cheng K, Mao J, Wang F, Liu W, Ye M, Zhao ZK, Zou H. Proteomic analysis of protein methylation in the yeast Saccharomyces cerevisiae [Internet] J. Proteomics. 2015;114:226–233. doi: 10.1016/j.jprot.2014.07.032. [DOI] [PubMed] [Google Scholar]
- 69.Bremang M, Cuomo A, Agresta AM, Stugiewicz M, Spadotto V, Bonaldi T. Mass spectrometry-based identification and characterisation of lysine and arginine methylation in the human proteome. [Internet] Mol. Biosyst. 2013;9:2231–2247. doi: 10.1039/c3mb00009e. [DOI] [PubMed] [Google Scholar]
- 70.Uhlmann T, Geoghegan VL, Thomas B, Ridlova G, Trudgian DC, Acuto O. A method for large-scale identification of protein arginine methylation. [Internet] Mol. Cell. Proteomics. 2012;11:1489–1499. doi: 10.1074/mcp.M112.020743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cao XJ, Arnaudo AM, Garcia BA. Large-scale global identification of protein lysine methylation in vivo. Epigenetics. 2013;8:477–485. doi: 10.4161/epi.24547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ong S-E, Mittler G, Mann M. Identifying and quantifying in vivo methylation sites by heavy methyl SILAC [Internet] Nat. Methods. 2004;1:119–126. doi: 10.1038/nmeth715. [DOI] [PubMed] [Google Scholar]
- 73.Lhoest J, Lobet Y, Costers E, Colson C. Methylated proteins and amino acids in the ribosomes of Saccharomyces cerevisiae [Internet] Eur. J. Biochem. 1984;141:585–590. doi: 10.1111/j.1432-1033.1984.tb08233.x. [DOI] [PubMed] [Google Scholar]
- 74.Wessjohann LA, Keim J, Weigel B, Dippe M. Alkylating enzymes. Curr. Opin. Chem. Biol. 2013;17:229–235. doi: 10.1016/j.cbpa.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 75.Gantt RW, Peltier-Pain P, Thorson JS. Enzymatic methods for glyco(diversification/randomization) of drugs and small molecules. [Internet] Nat. Prod. Rep. 2011;28:1811–1853. doi: 10.1039/c1np00045d. [DOI] [PubMed] [Google Scholar]
- 76.Goff RD, Thorson JS. Neoglycosylation and neoglycorandomization: enabling tools for the discovery of novel glycosylated bioactive probes and early stage leads [Internet] Medchemcomm. 2014;5:1036–1047. doi: 10.1039/C4MD00117F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee BWKK, Sun HG, Zang T, Kim BJ, Alfaro JF, Zhou ZS, Ju-Kim B, Alfaro JF, Zhou ZS, Kim BJ, et al. Enzyme-catalyzed transfer of a ketone group from an S-adenosylmethionine analogue: A tool for the functional analysis of methyltransferases [Internet] J. Am. Chem. Soc. 2010;132:3642–3643. doi: 10.1021/ja908995p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bishop a, Buzko O, Heyeck-Dumas S, Jung I, Kraybill B, Liu Y, Shah K, Ulrich S, Witucki L, Yang F, et al. Unnatural ligands for engineered proteins: New tools for chemical genetics. [Internet] Annu. Rev. Biophys. Biomol. Struct. 2000;29:577–606. doi: 10.1146/annurev.biophys.29.1.577. [DOI] [PubMed] [Google Scholar]
- 79.Shokat KM. Tyrosine kinases: Modular signaling enzymes with tunable specificities [Internet] Chem. Biol. 1995;2:509–514. doi: 10.1016/1074-5521(95)90183-3. [DOI] [PubMed] [Google Scholar]
- 80.Lin Q, Jiang F, Schultz PG, Gray NS. Design of allele-specific protein methyltransferase inhibitors [Internet] J. Am. Chem. Soc. 2001;123:11608–11613. doi: 10.1021/ja011423j. [DOI] [PubMed] [Google Scholar]
- 81.Li J, Wei H, Zhou MM. Structure-guided design of a methyl donor cofactor that controls a viral histone H3 lysine 27 methyltransferase activity. J. Med. Chem. 2011;54:7734–7738. doi: 10.1021/jm201000j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Prescher JA, Bertozzi CR. Chemistry in living systems [Internet] Nat. Chem. Biol. 2005;1:13–21. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- 83.Kim HJ, Balcezak TJ, Nathin SJ, McMullen HF, Hansen DE. The use of a spectrophotometric assay to study the interaction of S-adenosylmethionine synthetase with methionine analogues. Anal. Biochem. 1992;207:68–72. doi: 10.1016/0003-2697(92)90501-w. [DOI] [PubMed] [Google Scholar]
- 84.Schlenk F, Dainko JL. The S-n-propyl analogue of S-adenosylmethionine [Internet] Biochim. Biophys. Acta - Gen. Subj. 1975;385:312–323. doi: 10.1016/0304-4165(75)90359-1. [DOI] [PubMed] [Google Scholar]