Abstract
Type III secretion systems (T3SSs) contribute to microbial pathogenesis of Vibrio species, but the regulatory mechanisms are complex. We determined if the classic ExsACDE protein-protein regulatory model from Pseudomonas aeruginosa applies to Vibrio alginolyticus. Deletion mutants in V. alginolyticus demonstrated that, as expected, the T3SS is positively regulated by ExsA and ExsC and negatively regulated by ExsD and ExsE. Interestingly, deletion of exsE enhanced the ability of V. alginolyticus to induce host-cell death while cytotoxicity was inhibited by in trans complementation of this gene in a wild-type strain, a result that differs from a similar experiment with Vibrio parahaemolyticus ExsE. We further showed that ExsE is a secreted protein that does not contribute to adhesion to Fathead minnow epithelial cells. An in vitro co-immunoprecipitation assay confirmed that ExsE binds to ExsC to exert negative regulatory effect on T3SS genes. T3SS in V. alginolyticus can be activated in the absence of physical contact with host cells and a separate regulatory pathway appears to contribute to the regulation of ExsA. Consequently, like ExsE from P. aeruginosa, ExsE is a negative regulator for T3SS gene expression in V. alginolyticus. Unlike the V. parahaemolyticus orthologue, however, deletion of exsE from V. alginolyticus enhanced in vitro cytotoxicity.
Keywords: ExsE, negative regulator, T3SS, Vibrio alginolyticus, gene expression, ExsACDE
Introduction
The type III secretion system (T3SS) is an important virulence-associated surface structure of many Gram-negative pathogens, where it functions to translocate bacterial effector proteins across the bacterial and host membranes directly into the cytosol of host cells (Tseng et al., 2009). T3SSs are composed of a secretion apparatus, translocation apparatus and effector proteins (Hueck, 1998). Normally the genes encoding these proteins are not expressed unless cultured in defined media or in contact with host cells (Hueck, 1998). The regulatory pathway can be remarkably complex and usually involves a series of interacting proteins (Yahr and Wolfgang, 2006; Hauser, 2009).
Transcription of T3SS genes in Pseudomonas aeruginosa is controlled by ExsA, which is a member of the AraC/XyIS family of transcriptional regulators (Yahr and Wolfgang, 2006; Hauser, 2009). The transcriptional activity of ExsA is regulated by three additional interacting proteins: ExsC, ExsD, and ExsE (Yahr and Wolfgang, 2006). ExsD is an “anti-activator” that binds ExsA to prevent ExsA-dependent binding to T3SS promoter sequences (Mccaw et al., 2002). ExsC functions as an “anti-anti-activator” by binding directly to ExsD thereby preventing ExsD-ExsA interactions (Dasgupta et al., 2004). ExsC interacts with ExsE, a protein to which ExsC binds with greater affinity than ExsD (Rietsch et al., 2005). Under inducing conditions, ExsE is exported through T3SSs into either medium or host cells thereby allowing a cascade of interactions that frees ExsA to initiate T3SS transcription (Rietsch et al., 2005; Urbanowski et al., 2007).
Two different Vibrio T3SSs were originally described (T3SS1 and T3SS2) from a clinical strain of V. parahaemolyticus and the T3SS1 shares many characteristics with that of Yersinia and Pseudomonas (Makino et al., 2003; Troisfontaines and Cornelis, 2005). Subsequently, the mechanism of transcriptional control of T3SS1 genes in V. parahaemolyticus was shown to be similar to the P. aeruginosa T3SS regulatory pathway (Zhou et al., 2008, 2010; Kodama et al., 2010; Erwin et al., 2012). For example, T3SS1 genes in V. parahaemolyticus are positively regulated by ExsA and negatively regulated by ExsD (Zhou et al., 2008) while ExsC can directly bind ExsD to free ExsA and permit the expression of T3SS1 genes (Zhou et al., 2010). VP1702 is a functional equivalent of P. aeruginosa ExsE and this orthologue exerts a negative regulatory effect on the production of T3SS1-related proteins (Kodama et al., 2010). Interestingly, a later study revealed that deletion of exsE in a different V. parahaemolyticus strain, NY-4, showed no apparent impact on the synthesis of T3SS1 proteins and the ΔexsE strain was not cytotoxic based on a host-cell infection model (Erwin et al., 2012). The differences in transcriptional regulation of T3SS1 for different V. parahaemolyticus strains suggests that regulation of T3SSs may have diverged between genetic lineages of V. parahaemolyticus and may be divergent between Vibrio species as well.
Vibrio alginolyticus is widely distributed as the part of the normal microbial flora in marine environments (Zhao et al., 2010). It is also an opportunistic pathogen to people and causes otitis, conjunctivitis, superficial pyodermatitis, gastroenteritis, and life-threating infections in immunocompromised patients (Chien et al., 2002; Campanelli et al., 2008). V. alginolyticus is prevalent in coastal waters of southern China and it is commonly associated with costly disease of aquatic animals (Austin, 2010). Previous studies demonstrated that V. alginolyticus induces apoptosis, cell rounding and osmotic lysis in fish cells and autophagy in mammalian cell lines in a T3SS-dependent manner (Zhao et al., 2010, 2011). Although the T3SS of V. alginolyticus is similar to T3SS1 of V. parahaemolyticus with respect to gene synteny (Zhao et al., 2010), it is unclear if the same regulatory mechanism is employed by V. alginolyticus.
In this study, we found that expression of T3SS genes in V. alginolyticus, like that in V. parahaemolyticus, is positively regulated by ExsA and ExsC, and negatively regulated by ExsD. ExsE also functions as a negative regulator, which is consistent with previous studies (Rietsch et al., 2005; Kodama et al., 2010; Erwin et al., 2012). One important difference, however, is that the deletion of exsE enhanced the in vitro cytotoxicity of V. alginolyticus, an outcome that is distinct from the outcome of deleting exsE from V. parahaemolyticus strain NY-4 (Erwin et al., 2012).
Materials and methods
Bacterial strains, plasmids, and growth conditions
Vibrio alginolyticus strains, including wild-type strain ZJO and associated deletion mutants (Table 1), were routinely grown in trypticase soy broth (TSB) or on 1.5% TSB agar plates (TSA) at 30°C. E. coli S17λpir was used in gene deletion experiments and was cultured in LB medium. Expression vector pMMB207 (Morales et al., 1991) was used in complementation experiments and suicide plasmid pDM4 (Milton et al., 1996) was used to generate gene knockouts. Vector pACYCDuet-1 (Novagen) was used to examine protein-protein interactions. When appropriate, ampicillin (100 μg mL−1) or chloramphenicol (34 μg mL−1) was added.
Table 1.
Strains and plasmids used in this study.
| Strains and plasmids | Relevant characteristics | Sources |
|---|---|---|
| E. COLI | ||
| S17-1λpir | thi pro hsdR hsdM+ recA RP4-2-Tc::Mu-Km::Tn7λpir | Milton et al., 1992 |
| BL21 (DE3) | F−ompT hsdSB () gal dcm (DE3) | Novagen |
| S17_pDM4_exsA_A1F+A2R | S17 carrying pDM4_exsA_A1F+A2R | This study |
| S17_pDM4_exsC_A1F+A2R | S17 carrying pDM4_exsC_A1F+A2R | This study |
| S17_pDM4_exsD_A1F+A2R | S17 carrying pDM4_exsD_A1F+A2R | This study |
| S17_pDM4_exsE_A1F+A2R | S17 carrying pDM4_exsE_A1F+A2R | This study |
| S17_pMMB207_exsA_His | S17 carrying pMMB207_exsA_His | This study |
| S17_pMMB207_exsC_His | S17 carrying pMMB207_exsC_His | This study |
| S17_pMMB207_exsD_His | S17 carrying pMMB207_exsD_His | This study |
| S17_pMMB207_exsE_His | S17 carrying pMMB207_exsE_His | This study |
| S17_pDM4_exsA_HA_Insertion | S17 carrying pDM4_exsA_HA_Insertion | This study |
| BL21_pACYCDuet-1_exsA | BL21 carrying pACYCDuet-1_exsA | This study |
| BL21_pACYCDuet-1_exsC | BL21 carrying pACYCDuet-1_exsA | This study |
| BL21_pACYCDuet-1_exsD | BL21 carrying pACYCDuet-1_exsD | This study |
| BL21_pACYCDuet-1_exsE | BL21 carrying pACYCDuet-1_exsE | This study |
| V. ALGINOLYTICUS | ||
| ZJO | Opaque variant of wild strain ZJ51; isolated from diseased grouper fish off the Southern China coast; Apr | Chen et al., 2009 |
| ΔvscC:pexsE | In trans complementation of exsE in a T3SS dysfunctional strain (ΔvscC); Apr | Zhao et al., 2010 |
| ΔexsA | exsA deletion mutant; Apr | This study |
| ΔexsA: pexsA | ΔexsA complemented in trans with wild-type exsA gene located in the plasmid of pMMB207; Apr | This study |
| ZJO: pexsA | Wild-type strain complemented in trans with wild-type exsA gene located in the plasmid of pMMB207; Apr | This study |
| ΔexsC | exsC deletion mutant; Apr | This study |
| ΔexsC: pexsC | ΔexsC complemented in trans with wild-type exsC gene located in the plasmid of pMMB207; Apr | This study |
| ZJO: pexsC | Wild-type strain complemented in trans with wild-type exsC gene located in the plasmid of pMMB207; Apr | This study |
| ΔexsD | exsD deletion mutant; Apr | This study |
| ΔexsD: pexsD | ΔexsD complemented in trans with wild-type exsD gene located in the plasmid of pMMB207; Apr | This study |
| ZJO: pexsD | Wild-type strain complemented in trans with wild-type exsD gene located in the plasmid of pMMB207; Apr | This study |
| ΔexsE | exsE deletion mutant; Apr | This study |
| ΔexsE: pexsE | ΔexsE complemented in trans with wild-type exsE gene located in the plasmid of pMMB207; Apr | This study |
| ZJO: pexsE | Wild-type strain complemented in trans with wild-type exsE gene located in the plasmid of pMMB207; Apr | This study |
| ZJO_ pDM4_exsA_HA_Insertion | Wild-type strain was incorporated a exsA gene with a HA tag at the 3′ terminus in the chromosome; AprCmr | This study |
| ΔexsC_ pDM4_exsA_HA_Insertion | ΔexsC mutant was incorporated a exsA gene with a HA tag at the 3′ terminus in the chromosome; AprCmr | This study |
| V. PARAHAEMOLYTICUS | ||
| NY4 | Clinical isolate O3: K6 | Zhou et al., 2008 |
| PLASMIDS | ||
| pMMB207 | RSF1010 derivative, IncQ lacI q Cmr Ptac oriT | Morales et al., 1991 |
| pDM4 | A suicide vector with ori R6K sacB; Cmr | Milton et al., 1996 |
| pACYCDuet-1 | Prokaryotic expression vector with two MCS, each of which is preceded by a T7 promoter/lac operator; Cmr | Novagen |
| pDM4_exsA_A1F +A2R | Flanking region sequences of exsA cloned into pDM4 | This study |
| pDM4_exsC_A1F +A2R | Flanking region sequences of exsC cloned into pDM4 | This study |
| pDM4_exsD_A1F +A2R | Flanking region sequences of exsD cloned into pDM4 | This study |
| pDM4_exsE_A1F +A2R | Flanking region sequences of exsE cloned into pDM4 | This study |
| pDM4_exsA_HA_Insertion | exsA coding sequence and sequences for HA tag at the C-terminus cloned into pDM4 | This study |
| pMMB207_exsA_His | exsA coding sequence and sequences for 6 His amino acids at the C-terminus cloned into pMMB207 | This study |
| pMMB207_exsC_His | exsC coding sequence and sequences for 6 His amino acids at the C-terminus cloned into pMMB207 | This study |
| pMMB207_exsD_His | exsD coding sequence and sequences for 6 His amino acids at the C-terminus cloned into pMMB207 | This study |
| pMMB207_exsE_His | exsE coding sequence and sequences for 6 His amino acids at the C-terminus cloned into pMMB207 | This study |
| pACYCDuet-1_exsA | exsA coding sequence cloned into pACYCDuet-1 | This study |
| pACYCDuet-1_exsC | exsC coding sequence cloned into pACYCDuet-1 | This study |
| pACYCDuet-1_exsD | exsD coding sequence cloned into pACYCDuet-1 | This study |
| pACYCDuet-1_exsE | exsE coding sequence cloned into pACYCDuet-1 | This study |
Construction of deletion mutants
All deletions were made by allelic exchange following a method described previously (Milton et al., 1996). Briefly, primers ExsA_A1_F, ExsA_A1_R, ExsA_A2_F, and ExsA_A2_R (Table 2) were used to amplify two fragments flanking the coding sequence of exsA. Both amplified fragments incorporated 10-bp overlapping sequences in addition to BglII and SalI restriction site sequences, respectively. These products were used as templates for “splicing by overlap extension” (SOE) PCR to produce a hybrid product using primers ExsA_A1_F and ExsA_A2_R. The full-length fragment was digested with BglII and SalI and then ligated into the suicide vector pDM4 (digested with the same enzymes), generating pDM4_exsA_A1F+A2R. The resultant plasmid was electroporated into E. coli S17-1λpir and transferred into V. alginolyticus strain ZJO by conjugation. Successful transconjugants were selected on TSA plates with ampicillin and chloramphenicol. Secondary cross-over was detected by subsequent plating on agar containing 10% sucrose. For this latter selection, strains that successfully excised the plasmid sequence through secondary cross-over no longer propagate the plasmid-encoded sacB (Gay et al., 1985) and thus can grow in the presence of sucrose. The exsA deletion strain was confirmed by PCR with primers ExsA_int_F and ExsA_int_R and designated as ΔexsA. Construction of exsC, exsD and exsE deletion mutants were performed in the same manner using corresponding primers (Table 2).
Table 2.
Primers used in this study.
| Primer name | Sequences (5′-3′) |
|---|---|
| ExsA_A1_F | TGAAGATCTTATCTCGCTCCTTGAACAC |
| ExsA_A1_R | TAGCCACTTGTTTCTACCCTTCATTATTTTGA |
| ExsA_A2_F | AGGGTAGAAACAAGTGGCTATCGCGAAATGAA |
| ExsA_A2_R | AGCGTCGACCAGACGAGAGTTGATGTAGT |
| ExsA_int_F | TGTCGTTCACAATGGTCAG |
| ExsA_int_R | AGGCACATAATGGCATCAG |
| ExsC_A1_F | GTCAGAGCTCTCGACCCGTTAGGCTTCT |
| ExsC_A1_R | CTATGTCAGCGGTGTCTTTATGTCCAATGACA |
| ExsC_A2_F | TAAAGACACCGCTGACATAGGAATAGTCCC |
| ExsC_A2_R | GACTCTCGAGGAATAACCCAATAAAACC |
| ExsC_int_F | ACAATAACGCTTCCCACG |
| ExsC_int_R | TGTCAGCACGCCAAACTA |
| ExsD_A1_F | GTCAGAGCTCTGATGCCATTATGTGCCTAA |
| ExsD_A1_R | GAGGTGATTGTTTATGTTCGTCTCCGCAC |
| ExsD_A2_F | CGAACATAAACAATCACCTCAGCCAGAT |
| ExsD_A2_R | GACTCTCGAGCGTTCTTGTTCCAATAATGC |
| ExsD_int_F | GATAGCAGCACAATCACAAC |
| ExsD_int_R | GCACTTCCGAACACCAAT |
| ExsE_A1_F | GTCAGAGCTCTACATTCAGCCAACCATG |
| ExsE_A1_R | TTTATGTCCAGATATCACAATATAAGCAGG |
| ExsE_A2_F | TTGTGATATCTGGACATAAAGACACCTAAACTCTC |
| ExsE_A2_R | GACTCTCGAGCACTGCATCTAACGGAAA |
| ExsE_int_F | AGTTGCGGATCAAAGTCC |
| ExsE_int_R | ACACCAATTCATCGGTTC |
| ExsA-comp-F | AGGATAGAATTCATGGATGTGTCAGGCCAACTA |
| ExsA-comp-His-R | AGTTAGGGATCCTCAATGGTGATGGTGATGGTGTTTCGCGATAGCCACTTGA |
| ExsC-comp-F | AGGATAGAATTCATGTCAGCACGCCAAACTATC |
| ExsC-comp-His-R | AGTTAGGGATCCCTAATGGTGATGGTGATGGTGAACTCTCAAGTCTAAAGTTT |
| ExsD-comp-F | AGGATAGAATTCATGAAAAAGCAGCATTGGC |
| ExsD-comp-His-R | AGTTAGGGATCCTTAATGGTGATGGTGATGGTGGATCTGGCTGAGGTGATTGC |
| ExsE-comp-F | AGGATAGAATTCATGTCCAATGACATCCAATCCA |
| ExsE-comp-His-R | AGTTAGGGATCCTCAATGGTGATGGTGATGGTGGGAACGTTGAATTATCGCC |
| VscY_F | GGCGTGTTTACAAAGTGG |
| VscY_R | TGCCGAGTCAGGATGAAG |
| VseE_F | ATGAAGGCGAGACGAACA |
| VseE_R | GCACCCTAAATCCAACTGAC |
| 1687_F | ATGATTGTTGCCATCTACT |
| 1687_R | ACTCGGTTTATTACCTGAA |
| 1686_F | GCAAGCGGTGTTTGATAT |
| 1686_R | ATTGGTTACGCCACTTTT |
| VopB_F | AGAAGCGGGCGTAAATG |
| VopB_R | CACCACCAAACGTCACAAC |
| VopD_F | TCGGGTGTATTAGCGGGTGC |
| VopD_R | CTCGCCATTTCATTCTTGATTTCT |
| ExsA_F | AGCACTATGGCATTTCTC |
| ExsA_R | AACGACGACGGTAACTCT |
| ExsC_F | TAATCCAGTCGCCTAA |
| ExsC_R | CTCTATCGCTCTTTCTT |
| ExsD_F: | CGGAGTACACCTCTACAACC |
| ExsD_R | TCTTGAACCATTGCCATAC |
| ExsE_F | GCGTCATACTGCTTTCTG |
| ExsE_R | ACCAATTCATCGGTTCA |
| DnaK_F | TAAACCCTGACGAAGC |
| DanK_R | AGTCATCACGCCACCC |
| pACYC_ExsA_F | AGGATAGAATTCGATGGATGTGTCAGGCCAACTA |
| pACYC_ExsA_R | AGTTAGAAGCTTTCATTTCGCGATAGCCACTTGA |
| pACYC_ExsC_F | AGGATAGAATTCGATGTCAGCACGCCAAACTATC |
| pACYC_ExsC_R | AGTTAGAAGCTTCTAAACTCTCAAGTCTAAAGTTT |
| pACYC_ExsD_F | AGGATACGATCGATGAAAAAGCAGCATTGGC |
| pACYC_ExsD_R | AGTTAGCTCGAGTTAGGCGTAGTCAGGCACGTCGTAAGGATA GATCTGGCTGAGGTGATTGC |
| pACYC_ExsE_F | AGGATACGATCGATGTCCAATGACATCCAATCCA |
| pACYC_ExsE_R | AGTTAGCTCGAGTCAGGCGTAGTCAGGCACGTCGTAAGGATA GGAACGTTGAATTATCGCC |
| ExsA-insertion-F | AGGATAGAGCTCATGGATGTGTCAGGCCAACTA |
| ExsA-insertion-HA-R | AGTTAGTCTAGATCAGGCGTAGTCAGGCACGTCGTAAGGATATTTCGCGATAGCCACTTGA |
Complementation
To complement the exsA gene, primers ExsA-comp-F and ExsA-comp-His-R were used to amplify the complete exsA sequence with a 6X histidine sequence added at the C-terminus. The resulting amplicon was digested with EcoRI and BamHI and ligated into the expression vector pMMB207, which had been digested with the same enzymes to generate the plasmid pMMB207_exsA_His. This plasmid was then electroporated into E. coli S17-1λpir, resulting in the strain S17_pMMB207_exsA_His, and conjugated into the ΔexsA mutant strain and wild-type strain ZJO resulting in ΔexsA:pexsA and ZJO:pexsA, respectively. Construction of expression vectors for exsC, exsD and exsE was performed in the same manner with corresponding primers (Table 2).
Infection and lactate dehydrogenase (LDH) assay
Fathead minnow (FHM) epithelial cells were cultured in M199 medium (HyClone) supplemented with 10% (v/v) fetal bovine serum (HyClone) at 28°C. For LDH assays growth media (10% FBS) was replaced by fresh M199 supplemented with 1% FBS prior to infection to reduce background LDH activity. For complementation strains of V. alginolyticus, overnight culture was diluted 1:100 into fresh TSB with chloramphenicol and incubated at 30°C with shaking until an OD600 of ~0.6 was obtained. Isopropyl β-D-1-thiogalactopyranoside (IPTG; 1 mM) was then added to induce protein expression. After 3 h the induced bacteria were pelleted by centrifugation (3220 × g; 30 min) and resuspended in an equivalent volume of M199 with 1% FBS. Wild-type and deletion mutants were also treated in the same manner, but without antibiotics. Bacterial suspensions were added to cell monolayers in a 12-well plate at a multiplicity of infection (m.o.i.) of ~100. Plates were centrifuged at ~600 × g for 2 min to synchronize contact with host cells. Supernatants were collected at 1.5 h post infection and LDH activity was measured with a CytoTox 96 Non-Radioactive Cytotoxicity Assay (Promega). Maximum LDH release was achieved by lysis of cells using 10X Lysis Solution and spontaneous LDH release was measured from uninfected cells. Percent cytotoxicity was calculated as follows:
RNA isolation and quantitative RT-PCR (qPCR)
For bacteria grown in TSB, overnight culture was directly inoculated (1:100) into fresh TSB media with appropriate antibiotics and incubated for 3 h. An overnight bacterial culture was also used to infect FHM cell monolayers with an m.o.i. of 100 for 3 h. A total of 3 ml of each co-culture was collected and total RNA was isolated using a RiboPure-Bacteria kit (Ambion) followed by a secondary treatment of DNase using TURBO DNA-free kit (Ambion). Isolated RNA was reverse-transcribed into cDNA using an iScript Reverse Transcription Supermix (Bio-Rad). qPCR was performed with primer pairs (Table 2) to amplify internal fragments using SsoAdvanced SYBR Green Supermix (Bio-Rad) according to manufacturer's instructions. Cycling parameters were identical for all primer sets: 95°C for 30 s, 39 cycles of 95°C for 5 s, 55°C for 15 s, and 72°C for 30 s. Reactions were performed using the CFX 96 real-time PCR system (Bio-Rad) and relative expression was calculated using theΔΔCt method with dnaK serving as a house keeping gene (Livak and Schmittgen, 2001; Erwin et al., 2012; Nydam et al., 2014).
Adhesion assay
Adhesion assays were performed as described previously (Erwin et al., 2012). Briefly, FHM cell monolayers were grown on glass coverslips (Nunc; washed and sterilized) in six-well plates in the presence of M199 supplemented with 10% (v/v) FBS. Monolayers were then inoculated with indicated strains at an m.o.i. ~100. Importantly, V. alginolyticus strain ZJO does not adhere to glass coverslips unless host cells are present. We quantified the adherent bacteria by calculating the ratio of bacteria attached to coverslips to total bacteria as previously reported (Letourneau et al., 2011). Results were presented as average percentages from three independent replicates.
Swarming assay
We prepared semi-solid swarming motility agar (Niu et al., 2005) using LBS [2.5% (w/v) NaCl in LB] broth with the addition of 0.5% (w/v) agar, 0.04% (v/v) sterile Tween 80 and supplemented with ampicillin. A single isolate was selected for each strain and inoculated onto a freshly prepared swarm agar plate. Plates were incubated at 30°C for 8 h and images were obtained using ChemiDoc™MP System (Bio-Rad).
Protein interaction and purification
To examine the potential interactions between ExsA-ExsD, ExsA-ExsE, ExsC-ExsD, and ExsC-ExsE, recombinant proteins (ExsA-His, ExsC-His, ExsD-HA, and ExsE-HA) were induced and expressed in BL21 (DE3) with vector pACYCDuet-1. Overnight bacterial culture was diluted (1:100) into fresh LB media and IPTG (1 mM) was added when the OD600 reached ~0.6. After a 5 h incubation at 37°C, samples (~5 mL) were collected and centrifuged for 20 min at ~12,000 × g. Supernatant was decanted and the cell pellets were re-suspended in BugBuster Master Mix (1 g pellet in 5 mL reagent; Novagen) to prepare crude protein extracts. Whole-cell lysate was collected and loaded onto Ni2+ resin columns (Invitrogen) to allow protein binding overnight at 4°C. Columns were then washed six times with washing buffer (30 mM immidazole) and proteins were eluted with 500 μL elution buffer (300 mM immidazole). Presence of proteins was determined by western blot analysis with monoclonal anti-His antibody (1:2000; Invitrogen) and polyclonal anti-HA antibody (1:2000; Invitrogen). To control for the possibility that HA-tagged protein bound non-specifically to the resin column, ExsD-HA, and ExsE-HA proteins were expressed alone and passed through the Ni2+ column to serve as controls for further western blot analysis.
Trichloroacetic acid (TCA) precipitation assay
Fresh FHM cell culture was prepared 1 day prior to bacterial infection in a 75 cm2 cell culture flask (Nunc). Overnight bacterial culture (ΔexsE:pexsE or ΔvscC:pexsE) was diluted into fresh TSB media and IPTG (1 mM) was used to induce expression of exsE for 5 h. Induced bacteria were used to infect monolayers of FHM cells for 4 h and the bacterial cell pellet was analyzed for the presence of ExsE. Cell culture media (M199) was collected and filtered (0.8 μm, Millipore). TCA (100%) was added to the filtrates (1:4) and incubated for 1 h at 4°C. Supernatant was centrifuged (22,000 × g, 30 min) and the protein pellet was washed with cold acetone to remove residual TCA. Air-dried pellets were re-suspended with PBS and samples were analyzed by western blot.
Western blot
An equal volume of 2X Laemmli sample buffer (Bio-Rad) was added into each sample and all samples were boiled at 100°C for 5 min and loaded onto a 12% SDS-PAGE. After electrophoresis proteins were transferred to a PVDF membrane (Bio-Rad) and the membrane was blocked with 5% skimmed milk in PBS containing 0.05% (v/v) Tween 20 (PBS-T). After 1 h blocking, the membrane was probed with monoclonal anti-His or anti-HA antibody (1:2000; Invitrogen) for 1 h at room temperature. Secondary antibody (anti-mouse-DyLight 488; Thermo Scientific) was diluted 1:5000 in PBS-T with 1% milk for 1 h. Blots were washed 3X with PBS-T and images were obtained using a ChemiDoc™MP System (Bio-Rad).
Generation of HA-tagged ExsA in the chromosome
An HA-sequence was inserted at the C-terminus of the chromosomally encoded exsA as described previously (Zhou et al., 2010). Briefly, the coding sequence of exsA with a HA tag was amplified from wild-type V. alginolyticus by using primers ExsA-insertion-F and ExsA-insertion-HA-R (Table 2). The resulting amplicon was digested with XbaI and SacI and ligated into suicide vector pDM4 (digested with the same enzymes), resulting in plasmid pDM4_exsA_HA_Insertion. The plasmid was electroporated into E. coli S17-1λpir, generating the strain S17_pDM4_exsA_HA_Insertion. Plasmid pDM4_exsA_HA_Insertion was then conjugated into strain ZJO and ΔexsC resulting in strains ZJO_ pDM4_exsA_HA_Insertion and ΔexsC_pDM4_exsA_HA_Insertion, respectively. Synthesis of ExsA was determined by western blot analysis using anti-HA antibody as described above.
Statistical analysis
A paired t-test was used for paired comparisons across transcriptional profiles because we were interested in the overall pattern of difference rather than specific differences of individual genes (for deletion mutants, transcriptional results for the knockout gene were excluded for these comparisons). For phenotype comparisons, a Kruskal-Wallis one-way ANOVA was used to assess treatment effects in conjunction with a Tukey-Kramer test for multiple-comparison (α = 0.05). qPCR data were log-transformed for statistical analysis. Because log10(0) is undefined, 0.05 was added to zero values before transformation. Western blots were quantified using ImageJ (https://imagej.nih.gov/ij/index.html) for statistical comparison and Dnak was used to normalize band intensity data. Histograms were prepared by using SigmaPlot (ver. 12.5, Systat Software, Inc., San Jose, CA) and heat maps for qPCR data were prepared by using the gplots package in R (version 3.3.1).
Results
ExsE exhibits a negative regulatory effect on V. alginolyticus T3SS-induced cell death
To assess the applicability of the ExsACDE regulatory model in T3SS of V. alginolyticus, deletion mutants (ΔexsA, ΔexsC, ΔexsD, and ΔexsE) were co-cultured with FHM cells. Results from cytotoxicity assays were consistent with ExsA and ExsC functioning as positive regulators, while ExsD functions as a negative regulator for V. alginolyticus T3SS (Figure S1). LDH assays revealed that deleting exsE from V. alginolyticus enhanced cytotoxicity compared with the wild-type strain (Figure 1; P < 0.05), consistent with ExsE exhibiting a negative regulatory effect on V. alginolyticus T3SS-induced cell death. This conclusion is further supported when overexpression of exsE in a wild-type strain resulted in significantly reduced cytotoxicity (Figure 1; P < 0.05).
Figure 1.
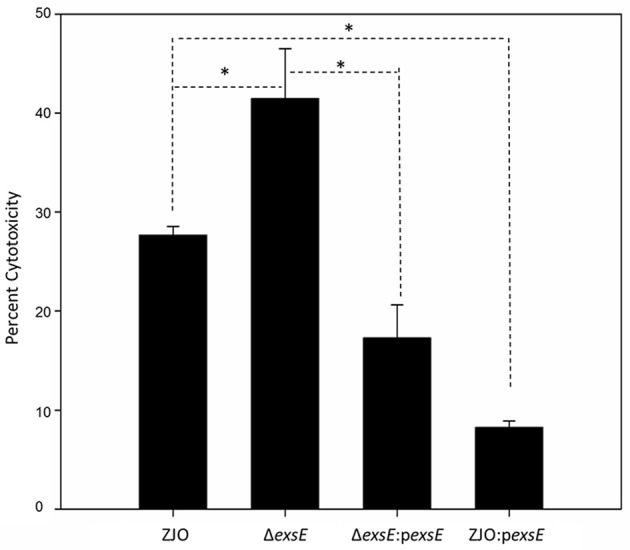
ExsE exerts a negative regulatory effect on in vitro cytotoxicity. LDH assay 1.5 h post infection. Deletion of exsE in V. alginolyticus led to increased T3SS-induced cell death while intrans expression of exsE inhibited cytotoxicity in the wild-type strain. Asterisk represents statistical significance (P < 0.05). Error bars represent SEM (n = three independent replicates).
ExsE is a negative regulator for T3SS gene transcription in V. alginolyticus
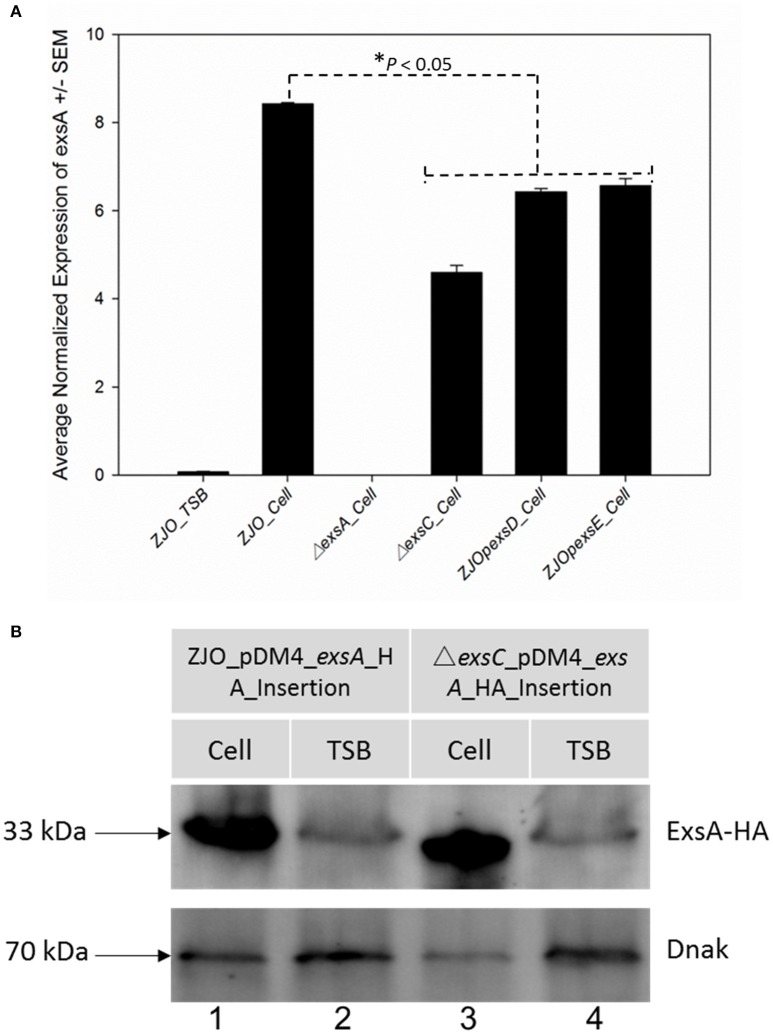
The type III secretion system (T3SS) genes in strain ZJO are transcribed when cultured with FHM cells, but are not transcribed when cultured in TSB (Figure 2, 20-fold mean increase, t = −3.12, 9 df, P = 0.012). Hereafter, we refer to FHM cells and TSB as “inducing” and “non-inducing” conditions, respectively. Deletion of exsE permitted transcription of T3SS genes under non-inducing conditions (ΔexsE, TSB) at levels comparable to the wild-type V. alginolyticus (ZJO, cell) strain when cultured under inducing conditions (Figure 2; t = −1.02, 8 df, P = 0.34). Presumably this occurs because without ExsE, ExsC is free to bind ExsD allowing any ExsA present in the cytoplasm to serve as a transcription regulator even in the absence of cell contact. Notably, under these conditions exsA expression was still nearly 15-fold less than what we observed when the wild-type strain was cultured under inducing conditions, which may indicate that protein turnover is relatively slow for ExsA. In trans expression of exsE (ZJOpexsE, cell) reduced overall gene expression by 10.8-fold (t = 2.47, 8 df, P = 0.039) compared to the wild-type strain under inducing conditions. This is to be expected if ExsE binds ExsC and allows ExsD to interfere with ExsA transcriptional regulator activity. Nevertheless, negative activity was clearly not complete because gene expression for the ZJOpexsE strain was still 8.8-fold greater than the wild-type strain under non-inducing conditions (ZJO, TSB; t = −3.1, 8 df, P = 0.015). Importantly, we also confirmed that knocking out exsA completely eliminates T3SS expression as predicted if it is the sole transcriptional regulator (Figure S2, t = −0.97, df = 8, P = 0.36). Knocking out exsC only partially reduced overall expression (Figure S2, mean 6-fold reduction, t = −5.46, 8 df, P < 0.001) while in trans expression of exsD in the presence of cells reduced expression relative to the wild-type strain with cells (Figure S2, 11.4-fold reduction, t = −2.46, 8 df, P = 0.04), but still exhibited significant overall expression relative to the wild-type strain when cultured in TSB alone (9.4-fold increase, t = 3.72, 8 df, P = 0.006). Collectively, these expression results are consistent with the ExsACDE model of transcriptional control, although positive regulation of exsA, and hence up-regulation of the entire T3SS, is probably controlled by a separate transcriptional regulation system as has been hypothesized for V. parahaemolyticus (Zhou et al., 2010).
Figure 2.
ExsE is a negative regulator for T3SS genes transcription in V. alginolyticus. 10 T3SS genes, including structural genes (vscY and vseE), effector genes (1687 and 1686), translocation genes (vopB and vopD) and regulatory genes (exsA, exsC, exsD and exsE), were examined using qPCR to show their transcription pattern in different V. alginolyticus strains (ZJO, ΔexsE and ΔexsE:pexsE) in TSB or contact with FHM cells. T3SS genes were transcribed after contact with host cells while transcription is limited under TSB only conditions. Deletion of exsE resulted in significantly more T3SS transcription under non-inducing conditions and transcription was inhibited by overexpression of exsE in wild-type strain even under inducing conditions. Average normalized expression data were added into individual box in the heatmap.
Evidence for Non-ExsACDE regulation of exsA
Loss of exsC or in trans expression of exsD or exsE are associated with reduced transcription of exsA (Figure 3A; P < 0.05), but the magnitude of exsA transcription was still well in excess of what was observed for non-inducing conditions (Figure 3A; P < 0.001). This could occur if exsA is at least partially autoregulated, but this also raises the possibility that other factors contribute to the regulation of T3SS in V. alginolyticus. To determine if protein levels were similarly affected, we quantified the synthesis of ExsA in both wild-type V. alginolyticus and the ΔexsC mutant strain by adding an in cis HA tag the 3′ terminus of ExsA. Western blot analysis indicated ExsA was produced at limited levels under non-inducing conditions (Figure 3B, lanes 2 and 4), but at higher concentrations by both wild-type and ΔexsC mutant strains when in contact with host cells (Figure 3B, lanes 1 and 3; P > 0.05). Thus, we surmise that a T3SS-independent regulatory pathway exists that can upregulate exsA expression independent of the ExsACDE regulon.
Figure 3.
exsA can be directly activated by alternative signaling pathway. (A) The transcription of exsA was not completely inhibited by deletion of exsC or overexpression of exsD and exsE in wild-type V. alginolyticus (B) exsA has similar expression pattern in ΔexsC mutant and wild-type strain when contacted with FHM cells (lane 1, 3) suggesting other factors may also contribute to the regulation of exsA. We detected limited expression of exsA in non-inducing conditions (lane 2, 4). Endogenous dnaK served as a loading control.
ExsE binds ExsC
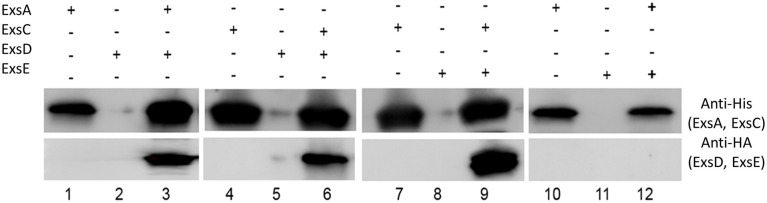
Co-immunoprecipitation experiments with His-tagged ExsC and HA-tagged ExsE demonstrated that these two proteins likely interact as expected (Figure 4, lane 9). Similar experiments indicated that ExsE does not directly bind to ExsA (Figure 4, lane 12). The expected interactions between ExsA and ExsD (Figure 4, lane 3) and ExsC and ExsD (Figure 4, lane 6) were both confirmed.
Figure 4.
ExsE interacts with ExsC. His-tagged ExsA, ExsC and HA-tagged ExsD, ExsE were expressed and incubated overnight at 4°C to examine protein-protein interaction. ExsA binds to ExsD (lane 3), ExsC binds to ExsD (lane 6) and ExsE binds ExsC (lane 9). Non-specific bindings were excluded in this experiment (lane 2, 5, 8, and11) and ExsE does not bind to ExsA (lane 12). + and − indicate the presence and absence of corresponding proteins, respectively.
ExsE is secreted under inducing conditions
Western blot analysis indicated that V. alginolyticus ExsE was secreted under inducing conditions (Figure 5, lane 4). VscC is a structural protein that is required for a functional T3SS in V. alginolyticus (Zhao et al., 2010). When vscC was knocked out and exsE was expressed in trans (ΔvscC:pexsE), ExsE was not detected in the supernatant (Figure 5, lane 6). Collectively, these results are consistent with T3SS-dependent secretion of ExsE. No proteins bands were detected when the wild-type V. alginolyticus was cultured in inducing conditions, confirming the specificity of the antibody that was used for the western blot (Figure 5, lane 1 and 2).
Figure 5.
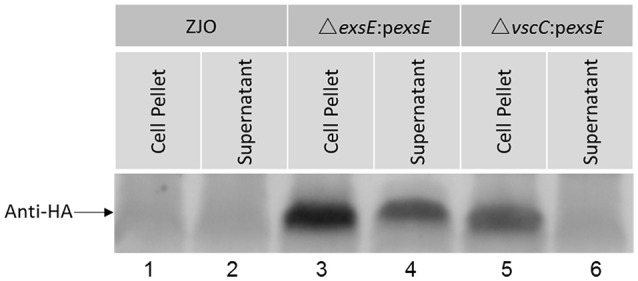
ExsE is secreted in a T3SS-dependent manner under inducing conditions. FHM cells were infected with V. alginolyticus strains (ZJO, ΔexsE:pexsE and ΔvscC:pexsE) and media was collected (4 h), filtered, precipitated and probed. Recombinant ExsE was detected in the supernatant (lane 4) unless a non-function T3SS was present (lane 6).
Physical contact with host cells is not required to trigger expression of T3SS in V. alginolyticus
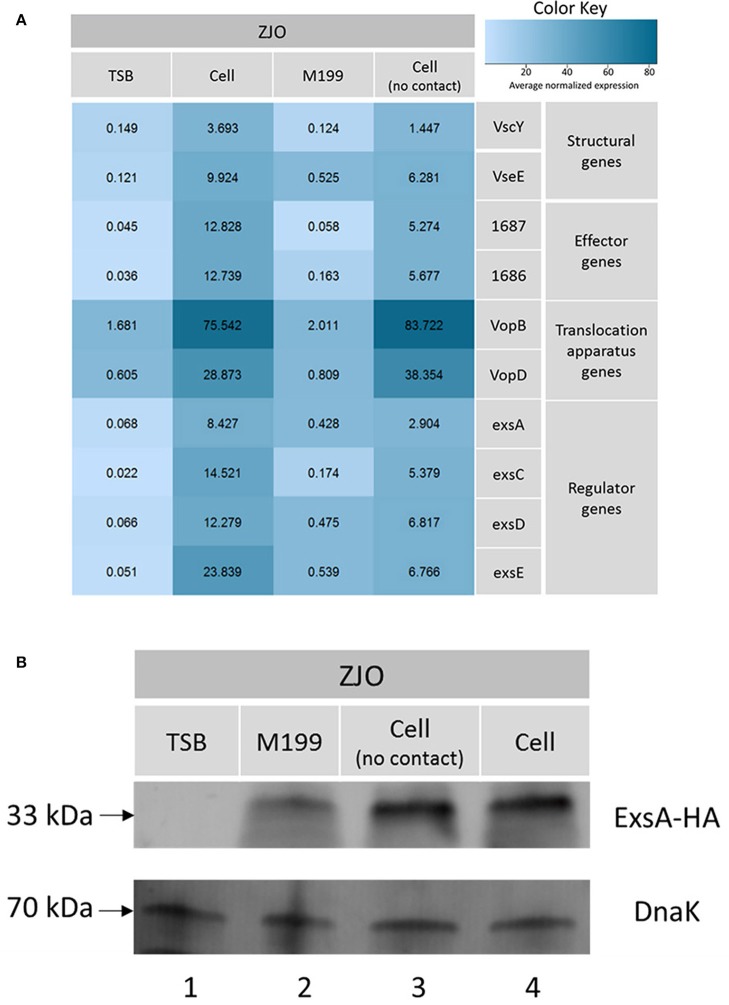
Vibrio alginolyticus was cultured in cell culture media (M199) on a six-well plastic plate (Nunc) with or without polycarbonate membrane inserts. Small molecules can pass through the membrane, but physical contact is blocked when host cells and V. alginolyticus are grown on either side of the membrane. Transcription of T3SS genes was upregulated >15-fold when host and bacterial cells were cultured in this manner (Figure 6A) although the effect was not statistically significant relative to M199 alone (t = −1.95, 9 df, P = 0.08). The lack of statistical significance is probably related to partial upregulation from the M199 media (relative to TSB; t = 4.39, 9 df, P = 0.002) and dilution of the presumptive soluble factor that signals for upregulation of exsA. Qualitatively, it appeared that the abundance of ExsA was similar when ZJO was grow in direct contact with cells or when separated by the membrane (Figure 6B; P > 0.05). ExsA was detected when ZJO was cultured in M199 compared to TSB (Figure 6B; P < 0.05), which is consistent with the upregulation of T3SS genes in the media alone (Figure 6A).
Figure 6.
Physical contact with host cells is not required to induce T3SS in V. alginolyticus. (A) Transcription of T3SS genes in V. alginolyticus was induced when cultured in cell culture media even in the absence of physical contact with eukaryotic cells. TSB and host cell contact served as negative and positive controls in this experiment. Average normalized expression data were added into individual box in the heatmap. (B) Without contacting with FHM cells, the expression of master regulator ExsA was induced to a high level (lane 3) which is comparable to host cell model (lane 4). Endogenous dnaK served as a loading control.
ExsE does not contribute to cell adhesion
Vibrio parahaemolyticus ExsE is required for adhesion to HeLa cells and a ΔexsE mutant strain exhibited a significant reduction in cyto-adherence compared with the wild-type strain (Erwin et al., 2012). We tested if V. alginolyticus ExsE contributes to bacterial adhesion to FHM cells following a standard adherence assay (Letourneau et al., 2011). After a 30 min incubation, 24.6% ± 0.03 of ΔexsE mutant cells adhered to FHM cells, which was comparable to 23.4% ± 0.06 of wild-type V. alginolyticus, indicating ExsE is not required for adhesion to FHM cells.
Discussion
The type III secretion system (T3SS) gene expression is induced by contact with eukaryotic cells or under specific environmental conditions. A regulatory cascade is usually involved in the transcriptional regulation of T3SS genes with a regulator that activates an AraC-like transcriptional activator. For example, in P. aeruginosa T3SS genes are upregulated under low-calcium growth condition (Finck-Barbancon et al., 1997; Vallis et al., 1999) and ExsE serves as a secreted regulator that indirectly contributes to the transcriptional activator ExsA by releasing ExsC to bind to ExsD (Rietsch et al., 2005). This model has been applied to Vibrio species and functional orthologues were identified in V. parahaemolyticus (Zhou et al., 2008, 2010; Kodama et al., 2010), but the mechanisms involved in the regulation of T3SS genes in Vibrio strains remain less clear (Erwin et al., 2012). In this study, we examined the transcription pattern of 10 T3SS genes from V. alginolyticus that encode presumptive structural proteins (n = 2), effector proteins (n = 2), translocation apparatus proteins (n = 2) and regulatory proteins (n = 4). As expected from P. aeruginosa and V. parahaemolyticus literature, deletion of exsE, or exsD resulted in significant increases in constitutive T3SS gene transcription and a similar up-regulation was detected from wild-type V. alginolyticus when complemented with either exsA or exsC (Figure 2 and Figure S2). These data indicated that both ExsA and ExsC are positive regulators while ExsD and ExsE are negative regulators for T3SS genes transcription in V. alginolyticus. Interestingly, the deletion of exsE enhanced the cytotoxicity of V. alginolyticus, which contrasts with the functional orthologue from V. parahaemolyticus (Erwin et al., 2012).
ExsE, a secreted repressor of the T3SS regulon that binds to ExsC, is involved in the ExsACDE regulatory cascade in P. aeruginosa (Rietsch et al., 2005). We confirmed that V. alginolyticus ExsE contributes to regulation of T3SS gene transcription in a manner similar to P. aeruginosa (i.e., via a specific interaction with ExsC; Figure 4). Kodama et al. (2010) indicated that V. parahaemolyticus ExsE exerted a negative regulatory effect on the production of T3SS1-related proteins. In contrast, Erwin et al. (2012) demonstrated that V. parahaemolyticus ExsE has no apparent impact on the synthesis of T3SS1 proteins although it was required for in vitro cytotoxicity using a HeLa cell model. To better characterize the role of ExsE in V. alginolyticus, we generated an exsE deletion mutant and overexpressed this gene in the wild-type V. alginolyticus. Consistent with the P. aeruginosa ExsACDE model, deletion of exsE from V. alginolyticus resulted in the greater cytotoxicity toward FHM cells while overexpression of the gene correspondingly inhibited host-cell death (Figure 1). We further examined the T3SS gene transcription and found that overall transcription is curtailed significantly when ExsE is overexpressed (Figure 2). Coupled with the lack of effect on adhesion relative to V. parahaemolyticus (Erwin et al., 2012), these differences may be species-specific differences, or they could be due to use of cell lines during infection experiments. The latter scenario is unlikely because in subsequent experiments we found that deletion of exsE from V. alginolyticus also enhanced the cytotoxicity toward HeLa cells (Figure S3). Erwin et al. (2012) reported reduced cyto-adhesion and swarming motility when exsE was deleted in V. parahaemolyticus and this is likely due to the loss of flagella biogenesis in the exsE-deficient strain. Herein we confirmed that deletion of exsE resulted in a reduced swarming phenotype in V. parahaemolyticus, but a non-swarming phenotype was observed in both exsE deletion mutant and wild-type V. alginolyticus (Figure S4). Thus, we surmise the species-specific differences account for these distinct phenotypes.
Pseudomonas aeruginosa ExsE is a secreted protein (Rietsch et al., 2005), and translocation of ExsE into Chinese Hamster Ovary cells is required for induction of T3SS gene expression (Urbanowski et al., 2007). We did not detect the translocation of ExsE into FHM cells based on an immunofluorescent assay (data not shown). Limited detection sensitivity of our assay or inherent difference among cell lines may explain this difference. It is also possible that translocation is not required for V. alginolyticus to trigger infection because its T3SS can be activated in the absence of direct contact with FHM cells (Figure 6). Indeed, even culture in M199 media is sufficient to induce some upregulation of the T3SS genes (Figure 6).
ExsA is the master transcriptional regulator of T3SS genes in V. alginolyticus and it is clearly required for in vitro cytotoxicity (Figures S1, S2). Our data demonstrated that ExsD binds to ExsA and ExsC binds to ExsD as expected (Figure 4). Deletion of exsC or overexpression of exsD or exsE did not completely suppress T3SS transcription (Figure 2 and Figure S2), and exsA transcription was still significantly higher than under non-inducing conditions (Figure 3A). This could occur if exsA is at least partially autoregulated, but also raises the possibility that exsA can be regulated by an alternative regulatory pathway that is independent of the T3SS. We further tested this hypothesis by examining the expression of exsA both in wild-type V. alginolyticus and ΔexsC mutant under inducing conditions, and our data revealed that deletion of exsC does not reduce the expression of exsA when in contact with FHM cells (Figure 3B). The observed phenotype in V. alginolyticus is consistent with previous data from V. parahaemolyticus (Zhou et al., 2010), but Dasgupta et al. (2004) reported diminished synthesis of ExsA from an ΔexsC strain in P. aeruginosa. In addition, compared to undetectable expression of exsA in LB containing 3% NaCl (non-inducing) from V. parahaemolyticus (Zhou et al., 2010), we observed the synthesis of ExsA under non-inducing conditions for V. alginolyticus (Figure 3B, lane 2 and 4). The apparent low-level presence of ExsA suggests that extant T3SS structures may be present at all times as has been reported for other pathogens (Cornelis, 2006). These T3SS could serve as “signaling” system for host-cell contact or for detection of soluble signal molecules (Figure 6).
In spite of the distinct differences between V. alginolyticus, V. parahaemolyticus, and P. aeruginosa, it is clear that the regulatory ExsACDE system is largely conserved for these T3SSs, although differences are apparent for ExsE beyond its contribution to the ExsACDE system. Loss of exsE in V. parahaemolyticus affects adhesion and cytotoxicity whereas loss of exsE in V. alginolyticus enhances cytotoxicity. ExsE is secreted by V. alginolyticus but its translocation is not required for upregulation of the T3SS as has been reported for P. aeruginosa. Indeed, co-culture but not physical contact is sufficient to up regulate transcription of exsA and thus an external sensing system appears to contribute to the regulation of the V. alginolyticus T3SS.
Author contributions
JL and ZZ conceived the experiments. JL, SL, LO, JA, and ZZ performed the experiments. JL, SL, LO, CR, CH, DC, and ZZ analyzed the results. JL, DC, and ZZ wrote the manuscript. CR, CH, and DC contributed the reagents.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by the National Natural Science Foundation of China (41276163), the Fundamental Research Funds for the Central Universities, the Project of Science and Technology New Star of Zhujiang in Guangzhou city (2013J2200094), and in part by the Science and Technology Planning Project of Guangdong Province, China (2014B030301064) and the Project of Chinese Academy of Sciences (KSCX2-EW-G-12B).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fcimb.2016.00177/full#supplementary-material
(A) ExsA is required for V. alginolyticus to induce in vitro cytotoxicity. (B) ExsC is a positive regulator for T3SS-induced host cell death in V. alginolyticus. (C) T3SS-induced host cell cytotoxicity is negatively regulated by ExsD in V. alginolyticus. Asterisk indicates statistically significant difference (*P < 0.05; **P < 0.01; ***P < 0.001).
Transcription pattern of T3SS genes in different V. alginolyticus strains. ExsA (A) and ExsC (B) exhibit positive regulatory effects on transcription of T3SS genes in V. alginolyticus and ExsD (C) is a negative regulator. Average normalized expression data were added into individual box in the heatmap.
Deletion of exsE in V. alginolyticus enhanced the cytotoxicity toward HeLa cells. NY4 is the wild-type V. parahaemolyticus and LDH assay was assessed after 4 h incubation with HeLa cells. Asterisk indicates statistically significant difference (P < 0.05).
Both exsE deletion mutant and wild-type V. alginolyticus were presenting a non-swarming phenotype. Swarm agar was inoculated with the indicated strains and incubated at 30°C for 8 h. The assay was repeated three times with similar results and a representative photograph is shown here.
References
- Austin B. (2010). Vibrios as causal agents of zoonoses. Vet. Microbiol. 140, 310–317. 10.1016/j.vetmic.2009.03.015 [DOI] [PubMed] [Google Scholar]
- Campanelli A., Sanchez-Politta S., Saurat J. H. (2008). Cutaneous ulceration after an octopus bite: infection due to Vibrio alginolyticus, an emerging pathogen. Ann. Dermatol. Venereol. 135, 225–227. 10.1016/j.annder.2007.04.010 [DOI] [PubMed] [Google Scholar]
- Chen C., Xie J., Hu C. Q. (2009). Phenotypic and genetic differences between opaque and translucent colonies of Vibrio alginolyticus. Biofouling 25, 525–531. 10.1080/08927010902964578 [DOI] [PubMed] [Google Scholar]
- Chien J. Y., Shih J. T., Hsueh P. R., Yang P. C., Luh K. T. (2002). Vibrio alginolyticus as the cause of pleural empyema and bacteremia in an immunocompromised patient. Eur. J. Clin. Microbiol. Infect. Dis. 21, 401–403. 10.1007/s10096-002-0726-0 [DOI] [PubMed] [Google Scholar]
- Cornelis G. R. (2006). The type III secretion injectisome. Nat. Rev. Microbiol. 4, 811–825. 10.1038/nrmicro1526 [DOI] [PubMed] [Google Scholar]
- Dasgupta N., Lykken G. L., Wolfgang M. C., Yahr T. L. (2004). A novel anti-anti-activator mechanism regulates expression of the Pseudomonas aeruginosa type III secretion system. Mol. Microbiol. 53, 297–308. 10.1111/j.1365-2958.2004.04128.x [DOI] [PubMed] [Google Scholar]
- Erwin D. P., Nydam S. D., Call D. R. (2012). Vibrio parahaemolyticus ExsE is requisite for initial adhesion and subsequent type III secretion system 1-dependent autophagy in HeLa cells. Microbiology 158, 2303–2314. 10.1099/mic.0.059931-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finck-Barbancon V., Goranson J., Zhu L., Sawa T., Wiener-Kronish J. P., Fleiszig S. M., et al. (1997). ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol. Microbiol. 25, 547–557. 10.1046/j.1365-2958.1997.4891851.x [DOI] [PubMed] [Google Scholar]
- Gay P., Le Coq D., Steinmetz M., Berkelman T., Kado C. I. (1985). Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J. Bacteriol. 164, 918–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser A. R. (2009). The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat. Rev. Microbiol. 7, 654–665. 10.1038/nrmicro2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueck C. J. (1998). Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62, 379–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama T., Yamazaki C., Park K. S., Akeda Y., Iida T., Honda T. (2010). Transcription of Vibrio parahaemolyticus T3SS1 genes is regulated by a dual regulation system consisting of the ExsACDE regulatory cascade and H-NS. FEMS Microbiol. Lett. 311, 10–17. 10.1111/j.1574-6968.2010.02066.x [DOI] [PubMed] [Google Scholar]
- Letourneau J., Levesque C., Berthiaume F., Jacques M., Mourez M. (2011). In vitro assay of bacterial adhesion onto mammalian epithelial cells. J. Vis. Exp. e2783 10.3791/2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Makino K., Oshima K., Kurokawa K., Yokoyama K., Uda T., Tagomori K., et al. (2003). Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V cholerae. Lancet 361, 743–749. 10.1016/S0140-6736(03)12659-1 [DOI] [PubMed] [Google Scholar]
- Mccaw M. L., Lykken G. L., Singh P. K., Yahr T. L. (2002). ExsD is a negative regulator of the Pseudomonas aeruginosa type III secretion regulon. Mol. Microbiol. 46, 1123–1133. 10.1046/j.1365-2958.2002.03228.x [DOI] [PubMed] [Google Scholar]
- Milton D. L., Norqvist A., Wolf-Watz H. (1992). Cloning of a metalloprotease gene involved in the virulence mechanism of Vibrio anguillarum. J. Bacteriol. 174, 7235–7244. 10.1128/jb.174.22.7235-7244.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milton D. L., O'toole R., Horstedt P., Wolf-Watz H. (1996). Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 178, 1310–1319. 10.1128/jb.178.5.1310-1319.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales V. M., Bäckman A., Bagdasarian M. (1991). A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97, 39–47. 10.1016/0378-1119(91)90007-X [DOI] [PubMed] [Google Scholar]
- Niu C., Graves J. D., Mokuolu F. O., Gilbert S. E., Gilbert E. S. (2005). Enhanced swarming of bacteria on agar plates containing the surfactant Tween 80. J. Microbiol. Methods 62, 129–132. 10.1016/j.mimet.2005.01.013 [DOI] [PubMed] [Google Scholar]
- Nydam S. D., Shah D. H., Call D. R. (2014). Transcriptome analysis of Vibrio parahaemolyticus in type III secretion system 1 inducing conditions. Front. Cell. Infect. Microbiol. 4:1. 10.3389/fcimb.2014.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietsch A., Vallet-Gely I., Dove S. L., Mekalanos J. J. (2005). ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 102, 8006–8011. 10.1073/pnas.0503005102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troisfontaines P., Cornelis G. R. (2005). Type III secretion: more systems than you think. Physiology (Bethesda). 20, 326–339. 10.1152/physiol.00011.2005 [DOI] [PubMed] [Google Scholar]
- Tseng T. T., Tyler B. M., Setubal J. C. (2009). Protein secretion systems in bacterial-host associations, and their description in the Gene Ontology. BMC Microbiol. 9(Suppl. 1), S2. 10.1186/1471-2180-9-S1-S2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowski M. L., Brutinel E. D., Yahr T. L. (2007). Translocation of ExsE into Chinese hamster ovary cells is required for transcriptional induction of the Pseudomonas aeruginosa type III secretion system. Infect. Immun. 75, 4432–4439. 10.1128/IAI.00664-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallis A. J., Yahr T. L., Barbieri J. T., Frank D. W. (1999). Regulation of ExoS production and secretion by Pseudomonas aeruginosa in response to tissue culture conditions. Infect. Immun. 67, 914–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahr T. L., Wolfgang M. C. (2006). Transcriptional regulation of the Pseudomonas aeruginosa type III secretion system. Mol. Microbiol. 62, 631–640. 10.1111/j.1365-2958.2006.05412.x [DOI] [PubMed] [Google Scholar]
- Zhao Z., Chen C., Hu C. Q., Ren C. H., Zhao J. J., Zhang L. P., et al. (2010). The type III secretion system of Vibrio alginolyticus induces rapid apoptosis, cell rounding and osmotic lysis of fish cells. Microbiology 156, 2864–2872. 10.1099/mic.0.040626-0 [DOI] [PubMed] [Google Scholar]
- Zhao Z., Zhang L., Ren C., Zhao J., Chen C., Jiang X., et al. (2011). Autophagy is induced by the type III secretion system of Vibrio alginolyticus in several mammalian cell lines. Arch. Microbiol. 193, 53–61. 10.1007/s00203-010-0646-9 [DOI] [PubMed] [Google Scholar]
- Zhou X., Konkel M. E., Call D. R. (2010). Regulation of type III secretion system 1 gene expression in Vibrio parahaemolyticus is dependent on interactions between ExsA, ExsC, and ExsD. Virulence 1, 260–272. 10.4161/viru.1.4.12318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Shah D. H., Konkel M. E., Call D. R. (2008). Type III secretion system 1 genes in Vibrio parahaemolyticus are positively regulated by ExsA and negatively regulated by ExsD. Mol. Microbiol. 69, 747–764. 10.1111/j.1365-2958.2008.06326.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) ExsA is required for V. alginolyticus to induce in vitro cytotoxicity. (B) ExsC is a positive regulator for T3SS-induced host cell death in V. alginolyticus. (C) T3SS-induced host cell cytotoxicity is negatively regulated by ExsD in V. alginolyticus. Asterisk indicates statistically significant difference (*P < 0.05; **P < 0.01; ***P < 0.001).
Transcription pattern of T3SS genes in different V. alginolyticus strains. ExsA (A) and ExsC (B) exhibit positive regulatory effects on transcription of T3SS genes in V. alginolyticus and ExsD (C) is a negative regulator. Average normalized expression data were added into individual box in the heatmap.
Deletion of exsE in V. alginolyticus enhanced the cytotoxicity toward HeLa cells. NY4 is the wild-type V. parahaemolyticus and LDH assay was assessed after 4 h incubation with HeLa cells. Asterisk indicates statistically significant difference (P < 0.05).
Both exsE deletion mutant and wild-type V. alginolyticus were presenting a non-swarming phenotype. Swarm agar was inoculated with the indicated strains and incubated at 30°C for 8 h. The assay was repeated three times with similar results and a representative photograph is shown here.