Abstract
Zoonotic potential of a rat-derived hepatitis E virus (HEV), designated as HEV-C1, remains unknown. To evaluate the risk for HEV-C1 infection in humans, paired sera of 208 hospitalized febrile patients collected from 2001 to 2003 in Hanoi, Vietnam, were examined for IgG antibodies to HEV-C1 and genotype 1 HEV (HEV-1), which is common in humans. IgG antibodies to virus-like particles (VLPs) of HEV-C1 and/or HEV-1 were detected from 99 of the 208 convalescent sera in enzyme-linked immunosorbent assay (ELISA). IgG antibody titers to HEV-C1 antigen in 3 of the 99 sera were more than 8-fold higher than those to HEV-1 antigen. IgM antibodies to HEV-C1 antigen were detected in acute sera from 2 of the 3 patients in ELISA and Western blotting. However, no HEV genome was detected. Clinical information was available for 1 of the 2 patients. Hepatic enzymes, aspartate aminotransferase and alanine aminotransferase, were mildly elevated (156 IU/l and 68 IU/l, respectively), and hepatomegaly was detected by ultrasonography. The patient recovered from the illness after 17 days. These results indicated that HEV-C1 or its variants infect humans in Vietnam and may cause acute febrile illness with mild liver dysfunction.
Keywords: antibody, hepatitis E virus, immunodiagnosis, rat, zoonosis
Hepatitis E virus (HEV) is one of the major causes of acute hepatitis. HEV is usually transmitted to humans via the fecal-oral route. There are more than 3 million cases worldwide every year with a fatality rate of 1.9%, and most cases occur in developing countries. Pregnant women are at higher risk for severe disease; the fatality rate is as high as 19.8% [15]. HEV is classified into the family Hepeviridae, genus Orthohepevirus, whose genome is single-stranded positive sense RNA containing 3 open reading frames (ORFs). ORF1 encodes polyprotein with putative functional domains of methyltransferase, protease, helicase and RNA-dependent RNA polymerase [6]. ORF2 encodes capsid protein, and ORF3 encodes phosphoprotein involved in virus release [2]. The genus contains 4 species: Orthohepevirus A to D [16]. To date, genotypes 1 to 4 HEV (HEV-1 to -4) of Orthohepevirus A, tentatively designated as human HEV, are known to cause disease in humans. However, recent discoveries of novel HEVs from various animals have raised suspicions about their potential risk for human health.
HEV-C1 of Orthohepevirus C, formerly called rat HEV, is a novel HEV discovered from wild rodents in Germany in 2009 [5]. Following that report, we and other groups have also found HEV-C1 in Vietnam, U.S.A., Indonesia, Denmark and China [10, 12, 13, 14, 17], suggesting a worldwide distribution of HEV-C1. HEV-C1 is genetically distant from human HEV: nucleotide sequence identity of entire genome is about 55%, and amino acid sequence identities of ORF1, ORF2 and ORF3 proteins are about 50%, 55% and 30%, respectively [4, 7]. Despite the low genetic identity, antiserum to human HEV cross-reacts to HEV-C1 antigen and vice versa [4, 5, 10]. The cross-reactivity could impede detection of specific antibody against HEV-C1 in serum specimens, while the low genetic identify could hinder specific detection of the viral genomic RNA by an RT-PCR method.
Zoonotic potential of HEV-C1 is a controversial issue. Purcell et al. [14] reported that rhesus monkeys, which are highly susceptible to HEV-3, did not develop viremia or antibodies even after intravenous inoculation of 105.2 50% infectious dose of HEV-C1. On the other hand, Dremsek et al. [1] reported that some sera of healthy forestry workers in Germany reacted more strongly to HEV-C1 antigen than to HEV-3 antigen. To determine whether HEV-C1 causes disease in humans, it is important to find patients with acute HEV-C1 infection. To this end, in this study, we examined sera of patients in Vietnam, where HEV-C1 is prevalent in rodents [10], for evaluation of the risk for HEV-C1 infection in humans.
MATERIALS AND METHODS
Samples: Tested samples were acute and convalescent sera collected from 208 hospitalized patients with febrile illness suspected to be caused by rickettsioses at Bach Mai Hospital in Hanoi, Vietnam, from 2001 to 2003. This is the largest referral medical center with approximately 1,900 beds, covering residents in all provinces of northern Vietnam.
Enzyme-linked immunosorbent assay (ELISA): Anti-HEV-1 and -HEV-C1 antibodies were detected by ELISA with some modifications from previous reports [11, 12]. Briefly, 96-well EIA/RIA plates (Corning, Corning, NY, U.S.A.) were each coated with 1 µg/ml of virus-like particles (VLPs) of HEV-1 and HEV-C1, which were generated by a recombinant baculovirus system [9, 10], at 4°C overnight. After blocking with phosphate buffered saline (PBS) containing 3% bovine serum albumin (BSA) (Sigma-Aldrich, St. Louis, MO, U.S.A.) at 37°C for 2 hr, the plates were incubated with sera (1:200) at 37°C for 1 hr. Then, the plates were incubated with horseradish peroxidase (HRP)-conjugated goat anti-human IgG (KPL, Gaithersburg, MD, U.S.A.) (1:10,000) or HRP-conjugated goat anti-human IgM (KPL) (1:100,000) at 37°C for 1 hr. After that, the plates were incubated with o-phenylenediamine dihydrochloride solution at room temperature (RT) for 30 min. The reaction was stopped with 10% H2SO4, and optical density (OD) at a wavelength of 492 nm was measured by a spectrometer. Plates were washed with PBS containing 0.05% Tween 20 (PBST) three times at the end of each step before coloring step. The cutoff value was tentatively set at OD=0.8, which was determined by histogram analysis (data not shown). At the titration of IgG, serial two-fold dilutions of sera (1:200 to 1:51,200) were subjected to ELISA in the same way. IgG titer was defined as the reciprocal of the highest dilution showing an OD value of more than 0.8.
Western blotting: VLPs of HEV-1 and HEV-C1 were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to an Immobilon-P membrane (Merck, Darmstadt, Germany) by conventional methods. After blocking with Block Ace (DS Pharma Biomedical, Osaka, Japan) at RT for 1 hr, the membrane was incubated with diluted acute sera (1:2,000 to 1:64,000) at RT for 1 hr. After washing with PBS for 15 min three times, the membrane was incubated with HRP-conjugated goat anti-human IgM (KPL) (1:40,000) at RT for 1 hr and washed again. Chemiluminescent signal was developed by an ECL Prime Western Blotting Detection System (GE Healthcare, Little Chalfont, U.K.) and detected by ImageQuant LAS 4000 mini (GE Healthcare).
RNA extraction, reverse transcription and polymerase chain reaction (PCR): Total RNA was extracted from acute sera of patients with suspected acute HEV-C1 infection by using a QIAamp Viral RNA Mini Kit (QIAGEN, Venlo, Netherlands). cDNA was synthesized from the total RNA using a random hexamer (Thermo Fisher Scientific, Waltham, MA, U.S.A.) and SuperScript II Reverse Transcriptase (Thermo Fisher Scientific) according to the manufacturer’s instructions. cDNA was amplified by nested PCR using Platinum Taq DNA Polymerase High Fidelity (Thermo Fisher Scientific) according to the manufacturer’s instructions. Primer pairs, HEV-cs and -cas and HEV-csn and -casn, reported by Johne et al. [5] and primer pairs, HEV-F1 and -R2, HEV-F2 and -R1, rat-HEV-F10 and -R7, and rat-HEV-F11 and -R9, reported by Li et al. [8, 11] were used for nested PCR. We also designed the following primers based on HEV-C1 sequences on the database and used them in various combinations: Rat HEV F1S (5′-GGCCCTTGGTTTAGGGCCATAGAGAAGGC-3′, nt 4,037–4,101), Rat HEV F2S (5′-GCCAACCTGCCTGAGTGGTGCTTTTATGG-3′, nt 4,109–4,137), Rat HEV F3S (5′-GAGAAGAACTGGGGCCCCGTGAAAGAGCG-3′, nt 4,661–4,689), Rat HEV F4S (5′-TTTGGCCCTTGGTTYMGGGCMATAGAGAA-3′, nt 4,070–4,098), Rat HEV F5S (5′-GCCAACCTGCCYGARTGGTGYTTTTATGG-3′, nt 4,109–4,137), Rat HEV F6S (5′-TGTTATGGAAYACWGTCTGGAAYATGGC-3′, nt 4,398–4,425), Rat HEV R1S (5′-GCGGCACGAACAGCAAAAGCACGAGC-3′, nt 4,945–4,970), Rat HEV R2S (5′-GCTACAGCCCAGAGTGTTATTCCTTC-3′, nt 4,891–4,916), Rat HEV R3S (5′-GCTGTCAWYGGCGACTGCCCGGCATCGGG-3′, nt 5,201–5,229), Rat HEV R4S (5′-CAGCGGCACGAACAGCARAAGCASGAGC-3′, nt 4,945–4,972) and Rat HEV R5S (5′-CGCTCYTTCACGGGRCCCCARTTCTTCTC-3′, nt 4,661–4,689). The nucleotide numbers after primer sequences correspond to positions in the genome sequence of HEV-C1 strain Vietnam-105 [7].
Ethic statement: This study was approved by the Ethics Committee of Hokkaido University Graduate School of Medicine.
RESULTS
Screening of anti-HEV-C1 and -HEV-1 IgG antibodies by ELISA: First, we screened for IgG antibodies to HEV-C1 and HEV-1 in convalescent sera by ELISA [10, 11]. VLPs of HEV-C1 and HEV-1 generated by a recombinant baculovirus system [9, 10] were used as antigens. Since antigenicities of HEV-1 to -4, which cause human infection, are closely related to each other [18], VLPs of HEV-1 were used as a representative antigen for HEV-1 to -4. As a result, 70 and 82 of the 208 convalescent sera were judged to be positive for IgG antibodies to HEV-C1 and HEV-1 antigens, respectively (Fig. 1). In total, 99 of the 208 sera were positive for anti-HEV-C1 and/or anti-HEV-1 antibodies.
Fig. 1.
Reactivity of convalescent sera to HEV-1 and HEV-C1 antigens in enzyme-linked immunosorbent assay (ELISA). 200-fold dilutions of 208 convalescent sera were subjected to ELISA using virus-like particles (VLPs) of HEV-1 and HEV-C1 as antigens. The cutoff value was tentatively set at optical density (OD) of 0.8 and is shown as dashed lines.
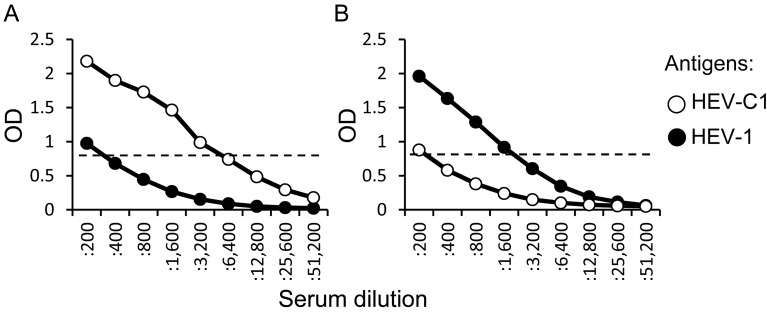
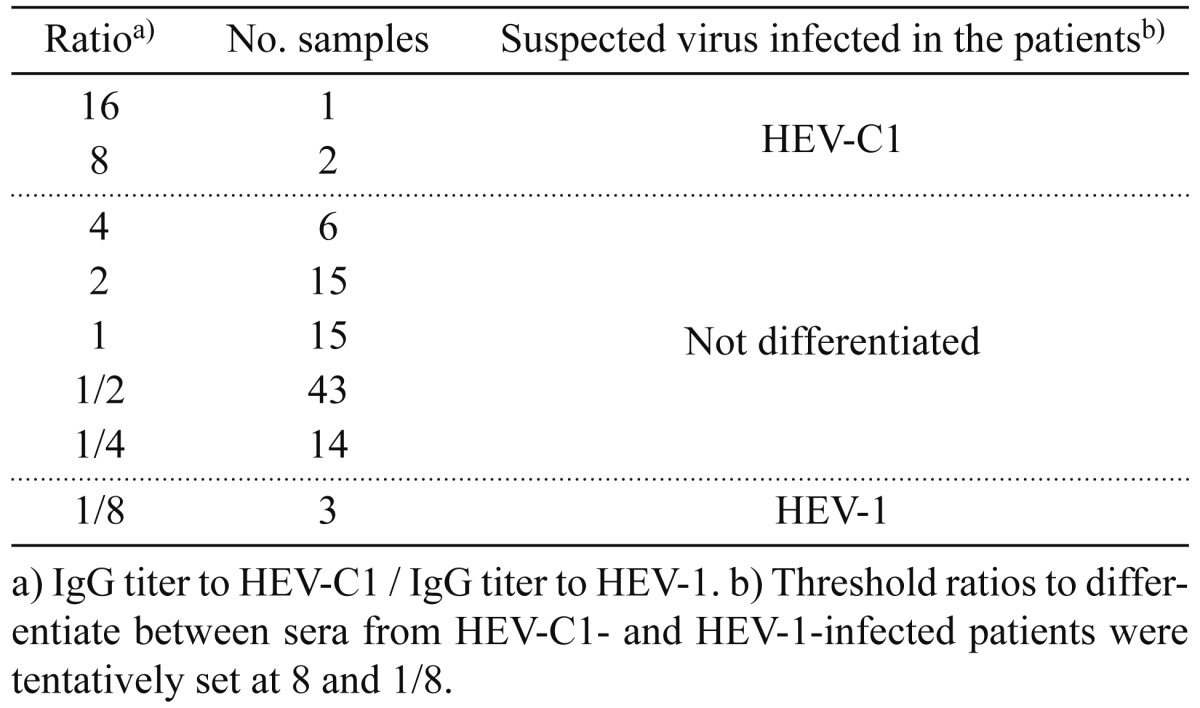
Comparison of IgG antibody titers to HEV-C1 and HEV-1 antigens: When reactivities of sera against HEV-C1 and HEV-1 antigens were compared, some sera showed higher reactivity against HEV-C1 antigen than HEV-1 antigen (Fig. 1). We then compared IgG antibody titers to HEV-C1 and HEV-1 antigens in the 99 sera. IgG antibody titer was defined as the reciprocal of the highest dilution showing more than the cutoff value. In most of sera, IgG titers to HEV-C1 and HEV-1 were similar level. However, some sera showed marked difference in the IgG titers (Fig. 2). In the case shown in Fig. 2A, IgG titers to HEV-C1 and HEV-1 were 3,200 and 200, respectively, and therefore, IgG titer to HEV-C1 was 16-fold higher than that to HEV-1. On the other hand, in the case shown in Fig. 2B, IgG titer to HEV-1 was 8-fold higher than that to HEV-C1. Ratio of IgG titer to HEV-C1 divided by IgG titer to HEV-1 was determined for each sample, and the number of samples at each ratio is listed in Table 1. Li et al. [10] reported that IgG titer to HEV-1 in a serum of HEV-1-infected patient was 16-fold higher than IgG titer to HEV-C1. Hence, we tentatively set up 8 and 1/8 as threshold ratios to differentiate between sera from HEV-C1- and HEV-1-infected patients. As a result, 3 patients each (designated as #1 to 3 and #4 to 6) were judged as patients suspected of having HEV-C1 and HEV-1 infection, respectively (Table 1 ).
Fig. 2.
Representative data of sera showing strong reactivity to HEV-C1 (A) or HEV-1 (B) antigen in IgG ELISA. Serial 2-fold dilutions of sera were subjected to ELISA using VLPs of HEV-C1 and HEV-1 as antigens. Open and filled circles indicate optical density (OD) values for HEV-C1 and HEV-1 antigens, respectively. The cutoff value was tentatively set at OD of 0.8 and is shown as dashed lines.
Table 1. Number of samples at each ratio of IgG titer to HEV-C1 to IgG titer to HEV-1 and suspected virus infected in the patients.
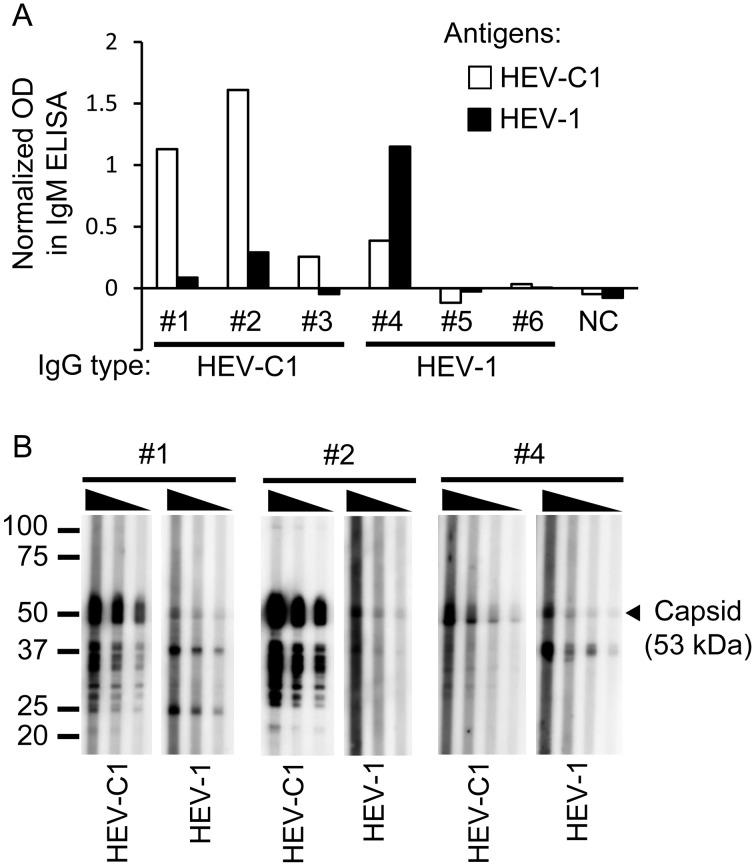
Detection of anti-HEV-C1 and anti-HEV-1 IgM antibodies: In order to find patients with acute HEV-C1 and HEV-1 infection, acute sera of the patients #1 to 3 and patients #4 to 6 suspected of having HEV-C1 and HEV-1 infection, respectively, were examined for IgM antibodies by ELISA. IgM antibodies to HEV-C1 were detected from 2 of the 3 acute sera of patients suspected of having HEV-C1 infection, while IgM antibodies to HEV-1 were detected from 1 of the 3 acute sera of patients suspected of having HEV-1 infection (Fig. 3A). In order to confirm the results, acute sera of patients #1, 2 and 4 were subjected to Western blotting for detecting IgM antibodies. As a result, acute sera of patients #1 and 2 showed obviously higher reactivity to recombinant capsid protein (about 53 kDa) constituting VLPs of HEV-C1 than that of HEV-1 (Fig. 3B). Acute serum of patient #4 reacted to the capsid protein of HEV-1 and also reacted to that of HEV-C1.
Fig. 3.
Detection of IgM antibodies in acute sera. (A) Detection by ELISA. Acute sera from patients #1 to 3 and patients #4 to 6 who were suspected of having HEV-C1 infection and HEV-1 infection, respectively, were diluted to 1:200 and subjected to ELISA using VLPs of HEV-C1 and HEV-1 as antigens. Open and filled bars indicate optical density (OD) values for HEV-C1 and HEV-1 antigens, respectively, which were normalized by subtracting OD values for null antigen. (B) Detection by Western blotting. Acute sera from 3 patients (#1, 2 and 4) were subjected to Western blotting using VLPs of HEV-C1 and HEV-1 as antigens. Acute sera of patients #1 and #4 were diluted to 1:4,000 to 1:16,000 and 1:2,000 to 1:16,000, respectively, and incubated with HEV-C1 and HEV-1 antigens. Acute serum of patient #2 was diluted to 1:16,000 to 1:64,000 or 1:4,000 to 1:16,000 and incubated with HEV-C1 or HEV-1 antigens, respectively. The arrowhead indicates the size of capsid protein constituting VLPs of HEV-C1 and HEV-1.
Clinical information: Some clinical information was available for the patient #1. Hepatic enzymes, aspartate aminotransferase (AST) and alanine aminotransferase (ALT), were mildly elevated (156 and 68 IU/l, respectively), and hepatomegaly was detected by ultrasonography. The patient recovered from the illness and was discharged after 17 days.
Detection of RNA of HEV-C1: In order to obtain direct evidence of HEV-C1 infection in humans, we tried to detect viral RNA from sera of the patients with IgM antibodies by reverse transcription and nested PCR. However, despite repeated trials, no HEV genome was detected.
DISCUSSION
In order to evaluate the risk for HEV-C1 infection in humans, we examined sera of 208 febrile patients in Vietnam where HEV-C1 is prevalent in rodents [10]. IgG antibodies to HEV-C1 antigen were detected from 70 of the 208 patient sera in ELISA. Although most of the 70 sera were also positive for IgG antibodies to HEV-1 antigen, IgG titers to HEV-C1 antigen in 3 sera were more than 8-fold higher than those to HEV-1 antigen (Table 1). IgM antibodies to HEV-C1 antigen were also detected from 2 of the 3 acute sera of patients in ELISA and Western blotting (Fig. 3). In addition, the reactivity of the IgM antibodies to HEV-C1 antigen was obviously higher than that to HEV-1 antigen. These results suggest that HEV-C1 infects humans.
Alternatively, there is a possibility that an HEV variant antigenically related to HEV-C1 infects humans. There are diverse members in the genus Orthohepevirus, which were discovered from a wide variety of animals. Orthohepevirus A contains isolates from human, pig, wild boar, deer, mongoose, rabbit and camel, Orthohepevirus B contains isolates from chicken, Orthohepevirus C contains isolates from rat, greater bandicoot, Asian musk shrew, ferret and mink, and Orthohepevirus D contains isolates from bat [16]. As in the case of HEV-C1 and HEV-1 [10], antigenic cross-reactivity between human and swine isolates of Orthohepevirus A and avian isolate of Orthohepevirus B was also reported [3], despite the low amino acid sequence identity of capsid proteins of the viruses (approximately 48 to 49%). These reports suggest the existence of viruses antigenically related to HEV-C1. Further studies are needed to determine antigenic relationship between HEV-C1 and various animal-derived HEVs.
Dremsek et al. have also found IgG antibodies to HEV-C1 antigen from some sera of healthy forestry workers in Germany [1]. However, they did not detect IgM antibodies and not investigate patient sera. We detected for the first time IgM antibodies to HEV-C1 antigen in patient sera. In addition, we found that one of the patients developed mild liver dysfunction. The patient was negative for IgG antibodies to Rickettsia japonica and Orientia tsutsugamushi. Although a low level of IgG antibodies to R. typhi was detected from the patient, the titer did not increase from acute to convalescent phase. These results suggest that HEV-C1 or its variants may cause disease in humans. However, despite repeated trials, no HEV genome was detected from the patient serum. This failure might be due to a low level of viral RNA in sera, heterogeneity of the virus and/or degradation of viral RNA during storage. Although whole blood or buffy coat may be suitable for detection of viral RNA, they were not available in this study. Genetic identification of the putative HEV-C1 is a future subject to be solved.
In conclusion, HEV-C1 or HEV variants that are antigenically related to HEV-C1 might infect humans. The patient with IgM antibodies to HEV-C1 antigen developed illness with mild liver dysfunction. However, no HEV genome was detected. Therefore, further studies are needed to elucidate the potential risk of HEV-C1 infection.
Acknowledgments
This study was supported in part by the Japan Initiative for Global Research Network on Infectious Diseases (J-GRID).
REFERENCES
- 1.Dremsek P., Wenzel J. J., Johne R., Ziller M., Hofmann J., Groschup M. H., Werdermann S., Mohn U., Dorn S., Motz M., Mertens M., Jilg W., Ulrich R. G.2012. Seroprevalence study in forestry workers from eastern Germany using novel genotype 3- and rat hepatitis E virus-specific immunoglobulin G ELISAs. Med. Microbiol. Immunol. (Berl.) 201: 189–200. doi: 10.1007/s00430-011-0221-2 [DOI] [PubMed] [Google Scholar]
- 2.Emerson S. U., Nguyen H. T., Torian U., Burke D., Engle R., Purcell R. H.2010. Release of genotype 1 hepatitis E virus from cultured hepatoma and polarized intestinal cells depends on open reading frame 3 protein and requires an intact PXXP motif. J. Virol. 84: 9059–9069. doi: 10.1128/JVI.00593-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haqshenas G., Huang F. F., Fenaux M., Guenette D. K., Pierson F. W., Larsen C. T., Shivaprasad H. L., Toth T. E., Meng X. J.2002. The putative capsid protein of the newly identified avian hepatitis E virus shares antigenic epitopes with that of swine and human hepatitis E viruses and chicken big liver and spleen disease virus. J. Gen. Virol. 83: 2201–2209. doi: 10.1099/0022-1317-83-9-2201 [DOI] [PubMed] [Google Scholar]
- 4.Johne R., Heckel G., Plenge-Bönig A., Kindler E., Maresch C., Reetz J., Schielke A., Ulrich R. G.2010. Novel hepatitis E virus genotype in Norway rats, Germany. Emerg. Infect. Dis. 16: 1452–1455. doi: 10.3201/eid1609.100444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johne R., Plenge-Bönig A., Hess M., Ulrich R. G., Reetz J., Schielke A.2010. Detection of a novel hepatitis E-like virus in faeces of wild rats using a nested broad-spectrum RT-PCR. J. Gen. Virol. 91: 750–758. doi: 10.1099/vir.0.016584-0 [DOI] [PubMed] [Google Scholar]
- 6.Koonin E. V., Gorbalenya A. E., Purdy M. A., Rozanov M. N., Reyes G. R., Bradley D. W.1992. Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: delineation of an additional group of positive-strand RNA plant and animal viruses. Proc. Natl. Acad. Sci. U.S.A. 89: 8259–8263. doi: 10.1073/pnas.89.17.8259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li T. C., Ami Y., Suzaki Y., Yasuda S. P., Yoshimatsu K., Arikawa J., Takeda N., Takaji W.2013. Characterization of full genome of rat hepatitis E virus strain from Vietnam. Emerg. Infect. Dis. 19: 115–118. doi: 10.3201/eid1901.121007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li T. C., Chijiwa K., Sera N., Ishibashi T., Etoh Y., Shinohara Y., Kurata Y., Ishida M., Sakamoto S., Takeda N., Miyamura T.2005. Hepatitis E virus transmission from wild boar meat. Emerg. Infect. Dis. 11: 1958–1960. doi: 10.3201/eid1112.051041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li T. C., Yamakawa Y., Suzuki K., Tatsumi M., Razak M. A., Uchida T., Takeda N., Miyamura T.1997. Expression and self-assembly of empty virus-like particles of hepatitis E virus. J. Virol. 71: 7207–7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li T. C., Yoshimatsu K., Yasuda S. P., Arikawa J., Koma T., Kataoka M., Ami Y., Suzaki Y., Mai T. Q., Hoa N. T., Yamashiro T., Hasebe F., Takeda N., Wakita T.2011. Characterization of self-assembled virus-like particles of rat hepatitis E virus generated by recombinant baculoviruses. J. Gen. Virol. 92: 2830–2837. doi: 10.1099/vir.0.034835-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li T. C., Zhang J., Shinzawa H., Ishibashi M., Sata M., Mast E. E., Kim K., Miyamura T., Takeda N.2000. Empty virus-like particle-based enzyme-linked immunosorbent assay for antibodies to hepatitis E virus. J. Med. Virol. 62: 327–333. doi: [DOI] [PubMed] [Google Scholar]
- 12.Li W., Guan D., Su J., Takeda N., Wakita T., Li T. C., Ke C. W.2013. High prevalence of rat hepatitis E virus in wild rats in China. Vet. Microbiol. 165: 275–280. doi: 10.1016/j.vetmic.2013.03.017 [DOI] [PubMed] [Google Scholar]
- 13.Mulyanto, Depamede S. N., Sriasih M., Takahashi M., Nagashima S., Jirintai S., Nishizawa T., Okamoto H.2013. Frequent detection and characterization of hepatitis E virus variants in wild rats (Rattus rattus) in Indonesia. Arch. Virol. 158: 87–96. doi: 10.1007/s00705-012-1462-0 [DOI] [PubMed] [Google Scholar]
- 14.Purcell R. H., Engle R. E., Rood M. P., Kabrane-Lazizi Y., Nguyen H. T., Govindarajan S., St Claire M., Emerson S. U.2011. Hepatitis E virus in rats, Los Angeles, California, U.S.A. Emerg. Infect. Dis. 17: 2216–2222. doi: 10.3201/eid1712.110482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rein D. B., Stevens G. A., Theaker J., Wittenborn J. S., Wiersma S. T.2012. The global burden of hepatitis E virus genotypes 1 and 2 in 2005. Hepatology 55: 988–997. doi: 10.1002/hep.25505 [DOI] [PubMed] [Google Scholar]
- 16.Smith D. B., Simmonds P., International Committee on Taxonomy of Viruses Hepeviridae Study Group,Jameel S., Emerson S. U., Harrison T. J., Meng X. J., Okamoto H., Van der Poel W. H. M., Purdy M. A., 2014. Consensus proposals for classification of the family Hepeviridae. J. Gen. Virol. 95: 2223–2232. doi: 10.1099/vir.0.068429-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolf S., Reetz J., Johne R., Heiberg A. C., Petri S., Kanig H., Ulrich R. G.2013. The simultaneous occurrence of human norovirus and hepatitis E virus in a Norway rat (Rattus norvegicus). Arch. Virol. 158: 1575–1578. doi: 10.1007/s00705-013-1646-2 [DOI] [PubMed] [Google Scholar]
- 18.Yang T., Kataoka M., Ami Y., Suzaki Y., Kishida N., Shirakura M., Imai M., Asanuma H., Takeda N., Wakita T., Li T. C.2013. Characterization of self-assembled virus-like particles of ferret hepatitis E virus generated by recombinant baculoviruses. J. Gen. Virol. 94: 2647–2656. doi: 10.1099/vir.0.056671-0 [DOI] [PubMed] [Google Scholar]