Abstract
Bats of the genus Eptesicus have several non-retroviral RNA virus-derived sequences in their genomes, among which an endogenous bornavirus-like L element, named eEBLL-1, was suggested to encode functional proteins in the hosts. However, the function of eEBLL-1 remains unclear due to a lack of appropriate investigation tools, such as cultured cells expressing eEBLL-1. Here, we established a continuous cell line, named HAMOI-EnK cells, from kidney of Eptesicus nilssonii. HAMOI-EnK cells are robust and could be passaged for at least 10 months. eEBLL-1 in the genomes of HAMOI-EnK cells retains an intact open reading frame. Additionally, eEBLL-1 is transcribed in the sense-orientation in cells. To our knowledge, this is the first report to demonstrate that eEBLL-1 is transcribed in cultured cells.
Keywords: cell line, eEBLL-1, endogenous bornavirus-like element, Eptesicus nilssonii, RNA-dependent RNA polymerase
Endogenous viral elements (EVEs) are heritable sequences that originate from ancient viruses in the eukaryotic genomes. The best-studied EVEs are endogenous retroviruses. Notably, some endogenous retrovirus-derived genes play essential roles in the hosts, such as placentation in mammalian species [9, 10]. Although non-retroviral viruses do not possess reverse transcriptase and/or integrase, recent studies have revealed that non-retroviral EVEs are also present in the genomes of mammals [1, 5, 7, 8, 11]. Notably, several non-retroviral EVEs contain relatively long open reading frames (ORFs) and have evolved under natural selection, suggesting that such EVEs encode functional proteins in the host cells [2, 6, 12]. Indeed, an endogenous bornavirus-like nucleoprotein (EBLN) element in Ictidomys tridecemlineatus showed anti-bornavirus activity in vitro [3]. These observations suggest that non-retroviral EVEs have been also co-opted by the hosts.
Bats (the order Chiroptera) have non-retroviral RNA virus-derived EVEs including EBL and endogenous filovirus-like (EFL) elements [7, 12, 13]. Taylor et al. [12] reported that an EFL element descended from filoviral VP35 gene has evolved under natural selection in Myotis bats. We also showed that an EBLL in bats of the genus Eptesicus (designated as eEBLL-1), derived from the L gene (encoding RNA-dependent RNA polymerase: RdRp) of an ancient bornavirus, has maintained a large and intact ORF for more than 11.8 million years and evolved under purifying selection in bats of the genus Eptesicus [6]. However, the function of eEBLL-1 is still unclear due to lack of proper research tools. To understand the biological significance of eEBLL-1 in the genus Eptesicus, cell lines from bats of the genus Eptesicus would be useful. There is one report describing establishment of cell lines from E. serotinus [4]. However, the expression profile of eEBLL-1 in these cell lines has not been determined. Therefore, it is required to establish and characterize cell lines from Eptesicus bats. Thus, we attempted to establish cell lines from Eptesicus nilssonii.
We first made primary cell cultures from E. nilssonii. E. nilssonii was caught in Obihiro, Hokkaido, and euthanized. Capture and handling of E. nilssonii was performed under a license from the Japanese Ministry of Environment (liscense No. 21-27-0213). The liver, kidney, spleen and lung were removed from the bat. Each of the tissue was minced, trypsinized and seeded on cell culture plates, which were incubated in a CO2 incubator (37°C in 5% CO2 in air) (full methods are available in Supplementary information). The cells were passaged every two to six days depending on the cell density: the cells were washed with PBS and detached with PBS containing 0.25% trypsin and 1 mM EDTA. The detached cells were mixed with growth medium and centrifuged at 1,000 ×g for 3 min. The supernatant was removed, and the cell pellet was resuspended in growth medium. Among them, kidney and spleen-derived cells grew well (data not shown). However, spleen-derived cells tended detached from culture dishes and could not propagate after 17 passages (approximately two months). On the other hand, kidney-derived cells were robust and could be passaged for at least 10 months without any immortalization process (as of May 24, 2016).
To confirm that the kidney-derived cells originated from E. nilssonii, we determined partial sequence of cytochrome b (cytb) gene from the cells by genomic PCR and direct sequencing (GenBank accession number LC122512) (Supporting information). We performed BLAST analysis using the determined sequence as a query, showing that the best hit is cytb gene from E. nilssonii (GQ272569). The nucleotide identity between the sequence we determined, and the best hit is 99.8%. We also conducted phylogenetic analysis using cytb sequences of the kidney-derived cell line and top 50 hits from the BLAST analysis. A Maximum Likelihood phylogenetic tree showed that the cytb gene from these cells was clustered with those of E. nilssonii (Supp. Fig. 1), indicating that the cells were indeed derived from E. nilssonii. We named this cell line as HAMOI-EnK after surnames of the investigators, species and tissue.
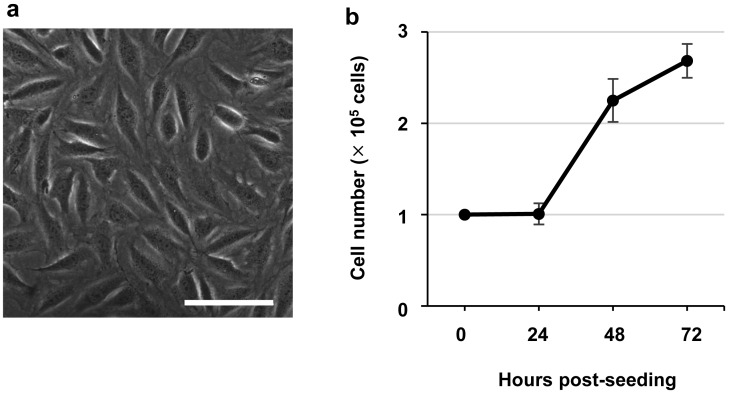
Fig. 1.
Characterization of HAMOI-EnK cells. (a) Morphology of HAMOI-EnK cells. Scale bar, 10 µm. (b) Growth kinetics of HAMOI-EnK cells. The means ± standard deviations of cell numbers from 3 independent wells are indicated.
HAMOI-EnK cells are adhesive and form monolayers (Fig. 1a). The cells are easily passaged using standard procedures (Supplementary information). To investigate the growth kinetics of HAMOI-EnK cells, 1 × 105 cells at the 64th passage were seeded into individual wells of a 12-well plate, and cell numbers were counted 24, 48 and 72 hr after plating (3 independent wells). The cell numbers at 24, 48 and 72 hr after plating were 1.01 × 105, 2.25 × 105 and 2.68 × 105 cells, respectively (Fig. 1b). We measured the doubling time in the exponential growth phase as approximately 23.4 −25.2 hr with DMEM containing 10% FCS.
Next, we analyzed eEBLL-1 in the genome of HAMOI-EnK cells. Although we demonstrated that eEBLL-1 is present in the genome of E. nilssonii in the previous study [6], eEBLL-1 might have been lost during long-term passage. Thus, we performed genomic PCR to detect eEBLL-1 in the genome (PCR conditions are available in Supporting information). We obtained a band of expected size (data not shown), which was subject to direct sequencing. The sequencing revealed that eEBLL-1 in HAMOI-EnK cells retains an intact ORF comprising 1,718 codons and all of the functional motifs in mononegavirus RdRps (see LC122511 in GenBank) that were reported in the previous study [6]. The nucleotide sequence of the putative ORF of eEBLL-1 in HAMOI-EnK cells showed 99.8% (5,146 of 5,157 nucleotides) identity to that of previously reported eEBLL-1 sequence in E. nilssonii (AB921210) [6]. Among the 11 nucleotide differences, 7 are synonymous, and 4 are non-synonymous substitutions. These differences might be due to genetic polymorphisms in bats. Alternatively, these mutations might have been introduced during the cell passage due to relaxed functional constraint. It would be interesting to determine eEBLL-1 sequences in several individuals of E. nilssonii.
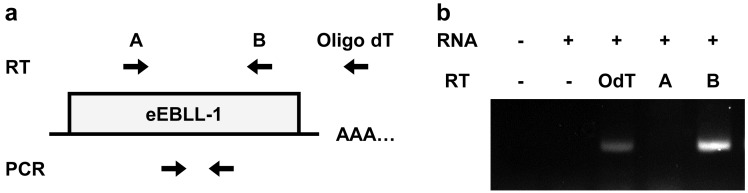
We also performed strand-specific RT-PCR (Fig. 2a) to investigate whether eEBLL-1 is transcribed in HAMOI-EnK cells. We synthesized cDNA with primer A (sense primer), primer B (antisense primer) or oligo dT primer, and then performed PCR (full methods and primer sequences are available in Supporting information). We detected expected bands using oligo dT or the antisense primer B for cDNA synthesis (Fig. 2b), demonstrating that sense-oriented eEBLL-1 transcripts are present in the cells. Unfortunately, we could not check the protein expression due to the lack of antibodies that detect eEBLL-1.
Fig. 2.
Expression of eEBLL-1 transcripts in HAMOI-EnK cells. (a) A schematic representation of strand-specific RT-PCR. A putative eEBLL-1 transcript is shown. Primers used for reverse transcription (RT) and PCR were indicated by arrows. (b) RT-PCR amplification of eEBLL-1 from RNA in HAMOI-EnK cells. Primers used for RT reactions are indicated: primer A, sense primer; primer B, antisense primer; OdT, oligo dT primer.
In this study, we established a cell line derived from kidney of E. nilssonii. Sequencing analyses showed that the cells were originated from E. nilssonii. The cells are robust and easy to passage. In addition, eEBLL-1 is present in the genome of HAMOI-EnK cells, which retains a large and intact ORF as previously reported [6]. Furthermore, we demonstrated the presence of sense-oriented eEBLL-1 transcripts in the cells. Although we detected eEBLL-1 transcripts in tissues of E. nilssonii and E. serotinus in the previous study [6], this is the first report demonstrating that eEBLL-1 is transcribed in cultured cells. Thus, HAMOI-EnK cells will be a powerful tool to investigate the biological significance of eEBLL-1, which could contribute to understand co-evolution of RNA viruses and their hosts.
Supplementary Material
Acknowledgments
This study was supported by JSPS KAKENHI grant number 26850208 (MH).
REFERENCES
- 1.Belyi V. A., Levine A. J., Skalka A. M.2010. Unexpected inheritance: multiple integrations of ancient bornavirus and ebolavirus/marburgvirus sequences in vertebrate genomes. PLoS Pathog. 6: e1001030. doi: 10.1371/journal.ppat.1001030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fort P., Albertini A., Van-Hua A., Berthomieu A., Roche S., Delsuc F., Pasteur N., Capy P., Gaudin Y., Weill M.2012. Fossil rhabdoviral sequences integrated into arthropod genomes: ontogeny, evolution, and potential functionality. Mol. Biol. Evol. 29: 381–390. doi: 10.1093/molbev/msr226 [DOI] [PubMed] [Google Scholar]
- 3.Fujino K., Horie M., Honda T., Merriman D. K., Tomonaga K.2014. Inhibition of Borna disease virus replication by an endogenous bornavirus-like element in the ground squirrel genome. Proc. Natl. Acad. Sci. U.S.A. 111: 13175–13180. doi: 10.1073/pnas.1407046111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He X., Korytař T., Schatz J., Freuling C. M., Müller T., Köllner B.2014. Anti-lyssaviral activity of interferons κ and ω from the serotine bat, Eptesicus serotinus. J. Virol. 88: 5444–5454. doi: 10.1128/JVI.03403-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horie M., Honda T., Suzuki Y., Kobayashi Y., Daito T., Oshida T., Ikuta K., Jern P., Gojobori T., Coffin J. M., Tomonaga K.2010. Endogenous non-retroviral RNA virus elements in mammalian genomes. Nature 463: 84–87. doi: 10.1038/nature08695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horie M., Kobayashi Y., Honda T., Fujino K., Akasaka T., Kohl C., Wibbelt G., Mühldorfer K., Kurth A., Müller M. A., Corman V. M., Gillich N., Suzuki Y., Schwemmle M., Tomonaga K.2016. An RNA-dependent RNA polymerase gene in bat genomes derived from an ancient negative-strand RNA virus. Sci. Rep. 6: 25873. doi: 10.1038/srep25873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horie M., Kobayashi Y., Suzuki Y., Tomonaga K.2013. Comprehensive analysis of endogenous bornavirus-like elements in eukaryote genomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368: 20120499. doi: 10.1098/rstb.2012.0499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horie M., Tomonaga K.2011. Non-retroviral fossils in vertebrate genomes. Viruses 3: 1836–1848. doi: 10.3390/v3101836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imakawa K., Nakagawa S., Miyazawa T.2015. Baton pass hypothesis: successive incorporation of unconserved endogenous retroviral genes for placentation during mammalian evolution. Genes Cells 20: 771–788. doi: 10.1111/gtc.12278 [DOI] [PubMed] [Google Scholar]
- 10.Jern P., Coffin J. M.2008. Effects of retroviruses on host genome function. Annu. Rev. Genet. 42: 709–732. doi: 10.1146/annurev.genet.42.110807.091501 [DOI] [PubMed] [Google Scholar]
- 11.Katzourakis A., Gifford R. J.2010. Endogenous viral elements in animal genomes. PLoS Genet. 6: e1001191. doi: 10.1371/journal.pgen.1001191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor D. J., Dittmar K., Ballinger M. J., Bruenn J. A.2011. Evolutionary maintenance of filovirus-like genes in bat genomes. BMC Evol. Biol. 11: 336. doi: 10.1186/1471-2148-11-336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor D. J., Leach R. W., Bruenn J.2010. Filoviruses are ancient and integrated into mammalian genomes. BMC Evol. Biol. 10: 193. doi: 10.1186/1471-2148-10-193 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.