Abstract
To investigate whether kokuvirus is present in Japanese dogs, we examined the fecal samples obtained from 94 diarrheal household dogs and 50 clinically healthy kenneled dogs by RT-PCR. The gene was detected in 37.2% and 48.0% in the former and the latter, respectively, suggesting that canine kobuvirus (CaKoV) is circulating among Japanese dogs. From the result of the latter, however, CaKoV may not be a primary pathogen. Furthermore, all gene-positive dogs were purebreds aged four months or younger. This finding suggests that CaKoV endemic is confined in multi-dog environments, and the dogs have a strong age-dependent resistance to CaKoV.
Keywords: canine kobuvirus, domestic dog, Japan, RNA
An epidemic outbreak of gastroenteritis in humans, possibly caused by consumption of raw oysters, occurred in Aichi Prefecture, Japan, in 1989, after which a small round virus was isolated from the feces of affected patients. Based on nucleotide and amino acid sequence analyses, the causal virus was classified as a new genus (the genus kobuvirus) in the family Picornaviridae and named the Aichi virus [30, 31]. Kobuviruses are non-enveloped, single-stranded and positive-sense RNA viruses, and have been isolated from samples obtained from a variety of mammals. Genetically, it is classified into three strains, termed Aichivirus A (formerly Aichi virus), B (formerly bovine kobuvirus) and C (formerly porcine kobuvirus) [6, 7, 13, 15, 16, 21, 22, 24, 29].
In 2011, Kapoor et al. [13] reported a virus related to the Aichi virus obtained from dog feces in the United States, which was the first report of its kind to suggest canine kobuvirus infection and was termed canine kobuvirus (CaKoV). CaKoV is classified as Aichivirus A species together with the Aichi virus, as well as kobuviruses isolated from cats and mice [7, 14]. Later, CaKoV was reported to be isolated from dogs in Italy, the United Kingdom, Korea, China and Tanzania [2, 4, 5, 10]. Furthermore, gene analysis showed the presence of viruses very closely related to CaKoV in canidae animals, such as jackal and fox, and the Hyaenidae [8, 18].
In Japan, viruses similar to CaKoV have been detected in abundance together with the Aichi virus in sludge from wastewater treatment plants, suggesting the presence of CaKoV [28]. However, to the best of our knowledge, there are no reports of CaKoV detection in Japanese dogs.
For the purpose of determining evidence for the circulation of CaKoV among domestic dogs in Japan, we attempted to isolate the CaKoV gene from fecal samples of household dogs and dogs being kept in pet shops.
In the period from 2012 to 2015, fecal samples were collected from a total of 94 household (privately owned) dogs with diarrhea (37, 6, 25 and 26 samples in 2012, 2013, 2014 and 2015, respectively), which visited veterinary clinics nationwide in Japan, in which the involvement of CPV-2 infection was ruled out by PCR (mentioned later). Furthermore, the fecal samples obtained from 50 clinically healthy kenneled dogs, aged 1–3 months, were collected from 4 pet shops in Japan. The sample was suspended in a sterile saline, the suspension was centrifuged, and nucleic acid was isolated from the supernatant using a QIAamp Viral RNA Mini Kit (Qiagen science, Germantown, MD, U.S.A.), following the manufacturer’s instruction. The extract was preserved at −45°C until used.
CaKoV was detected by RT-PCR with a primer pair (Forward; 5′-CTCCCCTCAGCTGCCTTCTC-3, Reverse; 5′-GAGGATCTGAAATTTGGAAG-3′) that was amplified at nucleotide positions 7,368–7,619 (a partial 3D gene) of CaKoV 12D049 (KF924623) [4], providing a 252-bp fragment, using a QIAGEN One Step RT-PCR kit (Qiagen science). Two point five µl of the extracted RNA was added to the 25 µl reaction mixtures containing 5 µl of 5 ×buffer (finally 1.5 mM MgCl2), 0.4 mM of each deoxynucleozide triphosphate, 10 U RNase inhibitor (Promega, Madison, WI, U.S.A.), 0.8 µM of the primers and 1.0 µl of enzyme mix. The RNA was reverse transcribed at 50°C for 30 min, followed by inactivation of reverse transcriptase and denaturation of cDNA template at 95°C for 15 min. The cDNA was amplified in 35 sequential cycles of denaturation at 94°C for 30 sec, annealing at 52°C for 30 sec and extension at 72°C for 40 sec, followed by a final extension of 72°C for 7 min. The PCR product was electrophoresed at 100 V for 35 min in a 2% agarose gel. Then, the gel was stained with ethidium bromide and was visualized under UV illumination.
Furthermore, 12 samples positive for the above mentioned RT-PCR were also submitted to RT-PCR with a primer pair which was amplified at nucleotide positions 6,982–7,485 (3D gene) of CaKoV 12D049 (KF924623) (Forward; 5′-CCCTGGAACACCCAAGGCCGCT-3′, Reverse; 5′- TCTGGTTGCCATAGATGTGGTG-3′) [5]. These PCR amplicons (504-bp) and above amplicons (252-bp) were purified using ExoSAP-IT and were submitted to direct sequencing on both strands by the dye-terminator cycle sequencing methodology (Bio Matrix Research, Chiba, Japan). The sequences (Nucleotide positions 6,982–7,619; 638-bp) were aligned with the reference strains in the GenBank Database using MEGA 6 software [27]. Phylogenetic trees were constructed employing the neighbor-joining method [23], and their homologies were analyzed using the BLAST search.
To investigate the involvement of other diarrheagenic viruses in the RT-PCR positive samples, canine parvovirus type 2 (CPV-2) [1], canine coronavirus types I (CCoV-I) and II (CCoV-II) [20], canine distemper virus (CDV) [9], canine adenovirus type 1 (CAdV-1) [12] and canine rotavirus (CRV) [11] were examined by PCR or RT-PCR.
The RT-PCR products (252-bp) were detected in 35 diarrheal household dogs (37.2%) and 24 healthy kenneled dogs (48.0%), with no significant differences (P>0.05). As shown in Table 1, grouped by age, the positivity of the RT-PCR in dogs, aged 2 months, was extremely high (63.9%) and was significantly different from the averaged positivity (37.2%) (P=0.0062, chi-squared=7.4845). In contrast, no positive PCR reaction was observed in all of 24 dogs aged 5 months or over. In addition, all the RT-PCR positive dogs were purebreds (Table 1). No significant differences were observed, grouped by year and region of Japan (P>0.05) (data not shown).
Table 1. Prevalence of canine kobuvirus RNA in diarrheal household and healthy kenneled dogs.
| Age | Positives/Samples tested | |||
|---|---|---|---|---|
| Diarreal household dogs | Healthy kenneled dogs | |||
| Purebred | Mixed breed | Total | ||
| A month | 2/6 | 0/1 | 2/7 (28.6%) | 19/36 (52.8%) |
| 2 months | 23/34 | 0/2 | 23/36 (63.9%) | 4/10 (40.0%) |
| 3 months | 7/17 | - | 7/17 (41.2%) | 1/4 (25.0%) |
| 4 months | 3/10 | - | 3/10 (30.0%) | - |
| 5 to 11 months | 0/5 | 0/1 | 0/6 | - |
| A year or older | 0/14 | 0/4 | 0/18 | - |
| Total | 35/86 (40.7%) | 0/8 | 35/94 (37.2%) | 24/50 (48.0%) |
When the 35 RT-PCR positive samples of the diarrheal household dogs were examined for other diarrheagenic viruses, CCoV-I, CCoV-II and CDV were detected in 21, 15 and 5 samples, respectively, while CAdV-1 and CRV were not detected. None of these 5 viruses were detected in 7 samples (20.0%) (data not shown).
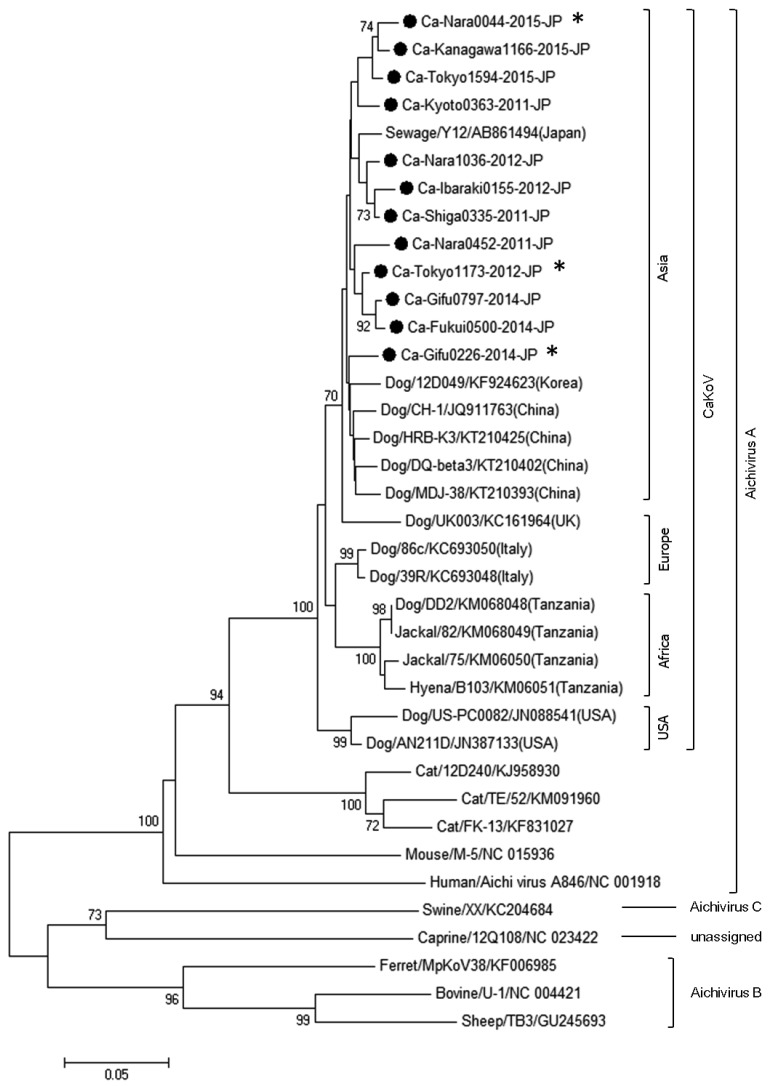
The partial 3D nucleotide sequences (638-bp) of the 12 present strains were compared with the reference strains in the GenBank database. Among the present strains, the homologies were 95.7–98.9%. The homologies of the present strains with known CaKoVs from dogs and canidae animals, and the Kobuvirus detected from a sewage sample (AB861494) were 94.5–97.4% and 96.1–97.7%, respectively. The present strains were clustered into the same clade with the known CaKoVs and the sewage strain in the phylogenetic tree (Fig. 1). The homologies of the present strains with the isolates from humans, cats and mice were relatively high (77.4–83.3%), showing that the present strains were classified into Aichivirus A species. In contrast, those with the isolates from other mammals were low (60.8–66.5%), which were clustered in different clades. Furthermore, the present strains and the sewage virus were clustered into the same clade (Asia), if the CaKoV clade was subdivided.
Fig. 1.
Phylogenetic tree based on the partial 3D nucleotide sequences of kobuviruses obtained by neighbor-joining method in the MEGA 6 software. Bootstrap analysis was performed with 1,000 replicates. Black dot marks represent 12 strains newly detected in this study. *Accession number: LC147654 (Ca-Nara0044-2015-JP), LC147655 (Ca-Gifu0226-2014-JP), LC147656 (Ca-Tokyo1173-2012-JP).
In the present study, kobuvirus RNA was detected at a high rate in the dog fecal samples. As a result of 3D nucleotide sequence analysis, it was revealed that the virus had a very high homology with previously reported CaKoV, suggesting that CaKoV has been transmitted among Japanese dogs. Generally, infectious viral diseases tend to be epidemic during the juvenile period [25, 26, 32], and we observed a similar tendency for CaKoV in the present study. Surprisingly, however, the frequency of viral RNA detection was confined in the dogs four months old or younger and was never found in those older than five months. These findings indicate that dogs have a strong age-dependent resistance to CaKoV and that the dog infected with CaKoV in the past has a high resistance to reinfection. Unfortunately, since the kenneled dogs of this study were aged 1–3 months, the positivity could not be analyzed by age. Furthermore, our findings showed a high rate of detection of viral RNA in kennels, similar to the finding of Choi et al. [4]. Thus, we speculated that CaKoV prevalence is confined to multi-dog environments.
Yamashita et al. [28] isolated a large amount of viral RNA similar to that of CaKoV from sludge obtained from a wastewater treatment plant. We found that the sewage virus had a very high homology with CaKoV detected in the dogs of the present study, indicating a possible canine origin. As noted above, CaKoV tends to be epidemic in an environment where several dogs are kept together. However, the risk of CaKoV infection even in a dog-free environment is understandable, since picornaviruses are highly stable in a normal environment.
Phylogenetic tree analysis of the 3D region showed that the viral strains isolated in Japan including CaKoV from sewage sludge could be classified into the same clade as those found in countries neighboring Japan, such as Korea and China, but into a different clade as compared to those isolated in Europe, the United States and Africa, if subdivided. Thus, CaKoV may be specific to various countries and regions, and may show independent transmission.
The diarrheal pathogenicity of CaKoV in dogs has not been fully elucidated, and it is still not clear whether CaKoV is diarrheagenic. [2, 5, 17]. Based on results of the present study showing that viral RNA was detected in approximately half of the examined healthy kenneled dogs, we suggest that it may not be a primary pathogen in dogs. However, all of the diarrheal dogs were found not to be infected with CPV-2, the most important diarrheal pathogen. Furthermore, 20.0% of the CaKoV-positive dogs were negative for other major diarrhea causative viruses, such as CDV, CAdV-1, CCoV and CRV. Recent epidemiological studies suggested possible involvement of porcine kobuvirus in diarrheal disease seen in piglets [3, 19]. Thus, it is possible that CaKoV might be involved in diarrheal disease seen in dogs as a mild or secondary infection factor, though various factors other than these viral infections can be related to an episode of diarrheal disease.
Further detailed epidemiological investigations including rearing environments and symptoms, as well as experimental infections, are considered necessary to clarify the pathogenicity of CaKoV.
REFERENCES
- 1.Buonavoglia C., Martella V., Pratelli A., Tempesta M., Cavalli A., Buonavoglia D., Bozzo G., Elia G., Decaro N., Carmichael L.2001. Evidence for evolution of canine parvovirus type 2 in Italy. J. Gen. Virol. 82: 3021–3025. doi: 10.1099/0022-1317-82-12-3021 [DOI] [PubMed] [Google Scholar]
- 2.Carmona-Vicente N., Buesa J., Brown P. A., Merga J. Y., Darby A. C., Stavisky J., Sadler L., Gaskell R. M., Dawson S., Radford A. D.2013. Phylogeny and prevalence of kobuviruses in dogs and cats in the UK. Vet. Microbiol. 164: 246–252. doi: 10.1016/j.vetmic.2013.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen L., Zhu L., Zhou Y. C., Xu Z. W., Guo W. Z., Yang W. Y.2013. Molecular and phylogenetic analysis of the porcine kobuvirus VP1 region using infected pigs from Sichuan Province, China. Virol. J. 10: 281. doi: 10.1186/1743-422X-10-281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi S., Lim S. I., Kim Y. K., Cho Y. Y., Song J. Y., An D. J.2014. Phylogenetic analysis of astrovirus and kobuvirus in Korean dogs. J. Vet. Med. Sci. 76: 1141–1145. doi: 10.1292/jvms.13-0585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Martino B., Di Felice E., Ceci C., Di Profio F., Marsilio F.2013. Canine kobuviruses in diarrhoeic dogs in Italy. Vet. Microbiol. 166: 246–249. doi: 10.1016/j.vetmic.2013.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Martino B., Di Profio F., Melegari I., Di Felice E., Robetto S., Guidetti C., Orusa R., Martella V., Marsilio F.2015. Molecular detection of kobuviruses in European roe deer (Capreolus capreolus) in Italy. Arch. Virol. 160: 2083–2086. doi: 10.1007/s00705-015-2464-5 [DOI] [PubMed] [Google Scholar]
- 7.Di Martino B., Di Profio F., Melegari I., Marsilio F., Martella V.2015. Detection of feline kobuviruses in diarrhoeic cats, Italy. Vet. Microbiol. 176: 186–189. doi: 10.1016/j.vetmic.2015.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Martino B., Di Profio F., Melegari I., Robetto S., Di Felice E., Orusa R., Marsilio F.2014. Molecular evidence of kobuviruses in free-ranging red foxes (Vulpes vulpes). Arch. Virol. 159: 1803–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frisk A. L., König M., Moritz A., Baumgärtner W.1999. Detection of canine distemper virus nucleoprotein RNA by reverse transcription-PCR using serum, whole blood, and cerebrospinal fluid from dogs with distemper. J. Clin. Microbiol. 37: 3634–3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geng Y., Guo D., Li C., Wang E., Wei S., Wang Z., Yao S., Zhao X., Su M., Wang X., Wang J., Wu R., Feng L., Sun D.2015. Co-circulation of the rare CPV-2c with unique Gln370Arg substitution, new CPV-2b with unique Thr440Ala substitution, and new CPV-2a with high prevalence and variation in Heilongjiang province, northeast China. PLOS ONE 10: e0137288. doi: 10.1371/journal.pone.0137288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gouvea V., Glass R. I., Woods P., Taniguchi K., Clark H. F., Forrester B., Fang Z. Y.1990. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J. Clin. Microbiol. 28: 276–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu R. L., Huang G., Qiu W., Zhong Z. H., Xia X. Z., Yin Z.2001. Detection and differentiation of CAV-1 and CAV-2 by polymerase chain reaction. Vet. Res. Commun. 25: 77–84. doi: 10.1023/A:1006417203856 [DOI] [PubMed] [Google Scholar]
- 13.Kapoor A., Simmonds P., Dubovi E. J., Qaisar N., Henriquez J. A., Medina J., Shields S., Lipkin W. I.2011. Characterization of a canine homolog of human Aichivirus. J. Virol. 85: 11520–11525. doi: 10.1128/JVI.05317-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khamrin P., Maneekarn N., Okitsu S., Ushijima H.2014. Epidemiology of human and animal kobuviruses. Virusdisease 25: 195–200. doi: 10.1007/s13337-014-0200-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee M. H., Jeoung H. Y., Lim J. A., Song J. Y., Song D. S., An D. J.2012. Kobuvirus in South Korean black goats. Virus Genes 45: 186–189. doi: 10.1007/s11262-012-0745-6 [DOI] [PubMed] [Google Scholar]
- 16.Li L., Victoria J. G., Wang C., Jones M., Fellers G. M., Kunz T. H., Delwart E.2010. Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses. J. Virol. 84: 6955–6965. doi: 10.1128/JVI.00501-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oem J. K., Choi J. W., Lee M. H., Lee K. K., Choi K. S.2014. Canine kobuvirus infections in Korean dogs. Arch. Virol. 159: 2751–2755. doi: 10.1007/s00705-014-2136-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olarte-Castillo X. A., Heeger F., Mazzoni C. J., Greenwood A. D., Fyumagwa R., Moehlman P. D., Hofer H., East M. L.2015. Molecular characterization of canine kobuvirus in wild carnivores and the domestic dog in Africa. Virology 477: 89–97. doi: 10.1016/j.virol.2015.01.010 [DOI] [PubMed] [Google Scholar]
- 19.Park S. J., Kim H. K., Moon H. J., Song D. S., Rho S. M., Han J. Y., Nguyen V. G., Park B. K.2010. Molecular detection of porcine kobuviruses in pigs in Korea and their association with diarrhea. Arch. Virol. 155: 1803–1811. doi: 10.1007/s00705-010-0774-1 [DOI] [PubMed] [Google Scholar]
- 20.Pratelli A., Decaro N., Tinelli A., Martella V., Elia G., Tempesta M., Cirone F., Buonavoglia C.2004. Two genotypes of canine coronavirus simultaneously detected in the fecal samples of dogs with diarrhea. J. Clin. Microbiol. 42: 1797–1799. doi: 10.1128/JCM.42.4.1797-1799.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reuter G., Boldizsár A., Kiss I., Pankovics P.2008. Candidate new species of Kobuvirus in porcine hosts. Emerg. Infect. Dis. 14: 1968–1970. doi: 10.3201/eid1412.080797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reuter G., Boros A., Pankovics P., Egyed L.2010. Kobuvirus in domestic sheep, Hungary. Emerg. Infect. Dis. 16: 869–870. doi: 10.3201/eid1605.091934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saitou N., Nei M.1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425. [DOI] [PubMed] [Google Scholar]
- 24.Smits S. L., Raj V. S., Oduber M. D., Schapendonk C. M., Bodewes R., Provacia L., Stittelaar K. J., Osterhaus A. D., Haagmans B. L.2013. Metagenomic analysis of the ferret fecal viral flora. PLoS ONE 8: e71595. doi: 10.1371/journal.pone.0071595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soma T., Ohinata T., Ishii H., Takahashi T., Taharaguchi S., Hara M.2011. Detection and genotyping of canine coronavirus RNA in diarrheic dogs in Japan. Res. Vet. Sci. 90: 205–207. doi: 10.1016/j.rvsc.2010.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soma T., Saito N., Kawaguchi M., Kawashima S., Osada H.2011. Prevalence of anti-canine distemper virus IgM antibodies during 1997 to 2007 in Japan. J. Enviro. Dis. 20: 1–6. [Google Scholar]
- 27.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S.2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. doi: 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamashita T., Adachi H., Hirose E., Nakamura N., Ito M., Yasui Y., Kobayashi S., Minagawa H.2014. Molecular detection and nucleotide sequence analysis of a new Aichi virus closely related to canine kobuvirus in sewage samples. J. Med. Microbiol. 63: 715–720. doi: 10.1099/jmm.0.070987-0 [DOI] [PubMed] [Google Scholar]
- 29.Yamashita T., Ito M., Kabashima Y., Tsuzuki H., Fujiura A., Sakae K.2003. Isolation and characterization of a new species of kobuvirus associated with cattle. J. Gen. Virol. 84: 3069–3077. doi: 10.1099/vir.0.19266-0 [DOI] [PubMed] [Google Scholar]
- 30.Yamashita T., Kobayashi S., Sakae K., Nakata S., Chiba S., Ishihara Y., Isomura S.1991. Isolation of cytopathic small round viruses with BS-C-1 cells from patients with gastroenteritis. J. Infect. Dis. 164: 954–957. doi: 10.1093/infdis/164.5.954 [DOI] [PubMed] [Google Scholar]
- 31.Yamashita T., Sakae K., Tsuzuki H., Suzuki Y., Ishikawa N., Takeda N., Miyamura T., Yamazaki S.1998. Complete nucleotide sequence and genetic organization of Aichi virus, a distinct member of the Picornaviridae associated with acute gastroenteritis in humans. J. Virol. 72: 8408–8412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zourkas E., Ward M. P., Kelman M.2015. Canine parvovirus in Australia: A comparative study of reported rural and urban cases. Vet. Microbiol. 181: 198–203. doi: 10.1016/j.vetmic.2015.10.009 [DOI] [PubMed] [Google Scholar]