Abstract
Reverse transcription quantitative-polymerase chain reaction (RT-qPCR) is a routine method for gene expression analysis, and reliable results depend on proper normalization by stable reference genes. Caloric restriction (CR) is a robust lifestyle intervention to slow aging and delay onset of age-associated diseases via inducing global changes in gene expression. Reliable normalization of RT-qPCR data becomes crucial in CR studies. In this study, the expression stability of 12 candidate reference genes were evaluated in inguinal white adipose tissue (iWAT), skeletal muscle (Sk.M) and liver of CR mice by using three algorithms, geNorm, NormFinder, and Bestkeeper. Our results showed β2m, Ppia and Hmbs as the most stable genes in iWAT, Sk.M and liver, respectively. Moreover, two reference genes were sufficient to normalize RT-qPCR data in each tissue and the suitable pair of reference genes was β2m-Hprt in iWAT, Ppia-Gusb in Sk.M and Hmbs-β2m in liver. By contrast, the least stable gene in iWAT or Sk.M was Gapdh, and in liver was Pgk1. Furthermore, the expression of Leptin and Ppar-γ were profiled in these tissues to validate the selected reference genes. Our data provided a basis for gene expression analysis in future CR studies.
Quantification of gene expression has become a staple in most molecular biological laboratories to understand the complex signaling networks that regulate different responses observed under various conditions. Reverse transcription quantitative real-time polymerase chain reaction (RT-qPCR) has become the most prevalent and powerful technique applied to quantify gene expression because of its simple, sensitive, accurate, reproducible and high-throughput features1. RT-qPCR gives a rapid means of understanding gene transcriptional level variation under various conditions. There are two methods of presenting quantitative gene expression - the absolute and relative quantification methods1,2. For researchers, it is often necessary to know not the actual copy number of gene but the expression change of gene, so that, relative quantification method is more common for gene expression assessment1,3. However, relative quantification presents qPCR data of target genes relying on internal control genes as reference4.
Reference genes with stable expression levels are essential as an internal control to normalize and monitor the variations of interested gene expression among different treatments or conditions5,6. The ideal reference genes should be expressed at constant levels regardless of experimental conditions, cell types, tissues, developmental stages or stress treatments4,7, and should have expression levels comparable to that of the target gene8. Nevertheless, increasing evidences suggest that the expression of reference genes often varies considerably under different experimental conditions as reviewed by Suzuki and Kozera9,10. Thus, identification of reliable reference genes is a prerequisite for qRT-PCR experiments. However, it has been ignored in most of the RT-qPCR studies that it is necessary to validate the stability of the reference gene under determined experimental condition to ensure proper normalization and a robust RT-qPCR analysis6,10,11.
Caloric restrictions (CR) is an efficient non-genetic intervention to slow aging, improve health, reduce most risk factors for chronic diseases and delay onset of age-associated diseases in a fashion that is evolutionarily conserved from yeast to primates and humans12,13,14,15,16. CR causes global changes in gene expressions which are the molecular basis of the beneficial effects of CR15,17,18. To decipher the mechanism of CR further and achieve the beneficial effects of CR easier, it’s necessary and important to analyze the changes of gene transcriptional level. The work in rat cortex and hippocampus showed that CR significantly influenced expression of 18S rRNA and cyclophilin B19. However, to date, appropriate reference genes have not been identified in case of CR in other tissues or organs, especially those tissues which are closely related to metabolism and have extensively response to CR, such as liver, skeletal muscle and white adipose tissue16,20,21. It has been reported that in adipose tissue or adipocyte, some of the commonly-used reference genes, for instance glyceraldehyde-3-phosphate dehydrogenase (GAPDH), can be regulated by some treatments, such as insulin or obesity22,23,24. Thus, it is essential to evaluate the stability of reference genes in these tissues after CR.
Several software packages have been established to identify reference genes with stable expression levels, such as geNorm7, NormFinder25, and BestKeeper26. With the help of these statistical algorithms, a number of studies on validation of reference genes have been reported27,28,29. In this study, we performed the first comprehensive study of CR effects in inguinal white adipose tissue (iWAT), skeletal muscle (Sk.M) and liver on twelve commonly-used reference genes involved in different biological functions9,27,30,31,32,33,34 (Table 1), including the formation of cellular cytoskeleton (beta-actin (Actb) and alpha-tubulin (Tuba)), protein biosynthesis (hydroxymethylbilane synthase (Hmbs), peptidylprolyl isomerase A (Ppia) and ribosomal protein L13A (Rpl13a)), immune response (beta-2 microglobulin (β2m)), metabolism (Gapdh, beta-glucuronidase (Gusb) and phosphoglycerate kinase 1(Pgk1)), nucleotide synthesis (hypoxanthine guanine phosphoribosyl transferase (Hprt)), transcription (TATA box binding protein (Tbp)) and signal transduction (tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta-polypeptide (Ywhaz)). This study provides a basis for the selection of reference genes and useful guidelines for future gene expression studies of CR and metabolism.
Table 1. Gene symbols, gene names, accession numbers and functions of the twelve reference genes investigated by RT-qPCR.
| Gene Symbol | Gene Name | Accession Number | Function |
|---|---|---|---|
| Actb | beta-actin | NM_007393.5 | Cytoskeletal structural protein |
| β2m | beta-2 microglobulin | NM_009735.3 | Beta-chain of MHC class I molecules |
| Gapdh | glyceraldehyde-3-phosphate dehydrogenase | NM_008084.3 | Involved in glycolysis and gluconeogenesis |
| Gusb | beta-glucuronidase | NM_010368.1 | Lysosomal exoglycosidase |
| Hmbs | hydroxymethylbilane synthase | NM_013551.2 | Heme synthesis and porphyrin metabolism |
| Hprt | hypoxanthine guanine phosphoribosyl transferase | NM_013556.2 | Purine synthesis through the purine salvage pathway |
| Pgk1 | phosphoglycerate kinase 1 | NM_008828.3 | A glycolytic enzyme |
| Ppia | peptidylprolyl isomerase A | NM_008907.1 | Protein coding, a cyclosporin binding-protein |
| Rpl13a | ribosomal protein L13A | NM_009438.5 | Sturcturalconstituent of ribosome |
| Tbp | TATA box binding protein | NM_013684.3 | Transcription initiation from RNA polymerase II, general transcription factor |
| Tuba | alpha-tubulin | NM_011653.2 | Microtubules of the eukaryotic cytoskeleton |
| Ywhaz | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta-polypeptide | NM_011740.3 | Protein domain in specific binding |
Materials and Methods
Ethics statement
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Biomedical Ethics Committee of Beijing Hospital and Beijing Institute of Geriatrics, Beijing, China. Staff veterinarians monitored mice on a regular basis, finding no pathogens. All surgeries were performed under 10% chloral hydrate anesthesia, and all efforts were made to minimize suffering.
Animals
Male C57BL/6 mice were purchased from the Vital River (Charles River China) at two months of age. After a one-week acclimation, all mice were randomly assigned to two groups and treated for 3 months: ad libitum (AL) fed mice were given uninhibited access to normal chow diet; caloric restriction (CR) mice were fed as 65~70% of the AL feeding rate15,35. They were housed individually. All the animals were housed at 21 °C in a 12-h light/12-h dark cycle. We recorded the body weight and food intake once a week during the study (Supplementary Fig. S1).
IPGTT and IPITT
For intra-peritoneal glucose tolerance test (IPGTT), following an overnight fast, the mice were injected intraperitoneally of glucose at a dose of 1 g/kg body weight (10% glucose in PBS, 100 ul/10 g body weight)36. For intra-peritoneal insulin tolerance test (IPITT), following a 4 h fasting, mice were injected intraperitoneally of insulin at a dose of 0.75 IU/kg body weight (0.075IU/ml in PBS, 100 ul/10 g body weight)36. Blood glucose levels were measured from tail blood using glucose strips at 0 (basal), 30, 60 and 120 min after injection by One Touch Ultra glucose strips (LifeScan, Inc, USA).
Tissue preparation
Mice were sacrificed according to the approved protocol. The liver, inguinal white adipose tissue (iWAT) and skeletal muscle (Sk.M) from hind legs were isolated, snap frozen in liquid nitrogen, ground into powder with mortar and pestle in liquid nitrogen and then stored at −80 °C.
Total RNA extraction
Total RNA was isolated from 10–80 mg tissues using TRIzol® reagent (Invitrogen Life Technologies, USA) following the manufacturer’s instructions. The residual DNA was removed by TURBO DNA free kit (Ambion Inc., UK). Yield and purity of RNA was determined by NanoDrop ND-1000 spectrophotometer (Nanodrop technologies, USA). RNA samples with an absorbance ratio OD 260/280 between 1.9–2.2 and OD 260/230 greater than 2.0 were used for further analysis. RNA integrity was assessed using agarose gel electrophoresis.
Reverse transcription cDNA synthesis
First-strand cDNA was synthesized from 2 μg of total RNA with random hexamer oligonucleotide primers using a 20 μl reverse transcription system (New England Biolabs, USA). cDNA was stored at −20 °C for future use. For qPCR analysis, each cDNA sample was diluted 20 times with nuclease free water.
Real-time PCR
Real-time PCRs were conducted in Bio-Rad iQ5 Real- Time System. For each reaction, the 20 μl mixture contained 1 μl of diluted cDNA, 5 pmol each of the forward and reverse primers, and 10 μl 2 × SYBR Premix Ex Taq II (Takara Bio Inc, Japan). The amplification program was as follows: 95 °C for 30 s, 40 cycles at 95 °C for 5 s and 60 °C for 30 s. After amplification, a thermal denaturing cycle was added to derive the dissociation curve of the PCR product to verify amplification specificity (Supplementary Fig. S1). Reactions for each sample were carried out in triplicate.
qPCR efficiencies in the exponential phase were calculated for each primer pair using standard curves (5 tenfold serial dilutions of pooled cDNA that included equal amounts from the samples set), the mean threshold cycle (Ct) values for each serial dilution were plotted against the logarithm of the cDNA dilution factor and calculated according to the equation E = 10(−1/slope) × 10011, where the slope is the gradient of the linear regression line. The linear dynamic range was determined by the standard curve and correlation coefficients (R2) for each gene as reported.
Primers
Twelve candidate reference genes were selected based on their common usage as endogenous control genes in previous studies. The candidate genes were Actb, β2m, Gapdh, Gusb, Hmbs, Hprt, Pgk1, Ppia, Rpl13a, Tbp, Tuba, Ywhaz. The primers were designed from nucleotide sequences identified using NCBI BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Primers were ordered from Life Technologies with their certificates of analysis. The primer characteristics of nominated reference genes are listed in Table 2. The primers for validation of selected reference genes are as follows: Leptin-F, GTGGCTTTGGTCCTATCTGTC and Leptin-R, CGTGTGTGAAATGTCATTGATCC; Ppar-γ (peroxisome proliferator-activated receptor gamma) -F, TGTCGGTTTCAGAAGTGCCTTG and Ppar-γ-R, TTCAGCTGGTCGATATCACTGGAG.
Table 2. List of reference genes investigated by RT-qPCR.
| Gene Symbol | Primer Sequence | Amplicon Size | Span Intron | Efficiency% | R2 |
|---|---|---|---|---|---|
| Actb | F:CCTTCTTGGGTATGGAATCCTGT R:CACTGTGTTGGCATAGAGGTCTTTAC | 101 | Y (E4-E5) | 98 | 0.9997 |
| β2m | F:CATGGCTCGCTCGGTGAC R:CAGTTCAGTATGTTCGGCTTCC | 135 | Y (E1-E2) | 101 | 0.9994 |
| Gapdh | F:TGCACCACCAACTGCTTAG R:GGATGCAGGGATGATGTTC | 177 | Y (E4-E5) | 101 | 0.9999 |
| Gusb | F:CCGACCTCTCGAACAACCG R:GCTTCCCGTTCATACCACACC | 169 | Y (E1-E2) | 104 | 0.9992 |
| Hmbs | F:ATGAGGGTGATTCGAGTGGG R:TTGTCTCCCGTGGTGGACATA | 134 | Y (E2-E4) | 99 | 0.9999 |
| Hprt | F:TGACACTGGCAAAACAATGCA R:GGTCCTTTTCACCAGCAAGCT | 95 | Y (E6-E7) | 107 | 1 |
| Pgk1 | F:ATGTCGCTTTCCAACAAGCTG R:GCTCCATTGTCCAAGCAGAAT | 164 | Y (E1-E3) | 99 | 0.9948 |
| Ppia | F:GGCAAATGCTGGACCAAAC R:CATTCCTGGACCCAAAACG | 149 | Y (E4-E5) | 105 | 0.9996 |
| Rpl13a | F:AGGGGCAGGTTCTGGTATTG R:TGTTGATGCCTTCACAGCGT | 120 | Y (E1-E3) | 101 | 0.9989 |
| Tbp | F:CCTTGTACCCTTCACCAATGAC R:ACAGCCAAGATTCACGGTAGA | 119 | Y (E3-E4) | 96 | 0.9913 |
| Tuba | F:TGTCCTGGACAGGATTCGC R:CTCCATCAGCAGGGAGGTG | 115 | Y (E3-E4) | 104 | 1 |
| Ywhaz | F:GAAAAGTTCTTGATCCCCAATGC R:TGTGACTGGTCCACAATTCCTT | 134 | Y (E3-E4) | 101 | 0.9999 |
Primers sequences, length of the RT-qPCR transcripts, efficiency of each pair of primers and the correlation coefficients (R2) of the standard curve are indicated.
Analysis of gene stability and minimum number of reference genes required
To assess the stability of candidate reference genes, three widely recognized reference gene normalization algorithms each with a unique advantage were used, geNorm7, NormFinder25, and BestKeeper26. Ct values were converted to non-normalized relative quantities according to the formula: 2−ΔCt (ΔCt = the corresponding Ct value – minimum Ct)7. geNorm and NormFinder calculations are based on these converted quantities; raw Cq values were directly analyzed by BestKeeper. The brief usage and characteristics of each algorithm are as follows:
GeNorm7 ranked the reference genes based on their M value. The lower the M value, the higher is the expression stability and the M value less than 1.5 is recommended to identify stably expressed gene. The two most stable reference genes are determined by step-wise exclusion of the least stable gene. It also compares the pairwise variation (V) of these genes with others. The V value of Vn/Vn + 1 between two sequential normalization factors was used to determine the optimal number of reference genes required for better normalization. A threshold value below 0.15 suggests the requirement of no additional reference gene for normalization.
NormFinder25 estimates both intra- and inter-group variations of gene expression and then combines the two to produce a stability value for all the samples in any number of groups. Genes with the lowest rank are considered to be most stably expressed and are ideal to select as reference gene(s) for that particular experimental condition.
BestKeeper is used to determine standard deviation and power of each reference gene, and then to select the best reference genes based on these variables26. The BestKeeper index is calculated from the geometric mean of the candidates Ct values for each specific sample. In BestKeeper, SDCt value < 1 indicates genes which are stably expressed. The most stable reference genes are the ones with the lowest SD values and highest coefficients of correlation with the BestKeeper index.
Validation of reference genes
To confirm the reliability of the reference genes, the relative expression profiles of Leptin and Ppar-γ genes were determined and normalized with the recommended gene pair, most and least stable gene, respectively. Relative fold changes in gene expression were calculated using the comparative 2−ΔΔCt method and normalized to the corresponding reference gene levels1,37.
Statistical analysis
Data are expressed as mean ± standard error (s.e.m). Means of different groups were compared and analyzed using the Student’s t-test. Differences were reported as statistically significant when p < 0.05. GraphPad Prism 5 (GraphPad Software, USA) was used for statistical procedures and graph plotting.
Results
Establishment of CR mice model
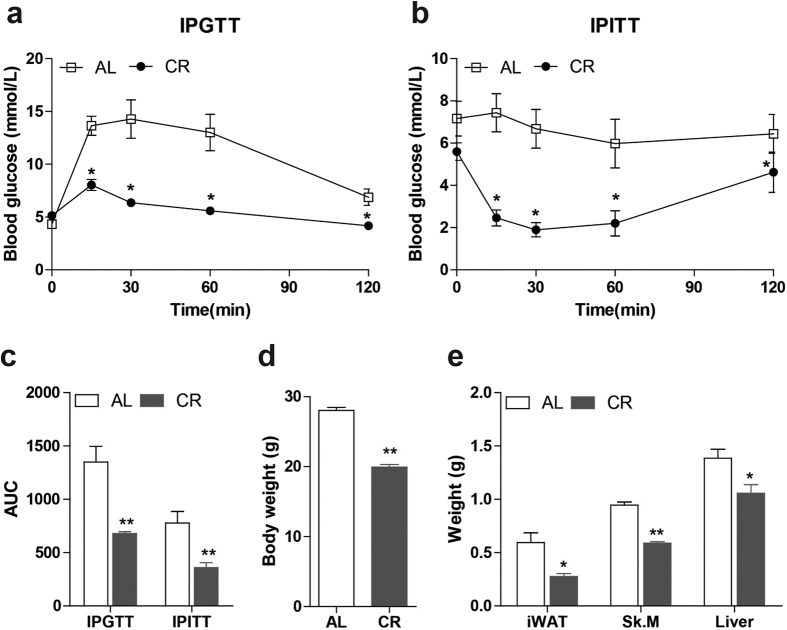
After treatment for 3 months, the body weight in CR group (19.94 ± 0.38 g) is significantly lower (p < 0.0001) than in AL group (28.03 ± 0.43 g) and is only 71.1% as in AL group (Fig. 1d). Since insulin sensitivity increase is one of the important effects of CR14,38,39, the establishment of CR was evaluated through assessing glucose and insulin homeostasis. We performed intraperitoneal glucose tolerance test (IPGTT) and insulin tolerance test (IPITT). Both glucose tolerance and insulin tolerance improved in CR mice relative to AL (Fig. 1a and b). Both of the area under the curve (AUC) measurements were significantly reduced in CR group (Fig. 1c. p = 0.002 and 0.008, respectively). Besides, the weight of iWAT, Sk.M and liver of CR mice reduced to 46.5% (p = 0.01), 62.2% (p < 0.0001) and 76.3% (p = 0.02) as AL mice, respectively. These results indicated that the CR model was established successfully.
Figure 1. The assessment of CR model establishment.
Glucose measurements (mmol/l) at 0, 30, 60 and 120 min after glucose (a) or insulin intraperitoneal injection (b) in mice after CR for 3 months. (c): AUC of IPGTT or IPITT was calculated accordingly. Weight of whole body (d), iWAT, Sk.M and liver (e) in both groups was measured after CR for 3 months. (*p < 0.05, **p < 0.01 vs AL. n = 5 in each group) iWAT: inguinal white adipose tissue, Sk. M: skeletal muscle, AUC: area under curve, IPGTT: intraperitoneal glucose tolerance test, IPITT: intraperitoneal insulin tolerance test, AL: ad libitum, CR: caloric restriction.
Expression profiles of candidate reference genes
Twelve candidate reference genes were selected for this study. They are widely used and recognized reference genes, which have been described in the literatures9,27,30,31,32,33,34 and represent several functional classes to minimize the possibility of co-regulation. The performance of each amplification primer set was tested by RT-qPCR. Their respective PCR amplification efficiencies were calculated as the first step. The amplification efficiency (E) of all reactions ranged from 96% to 107%; and the correlation coefficients (R2) of the standard curve varied from 0.9913 to 0.9999 (Table 2).
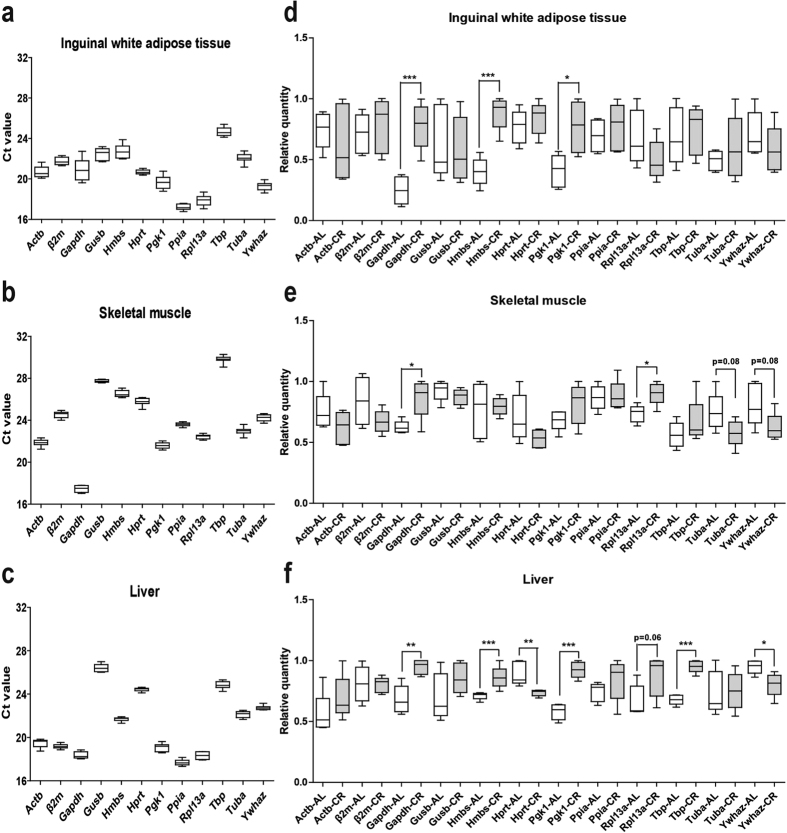
The expression profiles of candidate reference genes in all the three tissues of both groups were investigated (Fig. 2). The individual reference genes had different expression ranges across samples in each tissue. The expression levels of the candidate reference genes were determined by Ct values through RT-qPCR experiments. Ct is the amplification cycle number at which the fluorescence signal of the reporter dye reaches above an arbitrarily placed baseline threshold1. The distribution of the Ct values for each of the reference genes in different tissues is displayed by box plot. All the Ct values are in suitable range: 17.23 ± 0.10 to 24.67 ± 0.14 (standard deviation, SD is 0.30~1.07) in iWAT (Fig. 2a), 17.45 ± 0.11 to 29.81 ± 0.10 (SD is 0.18~0.35) in Sk.M (Fig. 2b) and 17.69 ± 0.08 to 26.42 ± 0.11 (SD is 0.18~0.38) in liver (Fig. 2c).
Figure 2. Ct values and relative quantities of 12 candidate reference genes in all samples of each tissue of CR mice.
Raw Ct values and relative quantities (without normalization to any reference genes) of ten samples of inguinal white adipose tissue (a and d), skeletal muscle (b and e) or liver (c and f), including five AL and five CR mice were described using a box and whiskers plot. The boxes encompass the 25th to 75th percentiles, and the line across the box is the median. Whisker caps denote the maximum and minimum values. The non-normalized relative quantities were calculated by dCt method with iQ5 software (*p < 0.05, **p < 0.01 and ***p < 0.001 vs AL).
We measured non-normalized relative quantity (the raw expression without normalization to any reference gene) of our 12 genes (Fig. 2d–f) in iWAT, Sk.M and liver. Among the tested genes, we observed variations between AL and CR mice within all three tissues: there are significant differences in three genes (Gapdh, Hmbs and Hprt (p < 0.05)) in iWAT (Fig. 2d) and two genes (Gapdh and Rpl13a (p < 0.05)) in Sk.M (Fig. 2e), and more candidate reference genes were changed by CR in liver, including Gapdh, Hmbs Hprt, Rpl13, Tbp and Ywhaz (p < 0.05) (Fig. 2f). These results clearly demonstrate variations in many commonly-used reference genes, but also differences between different tissues changed by CR. It also indicates that one reference gene is usually not enough, and normalization to multiple reference genes should be done for each tissue during CR. To address this question, we used three different methods: geNorm, Normfinder and BestKeeper algorithms.
Effect of CR on mRNA expression of candidate reference genes
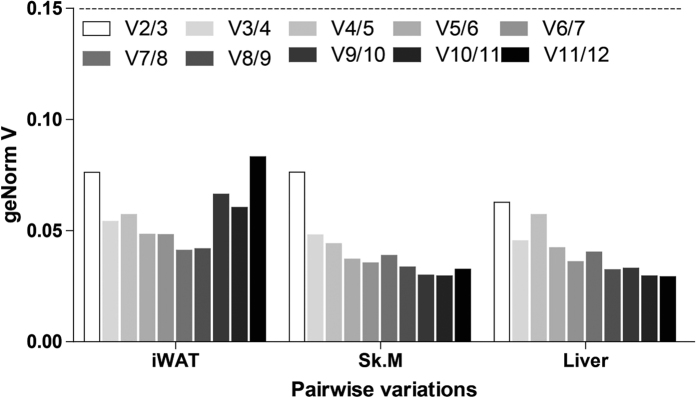
The candidate reference genes were ranked based on their gene expression stability between CR and AL treatments assessed by employing three statistical algorithms. All the geNorm V values generated by multiple analyses of pairwise variations between the twelve reference genes were under the 0.15 threshold in all these tissues (Fig. 3). It means that at least two reference genes among them are enough for normalization in RT-qPCR in each tissue. According to the geNorm algorithm, all candidate reference genes, regardless of which tissue, meet the programs default limit of M < 1.5 (0.251~1.024) (Table 3). Moreover, when considering the results of the BestKeeper algorithm, in which SD (±Ct) value < 1 (0.099~0.869) (Table 3) indicates that genes are stably expressed, these selected gene also were considered stable. However, the ranks order of the reference genes in each tissue showed similar trends with subtle variations by different software, which might be attributed to the differences across algorithms. For the purpose of clarity in depiction, we dealt separately the ranking of the reference genes in each tissue generated by different statistical method.
Figure 3. Pairwise variation (V) of candidate reference genes calculated by geNorm.
Pairwise variation (Vn/n + 1) was analyzed to determine the optimal number of reference genes. Two reference genes are enough in all the detected tissues to normalize RT-qPCR data as the pairwise variation V value for V 2/3 (n/n + 1) is under 0.15. iWAT: inguinal white adipose tissue, Sk. M: skeletal muscle.
Table 3. Candidate reference gene expression stability ranked by geNorm, NormFinder, and BestKeeper in iWAT, Sk.M and liver of CR mice.
| Tissue | Rank | geNorm |
NormFinder |
BestKeeper |
Comprehensive ranking by geoMean | ||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | M | Gene | Stability value | Gene | CV (% Ct) | SD (±Ct) | |||
| iWAT | 1 | Gapdh | 1.024 | Gapdh | 0.616 | Gapdh | 4.158 | 0.869 | Gapdh |
| 2 | Hmbs | 0.694 | Hmbs | 0.400 | Pgk1 | 2.498 | 0.492 | Hmbs | |
| 3 | Pgk1 | 0.653 | Rpl13a | 0.340 | Hmbs | 2.442 | 0.555 | Pgk1 | |
| 4 | Actb | 0.579 | Pgk1 | 0.323 | Actb | 2.330 | 0.483 | Actb | |
| 5 | Rpl13a | 0.557 | Actb | 0.321 | Gusb | 2.197 | 0.493 | Rpl13a | |
| 6 | Gusb | 0.547 | Ywhaz | 0.286 | Rpl13a | 2.173 | 0.389 | Gusb | |
| 7 | Ywhaz | 0.505 | Gusb | 0.278 | Ywhaz | 1.620 | 0.313 | Ywhaz | |
| 8 | Tuba | 0.490 | Tuba | 0.163 | Tbp | 1.542 | 0.380 | Tuba | |
| 9 | Hprt | 0.460 | Tbp | 0.134 | Ppia | 1.527 | 0.263 | Tbp | |
| 10 | Ppia | 0.447 | Hprt | 0.134 | Tuba | 1.505 | 0.332 | Ppia | |
| 11 | Tbp | 0.446 | Ppia | 0.129 | β2m | 1.484 | 0.323 | Hprt | |
| 12 | β2m | 0.419 | β2m | 0.104 | Hprt | 0.847 | 0.175 | β2m | |
| Sk.M | 1 | Gapdh | 0.437 | Gapdh | 0.232 | Gapdh | 1.657 | 0.290 | Gapdh |
| 2 | Hmbs | 0.380 | Hprt | 0.189 | Ywhaz | 1.187 | 0.288 | Hprt | |
| 3 | Hprt | 0.374 | Rpl13a | 0.172 | Tuba | 1.070 | 0.246 | Tuba | |
| 4 | Pgk1 | 0.347 | Tuba | 0.171 | Actb | 1.035 | 0.226 | Ywhaz | |
| 5 | β2m | 0.345 | Ywhaz | 0.169 | β2m | 1.018 | 0.250 | β2m | |
| 6 | Rpl13a | 0.343 | β2m | 0.158 | Hprt | 0.968 | 0.250 | Pgk1 | |
| 7 | Tuba | 0.334 | Pgk1 | 0.155 | Pgk1 | 0.953 | 0.206 | Rpl13a | |
| 8 | Tbp | 0.332 | Tbp | 0.153 | Hmbs | 0.855 | 0.227 | Hmbs | |
| 9 | Ywhaz | 0.327 | Actb | 0.152 | Rpl13a | 0.750 | 0.168 | Actb | |
| 10 | Actb | 0.324 | Hmbs | 0.151 | Tbp | 0.690 | 0.206 | Tbp | |
| 11 | Gusb | 0.268 | Gusb | 0.057 | Ppia | 0.511 | 0.121 | Gusb | |
| 12 | Ppia | 0.251 | Ppia | 0.040 | Gusb | 0.357 | 0.099 | Ppia | |
| Liver | 1 | Gusb | 0.414 | Pgk1 | 0.204 | Pgk1 | 1.776 | 0.341 | Pgk1 |
| 2 | Actb | 0.392 | Hprt | 0.204 | Rpl13a | 1.624 | 0.298 | Actb | |
| 3 | Hprt | 0.384 | Ywhaz | 0.200 | Actb | 1.483 | 0.288 | Gusb | |
| 4 | Tuba | 0.372 | Tuba | 0.158 | Gapdh | 1.405 | 0.258 | Hprt | |
| 5 | Pgk1 | 0.370 | Gapdh | 0.153 | Ppia | 1.090 | 0.193 | Tuba | |
| 6 | Ywhaz | 0.368 | Actb | 0.151 | Tuba | 1.069 | 0.236 | Rpl13a | |
| 7 | Rpl13a | 0.343 | Gusb | 0.150 | Gusb | 1.016 | 0.268 | Gapdh | |
| 8 | Gapdh | 0.325 | Rpl13a | 0.139 | Tbp | 0.983 | 0.245 | Ywhaz | |
| 9 | Ppia | 0.307 | Tbp | 0.132 | β2m | 0.767 | 0.147 | Ppia | |
| 10 | β2m | 0.306 | β2m | 0.115 | Hmbs | 0.611 | 0.133 | Tbp | |
| 11 | Tbp | 0.304 | Ppia | 0.083 | Ywhaz | 0.602 | 0.137 | β2m | |
| 12 | Hmbs | 0.276 | Hmbs | 0.059 | Hprt | 0.579 | 0.141 | Hmbs | |
The comprehensive ranking was based on the geometric mean of geNorm, NormFinder, and BestKeeper results. Candidates are listed from top to bottom in order of increasing expression stability. (SD (±Ct): standard deviation of the Ct; CV (% Ct): coefficient of variance expressed as a percentage of the Ct level; iWAT: inguinal white adipose tissue; Sk.M: skeletal muscle; geoMean: geometrical mean).
In inguinal white adipose tissue
In iWAT, it is showed that the best genes to use as reference genes were β2m > Tbp > Ppia recommended by geNorm, β2m > Ppia > Hprt by NormFinder and Hprt > β2m > Tuba by BestKeeper (Table 3). These genes display the same profile of expression between CR and AL (Fig. 2d). On the other hand, the least stable genes were Gapdh > Hmbs > Pgk1 showed by geNorm, Gapdh > Hmbs > Rpl13a by NormFinder and Gapdh > Pgk1 > Hmbs by BestKeeper (Table 3). While reference genes ranking varied slightly by algorithm, the overall ranking of the best candidate reference genes were calculated through geometric means of the three ranking numbers and the most stable reference genes had the smallest geometric mean32,40,41 (Table 3). The comprehensive ranking showed that β2m and Hprt appear to be the most suitable reference gene pair in iWAT of CR. Besides, the worst reference gene is Gapdh, the most stable one is β2m.
In skeletal muscle
The same method was applied in skeletal muscle. All the three algorithms showed that Ppia and Gusb is the best pair of reference genes to normalize RT-qPCR data in skeletal muscle after CR treatment (Table 3). There were some differences in the ranking of reference genes between algorithms. However, the comprehensive ranking based on the geometric mean of the ranks by these algorithms32,40,41 showed that Ppia is the most stable gene and Gapdh is the least stable one.
In liver
In liver of CR and AL mice (Table 3), according to the geNorm M value, the best three genes were Hmbs > Tbp > β2m. The results of Normfinder analysis showed that the three most stable reference genes were Hmbs > Ppia > β2m. The results of stable genes by BestKeeper were Hprt > Ywhaz > Hmbs > β2m. Integrating the results of all the three algorithms by geometric mean, the best pair of reference genes to normalize RT-qPCR data in liver after CR treatment appears to be Hmbs and β2m and the most stable gene may be Hmbs. On the other hand, the least stable genes were Gusb > Actb > Hprt analyzed by geNorm, Pgk1 > Hprt > Ywhaz by NormFinder and Pgk1 > Rpl13a > Actb by BestKeeper. The comprehensive ranking showed Pgk1 was the most unstable reference gene.
The overall stability of each gene in all the investigated tissues
To assess the overall stability of each gene in all the investigated tissues after CR, we calculated the comprehensive ranking by geometric mean of the ranks of each gene in all the tissues (Table 4). The results showed that after CR, Tbp and Ppia were relative stable, whereas Gapdh and Pgk1 were changed most in all these tissues.
Table 4. The comprehensive ranking of the candidate reference gene expression stability after CR.
| Gene | Comprehensive ranking by GeoMean | Rank in iWAT | Rank in Sk.M | Rank in liver |
|---|---|---|---|---|
| Gapdh | 1 | 1 | 1 | 7 |
| Pgk1 | 2 | 3 | 6 | 1 |
| Actb | 3 | 4 | 9 | 2 |
| Hprt | 4 | 11 | 2 | 4 |
| Tuba | 5 | 8 | 3 | 5 |
| Hmbs | 6 | 2 | 8 | 12 |
| Gusb | 7 | 6 | 11 | 3 |
| Rpl13a | 8 | 5 | 7 | 6 |
| Ywhaz | 9 | 7 | 4 | 8 |
| β2m | 10 | 12 | 5 | 11 |
| Tbp | 11 | 9 | 10 | 10 |
| Ppia | 12 | 10 | 12 | 9 |
The comprehensive ranking was based on the geometric mean of the gene rank in each tissue. Candidates are listed from top to bottom in order of increasing expression stability. (iWAT: inguinal white adipose tissue; Sk.M: skeletal muscle; geoMean: geometric mean).
Validation of reference genes selection on the target mRNA relative expression
To validate the reliability of selected reference genes, we decided to check them on the expression of genes which expression changed by CR. In previous studies, CR mice showed a marked increase in Ppar-γ16,42,43 and reduced circulating level of Leptin15,44,45. Leptin is mainly produced by adipocyte30,46,47. Studies have also shown leptin production by other tissues, including skeletal muscle48,49. Thus, we checked Leptin expression in Leptin- expressing tissues iWAT and Sk.M, and Ppar-γ expression in all the three tissues by normalized with the recommended gene pair, most and least stable gene, respectively (Fig. 4).
Figure 4.
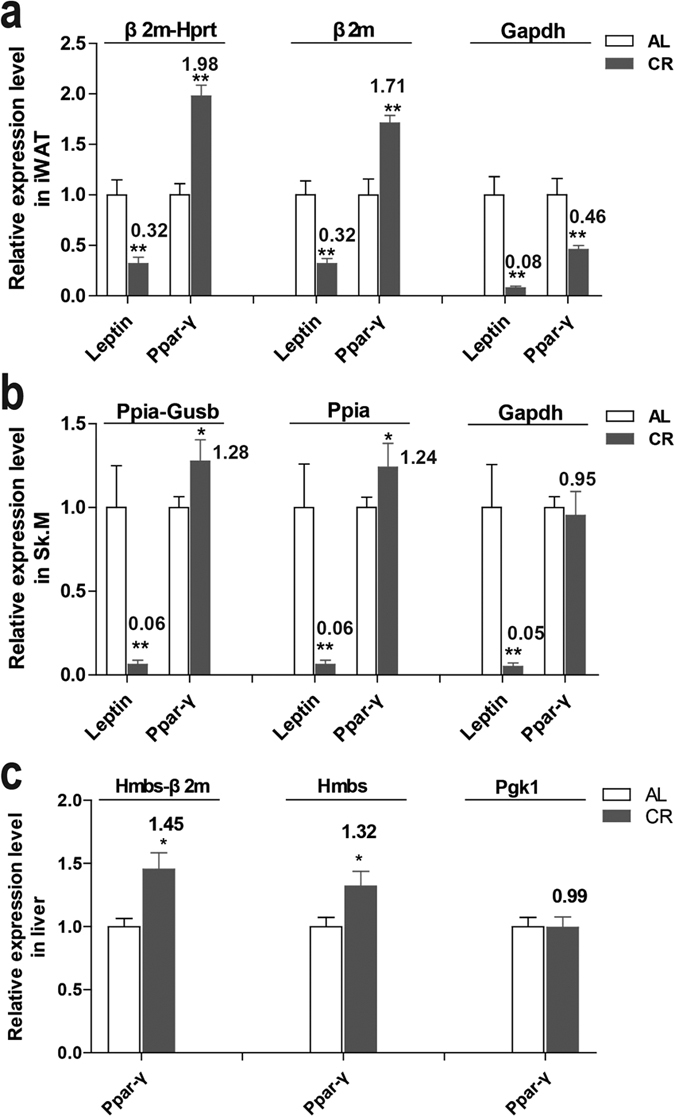
Relative expression level of Leptin and Ppar-γ in inguinal white adipose tissue (a), skeletal muscle (b) and liver (c) of CR and AL mice measured by RT-qPCR and normalized to the indicated reference gene or gene pair. The expression levels of AL mice were set at a relative expression of 1. The fold changes of CR mice were indicated near the bars. PCR reactions for each sample were carried out in triplicate. (*p < 0.05, **p < 0.01 vs AL. n = 5 in each group) AL: ad libitum, CR: caloric restriction.
By using the recommended reference gene pair by the above analysis - β2m-Hprt in iWAT, Ppia-Gusb in Sk.M and Hmbs-β2 m in liver - we observed the significant decrease of Leptin (p < 0.01) and increase of Ppar-γ (p < 0.01 in iWAT, p < 0.05 in Sk.M and liver) after CR (Fig. 4 left). These data confirmed the reliability of the recommended reference gene pair in each tissue.
The normalization results with the single most stable reference gene - β2m in iWAT, Ppia in Sk.M and Hmbs in liver - were similar to those with gene pair. However, the fold changes of Ppar-γ normalized with these single genes were less than with gene pairs (Fig. 4 middle). On the other hand, when using the least stable reference gene – Gapdh in iWAT and Sk.M and Pgk1 in liver (Fig. 4, right), which are also under the threshold of 1.5 on the geNorm M value - Ppar-γ expression were changed to the opposite direction in iWAT (p < 0.05, Fig. 4a) or had no obvious changes in Sk.M (Fig. 4b) or liver (Fig. 4c) (p > 0.05). These results highlight the importance of selection of suitable reference genes combination and efficient normalization for reliable results in gene expression studies in CR.
Discussion
In molecular biological research, gene expression analysis is one of the most frequently-used strategies in the field of gene study. RT-qPCR is the most classical technique in the field of gene expression study. This method requires an appropriate reference gene to normalize mRNA levels. It is vital to select appropriate reference genes for normalization of gene expression. Numbers of reference genes have been used, such as Gapdh and Actb, two commonly-used reference genes9. However, in many cases, these genes have been demonstrated to be variable in different cells, tissues and experimental conditions as reviewed by Suzuki9. For example, Gapdh is not stable in age-induced apoptosis in neurons27, in insulin stimulated adipocytes and hepatoma cell lines22 and in human omental and subcutaneous adipose tissue from obesity and type 2 diabetes patients50.
Therefore, for the first time, we evaluated CR effects in three metabolism related tissues (iWAT, Sk.M and liver) on the expression stability of twelve reference genes (Actb, β2m, Gapdh, Gusb, Hmbs, Hprt, Pgk1, Ppia, Rpl13a, Tbp, Tuba and Ywhaz) by using three different softwares (geNorm, NormFinder and BestKeeper). The most suitable combination of reference genes was recommended for each tissue. We showed that after CR, in all the detected tissues, 1) there are variations of expression for these twelve commonly-used reference genes, 2) at least two reference genes are necessary for good normalization of RT-qPCR data, and 3) each tissue needs its own paired reference genes.
The expression of reference genes can be changed by different experimental conditions or in different tissues9,10. Tanic and colleagues have detected five housekeeping genes, including Actb, Gapdh, Hprt, 18S rRNA and Cytb, in rat cortex and hippocampus after dietary restriction, a kind of CR. They found that the expression of these genes varied under that condition19. It indicates CR can alter some reference genes expression. Their study suggests that it is necessary to evaluate and select appropriate reference genes in CR studies. Furthermore, CR induces extensively responses in all the tissues or organs. There have been quantities of studies on CR, especially in those metabolism related tissues, such as adipose tissues, skeletal muscle and liver51,52. It has been reported that expression of reference genes in these tissues can also be altered under some conditions. For instance, the work by Zhang et al.53 found that four frequently-used reference genes have different expression stability in three types of adipose tissue from obese and lean rats. It was also observed by Svingen et al. that among the six rat tissues they checked following toxicological exposure, reference genes they detected were most unstable in liver54. In pig skeletal muscle, reference genes have variations at multiple developmental stages55. However, no study has been carried out to evaluate suitable reference genes in adipose tissues, skeletal muscle and liver after CR. In the present study, for the first time, we detected the effects of CR on reference genes in these tissues. We did find some unstable reference genes after CR in these tissues. Direct comparison of Ct or non-normalized relative quantity revealed significant difference in several genes in each tissue between CR and AL.
Since the instability of reference genes under different experimental conditions, the use of a single, unverified reference gene when normalizing experiments that attempt to demonstrate small differences in mRNA abundance has been shown to lead to unreliable conclusions6. By using the recommended pair of reference genes, we observed the same changes of Leptin and Ppar-γ after CR as previously reported15,16,42,43,44,45. It is also reported that CR downregulated56 or had no effects on hepatic Ppar-γ expression level57,58. These inconsistent results of Ppar-γ are possible because of the differences in strains, ages, energy restriction degree, duration, etc. as investigated by Mitchell et al.15. However, the results normalized with different reference genes can be discrepancy with different extend29, have no changes27 or even change to opposite direction59. In the current study, we observed all these situations when normalized the data with a single reference gene. Although the advantage of using multiple, validated reference genes was demonstrated as early as 20027, until recently, it is still widespread of using single, unverified reference genes6, an approach that has been demonstrated to cause biased results. The CR related studies are in the same situation. Our data show that it is important to apply at least two reference genes and to check their stability of expression when they are used to normalize RT-qPCR data.
It is interesting to note that there are some similarities among the three tissues: Gapdh and Pgk1, two metabolism related genes, were changed most in all these tissues, whereas Tbp and Ppia, which are not related with metabolism directly, were ranked low in all the three tissue. It might be due to that they have some similarities in functions or responses, such as they are all insulin target tissues and their insulin sensitivity can all be promoted by CR39,52. On the other side, it has been well known that gene expression profile has tissue specific. Tissue specificity also happens in reference genes as show in Table 4. Our study showed that each tissue needs its own specific reference genes. For example, β2m is the most stable gene in iWAT and second stable one in liver but relative unstable in Sk.M. Gapdh is most unstable in both iWAT and Sk.M but is ranked in the middle in liver. The facts that these tissues performed different functions and they are all heterogeneous tissues are two possibilities for the differences of reference genes in these tissues.
Considering that an algorithm is one-sided for evaluating the expression stability of reference genes, many statistical approaches are usually integrated to determine the best reference genes in different experimental conditions27,60. In the present study, we employed three common statistical programs, geNorm, NormFinder, and Bestkeeper. As expected, in each tissue, the stability ranking generated by distinct statistical algorithm is not exactly the same with each other. For example, for Sk.M, Ppia was ranked most stable reference gene by geNorm as well as NormFinder, whereas Gusb was recommended by Bestkeeper. These discrepancies reflect differences between the approaches7,25,26. The geNorm determines gene expression stability based on the principle that the expression ratio of two ideal internal control genes is identical among all test samples7. Co-regulated genes with similar expression profiles will obtain preferential stability ranking from geNorm, leading to an erroneous choice for normalization. On the contrary, the algorithms of NormFinder and Bestkeeper are less sensitive to co-regulation25,26. NormFinder combines both intra- and inter-group variation into a stability value. This model-based approach should provide a more precise and robust estimative of expression variation among subsets composed by different sample types14. BestKeeper15 software was applied as an expression standard of reference genes according to the ranking of the standard deviation (SD (±Ct)) and coefficient of variance (CV (%Ct)) of Ct values, which is inversely proportional to the stability of expression. However, when there are discrepancies among these methods, the recommended genes are always close to the most stable genes, reflecting their ability to be a good reference gene. It is interesting to note that the discrepancy among different methods is bigger in liver than in iWAT and Sk.M. In liver, Hprt is most stable gene generated by Bestkeeper, but second and third unstable by NormFinder and geNorm. Thus, it is advisable to evaluate reference genes by more than one method and integrate the data to generate final ranking, especially in tissues like liver. Here, we calculated the comprehensive ranking by geometric mean of results by all the software programs32,40,41. And validation by Leptin and Ppar-γ expression assessment confirmed the reliability of this strategy.
In this study we evaluated comprehensively and successfully the reference genes after CR in different tissues. At the same time, we acknowledge the limitations of the present study. First, our choice of reference genes analyzed was limited to twelve candidate genes. There are some other widely used genes, such as 18sRNA and Ubiquitin, which may warrant consideration in the future. However, the selected genes within the twelve candidates are suitable as validated by the expression of Leptin and Ppar-γ. Second, the results of this study may not be applicable to other tissues or treatments. Validation studies will need to be repeated in other tissues, cells, experimental conditions. Nevertheless, according to the similarities among these metabolism related tissues, our results are worthy of reference in metabolism related studies in these tissues. Third, there are some other statistical methods, such as qBasePlus61, comparative ΔCt62, and RefFinder63. We applied only three most commonly-used softwares. However, according to previous studies64,65, it’s possible that those methods could get similar results as in this study.
Conclusion
This is the first study specifically designed to evaluate effects of CR in iWAT, skeletal muscle and liver on a set of candidate reference genes for gene expression normalization using qRT-PCR in the C57BL6/J mice using the programs geNorm, NormFinder, and BestKeeper. We indicate that among 12 candidate reference genes investigated, each tissue needs its own combination of reference genes, which is β2m-Hprt for iWAT, Ppia-Gusb for Sk.M and Hmbs-β2m for liver. We also demonstrate the advantage of using more than one reference gene in combination for certain experimental conditions. Our study provides a basis for the selection of reference genes and useful guidelines for future gene expression studies of CR and metabolism.
Additional Information
How to cite this article: Gong, H. et al. Evaluation of candidate reference genes for RT-qPCR studies in three metabolism related tissues of mice after caloric restriction. Sci. Rep. 6, 38513; doi: 10.1038/srep38513 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported by National Natural Science Foundation of China to H.G. (No. 81300693) and L.S. (No. 81571385), and the Research Special Fund for Public Welfare Industry of Health to T.M.Z. (No. 201302008).
Footnotes
Author Contributions H.G., R.M.Q. and T.M.Z. designed the study. B.D.C., H.G., J.P., W.W. and Y.W.H. performed the experiments. H.G., L.S. and Y.W.H. analyzed the data. H.G., L.S. and T.M.Z. contributed reagents/materials/analysis tools and wrote the main part of the manuscript together. All authors reviewed the manuscript and added details to it.
References
- Schmittgen T. D. & Livak K. J. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3, 1101–1108 (2008). [DOI] [PubMed] [Google Scholar]
- Chen C. et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33, e179 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruijter J. M. et al. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37, e45 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M. L. & Medrano J. F. Real-time PCR for mRNA quantitation. Biotechniques 39, 75–85 (2005). [DOI] [PubMed] [Google Scholar]
- Huggett J., Dheda K., Bustin S. & Zumla A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun 6, 279–284 (2005). [DOI] [PubMed] [Google Scholar]
- Bustin S. A. et al. The need for transparency and good practices in the qPCR literature. Nat Methods 10, 1063–1067 (2013). [DOI] [PubMed] [Google Scholar]
- Vandesompele J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3, RESEARCH0034 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulson D. T. et al. Identification of valid reference genes for the normalization of RT qPCR gene expression data in human brain tissue. BMC Mol Biol 9, 46 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Higgins P. J. & Crawford D. R. Control selection for RNA quantitation. Biotechniques 29, 332–337 (2000). [DOI] [PubMed] [Google Scholar]
- Kozera B. & Rapacz M. Reference genes in real-time PCR. J Appl Genet 54, 391–406 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S. A. et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55, 611–622 (2009). [DOI] [PubMed] [Google Scholar]
- Fontana L., Partridge L. & Longo V. D. Extending healthy life span–from yeast to humans. Science 328, 321–326 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flurkey K., Astle C. M. & Harrison D. E. Life extension by diet restriction and N-acetyl-L-cysteine in genetically heterogeneous mice. J Gerontol A Biol Sci Med Sci 65, 1275–1284 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omodei D. & Fontana L. Calorie restriction and prevention of age-associated chronic disease. FEBS Lett 585, 1537–1542 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell S. J. et al. Effects of Sex, Strain, and Energy Intake on Hallmarks of Aging in Mice. Cell Metab 23, 1093–1112 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbiano S. et al. Caloric Restriction Leads to Browning of White Adipose Tissue through Type 2 Immune Signaling. Cell Metab (2016). [DOI] [PubMed] [Google Scholar]
- Wood S. H. et al. Transcriptome analysis in calorie-restricted rats implicates epigenetic and post-translational mechanisms in neuroprotection and aging. Genome Biol 16, 285 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gat-Yablonski G. et al. Quantitative proteomics of rat livers shows that unrestricted feeding is stressful for proteostasis with implications on life span. Aging (Albany NY) (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanic N., Perovic M., Mladenovic A., Ruzdijic S. & Kanazir S. Effects of aging, dietary restriction and glucocorticoid treatment on housekeeping gene expression in rat cortex and hippocampus-evaluation by real time RT-PCR. J Mol Neurosci 32, 38–46 (2007). [DOI] [PubMed] [Google Scholar]
- Cohen H. Y. et al. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science 305, 390–392 (2004). [DOI] [PubMed] [Google Scholar]
- Civitarese A. E. et al. Calorie restriction increases muscle mitochondrial biogenesis in healthy humans. PLoS Med 4, e76 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander-Bridges M. et al. Multiple insulin-responsive elements regulate transcription of the GAPDH gene. Adv Enzyme Regul 32, 149–159 (1992). [DOI] [PubMed] [Google Scholar]
- Dugail I., Quignard-Boulange A., Bazin R., Le Liepvre X. & Lavau M. Adipose-tissue-specific increase in glyceraldehyde-3-phosphate dehydrogenase activity and mRNA amounts in suckling pre-obese Zucker rats. Effect of weaning. Biochem J 254, 483–487 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthae S., May S., Hubersberger M., Hauner H. & Skurk T. Protein normalization in different adipocyte models and dependence on cell size. Horm Metab Res 45, 572–580 (2013). [DOI] [PubMed] [Google Scholar]
- Andersen C. L., Jensen J. L. & Orntoft T. F. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64, 5245–5250 (2004). [DOI] [PubMed] [Google Scholar]
- Pfaffl M. W., Tichopad A., Prgomet C. & Neuvians T. P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol Lett 26, 509–515 (2004). [DOI] [PubMed] [Google Scholar]
- Bruckert G., Vivien D., Docagne F. & Roussel B. D. Normalization of Reverse Transcription Quantitative PCR Data During Ageing in Distinct Cerebral Structures. Mol Neurobiol 53, 1540–1550 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z. J., Tian C., Jiang Q., Li X. H. & Zhuang J. Selection of suitable reference genes for qRT-PCR normalization during leaf development and hormonal stimuli in tea plant (Camellia sinensis). Sci Rep 6, 19748 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koramutla M. K., Aminedi R. & Bhattacharya R. Comprehensive evaluation of candidate reference genes for qRT-PCR studies of gene expression in mustard aphid, Lipaphis erysimi (Kalt). Sci Rep 6, 25883 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd G. T. et al. Leptin and insulin act on POMC neurons to promote the browning of white fat. Cell 160, 88–104 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita N. et al. Differential responses of white adipose tissue and brown adipose tissue to caloric restriction in rats. Mech Ageing Dev 133, 255–266 (2012). [DOI] [PubMed] [Google Scholar]
- Chen G. et al. Validation of reliable reference genes for real-time PCR in human umbilical vein endothelial cells on substrates with different stiffness. PLoS One 8, e67360 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissa N. et al. Stability of Reference Genes for Messenger RNA Quantification by Real-Time PCR in Mouse Dextran Sodium Sulfate Experimental Colitis. PLoS One 11, e0156289 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentile A. M. et al. RPL13A and EEF1A1 Are Suitable Reference Genes for qPCR during Adipocyte Differentiation of Vascular Stromal Cells from Patients with Different BMI and HOMA-IR. PLoS One 11, e0157002 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong H. et al. Histone modifications change with age, dietary restriction and rapamycin treatment in mouse brain. Oncotarget 6, 15882–15890 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jais A. et al. Heme oxygenase-1 drives metaflammation and insulin resistance in mouse and man. Cell 158, 25–40 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- Gong H. et al. The effects of energy intake of four different feeding patterns in rats. Exp Biol Med (Maywood) 241, 52–59 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cabo R., Carmona-Gutierrez D., Bernier M., Hall M. N. & Madeo F. The search for antiaging interventions: from elixirs to fasting regimens. Cell 157, 1515–1526 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira-Fantini P. M., Rajapaksa A. E., Oakley R. & Tingay D. G. Selection of Reference Genes for Gene Expression Studies related to lung injury in a preterm lamb model. Sci Rep 6, 26476 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Pan X., Xiao P., Farwell M. A. & Zhang B. Evaluation and identification of reliable reference genes for pharmacogenomics, toxicogenomics, and small RNA expression analysis. J Cell Physiol 226, 2469–2477 (2011). [DOI] [PubMed] [Google Scholar]
- Mitterberger M. C., Mattesich M. & Zwerschke W. Bariatric surgery and diet-induced long-term caloric restriction protect subcutaneous adipose-derived stromal/progenitor cells and prolong their life span in formerly obese humans. Exp Gerontol 56, 106–113 (2014). [DOI] [PubMed] [Google Scholar]
- Zhu M. et al. Circulating adiponectin levels increase in rats on caloric restriction: the potential for insulin sensitization. Exp Gerontol 39, 1049–1059 (2004). [DOI] [PubMed] [Google Scholar]
- Muller M. J. et al. Metabolic adaptation to caloric restriction and subsequent refeeding: the Minnesota Starvation Experiment revisited. Am J Clin Nutr 102, 807–819 (2015). [DOI] [PubMed] [Google Scholar]
- Maffei M. et al. Leptin levels in human and rodent: measurement of plasma leptin and ob RNA in obese and weight-reduced subjects. Nat Med 1, 1155–1161 (1995). [DOI] [PubMed] [Google Scholar]
- Zhang Y. et al. Positional cloning of the mouse obese gene and its human homologue. Nature 372, 425–432 (1994). [DOI] [PubMed] [Google Scholar]
- Sasaki N. et al. CDNA cloning of feline leptin and its mRNA expression in adipose tissue. J Vet Med Sci 63, 1115–1120 (2001). [DOI] [PubMed] [Google Scholar]
- Li X. et al. Establishment and Characterization of a Newly Established Diabetic Gerbil Line. PLoS One 11, e0159420 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamrick M. W. et al. The adipokine leptin mediates muscle- and liver-derived IGF-1 in aged mice. Exp Gerontol 70, 92–96 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalan V. et al. Validation of endogenous control genes in human adipose tissue: relevance to obesity and obesity-associated type 2 diabetes mellitus. Horm Metab Res 39, 495–500 (2007). [DOI] [PubMed] [Google Scholar]
- Ghosh S. et al. A systems biology analysis of the unique and overlapping transcriptional responses to caloric restriction and dietary methionine restriction in rats. FASEB J 28, 2577–2590 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speakman J. R. & Mitchell S. E. Caloric restriction. Mol Aspects Med 32, 159–221 (2011). [DOI] [PubMed] [Google Scholar]
- Zhang W. X. et al. Selection of Suitable Reference Genes for Quantitative Real-Time PCR Normalization in Three Types of Rat Adipose Tissue. Int J Mol Sci 17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svingen T., Letting H., Hadrup N., Hass U. & Vinggaard A. M. Selection of reference genes for quantitative RT-PCR (RT-qPCR) analysis of rat tissues under physiological and toxicological conditions. PeerJ 3, e855 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. et al. Evaluation of housekeeping genes for normalizing real-time quantitative PCR assays in pig skeletal muscle at multiple developmental stages. Gene 565, 235–241 (2015). [DOI] [PubMed] [Google Scholar]
- Mulligan J. D., Stewart A. M. & Saupe K. W. Downregulation of plasma insulin levels and hepatic PPARgamma expression during the first week of caloric restriction in mice. Exp Gerontol 43, 146–153 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masternak M. M. et al. Effects of caloric restriction and growth hormone resistance on the expression level of peroxisome proliferator-activated receptors superfamily in liver of normal and long-lived growth hormone receptor/binding protein knockout mice. J Gerontol A Biol Sci Med Sci 60, 1394–1398 (2005). [DOI] [PubMed] [Google Scholar]
- Rahman M., Halade G. V., Bhattacharya A. & Fernandes G. The fat-1 transgene in mice increases antioxidant potential, reduces pro-inflammatory cytokine levels, and enhances PPAR-gamma and SIRT-1 expression on a calorie restricted diet. Oxid Med Cell Longev 2, 307–316 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink T. et al. Instability of standard PCR reference genes in adipose-derived stem cells during propagation, differentiation and hypoxic exposure. BMC Mol Biol 9, 98 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petriccione M., Mastrobuoni F., Zampella L. & Scortichini M. Reference gene selection for normalization of RT-qPCR gene expression data from Actinidia deliciosa leaves infected with Pseudomonas syringae pv. actinidiae. Sci Rep 5, 16961 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans J., Mortier G., De Paepe A., Speleman F. & Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8, R19 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver N., Best S., Jiang J. & Thein S. L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol 7, 33 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F., Xiao P., Chen D., Xu L. & Zhang B. miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol Biol (2012). [DOI] [PubMed] [Google Scholar]
- Chen I. H. et al. Selection of suitable reference genes for normalization of quantitative RT-PCR in peripheral blood samples of bottlenose dolphins (Tursiops truncatus). Sci Rep 5, 15425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal M. F. et al. Comprehensive selection of reference genes for expression studies in meniscus injury using quantitative real-time PCR. Gene 584, 60–68 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.