Abstract
Neuronal inclusions of poly(GA), a protein unconventionally translated from G4C2 repeat expansions in C9ORF72, are abundant in patients with frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS) caused by this mutation. To investigate poly(GA) toxicity, we generated mice that exhibit poly(GA) pathology, neurodegeneration and behavioral abnormalities reminiscent of FTD and ALS. These phenotypes occurred in the absence of TDP-43 pathology and required poly(GA) aggregation. HR23 proteins involved in proteasomal degradation and proteins involved in nucleocytoplasmic transport were sequestered by poly(GA) in these mice. HR23A and HR23B similarly colocalized to poly(GA) inclusions in C9ORF72 expansion carriers. Sequestration was accompanied by an accumulation of ubiquitinated proteins and decreased xeroderma pigmentosum C (XPC) levels in mice, indicative of HR23A and HR23B dysfunction. Restoring HR23B levels attenuated poly(GA) aggregation and rescued poly(GA)-induced toxicity in neuronal cultures. These data demonstrate that sequestration and impairment of nuclear HR23 and nucleocytoplasmic transport proteins is an outcome of, and a contributor to, poly(GA) pathology.
A G4C2 repeat expansion located in an intronic region of chromosome 9 open reading frame 72 (C9ORF72) is the most common genetic cause of FTD and ALS1,2. FTD is characterized by abnormalities in behavior, language and personality, whereas ALS primarily affects motor systems. Nevertheless, in addition to a genetic overlap between FTD and ALS, there is considerable clinical and neuropathological overlap, indicating these two diseases may share common underlying pathological mechanisms.
Potential mechanisms by which C9ORF72 repeat expansions cause FTD and ALS, collectively referred to as c9FTD/ALS, include loss of C9ORF72 function consequent to decreased C9ORF72 mRNA expression3–6, and toxicity mediated by sense and/or antisense repeat-containing RNA7. For example, these transcripts form nuclear RNA foci believed to sequester several RNA-binding proteins and thereby cause abnormal RNA metabolism8–11. Recent evidence also suggests that sense G4C2 transcripts cause cellular injury by disrupting nucleocytoplasmic transport through nuclear pores12,13. Furthermore, both sense and antisense repeat-containing transcripts are susceptible to repeat-associated non-ATG (RAN) translation resulting in the synthesis of proteins of repeating dipeptides (GP, GA, GR, PA or PR), collectively referred to as c9RAN proteins14–18.
Several studies indicate that certain c9RAN proteins are toxic in cultured cells and/or Drosophila melanogaster models19–26. Arginine-rich poly(PR) and poly(GR) c9RAN proteins, found to induce nucleolar stress23,24,26 and implicated in impaired nucleocytoplasmic transport12,19, appear to be especially harmful. Nonetheless, their contribution to disease remains unclear, largely because of the relative paucity of poly(PR) and poly(GR) pathology in c9FTD/ALS14,16–18,27,28. In contrast, cytoplasmic poly(GA) inclusions are abundant in brains affected by c9FTD/ALS17,25,27. This is of particular interest given recent evidence that cytoplasmic protein aggregates interfere with nucleocytoplasmic transport of proteins and RNA29. In addition, several studies using cultured cells or primary neurons have demonstrated evidence of poly(GA) toxicity20,22,25. We and others have shown that poly(GA) proteins impair the ubiquitin-proteasome system (UPS)20,25, induce endoplasmic reticulum stress25 and cause loss of function of Unc119 proteins22. Ubiquitin- and p62-positive inclusions composed of c9RAN proteins are a neuropathological hallmark in c9FTD/ALS14–18, and mutations in p62 (refs. 30,31) and ubiquilin-2 (ref. 32) also result in ALS and FTD, indicating that UPS disruption may be a critical factor in c9FTD/ALS disease pathology.
No effective treatment for c9FTD/ALS exists, in part because of an incomplete understanding of the causative mechanisms of disease. To bridge this gap in our understanding and to gain much needed insight on the contribution of c9RAN proteins to c9FTD/ALS, with a particular focus on poly(GA), we used an adeno-associated virus (AAV) vector to mediate robust expression of poly(GA) proteins in the central nervous system (CNS) of mice.
RESULTS
Poly(GA) aggregation is necessary for its toxicity
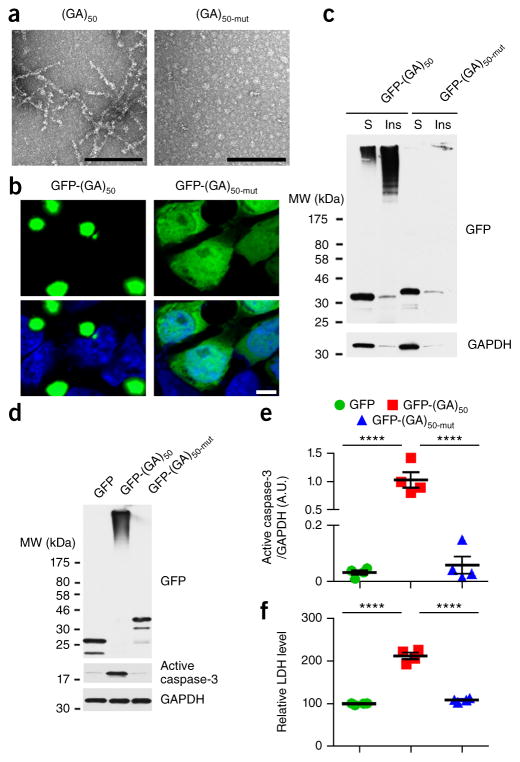
Using constructs that drive poly(GA) expression in the absence of other c9RAN proteins and foci, we previously reported that, in cultured cells and primary neurons, poly(GA) proteins form soluble and insoluble high-molecular-weight species, as well as inclusions composed of filaments similar to those observed in c9FTD/ALS brain tissues25. Here, to determine whether the conformation of poly(GA) proteins is important for their propensity to aggregate, we disrupted poly(GA) protein conformation by inserting proline residues among the repeating GA dipeptides. This strategy has been shown to disrupt the conformation and inhibit aggregation of other proteins associated with neurodegeneration (for example, polyglutamine33–35, amyloid peptides36 and tau37). We generated an ATG-initiated expression vector in which a proline residue was inserted after every 5 of the 50 GA repeats, referred to as (GA)50-mut. Consistent with our previous finding25, recombinant untagged (GA)50 proteins formed fibrils in vitro (Fig. 1a). In contrast, recombinant (GA)50-mut did not assemble into filaments (Fig. 1a). Whereas GFP-tagged (GA)50 formed cytoplasmic inclusions when expressed in cultured cells, and accumulated as insoluble high-molecular-weight species in cellular fractions, GFP-(GA)50-mut was diffusely distributed in cells and remained in a soluble, monomeric form (Fig. 1b,c). Expression of GFP-(GA)50 in primary neurons for 7 d resulted in the formation of high-molecular-weight poly(GA) species (Fig. 1d) and was neurotoxic, as evidenced by a significant increase in amounts of active caspase-3, a mediator of programmed cell death (Fig. 1d,e), and LDH activity (Fig. 1f) relative to cells expressing only GFP. The GFP-(GA)50-mut proteins, which could not aggregate, were not harmful to neurons, inducing neither caspase-3 activation nor LDH activity (Fig. 1d–f). These data suggest that the intrinsic conformation of poly(GA) proteins determines their propensity to aggregate and their toxicity.
Figure 1.
Disrupting the conformation of poly(GA) proteins inhibits poly(GA) protein aggregation and toxicity. (a) Transmission electron micrographs of purified recombinant untagged (GA)50 and (GA)50-mut proteins. Scale bars, 200 nm. (b) Confocal micrographs of GFP-(GA)50 and GFP-(GA)50-mut proteins expressed in HEK293T cells. Nuclei were counterstained with Hoechst 33258 (blue). Scale bar, 5 μm. (c) Immunoblot analysis of Triton X-100–soluble (S) and –insoluble (Ins) fractions of lysates from HEK293T cells expressing GFP-(GA)50 and GFP-(GA)50-mut. Blots were probed with antibodies specific for GFP. (d,e) Immunoblot (d) and densitometric analysis of immunoblots (e) indicating the levels of GFP-tagged proteins and active caspase-3 in neurons expressing GFP, GFP-(GA)50 or GFP-(GA)50-mut. GAPDH was used as a loading control in c and d. Full-length immunoblots are presented in Supplementary Figure 9. (f) Relative LDH activity, an indicator of cell toxicity, in the culture supernatant of primary neuronal cells expressing the indicated GFP-tagged proteins. Data are presented as mean ± s.e.m. from four separate experiments. P < 0.0001, as analyzed by one-way ANOVA. ****P < 0.0001, Tukey’s post hoc analysis. A.U., Arbitrary units
GFP-(GA)50 mice develop hallmark features of c9FTD/ALS
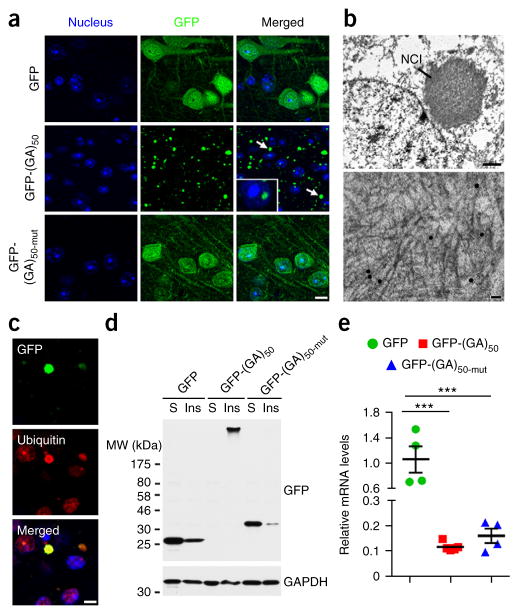
Given that poly(GA) expression was detrimental to primary neurons, we investigated the consequences of poly(GA) expression in vivo using AAV serotype 1 vectors to express GFP-(GA)50 or GFP in the CNS of mice. As above, we also generated GFP-(GA)50-mut–expressing mice to investigate whether poly(GA) toxicity depends on its ability to aggregate. Six months after intracerebroventricular (ICV) administration of AAV1 vectors at post-natal day 0, we assessed the mice to determine whether they exhibited pathological hallmarks associated with c9FTD/ALS. As in cultured cells, GFP-(GA)50 proteins, but not GFP or GFP-(GA)50-mut proteins, formed cytoplasmic and occasional nuclear inclusions in the cortex of mouse brains (Fig. 2a) and other brain regions, including the hippocampus, olfactory bulb, cerebellum and thalamus (Supplementary Fig. 1a,b and Supplementary Table 1). These inclusion were present mainly in MAP2-positive neurons rather than in GFAP-positive astrocytes (Supplementary Fig. 1c). Immunoelectron microscopy using anti-GA antibody labeled with gold particles showed neuronal cytoplasmic inclusions composed of fibrils of poly(GA) proteins (Fig. 2b). Immunofluorescence analysis revealed that nearly all GFP-(GA)50 inclusions were ubiquitin-positive (Fig. 2c). Ubiquitin-positive inclusions were not present in mice expressing GFP or GFP-(GA)50-mut (Supplementary Fig. 1d). Furthermore, GFP-(GA)50, but not GFP or GFP-(GA)50-mut, formed high-molecular-weight species in mouse brain lysates, as assessed by immunoblot analyses (Fig. 2d). Transgene mRNA levels were comparable between mice expressing GFP-(GA)50 or GFP-(GA)50-mut, but were significantly increased in GFP-expressing mice (Fig. 2e), ruling out the possibility that expression influenced poly(GA) aggregation.
Figure 2.
Expression of poly(GA) proteins in mouse brains results in the formation of ubiquitin-positive poly(GA) inclusions. (a) Confocal microscopy of mouse cortical tissue showing cell nuclei (blue) and the cellular distribution of the indicated GFP-tagged proteins 6 months after postnatal day 0 pups were ICV injected with AAV1 particles expressing GFP, GFP-(GA)50 or GFP-(GA)50-mut. Cytoplasmic (arrows) and intranuclear (inset) poly(GA) inclusions were observed. Scale bar, 10 μm. (b) Immunoelectron microscopy using an anti-GA antibody labeled with gold particles, showing neuronal cytoplasmic inclusion (NCI) composed of fibrils. Scale bar, 1 μm (top) and 50 nm (bottom). (c) Double immunofluorescence staining of cortex from GFP-(GA)50–transduced mice showing GFP- and ubiquitin-positive inclusions. Scale bar, 5 μm. (d) Immunoblot analysis of Triton X-100–soluble (S) and –insoluble (Ins) fractions from brain lysates of mice expressing GFP, GFP-(GA)50 or GFP-(GA)50-mut. Blots were probed with antibodies specific for GFP. GAPDH was used as a loading control. Full-length immunoblots are presented in Supplementary Figure 9. (e) Quantitative real-time PCR analysis of relative mRNA levels of the indicated GFP-tagged proteins in brains of mice expressing GFP, GFP-(GA)50 or GFP-(GA)50-mut (n = 4–5 per group). Data are presented as mean ± s.e.m. P = 0.0002, one-way ANOVA. ***P = 0.0003 (GFP mice versus GFP-(GA)50 mice) and ***P = 0.0007 (GFP mice versus GFP-(GA)50-mut mice), Tukey’s post hoc analysis.
Poly(GA) sequesters HR23 and nuclear pore proteins
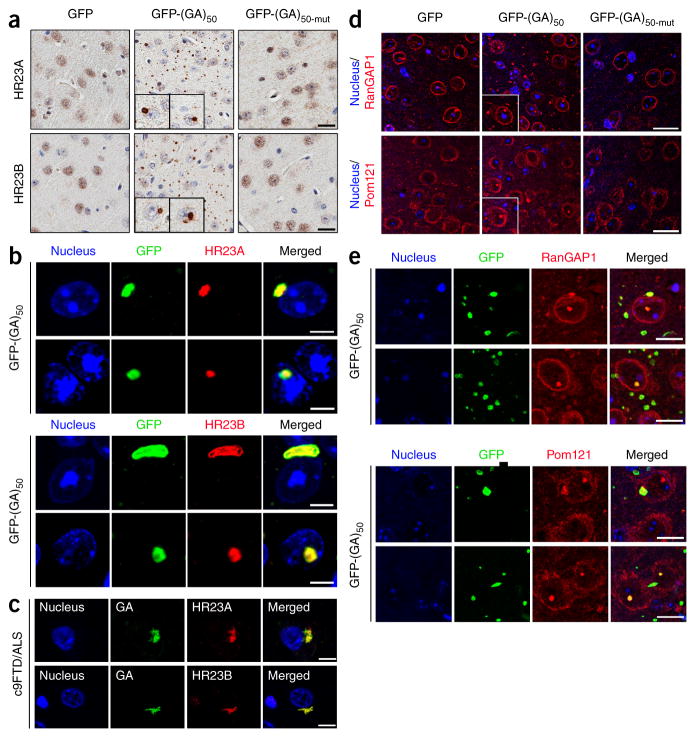
We and others reported that poly(GA) proteins cause UPS dysfunction20,25, and that poly(GA) interacts with components of the UPS and UPS-related proteins, such as HR23 proteins22. HR23A and HR23B, mammalian homologs of Saccharomyces cerevisiae Rad23, are nuclear proteins involved in the transfer of ubiquitinated proteins to the proteasome for degradation38. To investigate whether impairments in HR23A and HR23B underlie poly(GA)-mediated neurotoxicity, we examined the distribution of these proteins in 6-month-old GFP-(GA)50–expressing mice. Both HR23A and HR23B formed nuclear and, more frequently, cytoplasmic aggregates in the cortex (Fig. 3a), hippocampus (Supplementary Fig. 2a) and other regions (Supplementary Table 2) of mice expressing GFP-(GA)50 in contrast to their diffuse, predominantly nuclear distribution in mice expressing GFP or GFP-(GA)50-mut (Fig. 3a and Supplementary Fig. 2a). The redistribution of nuclear HR23 proteins to cytoplasmic aggregates in mice expressing GFP-(GA)50 was caused by sequestration of HR23 proteins into poly(GA) inclusions (Fig. 3b and Supplementary Fig. 2b). In fact, similar pathology was present in mice expressing (G4C2)66, another model of c9FTD/ALS in which c9RAN proteins, including poly(GA), are synthesized through RAN translation39. Compared to the concentration of poly(GA) in mice expressing GFP-(GA)50 (4,019 ± 206 ng/mg protein, mean ± s.e.m.), poly(GA) levels was approximately twofold lower in mice expressing (G4C2)66 (2,057 ± 129 ng/mg protein; P < 0.0001, unpaired t-test), but HR23B-positive inclusions were nonetheless present (Supplementary Fig. 2c). HR23 proteins formed cytoplasmic aggregates and co-localized with poly(GA) inclusions in hippocampal sections from patients with c9FTD/ALS but not from healthy controls (Fig. 3c and Supplementary Fig. 2d).
Figure 3.
Poly(GA) proteins sequester HR23 and nucleocytoplasmic transport proteins into inclusions. (a) Immunohistochemical analysis of HR23A and HR23B proteins in the cortex of mice expressing GFP, GFP-(GA)50 and GFP-(GA)50-mut. Insets, higher magnification examples of cytoplasmic and nuclear inclusions containing HR23 proteins. Scale bars, 20 μm. (b) Double immunofluorescence staining for GFP-(GA)50 and either HR23A or HR23B in the cortex of mice expressing GFP-(GA)50. Scale bars, 5 μm. (c) Double immunofluorescence staining for poly(GA) and either HR23A or HR23B in the hippocampus of c9FTD/ALS subjects. Scale bars, 10 μm. (d) Immunofluorescence staining of RanGAP1 and Pom121 in the cortex of mice expressing GFP, GFP-(GA)50 or GFP-(GA)50-mut. Insets, higher magnification examples of cytoplasmic and nuclear inclusions containing RanGAP1 or Pom121. Scale bars, 20 μm. (e) Double immunofluorescence staining for GFP-(GA)50 and either RanGAP1 or Pom121 in the cortex of mice expressing GFP-(GA)50. Scale bars, 10 μm.
Given that cytoplasmic poly(GA) inclusions are abundant in GFP-(GA)50 mice, that cytoplasmic protein aggregates interfere with the nuclear import and export of proteins29, and that defective nucleocytoplasmic transport is a putative pathomechanism of c9FTD/ALS12,13,19, we investigated whether nuclear pore complex defects occur in mice expressing GFP-(GA)50 as these could contribute to the mislocalization of nuclear HR23 proteins. We examined the distribution of RanGAP1, a key regulator of nucleocytoplasmic transport that is mislocalized in (G4C2)30-expressing flies as well as in c9ALS patient–derived neuronal cultures and brain tissues13, and of Pom121, an inner membrane nuclear pore protein that has a role in nuclear pore assembly and its anchoring to the nuclear envelope40,41. Six-month-old GFP-expressing mice had smooth perinuclear staining of RanGAP1, whereas in mice expressing GFP-(GA)50 we observed nuclear and cytoplasmic puncta of RanGAP1, which nearly always colocalized with poly(GA) inclusions (Fig. 3d,e and Supplementary Table 2). RanGAP1 puncta were occasionally immunopositive for HR23A and HR23B (Supplementary Fig. 2e). Pom121, which localized to the nuclear membrane in GFP mice, nearly always colocalized with poly(GA) inclusions in mice expressing GFP-(GA)50 (Fig. 3d,e and Supplementary Table 2). Both RanGAP1 and Pom121 were similarly mislocalized in 6-month-old mice expressing (G4C2)66 (Supplementary Fig. 2f), but we saw no such aberrations in mice expressing GFP-(GA)50-mut (Fig. 3d), indicating that poly(GA)-induced disruption of nuclear pore proteins is dependent on poly(GA) aggregation. These data suggest that expression of poly(GA) leads to defects in the nuclear pore complex, which could be detrimental to nucleocytoplasmic transport and may thus contribute, at least in part, to the mislocalization of HR23 proteins.
Loss of function of poly(GA)-sequestered HR23 proteins
To determine whether sequestration of HR23 proteins by poly(GA) hinders their function, we evaluated poly(GA)-induced HR23A and HR23B intracellular mislocalization. We observed an increased cytoplasmic accumulation of HR23A and HR23B, and a concomitant decrease in nuclear HR23A and HR23B in HEK293T cells expressing GFP-(GA)50 compared to GFP-expressing or GFP-(GA)50-mut–expressing controls (Supplementary Fig. 3a). Also, GFP-(GA)50, but not GFP or GFP-(GA)50-mut, precipitated with HR23A and HR23B in total cell lysates (Supplementary Fig. 3b). HR23 proteins bound both monomeric and high-molecular-weight forms of GFP-(GA)50 (Supplementary Fig. 3b), suggesting they interact with poly(GA) before its aggregation.
To determine whether HR23 proteins bind c9RAN proteins other than poly(GA), we carried out co-immunoprecipitation studies using lysates from cells expressing GFP-(GP)47, GFP-(GR)50, GFP-(GA)50 or GFP. As HR23A and HR23B both bind poly(GA) (Supplementary Fig. 3b), we focused on the potential interaction between HR23B and c9RAN proteins, which revealed that only poly(GA) interacted with HR23B (Supplementary Fig. 3c). We validated this in (G4C2)66 mice, which express poly(GA), poly(GP) and poly(GR)39; whereas the majority of HR23B inclusions in (G4C2)66 mice were immunopositive for poly(GA), HR23B inclusions were immunopositive for poly(GP) or poly(GR) much less frequently (Supplementary Fig. 3d). Staining of hippocampal sections from c9FTD/ALS patients for both HR23B and either poly(GA), poly(GP) or poly(GR) showed similar results. Whereas inclusions immunopositive for poly(GA) or poly(GP) outnumbered those positive for HR23B, the majority (85.0%) of HR23B-containing inclusions were positive for poly(GA). Less frequently, HR23B localized with poly(GP) or poly(GR) inclusions (9.2% and 4.5%, respectively) (Supplementary Fig. 3e).
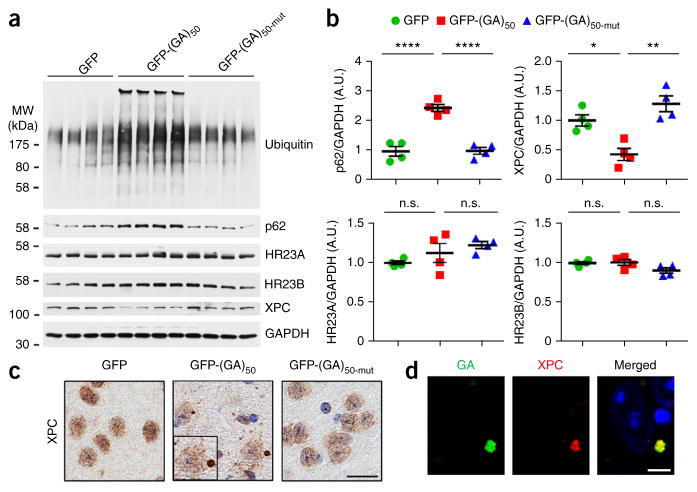
HR23 has a role in UPS function and also stabilizes xeroderma pigmentosum C (XPC)42, a DNA-binding protein involved in DNA-damage repair43,44. We observed an accumulation of p62 and ubiquitinated proteins in GFP-(GA)50 mice (Fig. 4a,b), which may result from loss of HR23 protein function. If this were the case, we might also expect to see enhanced XPC degradation. Indeed, we found that XPC protein levels were decreased by ~50% in the brains of mice expressing GFP-(GA)50 compared to mice expressing GFP or GFP-(GA)50-mut, while total HR23 protein levels remained unchanged (Fig. 4a,b). Consistent with the immunoblot data, immunohistochemistry showed a reduction in XPC levels in mice expressing GFP-(GA)50 relative to controls (Fig. 4c and Supplementary Fig. 4a). Of note, XPC proteins formed cytoplasmic inclusions and localized with poly(GA) in a subset of neurons in the cortex and hippocampus of mice expressing GFP-(GA)50 (Fig. 4c,d and Supplementary Fig. 4a,b) but remained mainly diffusely distributed in the nucleus of cells in control mice (Fig. 4c and Supplementary Fig. 4a). These data indicate that sequestration of HR23 by poly(GA) impairs HR23 function.
Figure 4.
Poly(GA) proteins cause ubiquitinated proteins to accumulate, decrease the stability of XPC proteins, and sequester XPC into inclusions. (a,b) Immunoblot (a) and densitometric analysis of immunoblots (b) for the indicated proteins to determine their expression in brains of mice expressing GFP, GFP-(GA)50 or GFP-(GA)50-mut (n = 4 per group). Data are presented as mean ± s.e.m. Top left: P < 0.0001, one-way ANOVA; ****P < 0.0001, Tukey’s post hoc analysis. Top right: P = 0.0012, one-way ANOVA; *P = 0.0129 and **P = 0.0010, Tukey’s post hoc analysis. Bottom left: P = 0.1599, one-way ANOVA. Bottom right: P = 0.0820, one-way ANOVA. n.s., not significant. Full-length immunoblots are presented in Supplementary Figure 9. (c) Immunohistochemical analyses of XPC in the cortex of GFP, GFP-(GA)50 or GFP-(GA)50-mut mice. Scale bar, 20 μm. (d) Double immunofluorescence staining for XPC and poly(GA) proteins in the cortex of mice expressing GFP-(GA)50. Scale bar, 5 μm.
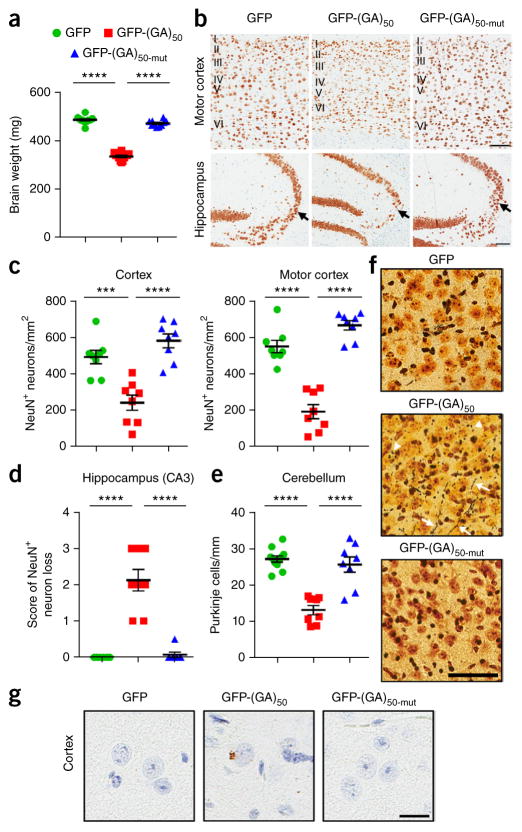
Poly(GA) mice develop brain atrophy and neurotoxicity
As poly(GA) aggregates are toxic to cultured cells (Fig. 1d–f), we evaluated whether poly(GA) aggregates are harmful in the mouse brain. Expression of GFP and GFP-(GA)50-mut had no effect on brain weight of mice. Conversely, mice expressing GFP-(GA)50 exhibited a significant decrease in brain weight (Fig. 5a) and size, the latter assessed by gross morphological analysis of hematoxylin and eosin–stained hemibrain sections (Supplementary Fig. 5a). We detected a decrease in body weight, but only in male mice expressing GFP-(GA)50 (Supplementary Fig. 5b). Consistent with the decrease in brain weight of mice expressing GFP-(GA)50, which was suggestive of atrophy, significantly fewer neurons were present in their cortex and hippocampus relative to mice expressing GFP or GFP-(GA)50-mut (Fig. 5b–d). The number of NeuN-positive neurons was significantly decreased in layer V of the motor cortex and in the CA3 region of the hippocampus, areas relevant to ALS and FTD (Fig. 5b–d and Supplementary Fig. 5c). The number of cerebellar Purkinje cells was also significantly reduced in GFP-(GA)50 mice (Fig. 5e). The evaluation of neurodegeneration by silver staining revealed argyrophilic neurites and neurons (Fig. 5f) in mice expressing GFP-(GA)50 but not in controls. Although rare, inclusions of phosphorylated TDP-43, another neuropathological hallmark of c9FTD/ALS, were specifically observed in mice expressing GFP-(GA)50 (~5–10 cells per sagittal hemibrain section were found to be positive for phosphorylated TDP-43; Fig. 5g).
Figure 5.
Poly(GA) mice develop brain atrophy, neuronal loss and neurodegeneration. (a) Mean brain weight of 6-month-old mice expressing GFP, GFP-(GA)50 or GFP-(GA)50-mut (n = 10–12 mice). (b) Representative images of NeuN-labeled cells in the motor cortex and hippocampus of GFP, GFP-(GA)50 or GFP-(GA)50-mut mice. Layers I to VI of the motor cortex are indicated, and the CA3 region of the hippocampus is indicated by an arrow. Scale bars, 100 μm. (c) Quantification of the number of NeuN-positive cells in the cortex (left) or motor cortex (right) of mice expressing GFP, GFP-(GA)50 or GFP-(GA)50-mut (n = 8 per group). (d) Semiquantitative analysis of NeuN-positive cells in the CA3 region of the hippocampus of mice (score 0: none; 1: mild; 2: modest; 3: severe) (n = 8 per group). (e) Quantification of the number of Purkinje cells in the cerebellum of mice expressing GFP, GFP-(GA)50 or GFP-(GA)50-mut (n = 8 per group). (f) Silver staining of cortex of GFP, GFP-(GA)50 or GFP-(GA)50-mut mice identifying argyrophilic degenerating neurites (arrows) and neuronal cell bodies (arrowheads). Scale bar, 60 μm. (g) Immunohistochemical analysis of phosphorylated TDP-43 in the brains of mice expressing GFP, GFP-(GA)50 or GFP-(GA)50-mut. Scale bar, 20 μm. Data are presented as mean ± s.e.m. In a: P < 0.0001, one-way ANOVA; ****P < 0.0001, Tukey’s post hoc analysis. In c–e: P < 0.0001, one-way ANOVA; ***P = 0.0005 and ****P < 0.0001, Tukey’s post hoc analysis.
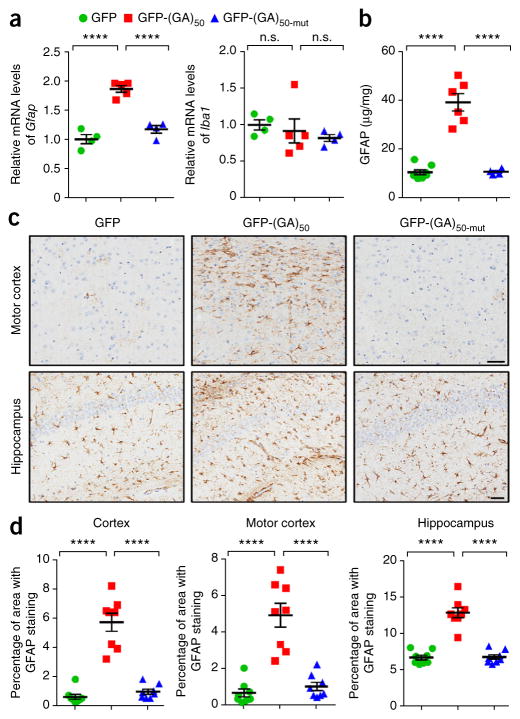
As neurodegeneration is often associated with the accumulation of reactive astrocytes and microglia, we analyzed the brains of mice for signs of these responses. Transcript levels of Gfap, a marker of astrogliosis, but not of Iba1, a marker of microgliosis, were significantly increased in the brains of GFP-(GA)50 mice compared to mice expressing GFP or GFP-(GA)50-mut (Fig. 6a). Likewise, increased GFAP protein expression was detected in the brains of mice expressing GFP-(GA)50, as assessed by immunoassay (Fig. 6b) and immunohistochemical analysis of cortex, motor cortex and hippocampus tissue (Fig. 6c,d). Taken together, these data indicate that expression of poly(GA) proteins induced neuronal loss, astrogliosis and brain atrophy in 6-month-old mice.
Figure 6.
Astrogliosis is observed in poly(GA) mouse brain. (a) Quantitative real-time PCR analysis of the relative mRNA levels of the astrocyte marker Gfap (left) and the microglial marker Iba1 (right) in brains of mice expressing GFP, GFP(GA)50 or GFP-(GA)50-mut (n = 4–5 per group). (b) GFAP protein levels in mice expressing the indicated GFP-tagged proteins, as assessed by ELISA (n = 4–8 mice per group). (c) Representative images of GFAP staining to identify reactive astrocytes in the motor cortex and hippocampus of mice encoding GFP, GFP-(GA)50 or GFP-(GA)50-mut. Scale bars, 50 μm. (d) Quantification of astrogliosis, analyzed by measuring GFAP-positivity in the cortex, motor cortex or hippocampus of mid-sagittal brain sections using a positive pixel count algorithm (n = 8 per group). Data are presented as mean ± s.e.m. In a (left), b and d: P < 0.0001, one-way ANOVA; ****P < 0.0001, as assessed by Tukey’s post hoc analysis. In a (right): P = 0.6106, one-way ANOVA. n.s., not significant.
To determine whether the aberrant features described above occurred progressively in mice expressing GFP-(GA)50 or were caused by developmental defects, we examined GFP-(GA)50–expressing mice between 4 and 6 weeks of age. Poly(GA) inclusions and ubiquitin-positive inclusions were detected in the brains of the young mice expressing GFP-(GA)50 but not in young mice expressing GFP or GFP-(GA)50-mut (Supplementary Fig. 6a,b). Nevertheless, we observed no significant changes in brain and body weight in these mice expressing GFP-(GA)50 (Supplementary Fig. 7a,b), nor was there a loss of NeuN-positive neurons in the cortex and hippocampus, or loss of cerebellar Purkinje cells (Supplementary Fig. 7c–e). Likewise, we detected no signs of astrogliosis in brains of mice expressing GFP-(GA)50 at this time point (Supplementary Fig. 7f). These data indicate that poly(GA) proteins did not influence early neuronal development and that poly(GA) inclusions formed before observable signs of neurodegeneration.
GFP-(GA)50 mice develop behavioral deficits
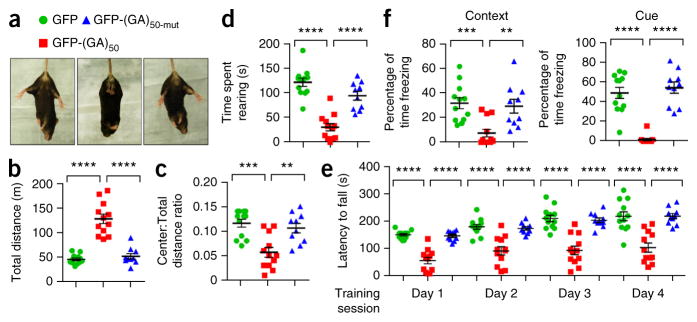
ALS patients display motor system deficits, and FTD is characterized by abnormalities in behavior, language and personality. To determine whether the combination of pathological features in poly(GA)-expressing mice is accompanied by c9FTD/ALS-relevant behaviors, we subjected 6-month-old mice to behavioral tasks. In the tail-suspension test, mice expressing GFP or GFP-(GA)50-mut exhibited a normal escape response by splaying their hindlimbs (Fig. 7a). In contrast, GFP-(GA)50 mice did not extend their hindlimbs and showed signs of forelimb clasping, the latter indicative of neurological abnormalities (Fig. 7a). To explore this further, we tested mice in the open-field assay to assess general locomotor activity, exploration and anxiety-like behavior. Mice expressing GFP-(GA)50 traveled a longer distance in comparison to control mice (Fig. 7b), indicative of hyperactivity, and exhibited a decreased tendency to explore the center of the open field (Fig. 7c), suggestive of anxiety-like behavior. Mice expressing GFP-(GA)50 also spent less time rearing (Fig. 7d), potentially indicative of a motor phenotype. We directly evaluated motor coordination and balance of mice using the rotarod test. During all training sessions, mice expressing GFP-(GA)50 spent significantly less time on the rotating rod compared to control mice (Fig. 7e). Finally, we used contextual and cued fear conditioning tests to measure associative learning and memory. In brief, we placed mice in a novel environment in which a conditioned stimulus (i.e., white noise) was paired with a foot shock. Upon returning mice to the same environment in the absence of the white noise (contextual condition), or re-exposing mice to a different environment but with the white noise (cued conditioning), we recorded the freezing behavior of mice as an indicator of memory associating the environment or auditory cue with the aversive foot shock. Both in the contextual and cued tests, mice expressing GFP-(GA)50 displayed a significant decrease in the amount of time freezing compared to mice expressing GFP or GFP-(GA)50-mut (Fig. 7f). Overall, mice expressing GFP-(GA)50 developed hyperactivity and anxiety-like behavior, as well as motor and cognitive deficits.
Figure 7.
Poly(GA) mice develop motor deficits, hyperactivity, anxiety and cognitive defects. (a) Response of mice expressing GFP, GFP-(GA)50 or GFP-(GA)50-mut after tail suspension. Splayed hind legs are indicative of a normal escape response. (b–d) Results from the open field test analyzing the behavior of mice expressing GFP, GFP-(GA)50 or GFP-(GA)50-mut (n = 10–12 per group), where hyperactivity is indicated by the total distance traveled (b), signs of anxiety are manifested by a decrease in the ratio of distance traveled in the center area to total distance traveled (c), and potential deficits in motor coordination are indicated by a decrease in rearing assessed using raised photobeams (d). (e) Results from a four day rotarod performance test used to determine motor deficits of mice expressing GFP, GFP-(GA)50 and GFP-(GA)50-mut (n = 10–12 per group) by evaluating latency to fall from a rotating rod. (f) In the fear conditioning test, associative learning and memory of mice expressing GFP, GFP-(GA)50 or GFP-(GA)50-mut (n = 10–12 per group) were evaluated by the percent of time freezing in response to an unconditioned (context) or conditioned (cued) stimulus. Data are presented as mean ± s.e.m. In b, d and e: P < 0.0001, one-way ANOVA; ****P < 0.0001, Tukey’s post hoc analysis. In c: P = 0.0001, one-way ANOVA; ***P = 0.0002 and **P = 0.0022, Tukey’s post hoc analysis. In f (left): P = 0.0005, one-way ANOVA; ***P = 0.0009 and **P = 0.0042, as assessed by Tukey’s post hoc analysis. In f (right): P < 0.0001, one-way ANOVA; ****P < 0.0001, Tukey’s post hoc analysis.
HR23 expression rescues poly(GA)-induced toxicity
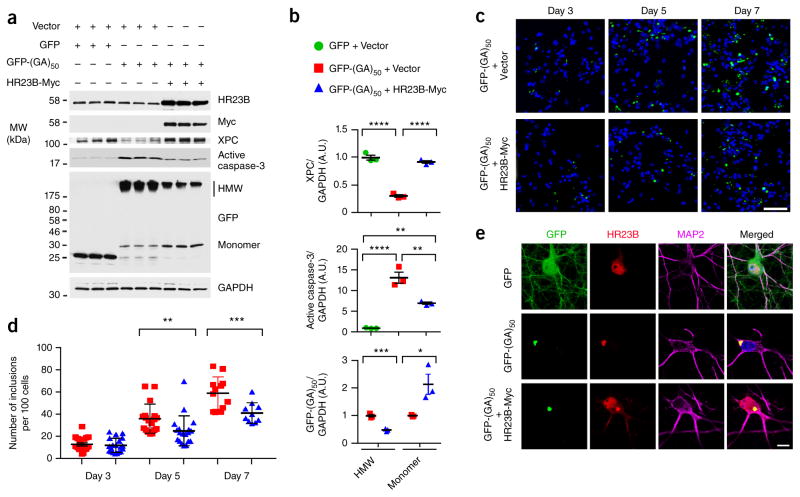
The above data indicate that poly(GA) proteins cause the sequestration and loss of function of HR23. To determine whether loss of HR23 protein function contributes to poly(GA) aggregation and toxicity, we sought to determine whether increasing HR23 protein levels in GFP-(GA)50–expressing primary neurons would provide neuroprotection. Previous studies have demonstrated that HR23B can completely compensate for the loss of HR23A activity but HR23A only partially compensates for the loss of HR23B42,45. We thus expressed GFP-(GA)50 proteins in primary neurons in the presence or absence of exogenous HR23B. Overexpressing HR23B restored HR23 protein function, as evidenced by a return of XPC to normal basal levels, and significantly decreased caspase-3 activation otherwise observed in neurons expressing GFP-(GA)50 (Fig. 8a,b). HR23B overexpression led to a significant decrease in high-molecular-weight GFP-(GA)50 and a concomitant increase in monomeric GFP-(GA)50 (Fig. 8a,b). In addition, significantly fewer poly(GA) inclusions were formed in GFP-(GA)50 primary neurons if exogenous HR23B was present (Fig. 8c,d) even though expression of mRNA encoding GFP-(GA)50 was not different between GFP-(GA)50–expressing cells without exogenous HR23B (1.00 ± 0.05) or with exogenous HR23B (0.93 ± 0.03, P = 0.32). Overexpressing HR23B in GFP-(GR)50–expressing primary neurons neither influenced levels of poly(GR) nor decreased activation of caspase-3 (Supplementary Fig. 8a,b), demonstrating that rescue of poly(GA)-induced toxicity by HR23B is specific and not the result of general neuroprotection.
Figure 8.
Exogenous HR23B attenuates poly(GA) aggregation and poly(GA)-induced neurotoxicity. (a,b) Immunoblot (a) and densitometric analysis of immunoblots (b) for the indicated proteins to determine their levels of expression in primary neurons transduced to express GFP-(GA)50 or GFP in the presence or absence of exogenous Myc-tagged HR23B. Note that HR23B overexpression restores XPC levels, attenuates poly(GA)-induced caspase-3 activation, as well as decreases high-molecular-weight (HMW) poly(GA) species while increasing monomeric poly(GA). Full-length immunoblots are presented in Supplementary Figure 9. (c) Confocal microscopy of GFP-(GA)50 in primary neurons with or without HR23B-Myc coexpression. Nuclei were counterstained with Hoechst 33258 (blue). Scale bar, 50 μm. (d) Graphs showing poly(GA) inclusions in primary neurons expressing GFP-(GA)50 in the presence or absence of exogenous HR23B at 3, 5 or 7 d after transduction. (e) Immunofluorescence staining for GFP, HR23B and MAP2 in primary neurons transduced to express the indicated proteins. Nuclei were counterstained with Hoechst 33258 (blue). Scale bar, 10 μm. Data are presented as mean ± s.e.m. of three separate experiments for b, or as mean ± s.e.m. of counts from 10–20 images for d. In b (top): P < 0.0001, one-way ANOVA; ****P < 0.0001, Tukey’s post hoc analysis. In b (middle): P = 0.0001, one-way ANOVA; ****P < 0.0001 (GFP + vector versus GFP-(GA)50 + vector), **P = 0.0041 (GFP + vector versus GFP-(GA)50 + HR23B-Myc) and **P = 0.0036 (GFP-(GA)50 + vector versus GFP-(GA)50 + HR23B-Myc), Tukey’s post hoc analysis. In b (bottom): ***P = 0.0006 (HMW) and *P = 0.0355 (monomer), unpaired t-test. In d: P < 0.0001, two-way ANOVA; **P = 0.0077 and ***P = 0.0006, Sidak post hoc analysis.
Given our findings suggesting that poly(GA) toxicity in cultured cells and mice is dependent on the ability of poly(GA) to aggregate, the HR23B-mediated attenuation of caspase-3 activation in neurons expressing GFP-(GA)50 likely resulted, at least in part, from the observed decrease in poly(GA) aggregation. Overexpressing HR23B may also have provided rescue from poly(GA) toxicity by restoring the intracellular localization, and consequently function, of HR23B. As endogenous HR23B becomes trapped in poly(GA) inclusions, primary neurons expressing only GFP-(GA)50 displayed little of the diffusely expressed nuclear and cytoplasmic HR23B seen in control cells expressing GFP (Fig. 8e). However, this loss of free HR23B in GFP-(GA)50 neurons was overcome by overexpressing HR23B despite the fact that a portion of HR23B remained sequestered in what poly(GA) inclusions remained (Fig. 8e). The recovery of nuclear HR23B likely accounted for the above-mentioned rescue of XPC levels (Fig. 8a). Taken together, these data implicate the involvement of impaired HR23 function in poly(GA) aggregation and neurotoxicity.
DISCUSSION
Here we provide evidence that (i) expression of poly(GA) proteins in the CNS of mice caused neurodegeneration and behavioral deficits; (ii) these abnormalities were dependent on the ability of poly(GA) to aggregate; (iii) poly(GA) inclusions sequestered various proteins, including those involved in nucleocytoplasmic transport and proteasomal protein degradation; (iv) HR23 protein pathology was present in c9FTD/ALS, (v) sequestration of HR23 led to its loss of function; and (vi) loss of HR23 protein function contributed to poly(GA) aggregation and toxicity.
It is well documented that various aggregation-prone proteins directly contribute to neurodegeneration, implicating c9RAN proteins in c9FTD/ALS pathogenesis. There is evidence that different c9RAN proteins induce cellular toxicity through a variety of pathways19–26. As poly(GA) pathology is particularly abundant in the brains of patients with c9FTD/ALS17,25,27, and because poly(GA) has proven toxic to cellular models20,22,25, we established a new poly(GA) mouse model to better understand the mechanisms underlying poly(GA) toxicity. We found that brains of mice expressing GFP-(GA)50 weighed less, contained abundant ubiquitin-positive poly(GA) inclusions composed of fibrils, and exhibited cortical, hippocampal and cerebellar cell loss. Mice expressing GFP-(GA)50 also developed behavioral, motor and memory deficits. The hippocampal neuronal loss in mice expressing GFP-(GA)50 likely accounted for their memory impairment. As we rarely detected poly(GA) proteins in the spinal cord (data not shown), deficits in motor coordination and balance likely stemmed from the loss of neurons in the motor cortex, particularly in layer V, and/or loss of Purkinje cells of the cerebellum, a region known to play a fundamental role in motor control. It is also possible that the hyperactivity observed in mice expressing GFP-(GA)50 influenced their performance in the rotarod test.
Unlike mice expressing GFP-(GA)50, mice expressing GFP-(GA)50-mut, a mutant form of (GA)50 that failed to aggregate, showed no signs of neurodegeneration nor of neuropathological and behavioral deficits. Thus, our data not only indicate that expression of poly(GA) is sufficient to induce neurodegeneration, but also that insoluble forms of the protein (i.e., oligomers or aggregates) are the toxic culprit. Given that the frequency of poly(GA) inclusions associates with cortical neurodegeneration only moderately27 if at all28,46, it seems likely that oligomers are the more likely effector, as is suspected to be the case for other aggregation-prone proteins in neurodegenerative disorders47.
Just as neurodegeneration was specific to mice expressing GFP-(GA)50, only GFP-(GA)50 and not GFP-(GA)50-mut caused the aberrant sequestration of HR23 proteins to poly(GA) inclusions, a pathological phenotype also observed in patients with c9FTD/ALS. These findings thus associate perturbations in HR23 proteins with neurodegeneration. Indeed, our data implicate poly(GA)-induced loss of HR23 protein function as a disease mechanism. HR23 proteins are responsible for transferring ubiquitinated proteins to the proteasome for degradation, and we discovered that ubiquitinated proteins and p62 accumulated in the brains of mice expressing GFP-(GA)50. We additionally observed a decrease in levels of XPC, a protein specifically stabilized by HR23 in the nucleus42. Prior reports indicate that loss of HR23 is detrimental; depletion of HR23B in mice results in a high rate of neonatal death with surviving mice displaying abnormalities including changes in behavior, such as jumping and circling45. It is also noteworthy that HR23 proteins form neuronal inclusions in patients with other repeat-expansion disorders, such as Huntington disease, spinocerebellar ataxias 3 and 7, and fragile X–associated tremor/ataxia syndrome48, suggesting HR23 impairment may be critical in various disorders.
We demonstrated that exogenous HR23B restored HR23 protein function and rescued the activation of caspase-3 that normally occurs upon GFP-(GA)50 expression in primary neurons. Exogenous HR23B decreased levels of high-molecular-weight poly(GA) and the formation of poly(GA) inclusions. These data suggest there is an interplay between HR23 proteins and poly(GA). Although this relationship requires further study, as does the mechanism by which HR23 proteins attenuate poly(GA) aggregation, several possible scenarios come to mind. For instance, because the decrease in high-molecular-weight poly(GA) was accompanied by an increase in monomeric poly(GA), HR23 proteins, perhaps upon binding to poly(GA) monomers, may stabilize the monomers and attenuate their oligomerization. Alternatively, given that we observed that soluble poly(GA) is recruited to poly(GA) aggregates, HR23 proteins may enhance proteasomal degradation of high-molecular-weight poly(GA) and thus reduce the sequestration of poly(GA) monomers to inclusions and their conversion to oligomeric species. Whether exogenous HR23B provided rescue primarily by preventing poly(GA) aggregation, or whether restoring nuclear HR23B function was also a key factor in mitigating poly(GA) toxicity, remains to be determined.
In addition to HR23 proteins, nuclear pore complex proteins, RanGAP1 and Pom121, localized with poly(GA) inclusions and formed abnormal puncta in mice expressing GFP-(GA)50. These data add to the growing body of evidence implicating nucleocytoplasmic defects as a key pathological mechanism of c9FTD/ALS. Unbiased screens to identify genetic modifiers or poly(PR) toxicity in yeast uncovered genes encoding nuclear import proteins, components of the nuclear pore complex, and regulators of the Ran-GTPase cycle needed to power nuclear import19. In a similar fashion, disrupted nucleocytoplasmic transport has been observed in Drosophila expressing (G4C2)58 (ref. 12) or (G4C2)30 (ref. 13). Further, cytoplasmic protein aggregates have been shown to interfere with the transport of proteins and RNA between the nucleus and cytoplasm29. Given that cytoplasmic poly(GA) inclusions are a prominent feature of c9FTD/ALS27, and given our findings of nuclear pore protein pathology in GFP-(GA)50 mice, poly(GA) may impair nucleocytoplasmic transport in c9FTD/ALS. Although this requires further study, it is also possible that nucleocytoplasmic transport defects might lead to an initial exclusion of HR23 proteins from the nucleus, leaving them vulnerable to cytoplasmic sequestration by poly(GA).
We recently generated mice that express 66 G4C2 repeats to study the consequences of the C9ORF72 repeat expansion in vivo39. By 6 months of age, mice expressing (G4C2)66 developed nuclear RNA foci and inclusions of c9RAN proteins, including poly(GA). Mice expressing (G4C2)66 were also marked by neuronal loss and behavioral deficits much like mice expressing GFP-(GA)50, suggesting that expression of poly(GA) may be the cause of these phenotypes. Likewise, that both mice expressing (G4C2)66 and mice expressing GFP-(GA)50 developed RanGAP1, Pom121 and HR23 protein pathology, suggests that poly(GA) expression causes or contributes to the development of these aberrant features in mice expressing (G4C2)66. However, because phosphorylated TDP-43 inclusions were rare in 6 month-old mice expressing GFP-(GA)50, poly(GA) expression is unlikely the cause of the TDP-43 pathology observed in mice expressing (G4C2)66 and in patients with c9FTD/ALS.
Overall, the generation of our mouse model led to the discovery of a new neuropathological feature of c9FTD/ALS: HR23 protein pathology. Our studies also provide compelling evidence that, through their high propensity to aggregate and by impairing HR23 protein function, as well as sequestering proteins involved in nucleocytoplasmic transport, poly(GA) proteins substantially contribute to the neurodegeneration in c9FTD/ALS. Finally, our findings indicate that small-molecule disruptors of poly(GA)’s conformation could offer a promising therapeutic strategy to inhibit poly(GA) aggregation and thus mitigate poly(GA)-mediated neurotoxicity.
ONLINE METHODS
Generation of (GA)50-mut plasmids
The plasmids encoding (GA)50-mut were generated using a previously described method25. In brief, a gene fragment that contains 50 GA dipeptide repeats interrupted by a proline every 5 repeats was synthesized by GeneArt, and then ligated to the HindIII and BamHI restriction sites of the pEGFP-C1 vector (Clontech Laboratories) and pAAV-EGFP fusion vector to generate GFP-(GA)50-mut and AAV-GFP-(GA)50-mut expression vectors, respectively, where expression is driven by the CMV enhancer-chicken beta-actin (CBA) promoter. To generate the GST-(GA)50-mut vector, PCR was performed to synthesize a cDNA fragment from (GA)50-mut, which was then inserted into a pGEX-6P-1 vector (GE Healthcare) using the BamHI and XhoI cloning sites. To generate the AAV-HR23B-Myc expression vector, a cDNA fragment encoding C-terminally Myc-tagged HR23B was synthesized by PCR using an HR23B plasmid as a template. This PCR product was then inserted into a pAAV vector using HindIII and XhoI cloning sites. The sequences of all plasmids was verified by sequence analysis.
Purification of recombinant proteins and transmission electron microscopy (TEM) analysis
Recombinant GST-(GA)50 and GST-(GA)50-mut proteins were purified according to methods we previously described25. Purified GST-(GA)50 or GST-(GA)50-mut proteins were dialyzed in Precision Protease Buffer and then incubated with PreScission Protease (GE Healthcare) for 16 h at 4 °C. The resulting cleaved products were extensively dialyzed in PBS. Next, the cleaved products were loaded onto a Glutathione Sepharose 4B column (GE Healthcare) and incubated for 10 min. Flow-through solutions containing untagged (GA)50 or (GA)50-mut proteins were collected and concentrated with Amicon spin filters (molecular weight cut-off (MWCO) of 3,500). To examine the filamentous structure of recombinant proteins by TEM, 10 μM untagged (GA)50 or (GA)50-mut were fixed to carbon grids, negatively stained with uranyl acetate, and imaged by TEM at 120,000× magnification.
Cell culture and preparation of cell lysates
HEK293T cells were grown in Opti-Mem plus 10% FBS and 1% penicillin-streptomycin. To examine the cellular distribution of GFP-(GA)50 or GFP-(GA)50-mut, cells grown on glass coverslips in 24-well plates were transfected with 0.3 μg of an expression vector encoding GFP-(GA)50 or GFP-(GA)50-mut using Lipofectamine 2000 (Life Technologies). After 48 h, cells were fixed with 4% paraformaldehyde in PBS for 15 min, and then permeabilized with PBS containing 0.5% Triton X-100 for 10 min. Afterward, Hoechst 33258 (1 μg/ml, Life Technologies) was used to stain cellular nuclei. Images were obtained on a Zeiss LSM 510 META confocal microscope.
To prepare the Triton X-100–soluble and insoluble fractions, cells grown in 6-well plates were transfected for 48 h with 1 μg of an expression vector (GFP-(GA)50 or GFP-(GA)50-mut). Then, cell pellets were lysed in co-immunoprecipitation (co-IP) buffer (50 mM Tris–HCl, pH 7.4, 300 mM NaCl, 1% Triton X-100, 5 mM EDTA) containing protease and phosphatase inhibitors. Lysates were sonicated on ice, and then centrifuged at 16,000g for 20 min. Supernatants were saved as the Triton X-100–soluble fraction. Pellets were dissolved in co-IP buffer plus 2% SDS, and both a protease and phosphatase inhibitor mixture. After sonication, lysates were centrifuged at 16,000g for 20 min, and the supernatants were saved as the Triton X-100–insoluble fraction. The protein concentration of the fractions was determined by BCA assay (Thermo Scientific), and samples were then subjected to immunoblot analysis.
To examine whether poly(GA) inclusions sequester HR23 proteins in the cytosol, cells grown in 6-well plates were transfected for 48 h with 2 μg of an expression vector (GFP, GFP-(GA)50 or GFP-(GA)50-mut). Then, fresh cell pellets were used to prepare nuclear and cytoplasmic fractions using a Nuclear/Cytosol Fractionation Kit (BioVision Inc.) according to the manufacturer’s instructions with some modifications. Briefly, fresh cell pellets were lysed in cytosol extraction buffer, and then centrifuged at 800g for 10 min at 4 °C. Supernatants were saved as the cytoplasmic fraction. The pellets were re-extracted with cytosol extraction buffer. After centrifugation, the resulting nuclear pellets were lysed in nuclear extraction buffer, and then centrifuged at 16,000g for 10 min at 4 °C. The supernatants were saved as the nuclear fraction. The protein concentration of the fractions was determined as described above, and samples were then subjected to immunoblot analysis.
Mouse studies
All procedures in this study using mice were performed in accordance with the National Institutes of Health Guide for Care and Use of Experimental Animals and approved by the Mayo Clinic Institutional Animal Care and Use Committee.
Primary neuronal cultures and their treatment
Primary neuronal cultures were prepared as previously described25 with slight modifications. In brief, fetal cortices were dissected in Hibernate A medium without calcium (BrainBits) and subsequently transferred into 10 ml of growth medium consisting of Neurobasal A (media Life Technologies) supplemented with B27, GMAX, gentamicin and bFGF (Life Technologies). Cells were dissociated in growth medium and strained through a 40 μm Falcon cell strainer (BD Bioscience) to remove cellular debris and to obtain a uniform, single-cell suspension. Neurons were seeded at a density of 7 × 105 cells/well in 6-well plates. After 4 d in culture, neurons were transduced with 1 × 1010 AAV1 genome particles of GFP, GFP-(GA)50 or GFP-(GA)50-mut. Seven days post-transduction, medium was collected for the LDH assay (Promega), and cell pellets were lysed in co-IP buffer with 2% SDS, as well as protease and phosphatase inhibitors. Lysates were sonicated and centrifuged at 16,000g for 20 min. Supernatants were saved for BCA assay to determine the protein concentration of samples before immuoblot. To examine whether overexpression of HR23B protects neurons from poly(GA)- or poly(GR)-induced toxicity, neurons were co-transduced with rAAV1 particles of GFP-(GA)50 or GFP-(GR)50 (0.75 × 1010 AAV1 genome particles) and either vector (0.5 × 1010 AAV1 genome particles) or HR23B-Myc (0.5 × 1010 AAV1 genome particles). Seven days post-transduction, neurons were harvested and cell lysates were prepared for immunoblot analysis as described above. To examine whether overexpression of HR23B inhibits formation of poly(GA) inclusions in mouse primary neurons, neurons were seeded at a density of 6 × 104 cells/coverslip in 24-well plates and co-transduced with rAAV1 virus of GFP-(GA)50 (0.3 × 1010 AAV1 genome particles) and either vector (0.2 × 1010 AAV1 genome particles) or HR23B-Myc (0.2 × 1010 AAV1 genome particles). Neurons were fixed at the indicated time-points for immunostaining as described below.
Intracerebroventricular injections of virus in neonatal mouse brain
ICV injections of AAV1 virus in mouse brain were performed according to previous reports49,50. In brief, at postnatal day 0 (P0) C57BL/6J pups were cryoanesthetized on ice until pups exhibited no movement. Next, a 32-gauge Hamilton needle attached to a 10 μl Hamilton syringe was inserted at approximately two-fifths the distance between the lambda and the eye of the pups. The needle was inserted perpendicular to the surface of the head and held at a depth of ~2 mm. Two microliters (1 × 1010 genomes/μl) of AAV1-GFP, GFP-(GA)50 or GFP-(GA)50-mut solution were manually injected into each cerebral ventricle. After injections, pups were placed on a heat pad until they completely recovered from anesthesia and were then placed back into their home cages.
Tissue processing
Sagittal half brains were immersion fixed in 10% formalin, embedded in paraffin, sectioned (5 μm thick), and then mounted on glass slides for immunofluorescence and immunohistochemistry staining. The cortex and hippocampus of the other half brain were dissected and frozen on dry ice. To further process the tissue, the frozen cortex and hippocampus were homogenized in ice-cold buffer (50 mM Tris pH 7.4, 50 mM NaCl and 1 mM EDTA) with 2× protease and phosphatase inhibitors. The resulting homogenates were used for RNA extraction and preparation of brain lysates. For immunohistochemical staining of XPC, mice were perfused with 0.9% saline followed by 4% paraformaldehyde. Afterward, brains were harvested and cut sagittally across the midline, placed in 4% paraformaldehyde, and then embedded in paraffin for sectioning.
Immunofluorescence staining
Paraffin sections of mouse and human brains were subjected to immunofluorescence staining, as we previously described51. Information on human subjects is provided in Supplementary Table 3. In brief, paraffin sections (5 μm) were deparaffinized and rehydrated in xylene and a graded series of alcohol (100%, 100%, 95% and 70%). Afterward, the sections were steamed for 30 min in Dako antigen retrieval solution or, when staining for RanGAP1 and Pom121, with HC-TekTM Epitope Retrieval Solution (IW-1100; IHC World; sodium citrate buffer). Sections were then blocked with Dako All Purpose Blocker for 1 h at room temperature (RT). To examine the cellular distribution of GFP, GFP-(GA)50 or GFP-(GA)50-mut, the sections were incubated overnight at 4 °C with a rabbit polyclonal anti-GFP antibody (A-6455, 1:1,000, Life Technologies) diluted in Dako Antibody Diluent. To determine whether poly(GA) inclusions are ubiquitin-positive, the sections were incubated with a rabbit polyclonal anti-GFP antibody (A-6455, 1:1,000, Life Technologies) and a mouse monoclonal anti-ubiquitin antibody (clone Ubi-1, 1:250, EMD Millipore). To examine what cell type has poly(GA) inclusions in mouse brain, sections were incubated with rabbit polyclonal anti-GFP antibody (A-6455, 1:1,000, Life Technologies), and mouse monoclonal anti-MAP2 (M1406, 1:200, Sigma) or mouse monoclonal anti-GFAP antibody (MU020-UC, 1:250, BioGenex). To determine if HR23 proteins co-localize with poly(GA) inclusions in mouse and human brain, sections were incubated with mouse monoclonal anti-HR23A (ab55725, 1:250, Abcam) or mouse monoclonal anti-HR23B antibody (ab88503, 1:250, Abcam), and rabbit polyclonal anti-GFP antibody (A-6455, 1:1,000, Life Technologies; used for mouse brain) or rabbit polyclonal anti-poly(GA) antibody (1:1,000; used for human brain). To examine the co-localization of HR23B proteins with c9RAN proteins in (G4C2)66 mouse and human brains, sections were incubated with mouse monoclonal anti-HR23B antibody (ab88503, 1:250, Abcam), and rabbit polyclonal anti-poly(GA) (1:1,000), rabbit polyclonal anti-poly(GP) (1:1,000) or rabbit polyclonal anti-poly(GR) (1:1,000). To examine whether XPC proteins co-localize with poly(GA) inclusions in mouse brains, the sections were incubated with rabbit polyclonal anti-XPC antibody (sc-30156, 1:50, Santa Cruz Biotechnology) and mouse monoclonal anti-GFP antibody (MAB3580, 1:250, EMD Millipore). To examine the distribution of RanGAP1 and Pom121, we used rabbit anti-RanGAP1 (SC-25630; 1:100; Santa Cruz) or rabbit anti-Pom121 (PA5-27623; 1:100; Thermo Fisher Scientific), respectively; these were coupled with mouse anti-GFP (MAB3580; 1:250; EMD Millipore) when investigating the co-localization of these proteins with GFP-(GA)50. After washing, sections were incubated with the corresponding Alexa Fluor 488–conjugated donkey or goat anti-mouse, Alexa Fluor 488–conjugated donkey anti-rabbit, Alexa Fluor 568–conjugated donkey anti-rabbit, or Alexa Fluor 555 goat anti-rabbit secondary antibody (1:500–1:1,000, Molecular Probes) at RT for 2 h. Hoechst 33258 (1 μg/ml, Life Technologies) was used to stain cellular nuclei or a mounting medium containing DAPI was used. Fixed primary neurons grown on glass coverslips were permeabilized with PBS and 0.5% Triton X-100 for 10 min. After blocking with 5% nonfat milk for 1 h at 37 °C, cells were incubated overnight at 4 °C with rabbit polyclonal MAP2 antibody (1:1,000, AB5622, EMD Millipore) and mouse monoclonal anti-HR23B antibody (ab88503, 1:250, Abcam). After washing, cells were incubated with the Alexa Fluor 568–conjugated donkey anti-mouse and Alexa 633-conjugated donkey anti-rabbit secondary antibody (1:500, Molecular Probes) at 37 °C for 2 h. Hoechst 33258 was used to stain nuclei. Images were obtained on a Zeiss LSM 510 META confocal microscope or a Zeiss LSM700 laser scanning confocal microscope.
Immunohistochemistry staining
Paraffin sections of mouse and human brain were subjected to immunohistochemistry staining as previously described48,51. In brief, following deparaffinization and rehydration, sections were pretreated by steaming for 30 min in dH2O for target retrieval, and endogenous peroxidase was blocked for 5 min with 0.03% hydrogen peroxide. Sections were then treated with 5% normal goat serum for 20 min at RT. Subsequently, sections were incubated in primary antibody for 45 min at RT using mouse monoclonal anti-ubiquitin antibody (clone Ubi-1, 1:60,000, EMD Millipore), rabbit polyclonal anti-GFP antibody (A-6455, 1:1,000, Life Technologies), rabbit polyclonal anti-GA antibody (1:50,000), rabbit polyclonal anti-GFAP antibody (PU020-UP, 1:2,500, Biogenex), rabbit polyclonal anti-Iba1 (019-19741, 1:3,000, Wako Chemicals), mouse monoclonal anti-NeuN (MAB377, 1:5.000, Chemicon International), rabbit polyclonal anti-phosphorylated TDP-43 (Serine 409/410) antibody39, mouse monoclonal anti-HR23A (ab55725, 1:250, Abcam), mouse monoclonal anti-HR23B antibody (ab88503, 1:250, Abcam) or rabbit polyclonal anti-XPC antibody (sc-30156, 1:50, Santa Cruz Biotechnology). After incubation in primary antibodies, sections were incubated for 30 min at RT in Dako Envision-Plus rabbit or mouse labeled polymer HRP. Peroxidase labeling was visualized with the chromogen solution 3, 3′-diaminobenzidine (DAB-Plus). Sections were then counterstained with Lerner 1 hematoxylin (Fisher Scientific) and coverslipped with Cytoseal mounting medium (Thermo Scientific).
Post-embedding immunoelectron microscopy (immunoEM)
To examine the filamentous structure of poly(GA) proteins in mouse brain, immunoEM was performed as previously described52. Thin sections were pretreated with sodium citrate buffer, pH 6 at 90–95 °C for 10 min before immunolabeling. Rabbit polyclonal antibody to poly(GA) (1:150 in PBS) was used as the primary antibody, and goat anti-rabbit IgG conjugated with 18 nm colloidal gold particles (1:20, Jackson ImmunoResearch Laboratories) was used as the secondary antibody. Thin sections stained with uranyl acetate and lead citrate were examined with a Philips 208S electron microscopy (FEI) fitted with a Gatan 831 Orius charge-coupled device (CCD) camera (Gatan). Digital images were processed with Adobe Photoshop CS5 software.
Quantification of neuropathology
Aperio ePathology technology (Leica Biosystems) was used to quantify the percent burden of immunohistochemical Iba1 and GFAP staining, as well as NeuN-positive neuronal density in a blinded fashion. High-resolution digitized images of immunostained slides were obtained using a ScanScope AT2 (Leica Biosystems). The whole cortex and the motor cortex were annotated on mid-sagittal serial sections of Iba1, GFAP and NeuN for each mouse using ImageScope software (v12.1; Leica Biosystems). Iba1-positive microglia and GFAP-positive astrocytes, labeled brown by chromagen 3,3′-diaminobenzine, were quantified using two custom-designed positive pixel count algorithms53. From the annotated area, the number of positively stained pixels as a proportion of all pixels was used as the output parameter for both Iba1 and GFAP. The rounded appearance and staining intensity of NeuN-positive neurons was used to design a custom-designed Nuclear Algorithm. A NeuN-positive neuronal density measure was calculated as the number of neurons per given square millimeter of the annotated area. The NeuN stain was additionally used to semi-quantify the severity of hippocampal CA3 neuronal loss using a four-point scale: none (0: no neuronal loss), mild (1: slightly disrupted bi-layer of the CA3 region that enters the dentate gyrus), moderate (2: extensive loss of neuronal bi-layer), severe (3: extensive loss of neurons throughout the CA3 region). To quantify the Purkinje cells in the cerebellum, hematoxylin and eosin (H&E)-stained slides were scanned as described above. The number of Purkinje cells was counted along a total of 10 mm of the Purkinje layer of the cerebellum for each mouse, and then calculated as the average number of Purkinje cells per millimeter.
Silver staining
Mice were anesthetized and perfused with saline for 1 min followed by 4% paraformaldehyde in 0.1 M phosphate buffer (PB, pH 7.4). Brains were removed and fixed in 4% paraformaldehyde in PB overnight at 4 °C. Afterward, brains were immersed in 0.1 M PB containing 20% sucrose for 48 h at 4 °C and then frozen for sectioning. Sections (40 μm thickness) were cut with a cryostat microtome (Leica Biosystems) and stored in 4% paraformaldehyde in PB buffer at 4 °C. After 5 d, the sections were subjected to silver staining using FD NeuroSilver kit II (PK301A, FD Neurotechnologies) according to the manufacturer’s instructions.
RNA extraction, reverse transcription and quantitative real-time PCR (qPCR)
To extract total RNA, one volume of the homogenate was immediately mixed with three volumes of Trizol LS Reagent (Life Technologies), and then frozen on dry ice. One day later, total RNA was extracted using the Direct-zol RNA MiniPrep kit (Zymo Research) according to the manufacturer’s instructions, combined with an in-column DNase I digestion step. Total RNA from primary neurons was extracted using the RNAeasy Plus Mini Kit (QIAGEN) as per the manufacturer’s instructions, combined with an in-column DNase I digestion step. cDNA was obtained after reverse transcription using 1,000 ng of RNA with random primers and the High Capacity cDNA Transcription Kit (Applied Biosystems) per the manufacturer’s instructions.
To quantify mRNA levels of GFP, GFP-(GA)50 or GFP-(GA)50-mut, and gliosis markers, GFAP and Iba1, in mouse brain and/or primary neurons, qPCR was conducted in triplicate for all samples using SYBR green assay (Life Technologies) on an ABI Prism 7900HT Fast Real-Time PCR System (Applied Biosystems). The primers used were: GFP: 5′-GAAG CGCGATCACATGGT-3′ and 5′-CCATGCCGAGAGTGATCC-3′; Gfap: 5′-GGAGAGGGACAACTTTGCAC-3′ and 5′-AGCCTCAGGTTGGTTTCATC-3′; Iba1: 5′-GGATTTGCAGGGAGGAAAAG-3′ and 5′-TGGGATCATCGAG GAATTG-3′; Gapdh: 5′-CATGGCCTTCCGTGTTCCTA-3′ and 5′-CCTGCTTCA CCACCTTCTTGAT-3. Relative mRNA expression of examined genes was normalized to GAPDH values, an endogenous transcript control.
Preparation of brain lysates
To prepare Triton X-100-soluble and insoluble fractions, Triton X-100 was added into the homogenate at a final concentration of 1%. After sonication, the lysates were centrifuged, and the soluble and insoluble fractions were prepared as described above. These soluble and insoluble fractions were used for immunoblot analysis to examine the solubility of GFP, GFP-(GA)50 or GFP-(GA)50-mut proteins. In addition, the soluble fraction was subjected to ELISA to measure protein levels of GFAP, as previously described54. To prepare total lysates, Triton X-100 and SDS were added to the homogenate at a final concentration of 1% and 2%, respectively. After sonication, the lysates were centrifuged at 16,000g for 20 min and the supernatants were saved for BCA assay (Thermo Scientific) to determine protein concentration.
Co-immunoprecipitation
Co-IP studies were performed as previously described55. In brief, HEK293T cells grown in 6-well plates were transfected for 48 h with 1 μg of an expression vector encoding GFP, GFP-(GA)50 or GFP-(GA)50-mut. Cell pellets were lysed in co-IP buffer containing protease and phosphatase inhibitors. After sonication on ice, the lysates were centrifuged at 16,000g for 20 min and the protein concentration of the resulting supernatants was determined by BCA assay. Supernatants containing 350 μg of total protein were precleared with 15 μl Dynabeads Protein G (Life Technologies), and then incubated with a rabbit polyclonal anti-GFP antibody (ab290, 1 μl, Abcam; used for GFP pulldown), mouse monoclonal anti-HR23A (ab55725, 1 μl, Abcam; used for HR23A pulldown) or mouse monoclonal anti-HR23B antibody (ab86781, 3 μl, Abcam; used for HR23B pulldown) overnight at 4 °C with gentle shaking. The antigen-antibody immuno-complex was captured by Dynabeads Protein G for 4 h, and then the beads were separated using a magnetic tube stand. After washing three times with co-IP buffer, captured proteins were eluted from the beads using loading buffer, and then the samples were subjected to immunoblot analysis.
Immunoblot analysis
Immunoblot analysis was performed as we previously described25,55. In brief, samples were heated in Laemmli’s buffer, and equal amounts of protein were loaded into 10-well 10% or 4–20% Tris-glycine gels (Novex). After transferring proteins to membranes, membranes were blocked with 5% nonfat dry milk in Tris-buffered saline containing 0.1% Triton X-100 (TBST) for 1 h, then incubated with a rabbit polyclonal anti-GFP antibody (A-6455, 1:4,000, Life Technologies), rabbit polyclonal anti-activated caspase-3 antibody (9661, 1:1,000, Cell signaling), mouse monoclonal anti-HR23A (ab55725, 1:1,000, Abcam), mouse monoclonal anti-HR23B antibody (ab88503, 1:2,000, Abcam), rabbit polyclonal anti-XPC antibody (sc-30156, 1:200, Santa Cruz Biotechnology), mouse monoclonal anti-Tubulin antibody (T5168, 1:2,000, Sigma), rabbit polyclonal anti Lamin A/C antibody (sc-6215, 1:100, Santa Cruz Biotechnology), rabbit polyclonal anti-poly(GR) (1:2,000), mouse monoclonal anti-Myc antibody (9E10, 1:2,000, Roche) or mouse monoclonal GAPDH antibody (H86504M, 1:10,000, Meridian Life Science) overnight at 4 °C. Membranes were washed in TBST and incubated with donkey anti-rabbit or anti-mouse IgG antibodies conjugated to horseradish peroxidase (1:5,000; Jackson ImmunoResearch) for 1 h. Protein expression was visualized by enhanced chemiluminescence treatment and exposure to film. The intensity of bands was quantified by Fuji Film MultiGauge Software, and then normalized to the corresponding controls. Full-length immunoblots are presented in Supplementary Figures 9 and 10.
Immunoassay analysis of poly(GA) proteins in mouse brain tissues
Mouse brain lysates were prepared as described above and poly(GA) levels in lysates were measured using a previously characterized sandwich immunoassay that specifically detects poly(GA) and other c9RAN proteins56. Lysates were diluted in Tris-buffered saline (TBS) and an equal amount of protein for all samples was tested in duplicate wells. Serial dilutions of recombinant (GA)50 in TBS were used to prepare the standard curve. Response values corresponding to the intensity of emitted light upon electrochemical stimulation of the assay plate using the MSD QUICKPLEX SQ120 were acquired and background corrected using the average response from lysates obtained from control mice (i.e., mice expressing (G4C2)2 or mice expressing only GFP) before interpolation of poly(GA) levels (presented as ng of poly(GA) per mg protein) using the standard curve.
Behavioral tests
Six-month-old mice expressing GFP (n = 12; 8 male, 4 female), GFP-(GA)50 (n = 12; 8 male, 4 female) or GFP-(GA)50-mut (n = 10; 6 male, 4 female) were subjected to behavioral analysis conducted in a blinded fashon on two consecutive weeks at the Mayo Mouse Behavior Core. On four consecutive days of week 1, the mice underwent a rotarod test. On 3 consecutive days of week 2, the mice were subjected to the open-field assay on day 1, and then to contextual and cued fear conditioning tests on days 2 and 3. All mice were acclimated to the room 1 h before testing, and then returned to their home cage and homeroom after each test.
Rotarod test
Motor performance of mice was assessed using an automated rotarod system (Med Associates, Inc). Before the training sessions, each mouse was habituated to stay on the spindle with slow constant speed for 1 min, and then the spindle was accelerated from 4 r.p.m. to 40 r.p.m. The latency to fall time was recorded when the mouse fell off the spindle, triggering a sensor that automatically stops the timer. Four consecutive trials were performed in a day and the test was repeated for 4 d.
Open-field assay
The open-field assay was performed as previously described57. The apparatus consists of a square Perspex box (40 cm × 40 cm × 30 cm, width × length × height) with side-mounted photobeams raised 7.6 cm above the floor to measure rearing and an overhead Fujinon camera to record behavioral activity. An imaginary 20 cm × 20 cm region in the center of the box and a corresponding perimeter region were defined using Anymaze software (Stoelting Co). For the test, each mouse was placed in a box and behavioral activity was recorded for 15 min and tracked using Anymaze software. Mice were analyzed for multiple measures, including rearing time to assess motor coordination, total distance traveled to measure hyperactivity, and center:total distance ratios to examine anxiety.
Context and cue fear conditioning assay
The context and cue fear conditioning assay was performed as previously described57. The apparatus consists of a sound attenuated chamber with a grid floor capable of delivering an electric shock, and freezing was recorded and measured with an overhead camera and FreezeFrame software (Actimetrics), respectively. On day 1, each mouse was placed into the chamber and baseline freezing behavior was recorded during the first two min. Then, mice were exposed to 80-dB white noise for 30 s as the conditioned stimulus (CS). During the final 2 s of this noise, mice received a mild foot shock (0.5 mA) as the unconditioned stimulus (US). After 1 min, mice were subjected to another CS-US pair stimulus, and then returned to their home cages and homeroom. On day 2, each mouse was returned to the test chamber and freezing behavior was recorded for 5 min (context test). Mice were returned to their home cage and placed in a different room with reduced lighting for at least 1 h. Moreover, environmental and contextual cues were changed. For the cued test, each mouse was placed in the modified chamber for 3 min, and then exposed to the auditory CS and freezing behavior was recorded for another 3 min (cue test). Baseline freezing behavior obtained during training was subtracted from the context or cue tests to control for variability in each animal.
Statistics
Data are presented as mean ± s.e.m., and were analyzed with one-way ANOVA followed by Tukey’s post hoc analysis, two-way ANOVA followed by Sidak post hoc analysis or unpaired t-test (Prism statistical software). Data distribution was assumed to be normal, but this was not formally tested. No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to those reported in previous publications39. For our mouse studies, all mice were subjected to the behavioral battery. Given our need for brain tissues for various biochemical and histological examinations, not all mice were included in each of these studies. If tissues from a subset of mice were used for a particular study, they were randomly chosen among the full group. With the exception of collecting data for behavioral studies, data collection and analysis were not performed blind to the conditions of the experiments.
Supplementary Material
Acknowledgments
We are grateful to all patients who agreed to donate post-mortem tissue. This work was supported by the US National Institutes of Health (NIH) National Institute on Aging (R01AG026251 (L.P.)); NIH National Institute of Neurological Disorders and Stroke (R21NS079807 (Y.-J.Z. and J.D.F.); R21NS089979 (T.F.G. and K.B.B.); F32NS087842 (J.J.); R01NS080882 (R.R.); R01NS085207 (J.D.R.); U54NS091046 (J.D.R.); R01NS063964 (L.P.); R01NS077402 (L.P.), R21NS084528 (L.P.); P01NS084974 (L.P., D.D., K.B.B. and R.R.); R01NS088689 (L.P.)); National Institute of Environmental Health Services (R01ES20395 (L.P.); Department of Defense (ALSRP AL130125 (L.P.)); Mayo Clinic Foundation (L.P.); Mayo Clinic Center for Individualized Medicine (L.P. and K.B.B.); Alzheimer’s Association (NIRP-14-304425 (Y.-J.Z.); NIRP-12-259289 (J.D.F.)); Amyotrophic Lateral Sclerosis Association (Y.-J.Z., T.F.G., K.B.B., D.W.C. and L.P.); Robert Packard Center for ALS Research at Johns Hopkins (J.D.R. and L.P.), Target ALS (C.L.-T., J.D.R. and L.P.); Brain Science Institute (J.D.R.); the Ludwig Institute for Cancer Research (D.W.C. and C.L.T.), and the European Union’s Seventh Framework Programme (FP7/2014-2019 grant 617198 (D.E.)). J.C.G. is the recipient of a National Science Foundation Graduate Research Fellowship Award, a Thomas Shortman Training Fund Graduate Scholarship and an Axol Science Scholarship.
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
L.P. and Y.-J.Z. contributed to the conception and design. Y.-J.Z. performed immunoblots, quantitative reverse-transcription PCR (qRT-PCR), co-immunoprecipitation and behavioral tests; T.F.G. completed anti-GA antibody generation and characterization, and performed poly(GA) assays with L.D.; J.C.G. and J.D.R. performed immunofluorescence staining for RanGAP1 and Pom121. H.S. performed ICV injection and behavioral tests; Y.-F.X. performed silver staining, immunofluorescence staining and primary neuronal cultures; Y.-F.X. and Z.S.W. quantified the Purkinje cells in cerebellum. M.E.M. and A.M.L. quantified neuronal loss and gliosis burden; M.S. and G.B. contributed to ELISA; W.-L.L. performed immunoEM; J.G. and A.G. performed immunofluorescence staining and immunoblotting; J.N.S. prepared primary neurons; K.J.-W. made plasmids; J.T. and M.Y. harvested mice and prepared brain lysates; E.A.P. produced AAV1; J.C. aided with ICV injections; M.C.-C. performed immunohistochemistry staining; A.K. and J.D.F. contributed to behavioral tests; J.D.B. and C.A.D. contributed to the purification of recombinant protein and the transmission electron microscopy study; J.J., C.L.-T., D.E. and D.W.C. characterized and provided anti-poly(GA) antibodies. R.R., K.B.B. and D.W.D. contributed to the tissue collection. C.D.L. analyzed data. L.P., Y.-J.Z., T.F.G. and R.B.K. analyzed data and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.DeJesus-Hernandez M, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Renton AE, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belzil VV, et al. Reduced C9orf72 gene expression in c9FTD/ALS is caused by histone trimethylation, an epigenetic event detectable in blood. Acta Neuropathol. 2013;126:895–905. doi: 10.1007/s00401-013-1199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu EY, et al. C9orf72 hypermethylation protects against repeat expansion-associated pathology in ALS/FTD. Acta Neuropathol. 2014;128:525–541. doi: 10.1007/s00401-014-1286-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xi Z, et al. Hypermethylation of the CpG island near the G4C2 repeat in ALS with a C9orf72 expansion. Am J Hum Genet. 2013;92:981–989. doi: 10.1016/j.ajhg.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Blitterswijk M, et al. Novel clinical associations with specific C9ORF72 transcripts in patients with repeat expansions in C9ORF72. Acta Neuropathol. 2015;130:863–876. doi: 10.1007/s00401-015-1480-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gendron TF, Belzil VV, Zhang YJ, Petrucelli L. Mechanisms of toxicity in C9FTLD/ALS. Acta Neuropathol. 2014;127:359–376. doi: 10.1007/s00401-013-1237-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sareen D, et al. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci Transl Med. 2013;5:208ra149. doi: 10.1126/scitranslmed.3007529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donnelly CJ, et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80:415–428. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper-Knock J, et al. Sequestration of multiple RNA recognition motif-containing proteins by C9orf72 repeat expansions. Brain. 2014;137:2040–2051. doi: 10.1093/brain/awu120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee YB, et al. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep. 2013;5:1178–1186. doi: 10.1016/j.celrep.2013.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freibaum BD, et al. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature. 2015;525:129–133. doi: 10.1038/nature14974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang K, et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015;525:56–61. doi: 10.1038/nature14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gendron TF, et al. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 2013;126:829–844. doi: 10.1007/s00401-013-1192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ash PE, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mori K, et al. Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol. 2013;126:881–893. doi: 10.1007/s00401-013-1189-3. [DOI] [PubMed] [Google Scholar]
- 17.Mori K, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339:1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- 18.Zu T, et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci USA. 2013;110:E4968–E4977. doi: 10.1073/pnas.1315438110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jovičić A, et al. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci. 2015;18:1226–1229. doi: 10.1038/nn.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamakawa M, et al. Characterization of the dipeptide repeat protein in the molecular pathogenesis of c9FTD/ALS. Hum Mol Genet. 2014;24:1630–1645. doi: 10.1093/hmg/ddu576. [DOI] [PubMed] [Google Scholar]
- 21.Mizielinska S, et al. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science. 2014;345:1192–1194. doi: 10.1126/science.1256800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.May S, et al. C9orf72 FTLD/ALS-associated Gly-Ala dipeptide repeat proteins cause neuronal toxicity and Unc119 sequestration. Acta Neuropathol. 2014;128:485–503. doi: 10.1007/s00401-014-1329-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wen X, et al. Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron. 2014;84:1213–1225. doi: 10.1016/j.neuron.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwon I, et al. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science. 2014;345:1139–1145. doi: 10.1126/science.1254917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang YJ, et al. Aggregation-prone c9FTD/ALS poly(GA) RAN-translated proteins cause neurotoxicity by inducing ER stress. Acta Neuropathol. 2014;128:505–524. doi: 10.1007/s00401-014-1336-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tao Z, et al. Nucleolar stress and impaired stress granule formation contribute to C9orf72 RAN translation-induced cytotoxicity. Hum Mol Genet. 2015;24:2426–2441. doi: 10.1093/hmg/ddv005. [DOI] [PubMed] [Google Scholar]
- 27.Mackenzie IR, et al. Quantitative analysis and clinico-pathological correlations of different dipeptide repeat protein pathologies in C9ORF72 mutation carriers. Acta Neuropathol. 2015;130:845–861. doi: 10.1007/s00401-015-1476-2. [DOI] [PubMed] [Google Scholar]
- 28.Schludi MH, et al. Distribution of dipeptide repeat proteins in cellular models and C9orf72 mutation cases suggests link to transcriptional silencing. Acta Neuropathol. 2015;130:537–555. doi: 10.1007/s00401-015-1450-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woerner AC, et al. Cytoplasmic protein aggregates interfere with nucleo-cytoplasmic transport of protein and RNA. Science. 2015;351:173–176. doi: 10.1126/science.aad2033. [DOI] [PubMed] [Google Scholar]
- 30.Fecto F, et al. SQSTM1 mutations in familial and sporadic amyotrophic lateral sclerosis. Arch Neurol. 2011;68:1440–1446. doi: 10.1001/archneurol.2011.250. [DOI] [PubMed] [Google Scholar]
- 31.Le Ber I, et al. French Clinical and Genetic Research Network on FTD/FTD-ALS. SQSTM1 mutations in French patients with frontotemporal dementia or frontotemporal dementia with amyotrophic lateral sclerosis. JAMA Neurol. 2013;70:1403–1410. doi: 10.1001/jamaneurol.2013.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng HX, et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477:211–215. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dougan L, Li J, Badilla CL, Berne BJ, Fernandez JM. Single homopolypeptide chains collapse into mechanically rigid conformations. Proc Natl Acad Sci USA. 2009;106:12605–12610. doi: 10.1073/pnas.0900678106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Popiel HA, et al. Disruption of the toxic conformation of the expanded polyglutamine stretch leads to suppression of aggregate formation and cytotoxicity. Biochem Biophys Res Commun. 2004;317:1200–1206. doi: 10.1016/j.bbrc.2004.03.161. [DOI] [PubMed] [Google Scholar]
- 35.Pelassa I, et al. Association of polyalanine and polyglutamine coiled coils mediates expansion disease-related protein aggregation and dysfunction. Hum Mol Genet. 2014;23:3402–3420. doi: 10.1093/hmg/ddu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiba T, et al. Amyloid fibril formation in the context of full-length protein: effects of proline mutations on the amyloid fibril formation of beta2-microglobulin. J Biol Chem. 2003;278:47016–47024. doi: 10.1074/jbc.M304473200. [DOI] [PubMed] [Google Scholar]
- 37.Mocanu MM, et al. The potential for beta-structure in the repeat domain of tau protein determines aggregation, synaptic decay, neuronal loss, and coassembly with endogenous Tau in inducible mouse models of tauopathy. J Neurosci. 2008;28:737–748. doi: 10.1523/JNEUROSCI.2824-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dantuma NP, Heinen C, Hoogstraten D. The ubiquitin receptor Rad23: at the crossroads of nucleotide excision repair and proteasomal degradation. DNA Repair (Amst) 2009;8:449–460. doi: 10.1016/j.dnarep.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 39.Chew J, et al. Neurodegeneration. C9ORF72 repeat expansions in mice cause TDP-43 pathology, neuronal loss, and behavioral deficits. Science. 2015;348:1151–1154. doi: 10.1126/science.aaa9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mitchell JM, Mansfeld J, Capitanio J, Kutay U, Wozniak RW. Pom121 links two essential subcomplexes of the nuclear pore complex core to the membrane. J Cell Biol. 2010;191:505–521. doi: 10.1083/jcb.201007098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Antonin W, Franz C, Haselmann U, Antony C, Mattaj IW. The integral membrane nucleoporin pom121 functionally links nuclear pore complex assembly and nuclear envelope formation. Mol Cell. 2005;17:83–92. doi: 10.1016/j.molcel.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 42.Ng JM, et al. A novel regulation mechanism of DNA repair by damage-induced and RAD23-dependent stabilization of xeroderma pigmentosum group C protein. Genes Dev. 2003;17:1630–1645. doi: 10.1101/gad.260003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Melis JPM, Luijten M, Mullenders LHF, van Steeg H. The role of XPC: implications in cancer and oxidative DNA damage. Mutat Res. 2011;728:107–117. doi: 10.1016/j.mrrev.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Rohde LH, Wu H. Involvement of nucleotide excision and mismatch repair mechanisms in double strand break repair. Curr Genomics. 2009;10:250–258. doi: 10.2174/138920209788488544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ng JM, et al. Developmental defects and male sterility in mice lacking the ubiquitin-like DNA repair gene mHR23B. Mol Cell Biol. 2002;22:1233–1245. doi: 10.1128/MCB.22.4.1233-1245.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davidson Y, et al. Neurodegeneration in frontotemporal lobar degeneration and motor neurone disease associated with expansions in C9orf72 is linked to TDP-43 pathology and not associated with aggregated forms of dipeptide repeat proteins. Neuropathol Appl Neurobiol. doi: 10.1111/nan.12292. Published online 5 November 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Todd TW, Lim J. Aggregation formation in the polyglutamine diseases: protection at a cost? Mol Cells. 2013;36:185–194. doi: 10.1007/s10059-013-0167-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bergink S, et al. The DNA repair-ubiquitin-associated HR23 proteins are constituents of neuronal inclusions in specific neurodegenerative disorders without hampering DNA repair. Neurobiol Dis. 2006;23:708–716. doi: 10.1016/j.nbd.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 49.Chakrabarty P, et al. Capsid serotype and timing of injection determines AAV transduction in the neonatal mice brain. PLoS One. 2013;8:e67680. doi: 10.1371/journal.pone.0067680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim JY, et al. Viral transduction of the neonatal brain delivers controllable genetic mosaicism for visualising and manipulating neuronal circuits in vivo. Eur J Neurosci. 2013;37:1203–1220. doi: 10.1111/ejn.12126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu YF, et al. Wild-type human TDP-43 expression causes TDP-43 phosphorylation, mitochondrial aggregation, motor deficits, and early mortality in transgenic mice. J Neurosci. 2010;30:10851–10859. doi: 10.1523/JNEUROSCI.1630-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin WL, Dickson DW, Sahara N. Immunoelectron microscopic and biochemical studies of caspase-cleaved tau in a mouse model of tauopathy. J Neuropathol Exp Neurol. 2011;70:779–787. doi: 10.1097/NEN.0b013e31822ac9c9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murray ME, et al. A quantitative postmortem MRI design sensitive to white matter hyperintensity differences and their relationship with underlying pathology. J Neuropathol Exp Neurol. 2012;71:1113–1122. doi: 10.1097/NEN.0b013e318277387e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shinohara M, Petersen RC, Dickson DW, Bu G. Brain regional correlation of amyloid-β with synapses and apolipoprotein E in non-demented individuals: potential mechanisms underlying regional vulnerability to amyloid-β accumulation. Acta Neuropathol. 2013;125:535–547. doi: 10.1007/s00401-013-1086-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang YJ, et al. Aberrant cleavage of TDP-43 enhances aggregation and cellular toxicity. Proc Natl Acad Sci USA. 2009;106:7607–7612. doi: 10.1073/pnas.0900688106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gendron TF, et al. Cerebellar c9RAN proteins associate with clinical and neuropathological characteristics of C9ORF72 repeat expansion carriers. Acta Neuropathol. 2015;130:559–573. doi: 10.1007/s00401-015-1474-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cook C, et al. Severe amygdala dysfunction in a MAPT transgenic mouse model of frontotemporal dementia. Neurobiol Aging. 2014;35:1769–1777. doi: 10.1016/j.neurobiolaging.2013.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.