Summary
Introduction.
Chlamydia trachomatis (Ct) is the most common bacterial cause of sexually transmitted infections (STI) and is associated with severe long-term sequelae in female populations.
In Italy Ct infections are not submitted to a screening programme, and its epidemiological profile is understudied. Even scarcer information is available about the genetic diversity on ompA gene, whose sequence defines 18 different genovars. This study aims at evaluating the prevalence of Ct infection in young sexually active asymptomatic women aged 18-25, and characterizing the molecular epidemiology of the different circulating genovars in this population.
Methods.
Cervical samples collected from 909 sexually-activeyoung women (mean age 21.5 years) were analyzed through molecular assay for the detection of Ct infection. Phylogenetic analysis on the ompA gene was performed on Ct positive samples to identify the circulating genovars.
Results.
The overall prevalence of Ct-infection was 4.4% (95%CI: 3.2-5.9%): 5.3% among women aged 18-21 years and 3.5% among those aged 22-25 years. Phylogenetic analysis has identified 5 different genovars: D, E, F, G, and H. The most common genovar was the E (46%), followed by genovar F and G (18.9% each), D (13.5%), and H (2.7%).
Conclusions.
This study underlines the high prevalence of asymptomatic Ct-infections among young women. Overall, about half of the asymptomatic infections is sustained by genovar E. The introduction in Italy of a systematic screening program should be considered to allow a better understanding of Ct spreading and providing women with an opportunity for early treatment to protect their sexual and reproductive health.
Key words: C. trachomatis infection, Sexually active asymptomatic young women, C. trachomatis genovars, Molecular epidemiology, Sexually transmitted diseases
Introduction
Chlamydia trachomatis (Ct) is an obligate intracellular Gram-negative bacterium that is the most commonly reported microorganism responsible for sexually transmitted infections (STI) in Europe and is the cause of considerable acute morbidity and long term reproductive health problems, particularly in young people [1, 2]. Many infections are asymptomatic and result in delayed diagnosis and uninterrupted transmission. This is of particular clinical relevance as untreated infections can ascend in the female genital tract and cause Pelvic Inflammatory Disease (PID), which includes any combination of endometritis, salpingitis, tubo-ovarian abscess and pelvic peritonitis. PID can also result in ectopic pregnancy, infertility and chronic pelvic pain [2-4].
The presence of few or no specific symptoms that, if untreated, could lead to an increase in female reproductive tract morbidity and the existence of an inexpensive treatment against all these infections make chlamydia screening widely recommended to all sexually active women aged 25 or less [5-7]. In fact, Ct is under epidemiological surveillance in several European regions and the reported prevalence in young sexually active women ranges from 5% to 10%. Infection rates are the highest in females below 20 years of age and decrease with increasing age [8, 9]; in 2007, the incidence of chlamydial infection was 4.5-fold higher in the age group 15 to 24 years than in the age group 25-44 years [9].
In Europe the incidence of genital Ct infections seems to have increased dramatically over the last 20 years. However, this is most likely the result of more extended testing rather than a true rise in the incidence. Targeted screening, opportunistic testing for asymptomatic infections, contact tracing and mandatory notification help to explain the high notification rates in the UK and the Scandinavian countries compared with other European states [10].
In Italy, a reporting system for Chlamydia infections is not in place and an organised screening in specific asymptomatic population groups is not available. Surveillance data are collected centrally by the Istituto Superiore di Sanità (ISS) by a sentinel network system consisting of 13 microbiology laboratories located throughout the Country. The last report, regarding data until 2012, described an overall prevalence of 2.4% in women aged between 15 and 45 years, with a significantly increased prevalence in lower age groups [11]. Moreover, a recent study identified 5.2% of Ct endocervical infection prevalence in a large population of sexually active women aged 15-55 years attending an outpatient service of cervico-vaginal pathology unit in Rome over a 10-year period [12]. Notwithstanding the existence of these studies, knowledge on the prevalence and molecular epidemiology of Chlamydial genital tract infections, particular in young asymptomatic women, remains modest.
Molecular epidemiology studies are currently based on the analysis of differences in the Ct major outer membrane protein (MOMP), whose coding gene (ompA) contains four spaced variable domains. Genetic variability on ompA gene reflects the existence of 18 genovars, classified according to their pathogenic potential [13]. Genovars A, B, Ba, and C have been commonly associated with trachoma, D-K, Da, Ia, and Ja with urogenital infections, and L1-L3 with lymphogranuloma venereum [13]. In addition, on the basis of amino acid similarities, these genovars have been grouped into the following groups or classes: group B (B, Ba, D, Da, E, L1, and L2), group C (A, C, H, I, Ia, J, Ja, K, and L3), and the intermediate group (F and G) [14]. These type of studies are useful to obtain new information about Ct genovars distribution, which can be further on translated into improved strategies for Ct infection management, for the traceability of sexual contacts, and in developing strategies for vaccine design [15, 16].
To improve our knowledge about Ct epidemiology and to expand the available Italian data, we evaluate here the prevalence of Ct infections and the molecular epidemiology of circulating genovars in young asymptomatic sexually active women aged 18-25 in Milan, Italy, over a period of 6 years (2008-2013).
Materials and methods
STUDY POPULATION
This was a cross-sectional study, performed on female subjects who spontaneously visited gynecological centers of the Local Health Units (LHUs) of Milan for medical consultations for contraception. No evidence of clinical symptoms related to Ct infection were described by the physician. Cervical cytological samples were collected for routine test among women aged 18 to 25 years, from September 2008 to June 2013. Written informed consent was obtained from young women so as to store their samples for further anonymous research testing. Due to its design, ethical approval was not required for this study, in compliance with the international policy [17] and with the current Italian legislation [18, 19]. The database was anonymised before the analysis.
SAMPLES COLLECTION
Cervical cytological samples were collected using a brush (Cytobrush Plus Medscand® Medical AB, Sweden), immersed and rinsed in a vial filled with 20 ml of PreservCyt® Solution (PreservCyt) and stored at room temperature (RT) until processing. A total of 10 ml of each PreservCyt® Solution containing cervical cells was centrifuged at 3800xg for 15 min at RT. After centrifugation the pellet was re-suspended in 1 ml of Phosphate Buffered Saline (PBS), transferred in a new 1.5 ml collection test tube, and stored at -20°C until nucleic acids extraction.
DNA EXTRACTION AND AMPLIFICATION
DNA was extracted with the EasyMAG kit (NucliSENS® easyMAG®, bioMérieux, France) from 500 μl of the resuspended pellet with a final elution volume of 100 μl. The concentration and purity of the extracted DNA was evaluated through a spectrophotometer (NanoDrop ND- 2000/200C, Euroclone®, Thermo Scientific, Wilmington, DE, USA). After DNA extraction molecular tests were promptly performed to limit the storage to a maximum of a week. DNA integrity was assessed by the amplification of a 268 bp fragment of the ubiquitous betaglobin gene using the primer pair GH20 and PCO4 [20].
A nested PCR targeting a Ct cryptic plasmid was performed to screen each extracted sample. The primers used for the nested PCR were previously described by Jalal et al. [21] and amplify a fragment of 150 nt. Amplifications were performed on 50 ng of extracted DNA in a 50 μlreaction mix containing 5x PCR Buffer, 200 μM dNTPs, 25 pmol of each primer, and 1 U Taq (GoTaq® DNA Polymerase 5U/μl Promega, Madison WI). Each run was accompanied by positive and negative control samples. The cycling conditions were as follow: 94°C for 5 min, followed by 30 cycles of amplification during the first step and 25 cycles during the nested step consisting of 94°C for 30 sec, 50°C for 30 sec, 72 °C for 30 sec, and a final 72°C for 7 minutes extension. The final amplification products were visualized using electrophoresis analysis on a 2% agarose gel containing ethidium bromide (0.5 mg/L) and compared with a standard (DNA Molecular Weight, Marker 100, SigmaAldrich, St. Louis, MO, USA).
All Ct-DNA positive samples were used to amplify a 395 bp fragment of the Ct ompA gene with previously described primer sets [21]. Amplifications were performed on 50 ng of extracted DNA in a 50 μl reaction mix containing 5x PCR Buffer, 200 μM dNTPs, 25 pmol of each primer, and 1 U Taq (GoTaq® DNA Polymerase 5U/μl Promega, Madison WI). The cycling conditions were: 94°C for 5 min, followed by 30 cycles during the first amplification round or 25 cycles during the second round of 94°C for 30 sec, 50°C for 30 sec, 72 °C for 30 sec, and a final 72°C step for 7 minutes.
Following the PCR of the ompA gene fragments, amplification products were purified using NucleoSpin® Extract II (Macherey-Nagel GmbH, Germany) and nucleotide sequences were obtained from automated DNA sequencing on the ABI PRISM 3100 genetic analyzer (Applied Biosystem, CA, USA).
PHYLOGENETIC ANALYSIS
Molecular characterization was performed by analysis of 395 bp amplicons of the ompA Ct gene (nucleotides 223-636 of Ct genovar A sequence, NC007429). All specimens presenting ompA variant sequences were confirmed by resequencing newly extracted DNA.
All sequences obtained in this study were deposited into NCBI GeneBank Database [27], under accession numbers: KX449367-KX449406. The reference genovars used for the construction of phylogenetic trees were obtained from the NCBI GenBank Database (genovar A: NC007429, M58938; genovar B: AF063194, U80075, M33636; genovar C: M17343; genovar D: NC000117, X62920, X62918, X62919; genovar E: X52557; genovar F: X52080; genovar G: AF063199; genovar H: X16007; genovar I: AF063200; genovar J: AF063202; genovar K: AF063204; genovar L1: M36533; genovar L2: M14738; genovar L3: X55700). Sequences were aligned using ClustalX 2.1 multiple aligner [22] and then used for phylogenetic inference. A model selection was performed to identify the best model for distance estimation. The evolutionary history was inferred using the maximum-likelihood method [23] based on the Tamura 3 parameter model [24], identified as the best fitting model after the model test analysis, using MEGA 6.06 [25]. A discrete Gamma distribution was used to model evolutionary rate differences among sites (G = 0.8952) and phylogenetic trees were constructed with MEGA 6.06. A bootstrap test [26] with 1,000 replicates was performed to test the robustness of the analyses.
STATISTICAL ANALYSIS
Data were expressed as mean (range) and percentages (95% confidence intervals, 95% CI) where appropriate. Comparisons between groups were performed using the chi-square test or Fisher's exact test. A p-value < 0.05 was considered statistically significant (two-tailed test). All statistical analyses were performed using OpenEPI software, version 2.3.1 [28].
Results
PREVALENCE OF CT INFECTION
A total of 909 samples collected from the same number of sexually active young women (mean age 21.5 years, range 18-25 years) were available for this study. All women were asymptomatic for Ct infection.
The beta-globin gene was successfully amplified from all 909 cervical samples collected, confirming the suitability of the extraction method for these biological samples (extracted DNA: mean 20.1 ng/μl, range [3.3-45.7 ng/μl]).
The overall prevalence of Ct infections was 4.4% (95% CI, 3.2-5.9%). The prevalence of Ct infection was the highest among 20-21 year-old women with a value of 5.5% (95% CI, 3.0-9.3%) and decreased to 3.5% (95% CI, 1.5-6.7%) and 3.6% (95% CI, 1.6-6.9%) in age groups 22-23 and 24-25 years (Tab. I). Differences between infection rates in the different age groups and during the different years of the study period were not significant.
Tab. I.
Prevalence of C. trachomatis infections in different age groups of sexually active young women (18-25 years old).
| Age group | Number of subjects N (%) |
Ct-DNA + N (%) |
Prevalence % |
95% CI |
|---|---|---|---|---|
| 18-19 | 220 (24.2%) | 11 (27.5%) | 5.0% | 2.5-8.8 |
| 20-21 | 235 (25.9%) | 13 (32.5%) | 5.5% | 3.0-9.3 |
| 22-23 | 230 (25.3%) | 8 (20.0%) | 3.5% | 1.5-6.7 |
| 24-25 | 224 (24.6%) | 8 (20.0%) | 3.6% | 1.6-6.9 |
| Total | 909 (100.0%) | 40 (100.0%) | 4.4% | 3.2-5.9 |
MOLECULAR CHARACTERIZATION
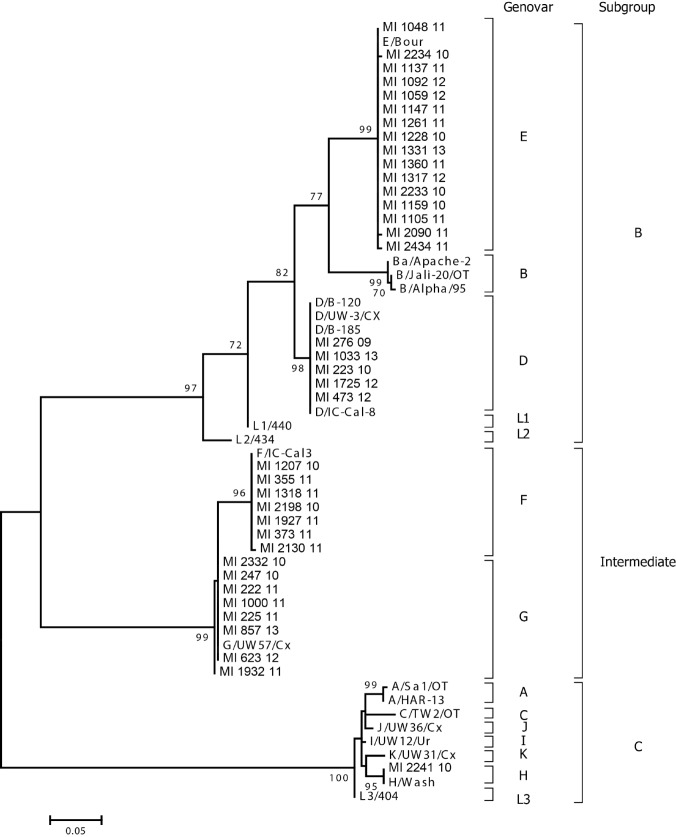
OmpA amplification was successful for 37 out of 40 positive samples (92.5%). Phylogenetic analysis of the ompA partial nucleotide sequences demonstrated 5 different genovars: D, E, F, G, and H (Fig. 1). In particular, 2 genovars (D and E) fell into subgroup B, 2 (F, and G) fell into the intermediate subgroup, and one, genovar H, fell into subgroup C. All circulating genovars are normally associated with infection of the urogenital tract.
Fig. 1.
Phylogenetic analysis of C. trachomatis partial ompA gene sequences (coded between nt 223-636 of C. trachomatis genovar A, accession number NC007429) obtained during this study and compared to reference sequences. The outcome of the bootstrap analysis is shown next to the nodes, and branch lengths are proportional to genetic distances as indicated by the scale bar. Genovars as well as subgroups are indicated on the right.
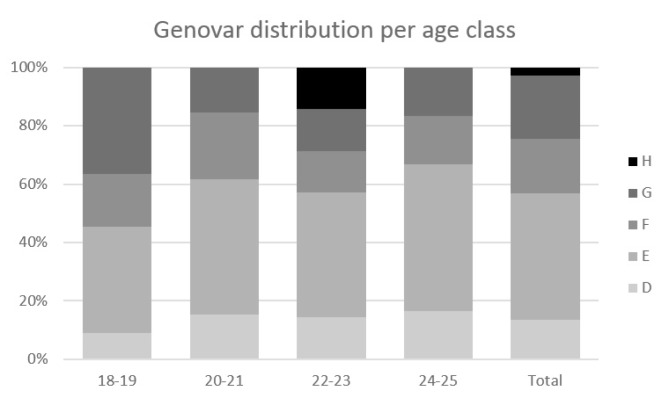
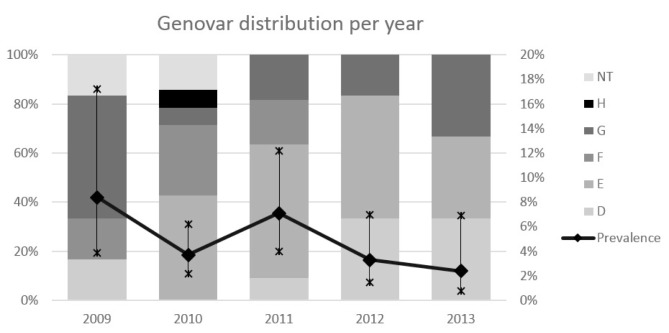
The most common genovar in our population was E (46%, 17/37), followed by genovars F and G (both 18.9%, 7/37), D (13.5%, 5/37), and H (2.7%, 1/37). No genovar distribution pattern was observed among the various age groups (Fig. 2), and during the different years of the study period (Fig. 3).
Fig. 2.
Genovar distribution among the studied age groups.
Fig. 3.
Overall Ct infection prevalence (line) with 95% CI (vertical lines) and genovar distribution (bars) during the different years of the study. The vertical axis on the left refers to the genovar distribution, while the one on the right refers to prevalence values. NT: not typed.
Discussion
Chlamydia trachomatis is the most common bacterium associated with STIs and genovars D-K cause genital tract infections in women (cervicitis and urethritis) and men (urethritis). These can also be responsible for sexually transmitted rectal and pharyngeal infections, be transmitted during labor, cause pneumonia and eye infections in infants, and be spread by close contact to cause eye infections in adults. Several data suggest that, even though young people aged 15-24 years represent only 25% of the sexually experienced population, they acquire nearly half of all new STIs [29]. Rates of reported chlamydial infection among persons aged 15-19 years and 20-24 years continue to increase. Overall, during 2009-2010, rates increased up to 2.8% and 7.5% in the age groups 15-19 and 20-24 years, respectively [30]. Compared with older women, sexually active adolescents aged 15-19 years and young adults aged 20-24 years are at higher risk of acquiring STIs for a combination of behavioural, biological, and cultural reasons [31].
This study focused on asymptomatic sexually active young women and describes the molecular epidemiology of Ct in Milan, Italy, over a period of 6 years. The prevalence of the infection was 4.4%, in line with what reported by other Italian studies [11, 12, 32, 33].
The most common genovars found in our populations were E (46%), F and G (both to 18.9%). Other recent studies described E (50-33%) and F (25-14%) as the most common genovars in symptomatic female populations [34-36]. A previous study, conducted in Italy in symptomatic and asymptomatic populations attending a Sexually Transmitted Disease (STD) outpatient clinic, described gerovar E as the most prevalent in female (37.2%), followed by genovar G (32.6%), and genovar F and J (both to 7%), regardless of presence or absence of related symptoms [37].
It has been demonstrated that genovars E and F have a biological advantage over the other genovars thanks to both their ability to escape the host immune response and the presence of specific virulence factors, which can facilitate the transmission and infectious processes [38, 39]. It is possible that genovars E and F are less immunogenic than other genovars and, therefore, remain the most prevalent strain in all populations, regardless of the presence or absence of clinical signs.
Although we could not detect any relationship between infection, related genovar, and absence of symptoms, the present study contributed to increase the current knowledge on genotype distribution of Ct in asymptomatic young women in northern Italy.
Conclusions
Our data indicate that Ct infections occur frequently in young sexually active women and that several different genovars are widely spread in Italy. Larger national and longitudinal studies are definitively required to better understand the spread of Ct infection and its impact on the young Italian population.
These results underscore the need to establish primary and secondary preventive measures and to allocate more resources for an adequate prevention of Ct infections. The introduction of an opportunistic screening program for Ct in Italy should be evaluated in order to achieve early treatment of infected subjects and reduce associated clinical manifestations. Finally, it seems essential to develop educational programs and information campaigns, particularly addressed to young people, about acquisition, risk factors and treatment of STIs with a particular focus on C. trachomatis infection.
The Authors have no conflict of interest to declare.
ACKNOWLEDGMENTS
The Authors thank Valentina Paladino for her helpful technical-scientific assistance. The Authors have no conflict of interest to declare.
References
- 1.Redmond SM, Alexander-Kisslig K, Woodhall SC, Broek IV, Bergen J, Ward H, Uusküla A, Herrmann B, Andersen B, Götz HM, et al. Genital Chlamydia prevalence in Europe and non-European high income countries: systematic review and meta-analysis. Plos One. 2015;10:e0115753–e0115753. doi: 10.1371/journal.pone.0115753. doi:10.1371/journal.pone.0115753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cates W, Wasseheit JN. Genital Chlamydia infections: epidemiology and reproductive sequelae. Am J Obstet Gynecol. 1994;164:1771–1781. doi: 10.1016/0002-9378(91)90559-a. [DOI] [PubMed] [Google Scholar]
- 3.Haggerty C, Gottlieb S, Taylor B, Low N, Xu F, Ness RB. Risk of sequelae after Chlamydia trachomatis genital infection in women. J Infect Dis. 2010;201:S134–S155. doi: 10.1086/652395. doi: 10.1086/652395. [DOI] [PubMed] [Google Scholar]
- 4.Oakeshott P, Kerry S, Aghaizu A, Atherton H, Hay S, Taylor-Robinson D, Simms I, Hay P. Randomised controlled trial of screening for Chlamydia trachomatis to prevent pelvic inflammatory disease: the POPI (prevention of pelvic infection) trial. BMJ. 2010;340:c1642–c1642. doi: 10.1136/bmj.c1642. doi: http://dx.doi.org/10.1136/bmj.c1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geisler WM. Diagnosis and management of uncomplicated Chlamydia trachomatis infections in adolescents and adults: summary of evidence reviewed for the 2010 Centers for Disease Control and Prevention Sexually Transmitted Diseases Treatment Guidelines. Clin Infect Dis. 2011;53:S92–S98. doi: 10.1093/cid/cir698. doi:10.1093/ cid/cir698. [DOI] [PubMed] [Google Scholar]
- 6.Paukku M, Kilpikari R, Puolakkainen M, Oksanen H, Apter D, Paavonen J. Criteria for selective screening for Chlamydia trachomatis. Sex Trans Dis. 2003;30:120–123. doi: 10.1097/00007435-200302000-00005. [DOI] [PubMed] [Google Scholar]
- 7.Low N, McCarthy A, Macleod J, Salisbury C, Campbell R, Roberts TE, Horner P, Skidmore S, Sterne JA, Sanford E, et al. Chlamydia Screening Studies Project Group, author. Epidemiological, social, diagnostic, and economic evaluation of population screening for genital chlamydial infection. Health Technol Assess. 2007;11:iii–iv. doi: 10.3310/hta11080. ix-xii, 1-165. [DOI] [PubMed] [Google Scholar]
- 8. European Centre for Disease Prevention and Control (ECDC) , author. Review of Chlamydia control activities in EU countries. Stockholm, May 2008. Technical Report. Stockholm: ECDC; 2008. Available at: http://ecdc.europa.eu/en/publications/publications/0805_ter_review_of_chlamydia_control_activities.pdf Accessed on: 29/06/2016. [Google Scholar]
- 9. European Centre for Disease Prevention and Control (ECDC) , author. Chlamydia control in Europe. ECDC Guidance. Stockholm: ECDC; 2009. Available at: http://ecdc.europa.eu/en/publications/Publications/0906_GUI_Chlamydia_Control_in_Europe.pdf Accessed on: 29/06/2016. [Google Scholar]
- 10. European Centre for Disease Prevention and Control (ECDC) , author. Sexually transmitted infections in Europe, 1990-2009. Stockholm: ECDC; 2011. Available at: http://ecdc.europa.eu/en/publications/Publications/110526_SUR_STI_in_Europe_1990-2009.pdf. Accessed on: 29/06/2016. [Google Scholar]
- 11.Salfa MC, Regine V, Ferri M. Suligoi B e la Rete Sentinella dei Centri Clinici e dei Laboratori di Microbiologia clinica. Le Infezioni sessualmente trasmesse: i dati dei due Sistemi di sorveglianza sentinella attivi in Italia. Notiziario ISS vol. 27, n° 4 - aprile 2014. Available at: www.iss.it/.../Notiziario_Istituto_Superiore_di_Sanit_2012.pdf_25_2_pp.3_10.pdf. Accessed on: 29/06/2016.
- 12.Marcone V, Recine N, Gallinelli C, Nicosia R, Lichtner M, Degener AM, Chiarini F, Calzolari E, Vullo V. Epidemiology of Chlamydia trachomatis endocervical infection in a previously unscreened population in Rome, Italy, 2000 to 2009. Euro Surveill. 2012;17(25) pii=20203. [PubMed] [Google Scholar]
- 13.Bandea CI, Kubota K, Brown TM, Kilmarx PH, Bhullar V, Yanpaisarn S, Chaisilwattana P, Siriwasin W, Black CM. Typing of Chlamydia trachomatis strains from urine samples by amplification and sequencing the major outer membrane protein gene (omp1) Sex Transm Infect. 2001;77:419–422. doi: 10.1136/sti.77.6.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Millman KL, Tavare S, Dean D. Recombination in the ompA gene but not the omcB gene of Chlamydia contributes to serovarspecific differences in tissue tropism, immune surveillance and persistence of the organism. J Bacteriol. 2001;183:5997–6008. doi: 10.1128/JB.183.20.5997-6008.2001. doi: 10.1128/JB.183.20.5997-6008.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morré SA, Rozendaal L, Valkengoed IG, Boeke AJ, Voorst Vader PC, Schirm J, Blok S, Hoek JA, Doornum GJ, Meijer CJ, et al. Urogenital Chlamydia trachomatis serovars in men and women with a symptomatic or asymptomatic infection: an association with clinical manifestations? Clin Microbiol. 2000;38:2292–2296. doi: 10.1128/jcm.38.6.2292-2296.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geisler WM, Suchland RJ, Whittington WL, Stamm WE. The relationship of serovar to clinical manifestations of urogenital Chlamydia trachomatis infection. Sex Transm Dis. 2003;30:160–165. doi: 10.1097/00007435-200302000-00013. [DOI] [PubMed] [Google Scholar]
- 17.Millum J, Menikoff J. Streamlining ethical review. Ann Intern Med. 2010;153:655–657. doi: 10.7326/0003-4819-153-10-201011160-00008. doi: 10.7326/0003-4819-153-10- 201011160-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Italian Data Protection Authority. Linee guida in tema di fascicolo sanitario elettronico e di dossier sanitario. [Guidelines on the Electronic Health Record and the Health File]. Gazzetta Ufficiale - Serie Generale. 3 Aug 2009;178. Rome: Istituto Poligrafico e Zecca dello Stato. Italian. Summary available at: www.garanteprivacy.it/web/guest/home/docweb/-/docwebdisplay/.../1634116; (English version available at: http://www.garanteprivacy.it/garante/doc.jsp?ID=1672821). [Google Scholar]
- 19. Italian Data Protection Authority. Autorizzazione generale al trattamento di dati personali effettuato per scopi di ricerca scientifica [General authorization to the processing of personal data for scientific research]. Gazzetta Ufficiale n. 72 del 26 mar 2012;85. Italian. Rome: Istituto Poligrafico e Zecca dello Stato. Available from: http://www.gazzettaufficiale.biz/atti/2012/20120072/12A03185.htm. [Google Scholar]
- 20.Puranen M, Saarikoski S, Syrjänen K, Syrjänen S. Polymerase chain reaction amplification of human papillomavirus DNA from archival, Papanicolaou-stained cervical smears. Acta Cytol. 1996;40:391–395. doi: 10.1159/000333842. [DOI] [PubMed] [Google Scholar]
- 21.Jalal H, Al-Suwaine A, Sonnex C, Sonnex C, Carne C. The superiority of polymerase chain reaction over an amplified enzyme immunoassay for the detection of genital chlamydial infections. Sex Transm Infect. 2006;82:37–40. doi: 10.1136/sti.2005.015362. doi:10.1136/sti.2005.015362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. doi:10.1093/bioinformatics/ btm404. [DOI] [PubMed] [Google Scholar]
- 23.Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 24.Tamura K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol Biol Evol. 1992;9:678–687. doi: 10.1093/oxfordjournals.molbev.a040752. [DOI] [PubMed] [Google Scholar]
- 25.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 27. NCBI GeneBank Database. Available at: http://www.ncbi.nlm.nih.gov/GenBank/index.html.
- 28.Dean AG, Sullivan KM, Soe MM. OpenEpi: Open Source Epidemiologic Statistics for Public Health, Version 2.3.1. http://www.OpenEpi.com.
- 29.Weinstock H, Berman S, Cates W., Jr Sexually transmitted diseases among American youth: incidence and prevalence estimates, 2000. Perspect Sex Reprod Health. 2004;36:6–6. doi: 10.1363/psrh.36.6.04. doi: 10.1363/psrh.36.6.04. [DOI] [PubMed] [Google Scholar]
- 30. Centers for Disease Control and Prevention , author. Sexually Transmitted Disease Surveillance 2010. Atlanta: U.S. Department of Health and Human Services; 2011. Available at http://www.cdc.gov/std/stats10/surv2010.pdf. [Google Scholar]
- 31.DiClemente RJ, Salazar LF, Crosby RA. A review of STD/HIV preventive interventions for adolescents: sustaining effects using an ecological approach. J Pediatr Psychol. 2007;32:888–906. doi: 10.1093/jpepsy/jsm056. doi: 10.1093/jpepsy/jsm056. Accessed on: 29/06/2016. [DOI] [PubMed] [Google Scholar]
- 32.Panatto D, Amicizia D, Bianchi S, Frati ER, Zotti CM, Lai PL, Domnich A, Colzani D, Gasparini R, Tanzi E. Chlamydia trachomatis prevalence and chlamydial/HPV co-infection among HPV-unvaccinated young Italian females with normal cytology. Hum Vaccin Immunother. 2015;11:270–276. doi: 10.4161/hv.36163. doi: 10.4161/hv.36163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Latino MA, Bello L, Lanza A, Leotta E, Tersiev P, Intinis G, Spagnolo E, Smirne C, Grio R. Chlamydia trachomatis infection among sexually active young women in Italy. Sex transm Infect. 2002;78:e4–e4. doi: 10.1136/sti.78.4.e4. [Google Scholar]
- 34.Gallo Vaulet L, Entrocassi C, Corominas AI, Rodríguez Fermepin M. Distribution study of Chlamydia trachomatis genotypes in symptomatic patients in Buenos Aires, Argentina: association between genotype E and neonatal conjunctivitis. BMC Res Notes. 2010;9:3–34. doi: 10.1186/1756-0500-3-34. doi: 10.1186/1756-0500-3-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lagergård T, Hadad R, Tunbäck P, Lindholm L, Löwhagen GB, Unemo M. Distribution of Chlamydia trachomatis ompA genovars and the new variant of C. trachomatis in the Göteborg area, Sweden. Eur J Clin Microbiol Infect Dis. 2010;29:609–611. doi: 10.1007/s10096-010-0887-1. doi: 10.1007/s10096-010-0887-1. [DOI] [PubMed] [Google Scholar]
- 36.Weill FX, Le Hello S, Clerc M, Scribans C, Barbeyrac B. Serological reactivity and bacterial genotypes in Chlamydia trachomatis urogenital infections in Guadeloupe, French West Indies. Sex Transm Infect. 2010;86:101–105. doi: 10.1136/sti.2009.037036. doi: 10.1136/ sti.2009.037036. [DOI] [PubMed] [Google Scholar]
- 37.Marangoni A, Foschi C, Nardini P, D'Antuono A, Banzola N, Francesco A, Ostanello F, Russo I, Donati M, Cevenini R. Chlamydia trachomatis serovar distribution and other sexually transmitted coinfections in subjects attending an STD outpatients clinic in Italy. New Microbiologica. 2012;35:215–219. [PubMed] [Google Scholar]
- 38.Mossman D, Beagley KW, Landay AL, Loewenthal M, Ooi C, Timms P, Boyle M. Genotyping of urogenital Chlamydia trachomatis in Regional New South Wales, Australia. Sex Transm Dis. 2008;35:614–616. doi: 10.1097/OLQ.0b013e31816b1b80. doi: 10.1097/OLQ.0b013e31816b1b80. [DOI] [PubMed] [Google Scholar]
- 39.Nunes A, Borrego MJ, Nunes B, Florindo C, Gomes JP. Evolutionary dynamics of ompA, the gene encoding the Chlamydia trachomatis key antigen. J Bact. 2009:7182–7192. doi: 10.1128/JB.00895-09. doi:10.1128/ JB.00895-09. [DOI] [PMC free article] [PubMed] [Google Scholar]