ABSTRACT
Polymorphisms in Fc-gamma-receptor (FCGR) genes as well as killer cell immunoglobulin-like receptor (KIR) and KIR ligand (KIRL) repertoires may influence antitumor effects of monoclonal antibodies (mAb). Here, we systematically analyzed high- and low-affinity FCGR2A and -3A genotypes as well as stimulating and inhibitory KIR/KIRL combinations in 53 neuroblastoma (NB) patients treated by long-term infusion (LTI) of anti-GD2 IgG1 Ab ch14.18/CHO using validated real-time PCR methods.
Patients with high-affinity FCGR2A and -3A genotypes showed a higher level of Ab-dependent cell-mediated cytotoxicity (ADCC) on day 8 after the start of ch14.18/CHO and superior event-free survival (EFS) compared to patients with low FCGR genotypes. Similar observations were made for patients with stimulatory KIR/KIRL haplotype B (combination of KIR genes including activating receptor genes) compared to inhibitory haplotype A (a fixed set of genes encoding for inhibitory receptors, except 2DS4) and stronger effects were found in patients when haplotype B and high-affinity FCGRs were combined. Surprisingly, independent analysis of KIRs showed a major role of activating KIR 2DS2 for high ADCC levels and prolongation of EFS. The greatest effect was observed in 2DS2-positive patients that also had high-affinity FCGR2A and -3A genotypes.
In summary, the presence of the activating KIR 2DS2 has a major effect on ADCC levels and survival in NB patients treated by LTI of ch14.18/CHO and may therefore be a useful biomarker in combination with FCGR polymorphisms for Ab-based immunotherapies.
KEYWORDS: Antibody-dependent cell-mediated cytotoxicity (ADCC), ch14.18/CHO, Fc-gamma-receptor polymorphisms (FCGR), killer-cell immunoglobulin-like receptor (KIR), KIR ligand (KIRL), neuroblastoma
Abbreviations
- Ab
antibody
- ADCC
antibody-dependent cell-mediated cytotoxicity
- A. dest.
Aqua destillata
- AM
acetoxymethyl ester
- CDC
complement-dependent cytotoxicity
- EFS
event-free survival
- FCGR
Fc-gamma-receptor polymorphisms
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- IL-2
interleukin-2
- KIR
killer-cell Immunoglobulin-like receptor
- KIRL
killer-cell Immunoglobulin-like receptor ligand
- LTI
long-term infusion
- mAb
monoclonal antibody
- NB
neuroblastoma
- RT
room temperature
- SD
standard deviation
- SEM
standard error of the mean
Introduction
Disialoganglioside GD2 is over expressed by neuroblastoma (NB) cells and passive immunotherapies based on administration of anti-GD2 monoclonal antibodies (mAb) have shown promising results.1,2 One major obstacle observed in patients treated with human/mouse chimeric anti-GD2 Ab ch14.18 given as a short-term infusion (8 h/d, 5 d, 100 mg/m2) is associated with induction of neuropathic pain3,4 that requires co-medication with intravenous morphine. Based on the clinical observation that pain intensity correlates with the infusion rate of anti-GD2 Ab, we applied a novel delivery method of ch14.18/CHO for the treatment of NB patients. 53 NB patients received the same cumulative dose of ch14.18/CHO given as a continuous long-term infusion (LTI; 24 h/d, 10 d, 100 mg/m2). In these patients, we observed a reduced pain toxicity profile as indicated by decreased morphine usage and low pain scores.5 We further showed that this novel delivery method of ch14.18/CHO induced Ab effector mechanisms (ADCC and CDC) over the entire treatment period of 6 mo and may therefore emerge as the preferred delivery method of anti-GD2 Ab in NB patients.6 We reported ch14.18/CHO Cmax concentrations of 12.56 ± 0.68 mg/mL and a CDC activity of 88 ± 2%, showing a low degree of variability in contrast to patient-specific ADCC levels of 21 ± 3% at this time point.6 This variation in ADCC may be associated with many factors including the cytotoxic activity of effector cells such as natural killer (NK) cells, monocytes, neutrophils and macrophages that play a crucial role in Ab-mediated antitumor effects.7,8 Therefore, the investigation of genes contributing to the cytotoxic activity of effector cells is of particular interest. The most important receptors involved in ADCC include FCGR3A (CD16) expressed primarily on NK cells and FCGR2A (CD32) expressed on monocytes, macrophages and neutrophils.7 Both receptors bind to the Fc fragment of human immunoglobulin (IgG) consequently inducing effector functions of leukocytes.9 It has been reported that the degree of effector cell-mediated cytotoxicity is also dependent upon genetic polymorphisms in FCGR genes as well as the repertoire of killer cell immunoglobulin-like receptors (KIRs) expressed by NK cells and KIR ligands (KIRL) expressed by target cells.8 Allotypic variants of FCGR2A and 3A have been reported to be of clinical significance, as they probably influence binding affinity to Fc-fragments of therapeutic Ab and, as a consequence, activation of effector cells.10 The first polymorphism of interest is a C/T nucleotide substitution in the extracellular domain of FCGR2A which changes the amino acid at position 131 from histidine to arginine (H131R) and influences binding affinity primarily to IgG2.10 The second is a G/T substitution in the extracellular domain 2 of FCGR3A which changes the amino acid at position 158 from valine to phenylalanine (V158F) and influences binding affinity to IgG1.10 In studies on therapeutic IgG1 mAb Rituximab, Trastuzumab and Cetuximab a better clinical response was reported in patients with high-affinity genotypes of FCGR3A.11-14 Similarly, patients with FCGR2A high-affinity genotype also showed a greater clinical benefit when treated with the same mAbs indicating an involvement of the FCGR2A H131R polymorphism in IgG1-mediated therapeutic effects.12,14,15
In addition to FCGR, NK-cell mediated ADCC depends on the KIR/KIRL repertoire. Models for KIR and KIRL related to NK cell activation were initially developed16 according to the “missing ligand” (or “KIR/KIRL mismatch”) concept, where absence of an inhibitory ligand might be sufficient to elicit NK cell-mediated cytotoxicity. This model was further detailed by the concept that NK cell cytotoxicity also requires the presence of an activating signal in addition to the absence of inhibitory KIRL on the target cells.16 Thus, the KIR/KIRL repertoire is of great relevance for NK cell-mediated antitumor cytotoxicity and consequently for mAb-based immunotherapies. In fact, several reports showed an association between KIR/KIRL genotypes and response to certain forms of immunotherapy.8 In treatment of leukemia, recipients missing KIRLs for the respective KIRs on the donor NK cells showed longer progression-free survival post-transplantation compared to those who had KIR/KIRL match. Furthermore, clinical studies with the murine anti-GD2 Ab 3F8 or humanized immunocytokine hu14.18-IL-2 demonstrated superior clinical outcome in patients missing inhibitory KIRL.17-19 Similar observations were made in lymphoma patients treated with rituximab.20 Extending on these results, interactions of activating KIR/KIRL has been recently studied in a number of malignancies.8 Underlying the diversity of KIR genomic regions, there are patterns that are conserved within the population suggesting conservation of distinct haplotypes that can be separated into two main groups described as haplotypes A and B.21,22
Haplotype A comprises a complement of KIR genes 3DL3, 2DL3, 2DL1, 2DL4, 3DL1, 2DS4 and 3DL2 that encode inhibitory receptors with the exception of 2DS4. In contrast, haplotype B consists of a more variable group of activating genes including 2DS1, 2DS2, 2DS3 and 2DS5.23 In leukemia, transplantation of haematopoietic cells from donors who had haplotype B led to prolonged relapse-free survival.23-25 In contrast, patients that received haematopoietic cells from donors with a set of inhibitory KIRs corresponding to haplotype A showed worse outcome indicating positive impact of activating KIRs on response to therapy.
Although in some reports involvement of activating KIR/KIRL interactions in clinical response to therapeutic Ab was shown, the specific role of activating KIRs in NB therapy is still unclear.8
Based on these considerations, we established validated methods and determined H131R and V158F polymorphisms in FCGR2A and -3A genes as well as six activating (2DS1-5 and 3DS1) and four inhibitory KIRs (2DL1-3 and 3DL1) and their ligands (HLA-C1, HLA-C2 and HLA-Bw4) in 53 high-risk NB patients treated with ch14.18/CHO given as a LTI. We examined the impact of distinct genotypes and their combinations on ADCC level and event-free survival (EFS).
Our data show for the first time a correlation of FCGR polymorphisms and KIR/KIRL repertoires with a functional immune parameter mediated by GD2-specific mAb ch14.18/CHO. We found that the patients combining high-affinity FCGR2A, -3A genotypes and genotype B/x or the presence of activating KIR 2DS2 showed the strongest anti-NB cellular cytotoxicity mediated by ch14.18/CHO and improved survival of NB patients indicating the synergistic effects of activating KIRs with high-affinity FCGRs. These data support that FCGR polymorphisms and KIR/KIRL genotypes may serve as biomarkers of response and outcome to immunotherapies with mAb in NB patients.
Results
Validation of FCGR polymorphism analysis
For validated detection of FCGR polymorphisms analysis, two control plasmids encoding for either FCGR2A-131H and FCGR3A-158V (“high-affinity plasmid”; Fig. 1A) or FCGR2A-131R and FCGR3A-158F alleles (“low-affinity plasmid”; Fig. 1B) were included in each analytical run instead of patient DNA. The GAPDH DNA sequence was also included into both plasmids as internal control allowing further assay precision (Figs. 1A and B). Only GAPDH-positive runs that showed amplification of the respective FCGR allele were used for further data analysis.
Figure 1.
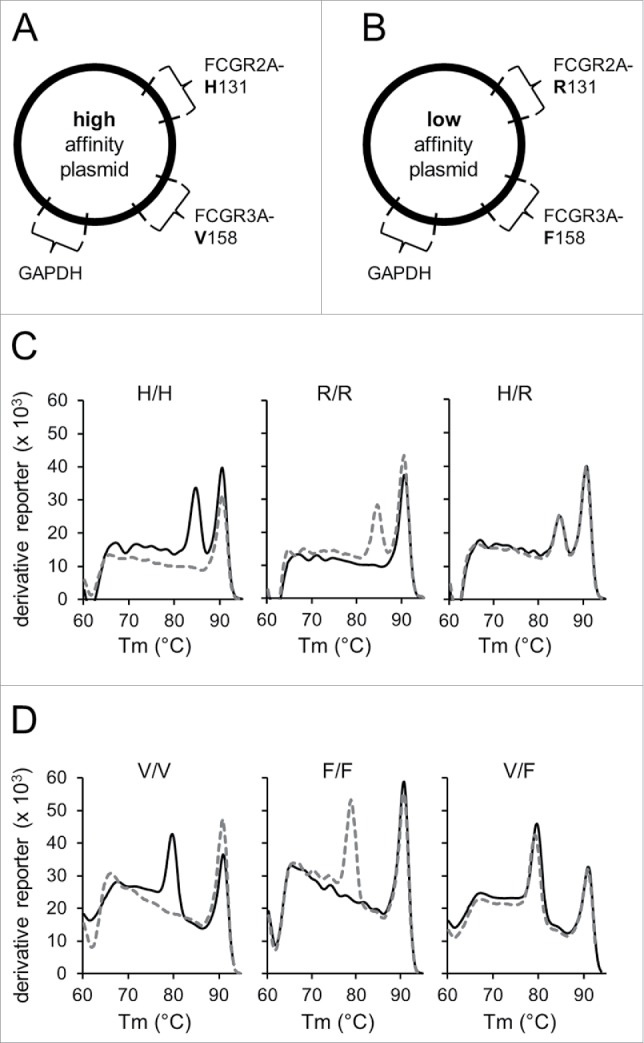
Establishment of the validated identification of FCGR polymorphisms. To validate analysis of FCGR polymorphisms, two control plasmids containing DNA sequences encoding for either FCGR2A-131H and FCGR3A-158V alleles (“high-affinity plasmid”; A) or FCGR2A-131R and FCGR3A-158F alleles (“low-affinity plasmid”; B) were generated and used as PCR templates in addition to patient samples. The plasmid encoding for high-affinity alleles was used for amplification with low- and high-affinity specific primers and thereby served as positive and negative control, respectively. Conversely, the plasmid carrying low-affinity alleles were utilized as positive and negative control for amplification with primers for low- and high-affinity polymorphisms, respectively. Due to similar Tm of allele-specific PCR products each allele was examined separately. Moreover, DNA sequences encoding for GAPDH were integrated into both plasmids and used as internal control for further assay precision. Only GAPDH-positive runs that showed amplification of the positive control and lack of amplification of the negative control of the respective allele-specific fragments were used for further data analysis. To determine FCGR polymorphisms in 2A (high-affinity allele 131H (H) and low-affinity allele 131R (R)) and 3A genes (high-affinity allele 158V (V) and low-affinity allele 158F (F)), allele-specific PCR followed by post-amplification melting-curve analysis was performed. To obtain melting-curves, the derivative reporter was plotted against the melting temperature (Tm) of PCR products. Representative melting-curves for possible genotypes of FCGR2A (C) and -3A (D) are shown in combination with GAPDH used as internal control. Black solid lines represent melting curves derived from reactions containing high affinity and gray dashed lines from reactions containing low-affinity primers. Tm of FCGR2A and -3A allele-specific amplicons were at 85°C and 80°C, respectively. GAPDH control amplicons showed a Tm of 91°C. Identification of FCGR genotypes H/H, R/R and H/R for 2A (C) and V/V, F/F and V/F for 3A (D) was realized using amplicon specific Tm profiles. Experiments were performed in duplicates. The derivative reporter was calculated as follows: change of SYBR Green I dye fluorescence/change of temperature.
Frequency of FCGR polymorphisms in the study population
We then applied the established procedure for genotyping of the 53 NB patients treated. In order to identify distinct FCGR genotypes, post-amplification melting-curve analysis was performed for each allele. We detected FCGR2A (Fig. 1C) and -3A allele-specific amplicons (Fig. 1D) using their specific melting temperatures (Tm) of 85°C and 80°C, respectively. The Tm of the internal control GAPDH was 91°C (Figs. 1C and D). With this method, we identified 14/53 (26%) patients homozygous for FCGR2A 131H, 15/53 (28%) patients homozygous for 131R allele and 24/53 (45%) heterozygous patients (H/R) (Fig. 2A). FCGR3A polymorphism analysis revealed 3/53 (5%) patients homozygous for 158V, 26/53 (49%) patients homozygous for 158F allele and 24/53 (45%) heterozygous patients (V/F) (Fig. 2A). Patients homozygous for low-affinity polymorphisms were assigned to the low-affinity cohort and those who were homozygous for high-affinity polymorphisms or heterozygous patients to the high-affinity cohort resulting in following genotypes: FCGR2A high (H/H or H/R; 38/53 patients (72%)) and low affinity (R/R; 15/53 patients (28%)) and FCGR3A high (V/V or V/F; 27/53 patients (51%)) and low affinity (F/F 26/53 patients (49%)) (Fig. 2A, numbers in parenthesis).
Figure 2.
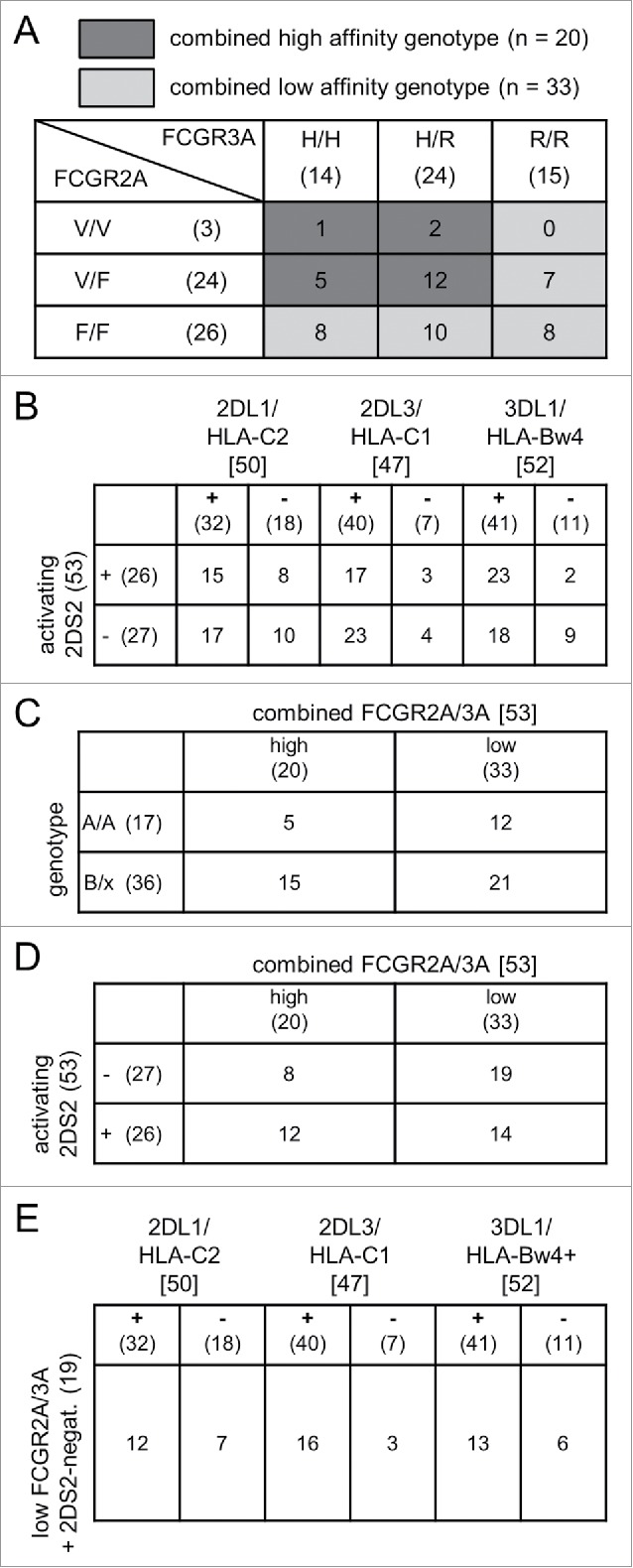
(A) Frequencies of FCGR2A/3A and KIR/KIRL genotypes. The total number of patients with distinct FCGR2A (lines) and 3A polymorphisms (columns) are shown in parentheses. Numbers without parentheses indicate patients with combinations of FCGR2A and 3A polymorphisms. Groups of patients with combined high- and low-affinity genotypes were defined as follows: combined high affinity (dark gray; FCGR2A H/H or H/R and FCGR3A V/V or V/F) and combined low affinity (light gray; FCGR2A R/R and/or F/F). (B) Frequencies of genotypes with inhibitory KIR/KIRL match and mismatch with or without the activating KIR 2DS2. The columns indicate the total number of patients with inhibitory KIR (2DL1, 2DL3, 3DL1) in square brackets. Numbers in round brackets reflect patient numbers with match (+) and mismatch (−) with the respective KIRL (HLA-C2, HLA-C1, HLA-Bw4). The lines show the number of patients in round brackets with presence (+) or absence (−) of activating KIR 2DS2. Numbers of patients without brackets indicate distinct combinations of inhibitory KIR/KIRL match- and activating KIR 2DS2 status. (C) Combination of FCGR2A/3A high and low genotype with genotype B/x and A/A. The columns indicate the total number of patients with combined high- and low-affinity FCGR2A/3A in round brackets. The lines show the number of patients with genotype A/A or B/x in round brackets. Numbers of patients without brackets indicate distinct combinations of combined high- and low-affinity FCGR2A/3A and genotype A/A or B/x. (D) Combination of FCGR2A/3A high and low genotype with or without the activating KIR 2DS2. The columns indicate the total number of patients with combined high- and low-affinity FCGR2A/3A in round brackets. The lines show the number of patients in round brackets with presence (+) or absence (−) of the activating KIR 2DS2. Numbers of patients without brackets indicate distinct combinations of combined high- and low-affinity FCGR2A/3A and 2DS2 status. (E) Frequencies of combined 2DS2-negative and FCGR2A/3A low-affinity genotype with or without KIR/KIRL match. Nineteen patients with combined low-affinity FCGR2A/3A phenotype and absent activating KIR 2DS2 were analyzed for inhibitory KIR/KIRL match and mismatch. The columns indicate the total number of patients with inhibitory KIR (2DL1, 2DL3, 3DL1) in square brackets. Numbers in round brackets reflect patient numbers with match (+) and mismatch (−) with the respective KIRL (HLA-C2, HLA-C1, HLA-Bw4). Numbers of patients without brackets indicate distinct combinations of inhibitory KIR/KIRL match- and absent activating KIR 2DS2 status.
Since a number of clinical studies showed strong deviations from Hardy–Weinberg equilibrium for gene polymorphisms detected indicating methodological problems, we also investigated the Hardy–Weinberg equilibrium for each FCGR genotype identified.10 Based on the observed frequencies of both alleles for FCGR2A and -3A genes, we calculated the expected genetic distribution of these polymorphisms on the Hardy–Weinberg principle.26 Statistical analysis using the Χ2 test did not reveal significant differences between the observed genotype distribution of FCGR2A and -3A and the expectations of the Hardy–Weinberg equilibrium (Table 1; p = 0.791 and 0.701, respectively). This data indicate that the patient cohort is representative clearly showing sensitivity and reliability of the established detection strategy and also the accuracy and validity of the concluded statements.
Table 1.
Frequencies of KIR, KIRL and KIR/KIRL combinations in neuroblastoma patients (n = 53).
| Patients |
|||
|---|---|---|---|
| Gene | N | % | |
| Activating KIR | 2DS1 | 20 | 38 |
| 2DS2 | 26 | 49 | |
| 2DS3* | 14 | 27 | |
| 2DS4 | 53 | 100 | |
| 2DS5* | 15 | 29 | |
| 3DS1 | 18 | 34 | |
| Inhibitory KIR | 2DL1 | 51 | 96 |
| 2DL2 | 26 | 49 | |
| 2DL3* | 47 | 90 | |
| 3DL1 | 53 | 100 | |
| KIRL* | HLA-C1 | 45 | 87 |
| HLA-C2 | 34 | 65 | |
| HLA-Bw4 | 41 | 79 | |
| 2DL1/HLA-C2** | match | 32 | 62 |
| mismatch | 18 | 35 | |
| 2DL2/HLA-C1 | match | 22 | 42 |
| mismatch | 3 | 6 | |
| 2DL3/HLA-C1 | match | 40 | 77 |
| mismatch | 7 | 13 | |
| 3DL1/HLA-Bw4** | match | 41 | 77 |
| mismatch | 11 | 21 | |
DNA samples of 52 patients were available for analysis.
Due to missing DNA of one patient for KIRL analysis, the total number of KIR/KIRL combinations is reduced by n = 1.
Finally, we defined two patient cohorts combining FCGR2A and -3A genotypes (Fig. 2A; light and dark gray). We included patients harboring at least one homozygous low-affinity genotype in the combined low-affinity cohort (R/R and/or F/F; 33/53 patients (62%)) (Fig. 2A, light gray). Patients who missed a homozygous low-affinity genotype were assigned to the combined high-affinity cohort (H/H or H/R and V/V or V/F; 20/53 patients (38%)) (Fig. 2A, dark gray).
Effect of FCGR polymorphism on ADCC levels
ADCC was analyzed in vitro against GD2 positive LA-N-1 NB cells using the effector cells and heat inactivated serum of each individual patient as previously described.27 Additionally, the level of ADCC increase achieved by ch14.18/CHO on day 8 of Ab infusion in cycle 1 was used to analyze correlations of ADCC levels with distinct FCGR polymorphisms. For this purpose, cytotoxicity levels of individual patients detected on day 1 prior to ch14.18/CHO application was subtracted from the absolute ADCC level determined on day 8. Overall, LTI of ch14.18/CHO on day 8 resulted in a 2.1-fold increased ADCC level compared to baseline in all patients analyzed, clearly showing ch14.18/CHO-dependent induction of effector mechanisms.
The approach to evaluate a role of FCGR on ADCC levels consisted of an initial analysis of FCGR2A and -3A polymorphisms separately followed by a combined assessment.
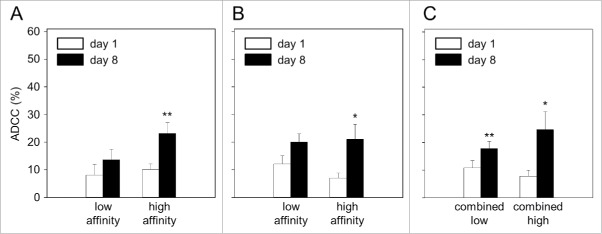
When patients were grouped according to high (38/53) and low (15/53) affinity in the FCGR2A polymorphism, analysis of ADCC revealed significantly higher levels on day 8 in patients with the high-affinity genotype (2.3-fold) compared to baseline (Fig. 3A). In contrast, ADCC observed in low-affinity genotype patients was only slightly increased compared to the baseline level (1.7-fold increase).
Figure 3.
Impact of FCGR2A and -3A genotypes on antibody-dependent cell-mediated cytotoxicity (ADCC). Patient blood samples were collected prior to start (baseline; day 1; white column) and on day 8 of long-term infusion of ch14.18/CHO (day 8; black column). ADCC was analyzed with Calcein-AM-based cytotoxicity assay using GD2-positive neuroblastoma cell line LA-N-1, patient-specific leukocytes and heat inactivated serum.27 ADCC in patients with low- (n = 15) and high-affinity (n = 38) polymorphisms of FCGR2A (8.05 ± 4% vs. 13.68 ± 4% and 10.12 ± 2% vs. 23.20 ± 4%, respectively) (A) and (B) FCGR3A low- (n = 26) and high-affinity polymorphisms (n = 27) (12.08 ± 3% vs. 20.08 ± 3% and 7.01 ± 2% vs. 21.03 ± 5%, respectively). (C) ADCC levels in patients with combined FCGR2A and -3A low (n = 33) and high affinity (n = 20) genotype (10.86 ± 3% vs. 17.80 ± 3% and 7.74 ± 2% vs.24.65 ± 6%, respectively). Data are shown as mean values ± SEM of experiments performed in triplicate. (A) Wilcoxon signed-rank test; **p = 0.084 and 0.002 vs. low- and high-affinity baseline, respectively. (B) Wilcoxon signed-rank test; *p = 0.011 and 0.023 vs. low- and high-affinity baseline, respectively. (C) Paired t-test; **p = 0.006 vs. low-affinity baseline. Wilcoxon signed-rank test; *p = 0.043 vs. high-affinity baseline.
A stronger effect was also observed for the FCGR3A polymorphism (Fig. 3B). ADCC levels increased in the FCGR3A high-affinity cohort (27/53 patients (51%)) by over a factor 3 compared to the low-affinity patients (26/53 (49%)) (3.1- vs. 1.7-fold, respectively).
Confounding factors in this analysis are ch14.18/CHO Ab serum concentration and NK-cell levels. To exclude effects of ch14.18/CHO levels on ADCC on day 8, we analyzed Ab serum levels between the high- and low-affinity cohorts for each FCGR cohort. Ab concentrations were determined as previously described.6 We found no difference in ch14.18/CHO levels between patient cohorts (data not shown, p = 0.782 and p = 0.642 for FCGR2A high vs. low and FCGR3A high vs. low cohorts, respectively). These data show that differences in ADCC levels observed are not related to varying ch14.18/CHO concentrations.
Moreover, we addressed the question whether differences in ADCC levels between the high- and low-affinity cohort are attributable to the NK cell numbers. Therefore, we compared NK cell numbers on day 8 between the high- and low-affinity patients for each FCGR genotype analyzed. Similar to results obtained for ch14.18/CHO levels, we did not find significant differences in NK cell numbers between the patient cohorts (data not shown, p = 0.132 and p = 0.324 for FCGR2A high vs. low and FCGR3A high vs. low cohorts, respectively).These data further show that observed differences in ADCC are not due to different NK cell numbers again emphasizing FCGR polymorphism-dependent impact on ADCC.
Next, we analyzed ADCC in patients combining the genotypes of the two FCGRs (2A and 3A). All patients with a homozygous low-affinity genotype in FCGR2A or -3A were combined to the low-affinity cohort (FCGR2A R/R and/or FCGR3A F/F; 33/53 patients (62%)) and compared to the high-affinity cohort (FCGR2A H/H or H/R and FCGR3A V/V or V/F; 20/53 patients (38%)) (Fig. 2A light and dark gray). We observed the highest increase of ADCC levels in the combined high-affinity group (Fig. 3C) compared to the combined low-affinity group (3.2- and 1.6-fold increase compared to baseline, respectively). In summary, these results clearly show correlation of high-affinity FCGR2A- and 3A genotypes with increased ch14.18/CHO-dependent cellular cytotoxicity levels and this effect translated into an a superior EFS as detailed below.
Frequency of distinct KIR/KIRL genotypes of treated neuroblastoma patients
To investigate the impact of stimulatory and inhibitory KIR/KIRL genotypes on ADCC levels and survival, six activating KIRs (2DS1-5 and 3DS1) and four inhibitory KIRs (2DL1-3 and 3DL1) were evaluated in 53 NB patients (Table 2). We limited our analysis to 2DL1-3 and 3DL1 based on the recently reported involvement of these KIRs on efficacy of Ab-dependent immunotherapy.7,28 Due to lack of expression on effector cells, we excluded the remaining known KIR pseudogenes 2DP1and 3DP1 from our analysis.29
Table 2.
Primer sequences. Allele-specific primer sequences for FCGR2A and -3A genes. The polymorphic sites are shown in bold. To increase PCR specificity, a mismatch was inserted at penultimate position of the respective allele-specific primer. GAPDH served as internal control.
| Gene | Primer | Polymorphism | Sequence (5′–3′) |
|---|---|---|---|
| FCGR2A | forward | 131H | ATGGAAAATCCCAGAAATTCTCACA |
| forward | 131R | ATGGAAAATCCCAGAAATTCTCACG | |
| reverse | H131R | AATGACCACAGCCACAATGA | |
| FCGR3A | forward | V158F | GGTCACATATTTACAGAATGGCAAACG |
| reverse | 158V | CAGTCTCTGAAGACACATTTTTACTCCCATC | |
| reverse | 158F | CAGTCTCTGAAGACACATTTTTACTCCCATA | |
| GAPDH | forward | ACTAGGCGCTCACTGTTCTCTC | |
| reverse | GGAGTAGGGACCTCCTGTTTCT |
For analysis of KIRLs HLA-C1, HLA-C2 and HLA-Bw4, DNA samples of 52 NB patients were available. After the analysis of inhibitory KIRs and their ligands, we then identified patients who were missing the KIRL for the respective KIR (KIR/KIRL mismatch) and those who had KIRL in the presence of its KIR (match) (Table 2). We found similar frequencies of KIR, KIRL and KIR/KIRL combinations in our patient cohort compared to previously published data30,31 clearly showing that the detection method applied is reliable as well as that the patient cohort is representative.
Based on our results of genotyping, we next assigned patients into haplotype A and B groups according to the following KIR status: haplotype A: absence of activating KIRs with the exception of 2DS4; haplotype B: presence of at least two activating KIRs. All patients homozygous for A haplotypes were assigned to the A/A genotype (17/53 patients (32%)) and those who had one or two B haplotypes to the B/x genotype (36/53 patients (68%)) (Fig. 2C).
Impact of KIR/KIRL genotypes on ADCC levels
According to the reported data showing that NK cell cytotoxicity could be influenced by the presence of activating signals and match or mismatch of inhibitory KIRs and the respective ligands,16 we analyzed ADCC in NB patients depending on their KIR/KIRL repertoires. Compared to baseline, we found about 2.4-fold increased ADCC in patients harboring haplotype B compared to almost unchanged ADCC in A/A patients (1.3-fold; Fig. 4A). Although the difference between these genotypes was not significant (p = 0.052), we observed a tendency toward increased ADCC in patients with B/x genotype compared to A/A patients.
Figure 4.
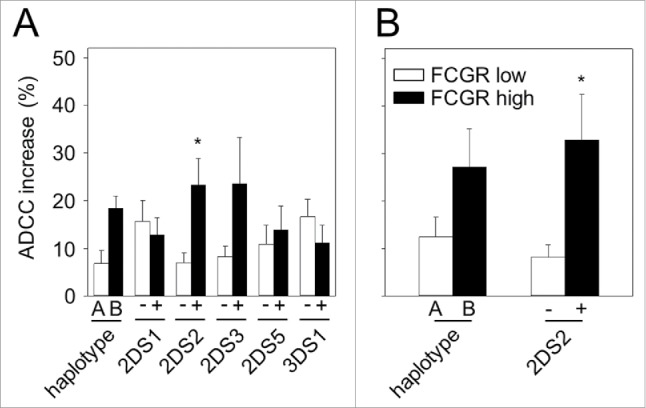
Impact of KIRs with or without FCGR2A and -3A genotypes on patient-specific ADCC. ADCC was analyzed prior to start (baseline; day 1) and on day 8 of long-term infusion of ch14.18/CHO. ADCC increase was calculated according to the formula: day 8 ADCC – baseline ADCC. (A) Patients harboring two haplotypes A (n = 17) or patients with the presence of the single activating KIR 2DS1 (n = 20), 2DS2 (n = 26), 2DS3 (n = 14), 2DS5 (n = 15) and 3DS1 genotype (n = 18) were compared to those who had haplotype B (n = 36) or missed the respective activating KIR (n = 33, 27, 38, 37 and n = 35, respectively). Patients with A/A genotype showed 1.3-fold increase (10.85 ± 3 vs. 8.19 ± 2%) and patients harboring haplotype B showed 2.4-fold increase (25.13 ± 3% vs. 10.26 ± 4% baseline). Following ADCC increases were found in patients with single activating KIR and those who were negative for respective activating KIR: 2.1- vs. 2.1-fold for 2DS1 (19.36 ± 3% vs. 9.13 ± 3% and 21.29 ± 5% vs. 10.17 ± 2%, respectively), 3.1- vs. 1.3-fold for 2DS2 (29.80 ± 5% vs. 9.70 ± 3% and 12.44 ± 2% vs. 9.50 ± 2%, respectively), 2.6- vs. 1.4-fold for 2DS3 (34.86 ± 10% vs. 13.63 ± 5% and 15.31 ± 2% vs. 10.62 ± 2%, respectively), 2.2- vs. 1.2-fold for 2DS5 (19.83 ± 5% vs. 8.86 ± 4% and 20.85 ± 5% vs. 17.05 ± 4%, respectively) and 1.5- vs. 2.7-fold for 3DS1(19.27 ± 3% vs. 12.82 ± 4% and 21.30 ± 5% vs. 7.93 ± 4%, respectively). (B) Impact of combined FCGR2A and -3A low- (white column) and high-affinity genotype (black column) with haplotype A or B (n = 12 and 15, respectively) and the presence or absence of the activating KIR 2DS2 on ADCC (n = 12 and 19, respectively). Increase of ADCC was evaluated in four patient cohorts: low-affinity FCGR2A and -3A + haplotype A, high-affinity FCGR2A and -3A + haplotype B (2.0- vs. 4.7-fold change (14.02 ± 4% vs. 7.06 ± % and 31.85 ± 8% vs. 6.78 ± 2%)), low-affinity FCGR2A and -3A + absence of 2DS2 and high-affinity FCGR2A and -3A + presence of 2DS2 (1.6- vs. 6.3-fold change (14.84 ± 3% vs. 9.11 ± 3% and 37.21 ± 9% vs. 5.91 ± 2%)). Data are shown as mean values ± SEM of experiments performed in triplicate. (A) Mann–Whitney rank sum test; *p = 0.011 vs. 2DS2-negative patients. (B) Mann–Whitney rank sum test; *p = 0.016 vs. combined FCGR2A and -3A low affinity + absence of 2DS2 group.
Then, we evaluated the impact of each activating KIR (2DS1-5- and 3DS1) on ADCC separately. For this, we divided patients in activating KIR-positive and -negative cohorts. KIR 2DS4 was detected in all patients and could therefore not be used to define distinct patient groups. Comparison of the increase of ADCC levels in patients with single activating KIR genotypes 2DS1-, 2DS3-, 2DS5- or 3DS1 did not reveal significant differences compared to the respective single KIR-negative cohort (Fig. 4A; 2.1- vs. 2.1-fold for 2DS1, 2.6- vs. 1.4-fold for 2DS3, 2.2- vs. 1.2-fold for 2DS5 and 1.5- vs. 2.7-fold for 3DS1). In contrast, 2DS2-positive patients (26/53 (49%)) showed significantly increased tumor cell lysis in comparison to patients missing 2DS2 (27/53 patients (51%)) (3.1- and 1.3-fold increase vs. baseline, respectively; Fig. 4A). Since all 2DS3-positive patients were also 2DS2 positive (14/26), the single impact of 2DS3 in this cohort could not be determined directly. To overcome this problem and to evaluate the single effects of 2DS2 independently of 2DS3, we analyzed ADCC levels in patients combining both genotypes (14/26 patients (54%)) with those who showed only the presence of 2DS2 (12/26 patients (46%)). Interestingly, the ADCC level increase was not significantly different between 2DS2- and 2DS3-positive patients and 2DS2-positive but 2DS3-negative patients. These results suggest that 2DS2 has a major impact on the level of ch14.18/CHO-mediated ADCC in NB patients.
To evaluate confounding factors, we found no difference in ch14.18/CHO concentrations (data not shown, p = 0.906) and NK cell numbers (data not shown, p = 0.840) in 2DS2-positive and -negative patients, further emphasizing a 2DS2-dependent effect on ADCC.
Next, we examined whether patients that are missing the KIRL (HLA-C1, HLA-C2 and HLA-Bw4) for the respective inhibitory KIR (2DL2 and 2DL3 for HLA-C1, 2DL1 for HLA-C2 and 3DL1 for HLA-Bw4) (KIR/KIRL mismatch) will have higher ADCC levels compared to those who have the KIRL present for its inhibitory KIR (match). Surprisingly, we did not observe significant differences between the matched and mismatched genotypes (2.2- and 1.7-fold increase for 2DL1/HLA-C2, 2.0- and 1.1-fold for 2DL3/HLA-C1, 2.5- and 1.3-fold for 3DL1/HLA-Bw4 compared to baseline, respectively). The impact of 2DL2/HLA-C1 mismatch could not be analyzed due to a small number of patients (3/52 patients (6%)). In summary, this finding underlines the importance of 2DS2 for ADCC.
Based on our results of significant lower ADCC levels in KIR 2DS2-negative patients (Fig. 4A), we addressed whether a combination of the absence of 2DS2 with match of inhibitory KIRs and their ligands (2DL1/HLA-C2, 2DL2/HLA-C1, 2DL3/HLA-C1 and 3DL1/HLA-Bw4) has an additional adverse effect on ADCC (Fig. 2B). Since 2DS2-negative patients also lack KIR 2DL2, analysis of this combination could not be performed. Statistical analysis of ADCC levels did not reveal significant differences between 2DS2-negative patients (1.3-fold increase) and those who additionally showed one unfavorable match (1.1-, 1.7- and 1.2-fold for 2DL1/HLA-C2, 2DL3/HLA-C1 and 3DL1/HLA-Bw4, respectively). This finding shows that a combination of two unfavorable KIR/KIRL genotypes (absence of stimulating but presence of inhibitory KIR) has no further adverse effect on ADCC levels, that again underscores the relevance of the KIR 2DS2 for ch14.18/CHO-mediated ADCC in treated patients.
Combined effect of FCGR2A, -3A and KIR/KIRL genotypes on ADCC levels
We evaluated the impact of FCGR2A and -3A in combination with KIR/KIRL genotype repertoires on ADCC using two approaches. First, we combined FCGR genotypes with KIR/KIRL A/A or B/x genotypes (Fig. 2C), second we assessed FCGR genotypes and the presence or absence of the activating KIR 2DS2 (Fig. 2D). For the first approach, we identified 17/53 (32%) homozygous for the inhibitory haplotype A (A/A genotype) patients of which 12/53 (23%) showed a combined low-affinity FCGR 2A and -3A genotype and 36/53 (68%) patients harboring haplotype B of which 14/53 (26%) had combined high-affinity FCGR 2A and -3A genotype.
For the second approach, we identified 12/52 (23%) patients with high-affinity FCGR2A and -3A polymorphisms and activating KIR2DS2 genotype and 19/52 patients (37%) with low-affinity FCGR2A and -3A polymorphisms and the absence of 2DS2. The remaining patients (22/53 patients (42%)) formed an intermediate genotype.
The result of the first approach revealed a 4.7-fold increased ADCC level over baseline in patients with high-affinity FCGR2A/3A and stimulatory B/x genotypes compared to the patients with low-affinity FCGRs and inhibitory A/A genotypes (2.0-fold increase over baseline) (Fig. 4B), however, this difference was not significant. The result of the second approach showed a strong increase of ADCC levels over baseline in 2DS2-positive and high-affinity FCGRs patients (6.3-fold) compared to 2DS2-negative patients with low-affinity FCGRs (1.6-fold over baseline) (Fig. 4B). Statistical analysis for the second approach revealed a clear correlation between these genotypes and ADCC levels (p = 0.016), with the highest effect size for ch14.18/CHO-mediated NB cell lysis in patients with high-affinity FCGR2A and -3A polymorphisms who were also positive for the activating KIR 2DS2, that again underlines the relevance of 2DS2.
Moreover, we examined whether additional unfavorable genotypes could further negatively affect ADCC levels. For this, we identified three patient cohorts with four unfavorable genotypes showing a match of inhibitory KIRs and their ligands (2DL1/HLA-C2 (12/52 patients (23%)), 2DL3/HLA-C1 (16/52 patients (31%)) and 3DL1/HLA-Bw4 (13/52 patients (25%)) in addition to the combination of three unfavorable genotypes described above (FCGR2A and 3A low-affinity patients missing 2DS2 genotype) (Fig. 2E). The remaining patients showing three unfavorable genotypes in combination with one favorable KIR/KIRL mismatch (Fig. 2E), were excluded from the analysis. The analysis of the four unfavorable genotype combinations did not reveal further decrease in ADCC compared to the three unfavorable genotypes showing a similar ADCC increase of about 1.6-fold in all groups analyzed (data not shown).
In summary, these findings show an association of high-affinity FCGR2A, -3A and KIR/KIRL genotypes in particular of 2DS2 with higher levels of anti-NB cellular cytotoxicity mediated by ch14.18/CHO in NB patients and thereby provide a clear genotype–phenotype correlation.
Effect of FCGR polymorphism on event-free survival
Based on the correlation of ADCC levels with FCGR genotypes, we addressed the question whether this functional observation also correlates with survival parameters. We first evaluated the impact of polymorphisms in FCGR 2A and 3A genes on EFS separately in our cohort of 53 NB patients treated. For FCGR2A, we found higher EFS rates in patients harboring high (H/H or H/R) (38/53 patients) compared to those who had low-affinity polymorphisms (R/R) (15/53 patients) (34% vs. 20 %). In line with this, the mean EFS time was longer in 2A high (EFS: 2.7 ± 0.4 y (95% CI [1.9, 3.5]) compared to low-affinity patients (EFS: 1.7 ± 0.5 y (95% CI [0.6, 2.7]). Although we found improved EFS probabilities in high-affinity patients compared to low-affinity, statistical analysis did not reveal significant difference between these groups (Fig. 5A; p = 0.141). These results suggest favorable effects of high-affinity polymorphism in FCGR2A on ch14.18/CHO-based immunotherapy.
Figure 5.
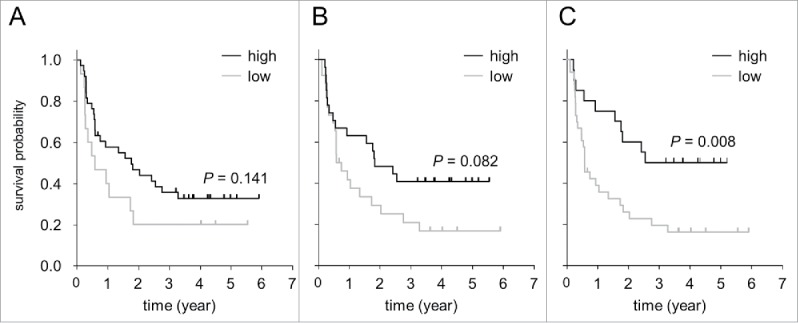
Impact of polymorphisms in FCGR2A and -3A genes on event-free survival of neuroblastoma patients (n = 53) treated by long-term infusion of ch14.18/CHO. Validated detection of FCGR2A and -3A polymorphisms was performed to identify high- and low-affinity patient cohorts. (A) Impact of FCGR2A high (H/H or H/R genotype; black line) and low affinity (R/R; gray line) on EFS. (B) Impact of FCGR3A high (V/V or V/F genotype; black line) and low affinity (F/F; gray line) on EFS. (C) Analysis of combined FCGR2A and -3A high (FCGR2A H/H or H/R and FCGR3A V/V or V/F; black line) and low-affinity genotypes (FCGR2A R/R or FCGR3A F/F; gray line) on EFS. Statistical analysis was performed using LogRank test.
Patients with FCGR3A high (V/V or V/F) (27/53 patients) compared to those who had low-affinity polymorphisms (F/F) (26/53 patients) did not reveal significant differences in EFS probabilities (Fig. 5B; p = 0.082). There was a tendency toward increased EFS in FCGR3A high- (V/V or V/F) compared to low-affinity patients (F/F) (Fig. 5B).
Similar to the correlation analysis for ADCC levels, we also investigated effects of combined FCGR2A and -3A genotypes (Fig. 2A). Higher EFS rates in the patient cohort with FCGR high affinity (H/H or H/R and V/V or V/F) (20/53 patients (38%)) compared to FCGR low affinity (R/R and/or F/F) (33/53 patients (62%)) (50% vs. 18%) was demonstrated. The mean EFS was 3.2 ± 0.5 y (95% CI [2.6, 4.2]) and 1.7 ± 0.4 y (95% CI [1.0, 2.4]) in the combined FCGR high- and low-affinity cohort, respectively. Now, the LogRank analysis revealed a clearly significant difference in EFS probabilities between both cohorts (Fig. 5C; p = 0.008).
Impact of KIR/KIRL genotype on event-free survival
Analysis of survival parameters in patients harboring A or B haplotypes revealed improved EFS in B/x genotype cohort compared to A/A patients (Fig. 6A), but this difference was not significant (p = 0.059). In line with our results showing a correlation of the activating KIR 2DS2 genotype on ADCC level increase, we observed higher EFS rates in 2DS2-positive patients (26/53 (49%)) compared to patients missing 2DS2 (27/53 patients) (42% vs. 19%). The mean EFS time was 2.9 ± 0.5 (95% CI [2, 3.8]) and 1.8 ± 0.4 (95% CI [1, 2.6]), for 2DS2-positive and -negative patients, respectively. Although the differences between EFS probabilities were found to be not significant, we observed a trend toward higher probabilities in patients with the presence of 2DS2 (Fig. 6B p = 0.055). Moreover, we did not find significant differences in EFS between patients with or without activating 2DS1, 2DS3, 2DS5 or 3DS1 confirming our ADCC data. Based on the fact, that all 2DS3-positive patients also presented 2DS2 (14/26 patients (54%)), we additionally compared EFS in these patients with those who harbored 2DS2 without 2DS3 (12/26 patients (46%)). We found similar survival probabilities in both groups (data not shown, p = 0.782) confirming our ADCC results and again emphasizing the crucial role of 2DS2 in EFS of patients under ch14.18/CHO-based immunotherapy.
Figure 6.
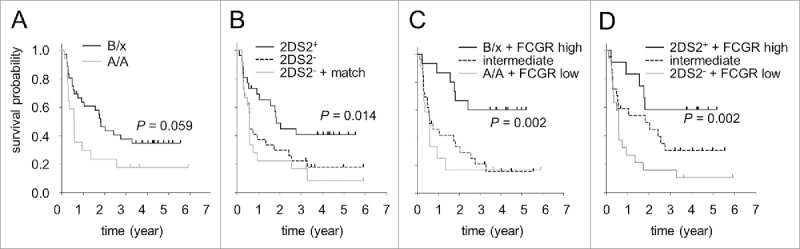
Impact of KIR haplotypes (A), KIR/KIRL repertoires (B) and their combination with FCGR2A and -3A polymorphisms (C) on event-free survival of neuroblastoma patients (n = 53) treated by long-term infusion of ch14.18/CHO. (A) EFS comparison of patients harboring two haplotypes A (genotype A/A; gray solid line) and patients with one or two haplotypes B (genotype B/x; black solid line). (B) EFS probabilities of patients with activating KIR2DS2 genotype (2DS2+; black solid line) were compared to KIR2DS2-negative patients with or without inhibitory KIR3DL1/HLA-Bw4 match (2DS2− + match; gray line and 2DS2−, black dashed line, respectively). (C) EFS in patients with combined high-affinity FCGR2A and -3A and B/x genotype (B/x + FCGR high; black solid line), low-affinity FCGR2A and -3A and A/A genotype (A/A + FCGR low; gray solid line) and in remaining patients with the intermediate genotype (intermediate; black dashed line). (D) Comparison of EFS probabilities of patients with high-affinity FCGR2A and -3A genotype in combination with the activating KIR 2DS2 (2DS2+ + FCGR high; black solid line), low-affinity FCGR2A and -3A genotype and the absence of 2DS2 (2DS2− + FCGR low; gray line) and intermediate patients (intermediate; black dashed line). Statistical analysis was performed using LogRank test.
Next, we evaluated the impact of the presence or absence of an inhibitory KIR/KIRL match (2DL1/HLA-C2, 2DL3/HLA-C1and 3DL1/HLA-Bw4) on EFS. In line with our ADCC data, statistical analysis did not reveal significant correlation between EFS probabilities and match or mismatch of KIRs and their ligands (data not shown).
Finally, we examined whether combination of two unfavorable genotypes (absence of activating KIR2DS2 and presence of inhibitory KIR/KIRL match 2DL1/HLA-C2, 2DL3/HLA-C1 or 3DL1/HLA-Bw4) (Fig. 2B) could negatively affect EFS probabilities. This combination revealed a significant difference in EFS between patients showing inhibitory 3DL1/HLA-Bw4 match in combination with the absence of 2DS2 (18/53 patients) compared to those who presented activating KIR2DS2 (26/53 patients) (Fig. 6B, p = 0.014). Analysis of EFS rates revealed 42% in the single 2DS2-positive cohort compared to 11% in the combined unfavorable cohort. The mean EFS time was 2.9 ± 0.5 y (95% CI [2, 3.8]) and 1.3 ± 0.4 y (95% CI [0.5, 2.1]) in 2DS2-positive and combined cohort, respectively.
Combined effect of FCGR2A, -3A and KIR/KIRL genotypes on event-free survival
In analogy to the stepwise approach selected for ADCC analysis, we evaluated the impact of polymorphisms in FCGR2A and -3A in combination with either A/A and B/x genotypes or presence and absence of the activating KIR 2DS2 (Figs. 2C and D) on EFS.
We grouped patients with high-affinity FCGRs and genotype B/x (15 patients (28%)), low-affinity FCGRs and genotype A/A (12 patients (23%)), 2DS2-positive and high-affinity FCGRs (12 patients (23%)) as well as 2DS2-negative and low-affinity FCGRs (19 patients (36%)) (Figs. 2C and D). Additionally, we defined intermediate groups including patients combining favorable FCGR with unfavorable KIR genotypes and the respective vice versa combinations (high-affinity FCGRs + genotype A/A (five patients (9%)), low-affinity FCGRs + genotype B/x (21 patients (40%)), high-affinity FCGRs + absence of 2DS2 (eight patients (15%)), low-affinity FCGRs + presence of 2DS2 (14 patients (26%)) (Figs. 2C and D).
Analysis of the FCGR+B/x and FCGR + A/A groups revealed a strong correlation between combined favorable genotypes and survival showing highly significant improved survival in patients with high-affinity FCGR2A and -3A in combination with B/X genotype (Fig. 6C). In contrast, patients with unfavorable genotypes (low-affinity FCGR2A and -3A in combination with genotype A/A) showed poor survival compared to the combined favorable group (Fig. 6C), clearly indicating genotype-dependent effects on outcome under ch14.18/CHO-based immunotherapy. Finally, patients with the intermediate genotype combination showed similar EFS compared to the unfavorable genotype. The mean EFS time of the favorable cohort was 3.7 ± 0.5 y (95% CI [2.7, 4.8]) compared to 1.4 ± 0.6 y (95% CI [0.2, 2.6]) in the unfavorable cohort. EFS probabilities of the favorable cohort were found to be significantly higher compared to the unfavorable cohort (Fig. 6C; p = 0.002). Analysis of EFS probabilities of the intermediate cohort revealed significant differences compared to the favorable cohort (p = 0.005 vs. favorable and p = 0.619 vs. unfavorable group) (Fig. 6C).
In line with our observations of the impact of FCGR and genotype A or B combinations, analysis of the FCGR + 2DS2+ and FCGR + 2DS2− combined groups showed highly significant increase in EFS of 2DS2-positive patients with high-affinity FCGR2A and -3A compared to the combined unfavorable genotype (low-affinity FCGRs and lack of 2DS2) (58% vs.11%) (Fig. 6D). The mean EFS time of the favorable cohort was 3.6 ± 0.6 y (95% CI [2.3, 4.8]) compared to 1.3 ± 0.4 y (95% CI [0.5, 2.1]) in the unfavorable cohort. Similar to the results of FCGR polymorphism and KIR/KIRL analysis, EFS probabilities of the favorable cohort were found to be significantly higher compared to the unfavorable cohort (Fig. 6D; p = 0.002). Analysis of EFS probabilities of the intermediate cohort did not reveal significant differences compared to the favorable and unfavorable cohorts (p = 0.132 vs. favorable and p = 0.140 vs. unfavorable group) (Fig. 6D).
Similar to ADCC analysis, we additionally evaluated the impact of four unfavorable genotypes on survival (Fig. 2E). For this, we identified patients who had an inhibitory match in addition to the three unfavorable genotypes described above (FCGR2A and -3A low-affinity patients missing 2DS2 genotype) (13/52 patients (25%)). Analysis revealed similar EFS probabilities compared to the patients of unfavorable genotypes without inhibitory match. Statistical analysis of patients with four unfavorable genotypes and those who had combined favorable genotypes (Fig. 2D) showed significant differences in EFS (p = 0.005 for 2DL3/HLA-C1, p = 0.011 for 2DL1/HLA-C2, and p = 0.002 for 3DL1/HLA-Bw4 matched unfavorable cohort) (data not shown), underlining the impact of FCGR2A and -3A polymorphisms and KIR/KIRL repertoires on ADCC level and outcome of immunotherapy with LTI of ch14.18/CHO. These data clearly show genotype–phenotype correlation between genotypes, Ab-dependent functional effector cell activity and treatment outcome.
Discussion
Immunotherapies based on administration of mAb directed against tumor antigens have revolutionized the treatment of cancer. One important mechanism of action of this class of cancer therapeutics is the elimination of tumor cells by host immune effector cells. Besides FCGR genotypes, expression repertoires of activating and inhibitory KIRs on NK cells as well as their ligands on target cells have been shown to affect Ab-dependent antitumor effects.16
Here, we report for the first time, that the patients combining high-affinity FCGR2A, -3A genotypes and stimulating genotype B/x or the presence of activating KIR 2DS2 showed the strongest anti-NB cellular cytotoxicity mediated by ch14.18/CHO and improved survival of NB patients indicating the synergistic effects of activating receptors.
A number of clinical studies have already demonstrated that the therapeutic efficacy of antitumor Ab correlates with FCGR polymorphisms in various cancers.10 Patients harboring FCGRs with high affinity for IgG have a better outcome following Ab-based immunotherapy probably due to a greater capacity to induce ADCC that is a major mechanism of action of mAbs.10 In line with this, we found a strong correlation between FCGR2A and -3A polymorphism and survival under ch14.18/CHO-based immunotherapy and, importantly we could show that the observed effects are associated with the ability of effector cells to mediate tumor cell lysis. Interestingly, our data show polymorphism-dependent effects of FCGR2A on ADCC and survival of NB patients under IgG1-based immunotherapy. These observations emphasize a role of macrophages, monocytes and granulocytes that express FCGR2A in addition to NK cells predominantly expressing FCGR3A in the ch14.18/CHO mediated anti-NB immune response.
Since ADCC was determined using white blood cells of ch14.18/CHO treated patients, the resulting cytotoxicity is a combined effect of all FCGR bearing effector cells including NK-cells, macrophages, monocytes and granulocytes. Here, we addressed whether a combination of favorable genotypes in both FCGR genes 2A and 3A may be associated with further improvement of patient-specific ADCC and survival. The combined effects of FCGR2A and -3A polymorphisms have been already reported in patients with metastatic colorectal cancer under cetuximab therapy.33 It has been shown that homozygous patients with high-affinity genotypes (131H/H and/or 158V/V) had a longer progress-free survival compared to patients missing favorable genotypes (131R/R and 158F/F).33 We extended these investigations and analyzed the impact of combined FCGR2A and -3A genotypes on ADCC during ch14.18/CHO immunotherapy. Importantly, we observed the strongest increase of ADCC in patients with combined high affinity compared to patients who had combined low-affinity FCGR genotypes. In line with our ADCC data, we found the greatest difference in EFS between the patients combining favorable and unfavorable genotypes. Thus, our observations extend previous studies with functional data and underline the importance of both polymorphisms in FCGR3A and -2A genes for therapeutic regimes based on administration of IgG1 mAb.
Based on the recent data showing that NK cell function and ADCC are associated with individual's KIR/KIRL genotypes and that these differences in genotypes correlate with the individuals' outcome in response to Ab-based immunotherapies,8 we further analyzed KIR/KIRL genotypes in our patient cohort. We first evaluated the impact of two distinct well-established KIR/KIRL combinations, inhibitory genotype A/A and activating genotype B/x, on ADCC and survival of NB patients under ch14.18/CHO immunotherapy. Although a positive impact of B/x genotype on survival has been previously reported in leukemia patients,23-25 the role of A/A and B/x genotypes in NB patients under ch14.18/CHO-based immunotherapy has not been shown yet. Here, we observed a strong tendency toward increased ADCC and improved survival in patients with B/x genotype compared to A/A patients suggesting an important role of activating KIRs in ch14.18/CHO immunotherapy.
Since there is a high genetic diversity in KIR haplotypes among the individuals, we further investigated immunotherapy-dependent impact of single KIRs on ADCC and EFS. In contrast to inhibitory KIRs that have been investigated for over a decade, effects of activating KIRs on NK cell cytotoxicity and clinical response to immunotherapies have only recently been addressed.8 From all activating KIRs analyzed in this study, the presence of only 2DS2 was associated with higher ADCC levels and improved survival compared to 2DS2-negative patients. Interestingly, a higher frequency of 2DS2 was shown in NB patients compared to healthy controls, indicating that 2DS2 in absence of immunotherapy has no protective effect against NB.31 This could be explained by the simultaneous presence of the inhibitory KIR 2DL2 in NB patients that may override effects of 2DS2 in absence of immunotherapy with ch14.18/CHO. The effect of 2DS2 is clearly reversed following ch14.18/CHO-based immunotherapy as could be indicated by a significantly increased ADCC in 2DS2-positive patients compared to those who missed 2DS2. Since 2DS2-positive patients were also positive for the inhibitory KIR 2DL2, which is in line with the previously reported data,34 we suggest that a potential inhibitory effect of 2DL2 might be compensated by activating signals of 2DS2 in the presence of ch14.18/CHO-mediated ADCC. Our data show that the reported increased frequency of 2DS2 in NB patients contributes to an improved anti-NB immune response following ch14.18/CHO-based immunotherapy, which underlines the usefulness of this type of immunotherapy in particular for NB patients.
In addition, we found also increased ADCC in patients with the 2DS3 genotype that has been reported as a protective factor in patients with leukemia.35-37 Since all our 2DS3-positive patients presented also 2DS2, we compared ADCC levels in this combined cohort with 2DS2-positive patients missing 2DS3. Importantly, we did not detect a difference in ADCC levels and EFS between patients with only 2DS2 compared to 2DS2- and 2DS3-positive patients. We also analyzed ch14.18/CHO serum concentrations and NK cell numbers in 2DS2-positive and -negative patients. Both ch14.18/CHO serum levels and NK cell numbers were not different between the patient groups (data not shown) underlining that differential effects observed are independent on NK-cell- and ch14.18/CHO-levels.
In summary, these data underline a major role of 2DS2 in mediating ch14.18-CHO-induced antitumor effects in NB patients.
The impact of inhibitory KIRs as well as match or mismatch combinations with their cognate ligands has been previously reported.8 In treatment of NB with the murine anti-GD2 Ab 3F8 or humanized immunocytokine hu14.18-IL-2, mismatch of inhibitory KIRL was associated with improved clinical outcome.17-19 These observations were also made in lymphoma patients treated with rituximab suggesting KIR/KIRL-dependent effects in NK-mediated tumor cell lysis.20 Surprisingly, in the present study we did not find significant differences in ADCC and EFS between matched and mismatched inhibitory KIR/KIRL genotypes in our patient cohort (data not shown), indicating only a minor role for exclusive consideration of the 2DL3/HLA-C1 and 3DL1/HLA-Bw4 KIR/KIRL repertoire. Interestingly, analysis of patients with combined unfavorable genotypes revealed significantly decreased EFS rates in 2DS2-negative missing patients with the inhibitory 3DL1/HLA-Bw4 match, again emphasizing the important contribution of 2DS2 to the efficacy of immunotherapy with ch14.18/CHO. The negative effect of the 3DL1/HLA-Bw4 matched combination in NB patients has been recently reported in one study on the murine anti-GD2 Ab 3F8.38 In line with these data, the greatest survival could be observed in patients with 3DL1/HLA-Bw4 mismatch underlining the role of the inhibitory KIR 3DL1 in the clinical response to anti-GD2 mAb-based immunotherapies.
Based on our data evaluating combined effects of KIR/KIRL on ADCC and survival, we hypothesized that the antitumor clinical response to ch14.18/CHO treatment will be higher in patients with combined favorable FCGR and KIR/KIRL genotypes. Recent studies showed a combined impact of multiple genotypes only in FCGR or KIR/KIRL repertoires separately.8,10
In summary, we showed that FCGR2A and -3A polymorphisms and KIR/KIRL repertoires, especially the presence of the activating KIR 2DS2, synergize with antitumor efficacy of the anti-GD2 Ab ch14.18/CHO in NB patients (ADCC and EFS) and may therefore be useful as biomarkers to predict treatment response.
Materials and methods
Ethic statement
All procedures involving human participants were in accordance with the ethical standards of the institutional and national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Treatment conducted under a compassionate use program and analysis protocols were approved by the ethical committee of the University Medicine Greifswald (Reg. No.: BB 179/15). Informed consent was obtained from all individual participants or their parents or legal guardians.
Patient characteristics
53 NB patients enrolled in this program were treated with ch14.18/CHO given as LTI in combination with interleukin-2 (IL-2) given subcutaneously. Only patients with biopsy-proven high-risk NB were treated. Patients after first-line therapy had to have evaluable disease. Following second-line chemotherapy, patients were allowed to be treated without evidence of disease; previous treatment had to be discontinued 3 weeks prior to the start of ch14.18/CHO. Treatment with isotretinoin, growth factor or other immunomodulatory therapy needed to be completed at least 7 d before treatment start. A performance score above 70% and a life expectancy of at least 12 weeks were required for inclusion.
Following treatment protocol was used: cytokine IL-2 was given for 5 d (s.c., day 1–5, 6 × 106 IU/m2/day), followed by a combined application of IL-2 (s.c., day 8–12, 6 × 106 IU/m2/day) with ch14.18/CHO (i.v., day 8–18, 10 mg/m2/day) and 13-cis-retinoic acid (p.o., day 22–35). At initial diagnosis, 46 patients had stage 4, four stage 3, one stage 2 and two stage 1 disease. Patients with lower stage than four had disseminated relapse. Prior treatment included chemotherapy (53 patients), surgery (51 patients), radiotherapy (34 patients) and high-dose therapy followed by peripheral blood stem cell rescue (51 patients), 32 patients received meta-iodo-benzyl-guanidine (mIBG) therapy preceding high-dose therapy.
The median of the patient age at diagnosis and at start of treatment was 4.4 y (range 0.5–24.1) and 6.0 y (1.9–25.5), respectively. The median of time interval from diagnosis to start of ch14.18/CHO treatment was 25 mo (range 8–111 mo). 28.3% of 53 patients showed MYCN amplification and 42.1% 1p deletion or imbalance. According to the International Neuroblastoma Staging System (INSS), 86.8% of 53 patients were stratified by stage 4 and classification of patients using International Neuroblastoma Risk Group (INRG) stratification system identified 92.5% as stage M patients. At treatment start, the performance scores were ≥ 90 (Lansky or Karnofsky).
Sampling
Patient-specific DNA was isolated from whole blood samples. For analysis of ADCC, patient sodium-heparin blood and serum samples were collected prior to start of immunotherapy (baseline) and on day 8 of ch14.18/CHO infusion. Increase of ADCC compared to baseline was available in 47 of 53 patients.
Analysis of antibody-dependent cell-mediated cytotoxicity (ADCC)
To evaluate ADCC, patient-specific effector cells and heat-inactivated serum collected at the same time point were used. Cytotoxicity was analyzed using a calcein-acetoxymethyl ester (AM)-based cytotoxicity assay as previously described.27 Briefly, patient leukocytes and heat-inactivated serum (12.5% final concentration) were incubated with 5,000 calcein-AM-labeled human GD2-positive NB cells LA-N-1 for 4 h (effector-to-target cell ratio: 40:1). Finally, supernatants of each well were transferred to black 96-well plates for determination of fluorescence at 495 nm excitation and 515 nm emission wavelengths using Synergy HT multimode microplate reader (BioTek Germany). Patient samples as well as samples for spontaneous (target cells only) and maximum release (target cells disrupted using an ultrasonic homogenizer) were analyzed in at least six replicates. ADCC was calculated according to the formula: (experimental release − spontaneous release)/(maximum release − spontaneous release) × 100%.
DNA isolation
53 DNA samples could be isolated using QIAamp® Blood Mini Kit (Qiagen GmbH, 51104) according to the manufacturer's guidelines. For analysis of FCGR polymorphisms and KIR/KIRL genotypes, DNA samples were diluted to achieve concentration of 2.5 and 30 ng/µL, respectively.
Analysis of FCGR polymorphisms
For analysis of FCGR2A high- and low-affinity alleles 131H and 131R as well as FCGR3A high- and low-affinity alleles 158V and 158F, respectively, four allele-specific PCRs were established. Polymorphisms in FCGR genes 2A and 3A were determined using post-amplification melting-curve analysis. First, allele-specific primers (Table 3, Eurofins MWG Operon) were designed to distinguish between allele-specific nucleotide combinations encoding for the respective amino acids (histidine or arginine and valine or phenylalanine for FCGR2A and -3A, respectively). Moreover, as DNA sequences encoding for FCGR3A and FCGR3B are highly homologous, we designed primers for the detection of FCGR3A polymorphisms allowing specific amplification of FCGR3A without amplification of the FCGR3B gene. Since the 3′ ends of the allele-specific primers overlap by only one single mismatched nucleotide (the polymorphic site of interest), we next included a mismatch in the primer sequence at the penultimate position to further increase PCR specificity. For detection of FCGR2A polymorphisms, two allele-specific forward primers and one common reverse primer were selected. For FCGR3A polymorphism analysis, one common forward primer and two allele-specific reverse primers were designed. Glyceraldehyde 3-phosphate dehydrogenase- (GAPDH) served as internal control.
Table 3.
Genotype distribution of FCGR2A and -3A in 53 neuroblastoma patients. Observed frequencies of FCGR2A and -3A genotypes were compared with the expectations of the Hardy–Weinberg equilibrium using Χ2 test. p < 0.05 was considered significant for statistical inference.
| Gene | Genotype | Observed frequency (%) | Expected frequency (%) | Χ2 | p |
|---|---|---|---|---|---|
| FCGR2A | H/H | 26 | 23 | ||
| H/R | 45 | 50 | 0.47 | 0.791 | |
| R/R | 28 | 26 | |||
| FCGR3A | V/V | 6 | 9 | ||
| V/F | 45 | 41 | 0.71 | 0.701 | |
| F/F | 49 | 50 |
Allele-specific PCR for both FCGR genes were performed using a StepOnePlus™ real-time PCR System (Thermo Fisher Scientific, 4376600) in a final volume of 10 µL containing 5 ng of genomic DNA, 5 µL of SensiMix™ SYBR® Hi-ROX 2.0x Mastermix (Bioline, QT605), 0.5 µL A. dest., 20 pg of the respective allele-specific primer pair as well as 2.5 pg of each GAPDH-specific primer. The PCR profile was: 10 min for initial DNA denaturation at 95°C followed by 30 cycles of 95°C for 15 s (denaturation), 61°C for FCGR2A and 56°C for FCGR3A for 15 s (annealing) and 72°C for 60 s (elongation). After the amplification cycles were completed, additional three steps allowing melt curve production were included (95°C for 15 s, 60°C for 60 s and 95°C for 15 s). Due to similar Tm of allele-specific PCR products each allele was examined separately. Using the established PCR method, allele-specific PCR products could be clearly detected by their melting characteristics. To obtain melting-curves, the derivative reporter was calculated as ratio of the change of SYBR Green I dye fluorescence to the change of temperature and was plotted against the melting temperature (Tm) of PCR products.
Validation of FCGR polymorphism analysis
To ensure reliable genotyping of patients, analysis of FCGR polymorphisms was validated. We generated two control plasmids containing DNA sequences encoding for either FCGR2A-131H and FCGR3A-158V alleles (“high-affinity plasmid”; Fig. 1A) or FCGR2A-131R and FCGR3A-158F alleles (“low-affinity plasmid”; Fig. 1B). Additionally, DNA sequences encoding for internal control GAPDH were integrated into both plasmids for further assay precision. These control plasmids were used as PCR template for polymorphism analysis instead of patient DNA to control for the amplification of the respective allele. Plasmids encoding for high-affinity polymorphisms were used for high- and low-affinity PCR as positive and negative control, respectively. Conversely, plasmids containing low-affinity genotypes were utilized as positive and negative control for low- and high-affinity PCR, respectively. Due to similar Tm of allele-specific PCR products each allele was examined separately. Only GAPDH-positive runs that showed amplification of positive control and lack of amplification of negative control of the respective allele-specific fragments were used for further data analysis.
Analysis of KIR/KIRL genotypes
Six activating KIRs (2DS1-5 and 3DS1) and four inhibitory KIRs (2DL1-3 and 3DL1-3) KIRs were determined in patient DNA samples using Olerup SSP® KIR Genotyping Kit incl. Taq polymerase (Olerup GmbH, 104.101-12). For KIRL analysis (HLA-C1; HLA-C2; HLA-Bw4), Olerup SSP® KIR HLA Ligand Kit incl. Taq polymerase (Olerup GmbH, 104.201-12) was used. A Olerup SSP® HLA-Negative Control SSP (Olerup GmbH, 102.102-01) was additionally included in each analytical run. Analysis was performed in a final reaction volume of 10 µL containing 60 ng of patient-specific genomic DNA according to the manufacturer's protocol. PCR products were finally visualized using 2% agarose gel electrophoresis.
Statistics
Statistical analysis was performed using Sigma Plot software (Version 13.0, Jandel Scientific Software). After testing for normality and equal variance across groups, differences between groups were assessed using either the Mann–Whitney rank sum test or Wilcoxon signed-rank test or one way ANOVA test followed by appropriate post hoc comparison. A p value of < 0.05 was considered significant and < 0.01 very significant. All data are presented as mean ± SD (standard deviation) or mean ± SEM (standard error of the mean).
To show whether the observed patterns of FCGR2A and -3A genotype frequencies were consistent with the expectations of the Hardy–Weinberg equilibrium, chi square (χ2) test was used.26 The null hypothesis was rejected at the 95% confidence (p < 0.05). EFS probabilities were estimated using Kaplan Meier analysis39 and compared using LogRank statistics.40 EFS was given as means ± SEM in years. Confidence interval (95% CI) was given in parenthesis.
Disclosure of potential conflicts of interest
No potential conflicts of Interest.
Acknowledgments
The authors would like to thank clinical stuff for support in collecting clinical specimens, Maria Asmus, Manuela Brueser and Theodor Koepp (University Medicine Greifswald, Pediatric Hematology and Oncology, Greifswald, Germany) for excellent technical assistance.
Funding
Financial support was provided by the Hector-Stiftung, Germany, Apeiron Biologics, Vienna, Austria, University Medicine Greifswald, Germany and Deutsche Kinderkrebsstiftung, Germany.
ORCID
Christian Jensen http://orcid.org/0000-0003-0577-1624
Sascha Troschke-Meurer http://orcid.org/0000-0002-6185-462X
Madlen Jüttner http://orcid.org/0000-0003-2040-0514
Holger N. Lode http://orcid.org/0000-0002-1201-208X
References
- 1.Modak S, Cheung NK. Disialoganglioside directed immunotherapy of neuroblastoma. Cancer Invest 2007; 25:67-77; PMID:17364560; http://dx.doi.org/ 10.1080/07357900601130763 [DOI] [PubMed] [Google Scholar]
- 2.Yang RK, Sondel PM. Anti-GD2 strategy in the treatment of neuroblastoma. Drugs Future 2010; 35:665; PMID:21037966; http://dx.doi.org/ 10.1358/dof.2010.035.08.1513490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ladenstein R, Weixler S, Baykan B, Bleeke M, Kunert R, Katinger D, Pribill I, Glander P, Bauer S, Pistoia V et al.. Ch14.18 antibody produced in CHO cells in relapsed or refractory Stage 4 neuroblastoma patients: a SIOPEN Phase 1 study. MAbs 2013; 5:801-9; PMID:23924804; http://dx.doi.org/ 10.4161/mabs.25215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu AL, Gilman AL, Ozkaynak MF, London WB, Kreissman SG, Chen HX, Smith M, Anderson B, Villablanca JG, Matthay KK et al.. Anti-GD2 antibody with GM-CSF, interleukin-2, and isotretinoin for neuroblastoma. N Engl J Med 2010; 363:1324-34; PMID:20879881; http://dx.doi.org/ 10.1056/NEJMoa0911123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lode HN, Jensen C, Endres S, Pill L, Siebert N, Kietz S, Ehlert K, Valteau-Couanet D, Janzek E, Loibner H et al.. Immune activation and clinical responses following long-term infusion of anti-GD2 antibody ch14.18/CHO in combination with interleukin-2 in high-risk neuroblastoma patients. J Clin Oncol 2015; 32:5. [Google Scholar]
- 6.Siebert N, Eger C, Seidel D, Juttner M, Zumpe M, Wegner D, Kietz S, Ehlert K, Veal GJ, Siegmund W et al.. Pharmacokinetics and pharmacodynamics of ch14.18/CHO in relapsed/refractory high-risk neuroblastoma patients treated by long-term infusion in combination with IL-2. MAbs 2016; 8:604-16; PMID:26785755; http://dx.doi.org/ 10.1080/19420862.2015.1130196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koehn TA, Trimble LL, Alderson KL, Erbe AK, McDowell KA, Grzywacz B, Hank JA, Sondel PM. Increasing the clinical efficacy of NK and antibody-mediated cancer immunotherapy: potential predictors of successful clinical outcome based on observations in high-risk neuroblastoma. Front Pharmacol 2012; 3:91; PMID:22623917; http://dx.doi.org/ 10.3389/fphar.2012.00091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang W, Erbe AK, Hank JA, Morris ZS, Sondel PM. NK Cell-mediated antibody-dependent cellular cytotoxicity in cancer immunotherapy. Front Immunol 2015; 6:368; PMID:26284063; http://dx.doi.org/ 10.3389/fimmu.2015.00368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carvalho S, Levi-Schaffer F, Sela M, Yarden Y. Immunotherapy of cancer: from monoclonal to oligoclonal cocktails of anti-cancer antibodies: IUPHAR Review 18. Br J Pharmacol 2016; 173:1407-24; PMID:26833433; http://dx.doi.org/ 10.1111/bph.13450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mellor JD, Brown MP, Irving HR, Zalcberg JR, Dobrovic A. A critical review of the role of Fc gamma receptor polymorphisms in the response to monoclonal antibodies in cancer. J Hematol Oncol 2013; 6:1; PMID:23286345; http://dx.doi.org/ 10.1186/1756-8722-6-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cartron G, Dacheux L, Salles G, Solal-Celigny P, Bardos P, Colombat P, Watier H. Therapeutic activity of humanized anti-CD20 monoclonal antibody and polymorphism in IgG Fc receptor FcgammaRIIIa gene. Blood 2002; 99:754-8; PMID:11806974; http://dx.doi.org/ 10.1182/blood.V99.3.754 [DOI] [PubMed] [Google Scholar]
- 12.Musolino A, Naldi N, Bortesi B, Pezzuolo D, Capelletti M, Missale G, Laccabue D, Zerbini A, Camisa R, Bisagni G et al.. Immunoglobulin G fragment C receptor polymorphisms and clinical efficacy of trastuzumab-based therapy in patients with HER-2/neu-positive metastatic breast cancer. J Clin Oncol 2008; 26:1789-96; PMID:18347005; http://dx.doi.org/ 10.1200/JCO.2007.14.8957 [DOI] [PubMed] [Google Scholar]
- 13.Taylor RJ, Chan SL, Wood A, Voskens CJ, Wolf JS, Lin W, Chapoval A, Schulze DH, Tian G, Strome SE. FcgammaRIIIa polymorphisms and cetuximab induced cytotoxicity in squamous cell carcinoma of the head and neck. Cancer Immunol Immunother 2009; 58:997-1006; PMID:18979096; http://dx.doi.org/ 10.1007/s00262-008-0613-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weng WK, Levy R. Two immunoglobulin G fragment C receptor polymorphisms independently predict response to rituximab in patients with follicular lymphoma. J Clin Oncol 2003; 21:3940-7; PMID:12975461; http://dx.doi.org/ 10.1200/JCO.2003.05.013 [DOI] [PubMed] [Google Scholar]
- 15.Zhang W, Gordon M, Schultheis AM, Yang DY, Nagashima F, Azuma M, Chang HM, Borucka E, Lurje G, Sherrod AE et al.. FCGR2A and FCGR3A polymorphisms associated with clinical outcome of epidermal growth factor receptor expressing metastatic colorectal cancer patients treated with single-agent cetuximab. J Clin Oncol 2007; 25:3712-8; PMID:17704420; http://dx.doi.org/ 10.1200/JCO.2006.08.8021 [DOI] [PubMed] [Google Scholar]
- 16.Benson DM Jr, Caligiuri MA. Killer immunoglobulin-like receptors and tumor immunity. Cancer Immunol Res 2014; 2:99-104; PMID:24592397; http://dx.doi.org/ 10.1158/2326-6066.CIR-13-0219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delgado DC, Hank JA, Kolesar J, Lorentzen D, Gan J, Seo S, Kim K, Shusterman S, Gillies SD, Reisfeld RA et al.. Genotypes of NK cell KIR receptors, their ligands, and Fcgamma receptors in the response of neuroblastoma patients to Hu14.18-IL2 immunotherapy. Cancer Res 2010; 70:9554-61; PMID:20935224; http://dx.doi.org/ 10.1158/0008-5472.CAN-10-2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tarek N, Le Luduec JB, Gallagher MM, Zheng J, Venstrom JM, Chamberlain E, Modak S, Heller G, Dupont B, Cheung NK et al.. Unlicensed NK cells target neuroblastoma following anti-GD2 antibody treatment. J Clin Invest 2012; 122:3260-70; PMID:22863621; http://dx.doi.org/ 10.1172/JCI62749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Venstrom JM, Zheng J, Noor N, Danis KE, Yeh AW, Cheung IY, Dupont B, O'Reilly RJ, Cheung NK, Hsu KC. KIR and HLA genotypes are associated with disease progression and survival following autologous hematopoietic stem cell transplantation for high-risk neuroblastoma. Clin Cancer Res 2009; 15:7330-4; PMID:19934297; http://dx.doi.org/ 10.1158/1078-0432.CCR-09-1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du J, Lopez-Verges S, Pitcher BN, Johnson J, Jung SH, Zhou L, Hsu K, Czuczman MS, Cheson B, Kaplan L et al.. CALGB 150905 (Alliance): rituximab broadens the antilymphoma response by activating unlicensed NK cells. Cancer Immunol Res 2014; 2:878-89; PMID:24958280; http://dx.doi.org/ 10.1158/2326-6066.CIR-13-0158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, Corliss B, Tyan D, Lanier LL, Parham P. Human diversity in killer cell inhibitory receptor genes. Immunity 1997; 7:753-63; PMID:9430221; http://dx.doi.org/ 10.1016/S1074-7613(00)80394-5 [DOI] [PubMed] [Google Scholar]
- 22.Valiante NM, Uhrberg M, Shilling HG, Lienert-Weidenbach K, Arnett KL, D'Andrea A, Phillips JH, Lanier LL, Parham P. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity 1997; 7:739-51; PMID:9430220; http://dx.doi.org/ 10.1016/S1074-7613(00)80393-3 [DOI] [PubMed] [Google Scholar]
- 23.Cooley S, Trachtenberg E, Bergemann TL, Saeteurn K, Klein J, Le CT, Marsh SG, Guethlein LA, Parham P, Miller JS et al.. Donors with group B KIR haplotypes improve relapse-free survival after unrelated hematopoietic cell transplantation for acute myelogenous leukemia. Blood 2009; 113:726-32; PMID:18945962; http://dx.doi.org/ 10.1182/blood-2008-07-171926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cooley S, Weisdorf DJ, Guethlein LA, Klein JP, Wang T, Le CT, Marsh SG, Geraghty D, Spellman S, Haagenson MD et al.. Donor selection for natural killer cell receptor genes leads to superior survival after unrelated transplantation for acute myelogenous leukemia. Blood 2010; 116:2411-9; PMID:20581313; http://dx.doi.org/ 10.1182/blood-2010-05-283051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cooley S, Weisdorf DJ, Guethlein LA, Klein JP, Wang T, Marsh SG, Spellman S, Haagenson MD, Saeturn K, Ladner M et al.. Donor killer cell Ig-like receptor B haplotypes, recipient HLA-C1, and HLA-C mismatch enhance the clinical benefit of unrelated transplantation for acute myelogenous leukemia. J Immunol 2014; 192:4592-600; PMID:24748496; http://dx.doi.org/ 10.4049/jimmunol.1302517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hardy GH. Mendelian proportions in a mixed population. Science 1908; 28:49-50; PMID:17779291; http://dx.doi.org/ 10.1126/science.28.706.49 [DOI] [PubMed] [Google Scholar]
- 27.Siebert N, Seidel D, Eger C, Juttner M, Lode HN. Functional bioassays for immune monitoring of high-risk neuroblastoma patients treated with ch14.18/CHO anti-GD2 antibody. PLoS One 2014; 9:e107692; PMID:25226154; http://dx.doi.org/ 10.1371/journal.pone.0107692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Terszowski G, Klein C, Stern M. KIR/HLA interactions negatively affect rituximab- but not GA101 (obinutuzumab)-induced antibody-dependent cellular cytotoxicity. J Immunol 2014; 192:5618-24; PMID:24795454; http://dx.doi.org/ 10.4049/jimmunol.1400288 [DOI] [PubMed] [Google Scholar]
- 29.Marsh SG, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA, Geraghty DE, Hansen JA, Mach B, Mayr WR et al.. Nomenclature for factors of the HLA system, 2002. Tissue Antigens 2002; 60:407-64; PMID:12492818; http://dx.doi.org/ 10.1034/j.1399-0039.2002.600509.x [DOI] [PubMed] [Google Scholar]
- 30.Jobim MR, Jobim M, Salim PH, Portela P, Jobim LF, Leistner-Segal S, Bittelbrunn AC, Menke CH, Biazus JV, Roesler R et al.. Analysis of KIR gene frequencies and HLA class I genotypes in breast cancer and control group. Hum Immunol 2013; 74:1130-3; PMID:23792055; http://dx.doi.org/ 10.1016/j.humimm.2013.06.021 [DOI] [PubMed] [Google Scholar]
- 31.Keating SE, Ni CC, Dring MM, Stallings RL, O'Meara A, Gardiner CM. Increased frequencies of the killer immunoglobulin-like receptor genes KIR2DL2 and KIR2DS2 are associated with neuroblastoma. Tissue Antigens 2015; 86:172-7; PMID:26202659; http://dx.doi.org/ 10.1111/tan.12608 [DOI] [PubMed] [Google Scholar]
- 32.Ferris RL, Jaffee EM, Ferrone S. Tumor antigen-targeted, monoclonal antibody-based immunotherapy: clinical response, cellular immunity, and immunoescape. J Clin Oncol 2010; 28:4390-9; PMID:20697078; http://dx.doi.org/ 10.1200/JCO.2009.27.6360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bibeau F, Lopez-Crapez E, Di FF, Thezenas S, Ychou M, Blanchard F, Lamy A, Penault-Llorca F, Frebourg T, Michel P et al.. Impact of Fc{gamma}RIIa-Fc{gamma}RIIIa polymorphisms and KRAS mutations on the clinical outcome of patients with metastatic colorectal cancer treated with cetuximab plus irinotecan. J Clin Oncol 2009; 27:1122-9; PMID:19164213; http://dx.doi.org/ 10.1200/JCO.2008.18.0463 [DOI] [PubMed] [Google Scholar]
- 34.Hsu KC, Liu XR, Selvakumar A, Mickelson E, O'Reilly RJ, Dupont B. Killer Ig-like receptor haplotype analysis by gene content: evidence for genomic diversity with a minimum of six basic framework haplotypes, each with multiple subsets. J Immunol 2002; 169:5118-29; PMID:12391228; http://dx.doi.org/ 10.4049/jimmunol.169.9.5118 [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, Wang B, Ye S, Liu S, Liu M, Shen C, Teng Y, Qi J. Killer cell immunoglobulin-like receptor gene polymorphisms in patients with leukemia: possible association with susceptibility to the disease. Leuk Res 2010; 34:55-8; PMID:19450876; http://dx.doi.org/ 10.1016/j.leukres.2009.04.022 [DOI] [PubMed] [Google Scholar]
- 36.Shahsavar F, Tajik N, Entezami KZ, Fallah RM, Asadifar B, Alimoghaddam K, Ostadali DM, Jalali A, Ghashghaie A, Ghavamzadeh A. KIR2DS3 is associated with protection against acute myeloid leukemia. Iran J Immunol 2010; 7:8-17; PMID:20371915; http://dx.doi.org/IJIv7i1A2 [PubMed] [Google Scholar]
- 37.Karabon L, Jedynak A, Giebel S, Wolowiec D, Kielbinski M, Woszczyk D, Kapelko-Slowik K, Kuliczkowski K, Frydecka I. KIR/HLA gene combinations influence susceptibility to B-cell chronic lymphocytic leukemia and the clinical course of disease. Tissue Antigens 2011; 78:129-38; PMID:21726204; http://dx.doi.org/ 10.1111/j.1399-0039.2011.01721.x [DOI] [PubMed] [Google Scholar]
- 38.Forlenza CJ, Boudreau JE, Zheng J, Le Luduec JB, Chamberlain E, Heller G, Cheung NV, Hsu KC. KIR3DL1 Allelic Polymorphism and HLA-B Epitopes Modulate Response to Anti-GD2 Monoclonal Antibody in Patients With Neuroblastoma. J Clin Oncol 2016; 34:2443-51; PMID:27069083; http://dx.doi.org/ 10.1200/JCO.2015.64.9558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc 1958; 53:457-80; http://dx.doi.org/ 10.1080/01621459.1958.10501452 [DOI] [Google Scholar]
- 40.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 1959; 22:719-48; PMID:13655060; http://dx.doi.org/ 10.1093/jnci/22.4.719 [DOI] [PubMed] [Google Scholar]