Abstract
Background and Objectives:
Macrolide, lincosamide and streptogramin type B (MLSB) antibiotics are important in the treatment of Staphylococcus aureus infections and existence of isolates with ability to resist against MLSB antibiotics is worrisome.
Materials and Methods:
In this cross sectional study, 101 S. aureus isolates were collected from patients of five selected hospitals in Tehran over a period of five months. Disk diffusion tests and differentiation between constitutive and inducible resistances were carried out by D-test. The presence of mecA, msrA, ermA and ermC genes were detected using PCR or multiplex PCR.
Results:
Out of 101 S. aureus isolates, 58 (57.4%) were methicillin resistant and 57 (56.4%) expressed resistance to erythromycin. The prevalence of constitutive MLSB (cMLSB), inducible MLSB (iMLSB) and MS (Negative) phenotype in all erythromycin resistant isolates were 71.9, 26.3 and 1.7%, respectively. Out of all the erythromycin resistant isolates, 57.8% harbored both ermA and ermC genes which possessed constitutive resistance. 8.7% of the isolates contained ermA gene alone which possessed inducible resistance with D phenotype and 5.2% of isolates just contained ermC gene which had inducible resistance with D+ phenotype. msrA gene was detected in 3.5% of the erythromycin resistant S. aureus isolates with constitutive resistance. None of the genes were detected among MS phenotypes.
Conclusion:
In this study, most of S. aureus isolates carried both ermA and ermC genes and there was a significant relationship (P value ≤ 0.05) between different resistance phenotypes and erm genes.
Keywords: Staphylococcus aureus, D-test, Erm A, ErmC, MsrA
INTRODUCTION
Staphylococcus aureus has been recognized as a significant pathogen among infectious diseases of human. S. aureus, particularly methicillin resistant isolate (MRSA), is an important cause of hospital and community acquired infection throughout the world.
The resistance to methicillin belongs to a penicillin-binding protein encoded by a mobile genetic element called the methicillin-resistant gene (mecA) (1). Increasing its resistance to antibiotics is limited to those available antibiotics which are prescribed for treatment of different infections caused by this bacterium (2).
Macrolide, lincosamide and streptogramin type B (MLSB) antibiotics such as erythromycin, clindamycin and streptogramin B are usually used in the treatment of infections, particularly skin and soft tissue infections. Clindamycin is a noteworthy choice in the treatment strategies for various reasons; 1) it has high tissue penetration (except for the central nervous system), 2) good oral absorption makes it appropriate for outpatient therapy, 3) clindamycin can be used as a choice antibiotic in patients suffer from allergy to penicillin. However, resistance to these antibiotics is significantly increasing all over the world (3, 4). Three different mechanisms of MLSB resistance have been described in Staphylococcus genus. One mechanism can occur through methylation of their ribosomal site which is mediated by the presence of erythromycin resistance methylase (erm) genes. Methylases reduce binding of MLSB antibiotics to the target site in the 50S ribosomal subunit. ermA and ermC are two common genes responsible for the MLSB resistance in S. aureus. Most of the bacteria targeted by macrolides and lincosamides, including Gram-positive species, spirochetes, and anaerobes, express Erm methylases. erm(A) genes are frequently spread in methicillin resistant isolates and are generated by Tn554 transposons while erm(C) genes are mostly responsible for erythromycin resistance in methicillin susceptible isolates and are carried by plasmids. The second mechanism can occur by drug efflux typically mediated by the ATP binding cassette (ABC) transporter msrA and the other mechanism is drug modification by two enzymes that confer resistance to macrolides and lincosamides such Mph(C) or Lnu(A), respectively (5–7). Despite the high incidence rate of MLSB resistant staphylococci, especially among MRSA (8–10), currently there is a little information available on the incidence and types of these resistant bacteria in Iran (11). The present study aimed to provide information regarding the prevalence of MLSB resistant S. aureus isolates in Tehran, Iran.
MATERIALS AND METHODS
Bacterial isolates.
In this descriptive study, a total of 101 clinical isolates of S. aureus were collected from hospitalized patients in five hospitals in Tehran, Iran (December 2012 to April 2013). Isolates were obtained from different clinical specimens including wounds, respiratory tract, urine, blood, sterile body fluids and abscesses. Only one isolate per patient was included. An informed consent was obtained from all subjects enrolled in this study. Isolates were characterized as S. aureus by standard microbiological methods including Gram staining, catalase test, slide and tube coagulase test, growth on mannitol salt agar and deoxy ribonuclease test (12). All detected S. aureus isolates were stored in nutrient broth plus 20% glycerol at −70 °C until study time.
Phenotypic determination of antibiotic resistance.
Determination of MLSB phenotypes was performed by the use of D-test as described previously (13) and according to the guidelines of Clinical and Laboratory Standards Institute (CLSI) (14). Three antibiotic resistance phenotypes were determined; isolates containing constitutive MLSB (cMLSB) which were resistant to both erythromycin and clindamycin, isolates containing inducible MLSB (iMLSB) which were resistant to erythromycin but sensitive to clindamycin and isolates containing the MS phenotype which showed resistance to merely macrolide and streptogramin B. A special disk diffusion procedure, D-test, was developed for discrimination of iMLSBs. In iMLSB resistant isolates, resistance to clindamycin was induced by diffusion of erythromycin through the agar which led to flattening of the clindamycin zone of inhibition closest to the erythromycin disk (A D-shaped zone) while MS phenotype contained isolates forming a circular zone around the clindamycin disk. Induction test in cMLSBs included two phenotypes; resistant (R) and hazy D zone (HD). In R phenotype, bacterial growth was seen in the presence of erythromycin and clindamycin. In contrast, in HD phenotype, in addition to the observed bacterial growth at the presence of erythromycin, two zones of growth could be observed around the clindamycin disk. Outer zone had a light and hazy growth extending to the clindamycin disk and a more dense bacterial growth in the inner zone which was blunted proximal to the erythromycin disk as observed in phenotype D. Erythromycin resistant isolates with flattening of the inhibition zone around the clindamycin disk were considered as positive for iMLSB. Induction test description in iMLSB included two phenotypes; D and D+. In D phenotype a D shaped clear zone was seen around the clindamycin disk proximal to the erythromycin disk while in D+ phenotype, in addition to the observed D shaped zone around the clindamycin disk, small colonies were grown at the inhibition zone of the clindamycin disk. Bacterial growth at the presence of erythromycin and circular clear zone around clindamycin disk was considered as MS positive and negative phenotype in induction test (13). Detection of MRSA was performed by cefoxitine disk (30 μg) on the Mueller Hinton agar (Merck, England) plate according to the CLSI guidelines (14). All antibiotic disks were purchased from Mast Co, UK and S. aureus ATCC 25923 was used as a standard strain.
Amplification of mecA, ermA, ermC, msrA genes.
Bacterial genomic DNA was extracted according to the method described previously (15). In Brief, five colonies from overnight incubated brain heart infusion agar plates (Merck, England) were suspended in 300 μl sterile distilled water and were heated at 100 °C for 15 min. After centrifugation at 14,000 rpm (10 min), the supernatant was used as the template DNA in PCR. Multiplex PCR was performed for detection of ermA and ermC genes whereas mecA and msrA genes were detected by single PCRs (15–17). Sequence of each primer and the reference for each PCR has been shown in Table 1. Amplification was performed in a final volume of 25 μl containing 0.5 μl of each primer (25 pmol), 1X PCR buffer, 1.5 mM MgCl2, 0.2 mM dNTP Mix, 5 μl of template DNA and 1.5 U of Taq DNA polymerase. PCR products were electrophoresed on 1% agarose gel (Roche, Germany) at 100 volts and later stained with ethidium bromide solution to see the amplified DNA fragments under gel documentation system (UV Tech, UK) with a molecular size marker (100 bp ladders, Fermentas, Lithuania). Resistant S. aureus isolates (ermA, ermC, and msrA gene positive strains, courtesy of Dr. Fereshteh Jabalameli) were used as the positive controls.
Table 1.
Primer sequences
| Genes | Sequence (5′-3′) | References |
|---|---|---|
| mecA | F: 5′-gtagaaatgactgaacgtccgataa-3′ R: 5′-ccaattccacattgtttcggtctaa-3′ |
15, 16 |
| msrA | F: 5′-ggcacaataagagtgtttaaagg-3′ R: 5′-aagttatatcatgaatagattgtcctgtt-3′ |
17 |
| ermA* | F: 5′-gttcaagaacaatcaatacagag-3′ R: 5′-ggatcaggaaaaggacattttac-3′ |
17 |
| ermC* | F: 5′-gctaatattgtttaaatcgtcaattcc-3′ R: 5′-ggatcaggaaaaggacattttac-3′ |
17 |
Multiplex PCR
Statistical analysis.
Data were summarized using mean ± standard deviation (SD) and assurance intervals for the microbiological and demographic characteristics. All analyses were performed using SPSS (v. 20, Chicago, USA) using Fisher exact or Chi-square tests and P values ≤ 0.05 were considered statistically significant.
RESULTS
S. aureus isolates were isolated from wound (n=29), urine (n=25), respiratory tract (n=14), sterile body fluids (n=16), blood (n=11) and abscesses (n=6). These isolates were obtained from the patients admitted to internal medicine (48.0%), intensive care units (28.0%), infectious diseases (16.5%) and surgery (7.5%). Demographic characteristics showed that there were 64 males and 37 females with the mean age of 52.57 (± 21.15) years. The distribution of MLSB resistance phenotypes based on the studied genes is illustrated in Table 2. Among 101 S. aureus isolates, 58 (57.4%) were resistant to methicillin and 57 (56.4%) were reported as erythromycin resistant. Forty nine (84.4%) of MRSA isolates and 8 (18.6%) methicillin susceptible S. aureus (MSSA) isolates were resistant to erythromycin, respectively. The prevalence of cMLSB, iMLSB and MS resistance phenotypes among erythromycin resistant isolates were 71.9, 26.3 and 1.7%, respectively. mecA gene was detected in all MRSA isolates. Among 41 isolates with cMLSB resistance phenotype, 38 isolates were MRSA and 10 out of 15 isolates with iMLSB resistant phenotypes were MRSA. Thirty three isolates in the cMLSB group were positive for both ermA and ermC genes aside from the presence or absence of msrA gene. Eight isolates of iMLSB category contained one of ermA or ermC genes and none of them had msrA gene. MS phenotype was only found in one MRSA isolate and this isolate did not harbor erm genes or msrA gene. The most common genes in erythromycin resistant isolates were ermA (66.6%), ermC (63.1%) and msrA (3.5%), respectively. None of the 8 MSSA erythromycin resistant isolates had ermA, ermC or msrA gene. Erythromycin susceptible isolates did not harbor ermA, ermC and msrA genes.
Table 2.
Distribution of resistance phenotypes of MLSB based on the studied genes (mecA, ermA, ermC and msrA)
| Genes | Resistance phenotypes of isolates (%) | |||||
|---|---|---|---|---|---|---|
| R* (n=40) | HD (n=1) | D (n=11) | D+ (n=4) | N (n=1) | Total (n=57) | |
| Negative PCR (for all 4 genes) | 3 (7.5) | - | 5 (45.4) | - | - | 8 (14) |
| mecA | 5 (12.5) | - | 1 (9) | 1 (25) | 1 (100) | 8 (14) |
| mecA + ermA | - | - | 5 (45.4) | - | - | 5 (8.7) |
| mecA + ermC | - | - | - | 3 (75) | - | 3 (5.2) |
| mecA + ermA + ermC | 30 (75) | 1 (100) | - | - | - | 31 (54.3) |
| mecA + ermA + ermC + msrA | 2 (5) | - | - | - | - | 2 (3.5) |
R*: resistant phenotype; HD: hazy D zone phenotype; D: D phenotype; D+: D+ phenotype; N: negative phenotype.
Among erythromycin resistant isolates, 40 isolates had constitutive resistance R phenotype and 32 of these isolates contained both ermA and ermC genes simultaneously with or without msrA gene. In this study only one isolate showed constitutive resistance HD phenotype which contained both ermA and ermC genes in the absence of msrA gene. The D-zone phenotype was observed in 11 isolates showing D-shaped clear zone around clindamycin disk. The D+ phenotype was observed in 4 isolates.
PCR amplifications (Fig. 1) revealed that five D phenotype isolates possess ermA gene alone and three isolates with D+ phenotype just harbored ermC gene. One negative phenotype isolate which showed resistance to erythromycin but was sensitive to clindamycin with a clear zone around clindamycin disk did not carry ermA, ermC or msrA genes (Table 2). The mecA gene was detected in all MRSA isolates. Among 41 cMLSB resistant phenotype isolates, 38 isolates were MRSA whereas 10 out of 15 iMLSB resistant phenotype isolates were MRSA. MS phenotype was only found in one MRSA isolate. None of 8 erythromycin resistant MSSAs had ermA, ermC or msrA genes. Erythromycin susceptible isolates did not harbor ermA, ermC or msrA genes.
Fig. 1.
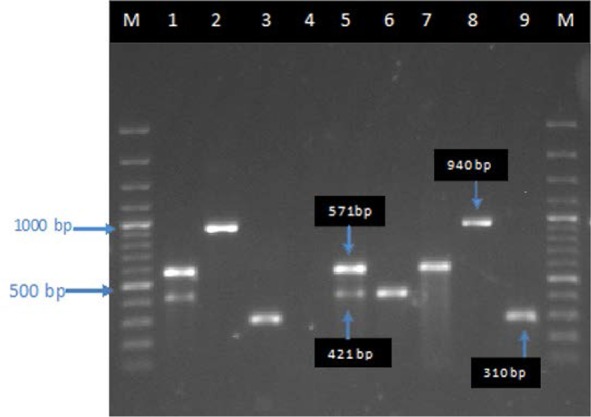
Agarose gel electrophoresis patterns from multiplex and single PCR on S. aureus isolates. Lane M, 1000 bp DNA ladder; lane 1, positive control PCR for ermA (421 bp) and ermC (571bp) genes; lanes 2–3, positive control PCRs for msrA (940 bp) and mecA (310 bp) genes; lane 4, negative control PCR; lane 5, ermA and ermC positive MRSA (a R phenotype); lane 6, ermA gene positive MRSA (a D pheno-type); lane 7, ermC gene positive MRSA (a D+ phenotype); lane 8, msrA gene positive MRSA (a R phenotype); lane 9, MRSA positive (a N phenotype).
In this study, most of the isolated S. aureus strains carried both of the ermA and ermC genes and a significant association was observed between different resistance phenotypes and erm genes (P ≤ 0.01).
DISCUSSION
Resistance to antimicrobial agents is an important problem in clinical issues and MRSA is now one of the most common nosocomial pathogens in many countries (18). In this study the rate of methicillin resistance among clinical isolates of S. aureus was 57.4% which is higher than reports from Tehran (29.7–52%) (19–22), other cities in the country (44.4–56.8%) (23–26) or other countries including Pakistan (48%), Australia (33.6%) and Turkey (25.9%) (27–29). On the other hand, it was lower than reports from China (72.8%) and India (59.3%) (30, 31). Geographic variations in the prevalence rate of MRSA among Iranian S. aureus isolates and variations from one hospital to the others may be due to the various factors such as efficacy of practices in controlling the infection, healthcare facilities and antibiotic usages that vary in the hospitals (32). Due to the changing pattern of antibiotic resistance among S. aureus isolates, it would be wise to have periodical surveillance of these changes every 3 to 4 years (33).
In the present study, erythromycin resistance (56.4%) was higher than published reports from Belgium (37.4%) (7), Iran (42%) (21) or India (51.7%) (34) and lower than Turkish (60.4%) (29) or Korean reports (77.5%) (35). In this study, the rates of constitutive, inducible and MS phenotypes among 57 erythromycin resistant isolates were 71.9, 26.3 and 1.7%, respectively. The observed higher rate of cMLSB than iMLSB resistance phenotype among erythromycin resistant S. aureus isolates was in accordance to the other reports (11, 36, 37). Rate of inducible phenotype in the erythromycin resistant S. aureus isolates was higher than other Iranian studies (6.4%, 14%) (11, 23) and lower than studies from all over the world (6, 34, 37). In this study, the MS resistance phenotype was slightly higher than what reported by another study from Iran (11) and lower than other studies from Greece and the US (6, 13). Increased rate of inducible resistant isolates is attributed to the increased usages of macrolides and clindamycin. It is crucial to perform the D-test to determine the erythromycin resistance (3). In our study 57.8% of erythromycin resistant S. aureus isolates had a coexistence of ermA and ermC genes which was higher than the other one conducted study from Iran (48.4%) (11) and two published studies from Turkey (37.5 and 18.6%) (38, 39) but lower than Greece (0.5%) and Belgium (3%) (6, 7). msrA gene was detected in 3.5% of erythromycin resistant S. aureus isolates which was higher than reports from Canada (1%) (40) and lower than other reports from Belgium hospitals (5%) (7) and Turkey (9.5%) (29). Coexistence of ermA and ermC genes in constitutive resistance phenotype was similar to the reports in Turkey (38, 39). We found 10 MRSA isolates having inducible phenotype among which 5 isolates harbored ermA and 3 other isolates only consisted ermC gene. It is notable that the former group had D phenotype while the latter ones were D+ similar to the findings of the study performed by Stward et al. (13). In our study msrA gene was seen in 2 constitutive resistant MRSA isolates in contrast with the study published by Spilopoulou et al. (6) and Steward et al. (13) in which all msrA isolates represented MS resistance phenotype.
CONCLUSION
It appears that the high prevalence of inducible resistance in this study may be due to the variable use of erythromycin and clindamycin in Iran. This is the first report from Iran that shows differentiation of MLSB resistance phenotypes according to their corresponding genes which can provide information to help characterization of isolates for epidemiologic studies in the communities. D-test should be an obligatory test in routine disk diffusion methods to detect inducible antibiotic resistance in treatment of infections. On the other hand, increasing of (ermA + ermC) and msrA genes in the MRSA emphasizes on the accurate use of these antibiotics to prevent any treatment failure.
ACKNOWLEDGEMENTS
This research has been supported by Tehran University of Medical Sciences, grant number 25177/93-02-30. It has also been supported by Research Chancellor of Shahed University.
REFERENCES
- 1.Nicholas BD, Bhargave G, Hatipoglu A, Heffelfinger R, Rosen M, Pribitkin EA. Preoperative prevalence of methicillin-resistant Staphylococcus aureus (MRSA) colonization in patients undergoing intranasal surgery. Med Sci Monit 2010; 16:CR365–368. [PubMed] [Google Scholar]
- 2.Weigel LM, Clewell DB, Gill SR, Clark NC, McDougal LK, Flannagan SE, et al. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science 2003; 302:1569–1571. [DOI] [PubMed] [Google Scholar]
- 3.Zelazny AM, Ferraro MJ, Glennen A, Hindler JF, Mann LM, Munro S, et al. Selection of strains for quality assessment of the disk induction method for detection of inducible clindamycin resistance in Staphylococci: a CLSI Collaborative Study. J Clin Microbiol 2005; 43:2613–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Emaneini M, Eslampour MA, Sedaghat H, Aligholi M, Jabalameli F, Shahsavan S, et al. Characterization of phenotypic and genotypic inducible macrolide resistance in Staphylococci in Tehran, Iran. J Chemother 2009; 21:595–597. [DOI] [PubMed] [Google Scholar]
- 5.Leclercq R. Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin Infect Dis 2002; 34:482–492. [DOI] [PubMed] [Google Scholar]
- 6.Spilopoulou I, Petinaki E, Papandreou P, Dimitracopoulos G. erm(C) is the predominant genetic determinant for the expression of resistance to macrolides among methicillin-resistance Staphylococcus aureus clinical isolates in Greece. J Antimicrob Chemother 2004; 53:814–817. [DOI] [PubMed] [Google Scholar]
- 7.Vandendriessche S, Kadlec K, Schwarz S, Denis O. Methicillin-susceptible Staphylococcus aureus ST398-t571 harbouring the macrolide-lincos-amide-streptogramin B resistance gene erm(T) in Belgian hospitals. J Antimicrob Chemother 2011; 66:2455–2459. [DOI] [PubMed] [Google Scholar]
- 8.Prabhu K, Rao S, Rao V. Inducible clindamycin resistance in Staphylococcus aureus isolated from clinical samples. J Lab Physicians 2011; 3:25–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Debdas D, Joshi S. Incidence of clindamycin resistance in clinical isolates of Staphylococcus aureus. J Infect Dev Ctries 2011; 5:316–317. [DOI] [PubMed] [Google Scholar]
- 10.Yilmaz G, Aydin K, Iskender S, Caylan R, Koksal I. Detection and prevalence of inducible clindamycin resistance in staphylococci. J Med Microbiol 2007; 56:342–345. [DOI] [PubMed] [Google Scholar]
- 11.Saderi H, Emadi B, Owlia P. Phenotypic and genotypic study of macrolide, lincosamide and streptogramin B (MLSB) resistance in clinical isolates of Staphylococcus aureus in Tehran, Iran. Med sci Monit 2011; 17:BR48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahon CR, Lehman DC, Manuselis G, with 49 contributors (2007). Text book of diagnostic microbiology, 3rd ed Philadelphia Saunders, US. [Google Scholar]
- 13.Steward C, Raney PM, Morrell AK, Williams PP, McDougal LK, Jevitt L, et al. Testing for induction of clindamycin resistance in erythromycin-resistant isolates of Staphylococcus aureus. J Clin Microbiol 2005; 43:1716–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.CLSI. Clinical and Laboratory Standards Institute Performance standards for antimicrobial susceptibility testing; twenty-first informational supplement. CLSI document M100-S21. Wayne, PA: Clinical and Laboratory Standards Institute; 2011. 31:M100-S21. [Google Scholar]
- 15.Perez-Roth E, Claverie-Martin F, Villar J, Méndez-Alvarez S. Multiplex PCR for simultaneous identification of Staphylococcus aureus and detection of methicillin and mupirocin resistance. J Clin Microbiol 2001; 39:4037–4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang K, Sparling J, Chow BL, Elsayed S, Hussain Z, Church DL, et al. New quadriplex PCR assay for detection of methicillin and mupirocin resistance and simultaneous discrimination of Staphylococcus aureus from coagulase-negative staphylococci. J Clin Microbiol 2004; 42:4947–4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lina G, Quaglia A, Reverdy ME, Leclercq R, Vandenesch F, Etienne J. Distribution of genes encoding resistance to macrolides, lincosamides and streptogramins among staphylococci. Antimicrob Agents Chemother 1999; 43:1062–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olarte NM, Valderrama IA, Reyes KR, Garzón MI, Escobar JA, Castro BE, et al. Methicillin-resistant Staphylococcus aureus colonization in a Colombian hospital intensive care unit: phenotypic and molecular characterization. Biomedica 2010; 30:353–361. [PubMed] [Google Scholar]
- 19.Rahimi F, Bouzari M, Katouli M, Pourshafie MR. Antibiotic resistance pattern of methicillin resistant and methicillin sensitive Staphylococcus aureus isolates in Tehran, Iran. Jundishapur J Microbiol 2013; 6:144–149. [Google Scholar]
- 20.Fatholahzadeh B, Emaneini M, Gilbert G, Udo E, Aligholi M, Modarressi MH, et al. Staphylococcal cassette chromosome mec (SCCmec) analysis and antimicrobial susceptibility patterns of methicillin-resistant Staphylococcus aureus (MRSA) isolates in Tehran, Iran. Microb Drug Resist 2008; 14:217–220. [DOI] [PubMed] [Google Scholar]
- 21.Farhadian A, Behzadian Nejad Q, Najar peerayeh SH, Rahbar M, Vaziri F. Determination of vancomycin and methicillin resistance in clinical isolates of Staphylococcus aureus in Iranian hospitals. Br Microbiol Res J 2014; 4:454–461. [Google Scholar]
- 22.Emaneini M, Aligholi M, Hashemi FB, Jabalameli F, Shahsavan S, Dabiri H, et al. Isolation of vancomycin-resistant Staphylococcus aureus in a teaching hospital in Tehran. J Hosp Infect 2007; 66:92–93. [DOI] [PubMed] [Google Scholar]
- 23.Motamedifar M, Seddigh Ebrahim Sarai H, Mansury D. Patterns of constitutive and inducible clindamycin resistance in Staphylococcus aureus isolated from clinical samples by D-test method, Shiraz, Southwest of Iran. GMJ 2014; 3:216–221. [Google Scholar]
- 24.Dibah S, Arzanlou M, Jannati E, Shapouri R. Prevalence and antimicrobial resistance pattern of methicillin resistant Staphylococcus aureus (MRSA) strains isolated from clinical specimens in Ardabil, Iran. Iran J Microbiol 2014; 6:163–168. [PMC free article] [PubMed] [Google Scholar]
- 25.Hasani A, Sheikhalizadeh V, Naghili B, Valizadeh V, Nikoonijad AR. Methicillin resistant and susceptible Staphylococcus aureus: Appraising therapeutic approaches in the Northwest of Iran. Iran J Microbiol 2013; 5:56–62. [PMC free article] [PubMed] [Google Scholar]
- 26.Sadeghi J, Mansouri S. Molecular characterization and antibiotic resistance of clinical isolates of methicillin-resistant Staphylococcus aureus obtained from Southeast of Iran (Kerman). APMIS 2014; 122:405–411. [DOI] [PubMed] [Google Scholar]
- 27.Ahmad MK, Asrar A. Prevalence of methicillin resistant Staphylococcus aureus in pyogenic community and hospital acquired skin and soft tissues infections. J Pak Med Assoc 2014; 64:892–895. [PubMed] [Google Scholar]
- 28.Nimmo GR, Pearson JC, Collignon PJ, Christiansen KJ, Coombs GW, Bell JM, et al. Antimicrobial susceptibility of Staphylococcus aureus isolated from hospital inpatients, 2009: report from the Australian Group on Antimicrobial Resistance. Commun Dis Intell Q Rep 2011; 35:237–243. [PubMed] [Google Scholar]
- 29.Duran N, Ozer B, Duran GG, Onlen Y, Demir C. Antibiotic resistance genes & susceptibility patterns in staphylococci. Indian J Med Res 2012; 135:389–396. [PMC free article] [PubMed] [Google Scholar]
- 30.Song Y, Du X, Li T, Zhu Y, Li M. Phenotypic and molecular characterization of Staphylococcus aureus recovered from different clinical specimens of inpatients at a teaching hospital in Shanghai between 2005 and 2010. J Med Microbiol 2013; 62:274–282. [DOI] [PubMed] [Google Scholar]
- 31.Tiwari HK, Sen MR. Emergence of vancomycin resistant Staphylococcus aureus (VRSA) from a tertiary care hospital from northern part of India. BMC Infect Dis 2006; 6:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Carvalho MJ, Pimenta FC, Hayashida M, Gir E, da Silva AM, Barbosa CP, et al. Prevalence of methicillin-resistant and methicillin-susceptible S. aureus in the saliva of health professionals. Clinics (Sao Paulo) 2009; 64:295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rahimi F, Bouzari M, Maleki Z, Rahimi F. Antibiotic susceptibility pattern among Staphylococcus spp. with emphasis on detection of mecA gene in methicillin resistant Staphylococcus aureus isolates. Arch Clin Infect Dis 2009; 4:143–150. [Google Scholar]
- 34.Lyall KDS, Gupta V, Chhina D. Inducible clindamycin resistance among clinical isolates of Staphylococcus aureus. J Mahatma Gandhi Inst Med Sci 2013; 18:112–115. [Google Scholar]
- 35.Jung YH, Kim KW, Lee KM, Yoo JI, Chung GT, Kim BS, et al. Prevalence and characterization of macro-lide-lincomycin-streptogramin B-resistant Staphylococcus aureus in Korean hospitals. J Antimicrob Chemother 2008; 61:458–460. [DOI] [PubMed] [Google Scholar]
- 36.Fiebelkorn KR, Crawford SA, McElmeel ML, Jorgensen JH. Practical disk diffusion method for detection of inducible clindamycin resistance in Staphylococcus aureus and coagulase-negative staphylococci. J Clin Microbiol 2003; 41:4740–4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saribas Z, Tunckanat F, Pinar A. Prevalence of erm genes encoding macrolide-lincosamide-streptogramin (MLS) resistance among clinical isolated of Staphylococcus aureus in a Turkish university hospital. Clin Microbiol Infect 2006; 12:797–799. [DOI] [PubMed] [Google Scholar]
- 38.Aktas Z, Aridogan A, Kayacan CB, Aydin D. Resistance to macrolide, lincosamide and streptogramin antibiotics in staphylococci isolated in Istanbul, Turkey. J Microbiol 2007; 45:286–290. [PubMed] [Google Scholar]
- 39.Gul HC, Kilic A, Guclu AU, Bedir O, Orhon M, Basustaoglu AC. Macrolide-lincosamide-streptogramin B resistant phenotypes and genotypes for methicillin-resistant Staphylococcus aureus in Turkey, from 2003 to 2006. Pol J Microbiol 2008; 57:307–312. [PubMed] [Google Scholar]
- 40.Martineau F, Picard FJ, Lansac N, Ménard C, Roy PH, Ouellette M, et al. Correlation between the resistance genotype determined by multiplex PCR assays and the antibiotic susceptibility patterns of Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob Agents Chemother 2000; 44:231–238. [DOI] [PMC free article] [PubMed] [Google Scholar]


