Abstract
Background and Objectives:
Citrobacter freundii is an opportunistic pathogen causing nosocomial infections and resistant to various antibiotics. We aimed to determine the clonal relationship of C. freundii isolates using Pulsed-Field Gel Electrophoresis (PFGE).
Materials and Methods:
Fifty clinical isolates of C. freundii were collected from the main hospital in Kermanshah. After antibiotic susceptibility testing and screening for extended-spectrum beta lactamase (ESBL), all isolates were genotyped by PFGE. The DNA fragment patterns were analysed using Gelcompar II version 6.6 software. The Dice coefficient was used to calculate similarities for cluster analysis.
Results:
The PFGE results of 12 (24%) and 38 (76%) ESBL positive and negative isolates, respectively, produced 39 clusters (X1–39) with different genotype patterns. The X1 and X2 clusters were the major clusters, each contained 3 isolates from different hospital wards. However, the majority of isolates showed a high genotypic diversity.
Conclusion:
Results revealed the genotypic diversity of C. freundii isolates indicating the various sources for the bacterial isolates. However, the presence of isolates with similar genotypes indicates the common origin for these strains and may reflect the strain dissemination within the hospital wards, in particular in infectious ward and intensive care unit.
Keywords: Clonal dissemination, Citrobacter freundii, PFGE
INTRODUCTION
The genus Citrobacter is a member of the Enterobacteriaceae family and ubiquitous in soil, sewage and water. This group of bacteria can also colonize human and animal gastrointestinal tract (1). Several species of Citrobacter have been identified and a few of them, including Citrobacter freundii, have been recognized as opportunistic pathogens (2). Although C. freundii is often considered as a commensal bacterium of the human intestinal microbiota, it can cause opportunistic infections in hospitalized patients (3). It has been associated with nosocomial infections of urinary, respiratory, biliary, gastritis, meningitis, brain abscesses and neonatal sepsis (2–4). Citrobacter spp. are bacteria with low virulence and can persist as a population for a long time and thus accumulate resistance determinants, which may make the treatment of their infections more difficult (4). Citrobacter infections can be fatal, with 33–48 % overall death rates, and 30% for neonates (4). Resistance to antibiotics may develop by several mechanisms in Citrobacter species, but the most important one is the production of β-lactamases that hydrolyze the β-lactam ring of antibiotics (5). Plasmid-mediated extended-spectrum β-lactamases (ESBLs) were first reported in the mid 1980s (6). There are now over 200 distinct ESBLs that have been characterized and classified on the basis of amino acid gene sequences (7).
The dissemination of nosocomial infection caused by C. freundii has been attributed to the various ways of bacterial spreading including medical staff hands and other objects shared in hospitals (8). Molecular typing of microorganisms is a tool to identify the sources of contamination and to determine the routes of transmission for pathogenic bacteria that could enable us to more accurately detect and limit the spread of hospital-acquired infections. The pulsed-field gel electrophoresis (PFGE) has been used with a good discriminatory power for investigation of outbreaks and sporadic cases of Citrobacter spp. (9, 10). Given the increased rate of nosocomial infection, we aimed to determine the clonal relationship of C. freundii isolates from the main hospital in Kermanshah city.
MATERIALS AND METHODS
Bacterial isolates.
Fifty isolates of C. freundii were collected during 2013 from the main hospital (Imam Reza) in Kermanshah (west of Iran). This is a major local hospital in the area and patients are referred to this center from cities and neighboring provinces. All isolates were from hospitalized patients. The clinical samples were included urine (n=24, 48%), blood (n=11, 22%), stool (n=8, 16%), and others (respiratory tract and wound secretions, ascitic fluid and sputum) (n=7, 14%). The patients were 23 females (46%) and 27 males (54%) with the average age of 36.8 year old. All bacterial isolates were identified by bacteriological and biochemical tests followed by confirming using API20E Kit (bio-Merieux, France).
Antibiotic susceptibility testing.
Antimicrobial susceptibility testing for six β-lactam antibiotics was carried out using disk diffusion method as recommended by Clinical and Laboratory Standard Institute (CLSI) (11). The antibiotics were penicillins (ampicillin, pipracilin-tazobactam), cephalosporins (cefazolin, cefotaxim), monobactam (azetronam) and carbapenem (meropenem) (Mast, England).
ESBL phenotypic confirmatory test.
Phenotypic confirmatory test (PCT) of ESBL was performed using the combined disk method on Mueller Hinton Agar (Merck, Germany) according to the CLSI recommendations. In this method, cefotaxime and ceftazidime alone and in combination with clavulanic acid were used. If the inhibited zone diameter increased ≥5 mm for either antimicrobial agents in combination with clavulanic acid it was considered phenotypic positive for ESBL (11).
Molecular typing by PFGE.
All fifty isolates were used for genotyping by PFGE according to a previously described protocol (12) with some modifications. C. freundii isolates and Salmonella enterica serovar Braenderup H9812 (As DNA marker) genome were digested with 20U XbaI (Fermentas, Lithuania). Following XbaI digestion, the genomes were loaded into a 1% Low electroendoosmosis agarose (Merck, Germany). Electrophoresis was performed using a CHEF MAPPER apparatus (Bio-Rad, USA) at 14 °C for 22 h under the following conditions: initial switch time, 6.8 s; final switch time, 34.8 s; included angle, 120°; voltage gradient, 6 V/cm; ramping factor, linear. Gels were stained with ethidium bromide and visualized under UV light using Gel Doc apparatus (Bio-Rad, USA).
Software analysis.
The DNA fragment patterns were analysed using Gelcompar II version 6.6 software (Applied maths, Belgium). The Dice coefficient was used to calculate similarities for cluster analysis. A cluster of isolates was defined to include all isolates with 80% cutoff for similarity of their DNA patterns according to the Tenover’s criteria (13).
Statistical Analysis.
Data were recorded and entered into an Excel file. Statistical analyses were performed using SPSS software (version 16). Variables were analysed by chi-square test. A p-value of < 0.05 was set as the statistical significance of all analyses. Simpson's Index of Diversity (D value) was calculated using equation.
RESULTS
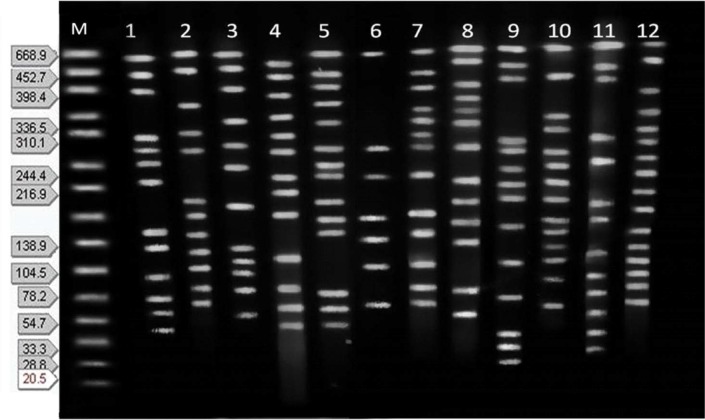
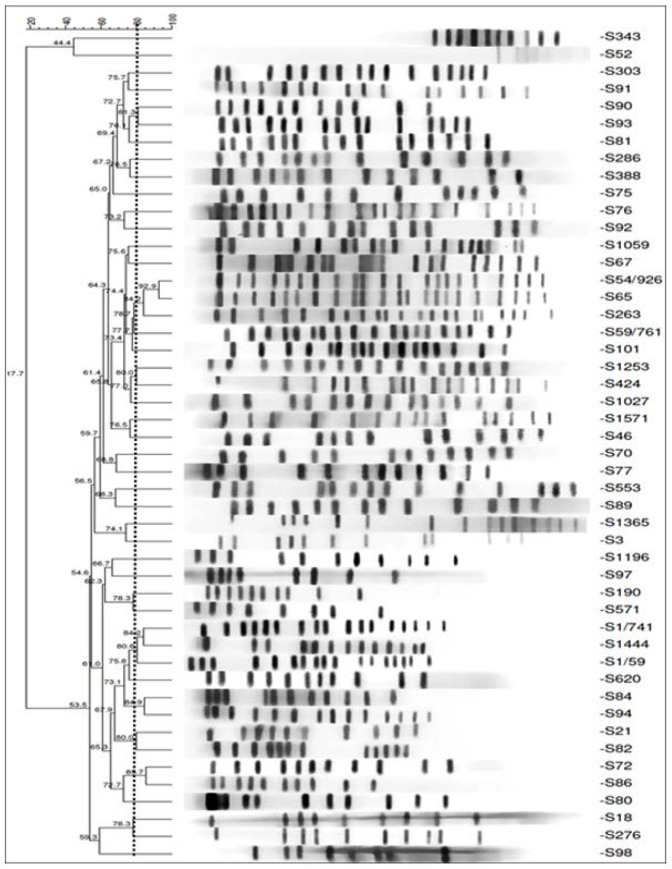
The antibiotic susceptibility of isolates presented in Table 1. Phenotypic ESBL production was detected in 12 (24%) isolates. PFGE showed the majority of isolates had 11 to 13 DNA bands (Figs 1, 2). According to the DNA fingerprints, 39 clusters (X1–39) were differentiated among isolates. The X1 and X2 clusters were the major clones, each with 3 isolates that were mostly urinary isolates. The next cluster (X3) with 2 isolates included 1 ESBL positive isolates resistant to all antibiotics tested, except carbapenems. The cluster X4 to X7 each with 2 isolates were from ICU and infectious wards (Table 2). The X8 to X39 clusters, each had a unique genotype and they were mostly from urine and blood samples with 8 (16%) ESBL positive isolates. They showed highly variable levels of resistance with diversely genotypic patterns. Finally, two strains of bacteria were nontypeble. The D value was 0.9902.
Table 1.
The antibiotic susceptibility of C.freundii isolates to β-lactam antibiotics (%)
| Antibiotics | Resistance | Intermedite | Sensetive |
|---|---|---|---|
| Ampicillin | 38(76) | 2(4) | 10(20) |
| Pipracilin-tazobactam | 3(6) | 8(16) | 39(78) |
| Cefazolin | 44(88) | 6(12) | 0 |
| Cefotaxim | 13(26) | 6(12) | 31(62) |
| Meropenem | 2(4) | 2(4) | 46(92) |
| Azetronam | 9(18) | 2(4) | 39(78) |
Fig 1.
CHEF profiles of 12 C. freundii isolates. M: Salmonella serotype Braenderup strain H9812 as a molecular size marker. Number 1 to 12 the profiles of isolates.
Fig 2.
Dendrogram for phylogenetic relationship among C.freundii isolates using PFGE data. The dotted line shows 80% cut off.
Table 2.
The distribution of clusters isolated among hospital wards
| Cluster number | Number of isolates | The number of isolates from hospital wards | ||
|---|---|---|---|---|
| ICU | Infectious | Surgery | ||
| X1 | 3 | 1 | 1 | 1 |
| X2 | 3 | 1 | 1 | 1 |
| X3 | 2 | - | 1 | 1 |
| X4 | 2 | 1 | 1 | - |
| X5 | 2 | 1 | 1 | - |
| X6 | 2 | 1 | 1 | |
| X7 | 2 | 1 | 1 | - |
DISCUSSION
Over recent years, the rate of nosocomial infections caused by C. freundii has been increased worldwide (4, 14). Research found the most frequent isolates of nosocomial infections among hospitalized neonates are Citrobacter spp. indicated the emergence of this bacteria as important nosocomial pathogens (4). In our study, resistance to cephalosporins, aztreonam and piperacillin was lower than some other studies. For example, a study in India has reported the resistance for cefotaxime 89%, ceftazidime 81.5% and aztreonam 74.7% (15). A research in Japan also showed 17.9% resistant to piperacillin(16). The results of previous studies, as well as our study, are compatible for the lowest resistance of C. freundii to carbapenems (16, 17).
The rate of ESBL production among our isolates was consistent with the findings of some previous studies. For instance, the prevalence of ESBLs in C. freundii isolates has been reported from 4.9 to 20.6 percent in South Korea (18, 19), 10.5 percent in Pakistan, 12.5 percent in Jordan and 29.5 percent in Poland (20–22). However, some other studies have reported lower or higher rates for ESBL in C. freundii. For example, the rates of 1.6 percent in France and 0.2 to 4.6% in Japan have reported which are lower than our results (15, 23). On the other hand, studies have reported higher rates including a study in India with 41.1% of ESBL (24). These variations may reflect the diversity of regional resistant strains and suggest that continuous surveillance is essential to detect the emergence and spread of ESBL-producing strains for this bacterium. Excessive antibiotic exposure (especially the extended spectrum cepholosporins) and extended hospital stay has been identified as the risk factors for the selection of the ESBL producing strains (25).
The molecular typing of C. freundii isolates revealed two features among genotypic patterns; the first was the isolates with genotypic diversity and the second, the isolates with more than 80% similar genotypes. The genotypic polymorphism can reflect the various origins of isolates. Given the spread of C. freundii in hospital environment, it is not surprising that the isolates may be acquired by patients from different sources. Similarly, the results of several studies in various countries on Citrobacter spp. indicated the genotype diversity among isolates. For example, a study on 57 strains of Citrobacter spp. found 36 different molecular types among the isolates (4). Research on 38, 26 and 12 of C. freundii isolates using PFGE, have reported 23, 20 and 11 genotypic patterns, respectively (26–28). A study in China on 51 Citrobacter strains (including 43 C. freundii, 4 C. youngae and 4 C. braakii) from different sources, revealed 48 different clusters by PFGE with similarity between 43.8% and 100% (29). Another study in Japan on 57 ESBL-producing isolates found 13 different clusters and18 unique PFGE types (23). The results of all above studies are compatiblie with our study for genenic diverisity of isolates.
On the other hand, a number of isolates with similar genotype in our study may indicate the dissemination of strains inside hospital environment, in particular in infectious ward and intensive care units. The spread of bacteria can happen in different ways, including by hospital staff or contaminated devices and equipments (8). Resistant isolates, in particular ESBL positive strains that cause nosocomial infections can be dominated population over time and this may limit the choice of effective antibiotics for their treatment (4).
CONCLUSION
Based on the results, the diversity patterns of C. freundii genotypes indicate the various origins of strains. However, the presence of strains with similar geno-types in clusters indicating a common origin for these strains. This fact may reflect the spread through hospital environments, especially in intensive care unit of the hospital. Given the clinical significance of ESBLs producing isolates, identification of these organisms is critical in order to get a better therapeutic response and reduce the risk of nosocomial outbreaks in the future.
REFERENCES
- 1.Smith A, Saiman L, Zhou J, Della-Latta P, Jia H, Graham PL., 3rd Concordance of gastrointestinal tract colonization and subsequent bloodstream infections with gram-negative bacilli in very low birth weight infants in the neonatal intensive care unit. Pediatr Infect Dis J 2010; 29:831–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whalen JG, Mully TW, English JC., 3rd Spontaneous Citrobacter freundii infection in an immunocompetent patient. Arch Dermatol 2007; 143:124–125. [DOI] [PubMed] [Google Scholar]
- 3.Gupta R, Rauf SJ, Singh S, Smith J, Agraharkar ML. Sepsis in a renal transplant recipient due to Citrobacter braakii. South Med J 2003; 96:796–798. [DOI] [PubMed] [Google Scholar]
- 4.Pepperell C, Kus JV, Gardam MA, Humar A, Burrows LL. Low-virulence Citrobacter species encode resistance to multiple antimicrobials. Antimicrob Agents Chemother 2002; 46:3555–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iredell J, Brown J, Tagg K. Antibiotic resistance in Enterobacteriaceae: mechanisms and clinical implications. BMJ 2016; 352:h6420. [DOI] [PubMed] [Google Scholar]
- 6.Cantón R, Oliver A, Coque TM, Varela Mdel C, Pérez-Díaz JC, Baquero F. Epidemiology of extended-spectrum beta-lactamase-producing Enterobacter isolates in a Spanish hospital during a 12-year period. J Clin Microbiol 2002; 40:1237–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacoby GA, Munoz-Price LS. The new beta-lactamases. N Engl J Med 2005; 352:380–391. [DOI] [PubMed] [Google Scholar]
- 8.Garcia-Cruz CP, Arguilar MJN, Arroyo-Helguera OE. Fungal and bacterial contamination on indoor surfaces of a hospital in Mexico. Jundishapur J Microbiol 2012; 5:460–464. [Google Scholar]
- 9.Hyytiä-Trees EK, Cooper K, Ribot EM, Gerner-Smidt P. Recent developments and future prospects in subtyping of foodborne bacterial pathogens. Future microbiol 2007; 2:175–185. [DOI] [PubMed] [Google Scholar]
- 10.Foley SL, Lynne AM, Nayak R. Molecular typing methodologies for microbial source tracking and epidemiological investigations of Gram-negative bacterial foodborne pathogens. Infect Genet Evol 2009; 9:430–440 [DOI] [PubMed] [Google Scholar]
- 11.CLSI. Clinical and Laboratory Standards Institute Performance standards for antimicrobial susceptibility testing; twenty-first informational supplement. CLSI document M100-S21. Wayne, PA: Clinical and Laboratory Standards Institute; 2011. 31:M100-S21. [Google Scholar]
- 12.Durmaz R, Otlu B, Koksal F, Hosoglu S, Ozturk R, Ersoy Y, et al. The optimization of a rapid pulsed-field gel electrophoresis protocol for the typing of Acinetobacter baumannii, Escherichia coli and Klebsiella spp. Jpn J Infect Dis 2009; 62:372–377. [PubMed] [Google Scholar]
- 13.Tenover FC, Gay EA, Frye S, Eells SJ, Healy M, McGowan JE., Jr. Comparison of typing results obtained for methicillin-resistant Staphylococcus aureus isolates with the DiversiLab system and pulsed-field gel electrophoresis. J Clin Microbiol 2009; 47:2452–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim PW, Harris AD, Roghmann MC, Morris JG, Jr, Strinivasan A, Perencevich EN. Epidemiological risk factors for isolation of ceftriaxone-resistant versus -susceptible Citrobacter freundii in hospitalized patients. Antimicrob Agents Chemother 2003; 47:2882–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohan S, Agarwal J, Srivastava R, Singh M. Observations on Citrobacter species from a tertiary care health center with special reference to multi-drug resistance and presence of CTX-M gene. Indian J Pathol Microbiol 2014; 57:439–441. [DOI] [PubMed] [Google Scholar]
- 16.Ishii Y, Tateda K, Yamaguchi K, Japan Antimicrobial Resistance Surveillance Participants Group (JARS) Evaluation of antimicrobial susceptibility for beta-lactams using the E-test method against clinical isolates from 100 medical centers in Japan (2006). Diagn Microbiol Infect Dis 2008; 60:177–183. [DOI] [PubMed] [Google Scholar]
- 17.Ishii Y, Ueda C, Kouyama Y, Tateda K, Yamaguchi K. Evaluation of antimicrobial susceptibility for β-lactams against clinical isolates from 51 medical centers in Japan (2008). Diagn Microbiol Infect Dis 2011; 69:443–448. [DOI] [PubMed] [Google Scholar]
- 18.Choi SH, Lee JE, Park SJ, Kim MN, Choo EJ, Kwak YG, et al. Prevalence, microbiology, and clinical characteristics of extended-spectrum beta-lactamase-producing Enterobacter spp., Serratia marcescens, Citrobacter freundii, and Morganella morganii in Korea. Eur J Clin Microbiol Infect Dis 2007; 26:557–561. [DOI] [PubMed] [Google Scholar]
- 19.Park YJ, Park SY, Oh EJ, Park JJ, Lee KY, Woo GJ, et al. Occurrence of extended-spectrum beta-lactamases among chromosomal AmpC-producing Enterobacter cloacae, Citrobacter freundii, and Serratia marcescens in Korea and investigation of screening criteria. Diagn Microbiol Infect Dis 2005; 51:265–269. [DOI] [PubMed] [Google Scholar]
- 20.Lartigue MF, Fortineau N, Nordmann P. Spread of novel expanded-spectrum beta-lactamases in Enterobacteriaceae in a university hospital in the Paris area, France. Clin Microbiol Infect 2005; 11:588–591. [DOI] [PubMed] [Google Scholar]
- 21.Hafeez R, Aslam M, Mir F, Tahir M, Javaid I, Ajmal AN. Frequency of extended spectrum beta lactamase producing gram negative bacilli among clinical isolates. Biomedica 2009; 25:112–115. [Google Scholar]
- 22.Empel J, Baraniak A, Literacka E, Mrówka A, Fiett J, Sadowy E, et al. Molecular survey of beta-lactamases conferring resistance to newer beta-lactams in Enterobacteriaceae isolates from Polish hospitals. Antimicrob Agents Chemother 2008; 52:2449–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanamori H, Yano H, Hirakata Y, Endo S, Arai K, Ogawa M, et al. High prevalence of extended-spectrum β-lactamases and qnr determinants in Citrobacter species from Japan: dissemination of CTX-M-2. J Antimicrob Chemother 2011; 66:2255–2262. [DOI] [PubMed] [Google Scholar]
- 24.Batchoun RG, Swedan SF, Shurman AM. Extended spectrum beta-lactamases among gram-negative bacterial isolates from clinical specimens in three major hospitals in northern Jordan. Int J Microbiol 2009; 2009:513874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shaikh S, Fatima J, Shakil S, Rizvi SM, Kamal MA. Risk factors for acquisition of extended spectrum beta lactamase producing Escherichia coli and Klebsiella pneumoniae in North-Indian hospitals. Saudi J Biol Sci 2015; 22:37–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nawaz M, Khan AA, Khan S, Sung K, Steele R. Isolation and characterization of tetracycline-resistant Citrobacter spp. from catfish. Food Microbiol 2008; 25:85–91. [DOI] [PubMed] [Google Scholar]
- 27.Bai L, Xia S, Lan R, Liu L, Ye C, Wang Y, et al. Isolation and characterization of cytotoxic, aggregative Citrobacter freundii. PLoS One 2012; 7:e33054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu CP, Weng LC, Tseng HK, Wang NY, Lee CM. Cefotaxime-resistant Citrobacter freundii in isolates from blood in a tertiary teaching hospital in Northern Taiwan. J Infect 2007; 55:363–368. [DOI] [PubMed] [Google Scholar]
- 29.Zhang XA, Bai XM, Ye CY, Ren ZH, Xu JG. Establishment and comparison of pulsed-field gel electrophoresis, multiple-locus variable number tandem repeat analysis and automated ribotyping methods for subtyping of Citrobacter strains. Biomed Environ Sci 2012; 25: 653–662. [DOI] [PubMed] [Google Scholar]