Dear Editor-in-Chief
Recently different methods are introduced for DNA extraction from bacteria. Bacterial DNA can be extracted using a commercial kit and conventional (e.g., boiling, phenol-chloroform and detergent methods). In the manual methods, chemically solution such as EDTA, SDS, NaCl, Tris hydrochloride, phenol-chloroform, proteinase K and lysozyme can be proposed (1).
The phenol-chloroform DNA extraction method is time-consuming and manipulation of toxic solvents may be hazardous to the environment and the technician. Furthermore, several washing and centrifugation steps increases the risk of sample contamination (2). Therefore, several methods have been introduced as alternative techniques to the phenol-chloroform method including boiling and detergent methods. These approaches are convenient and low-cost, but due to remaining of some residual protein with the DNA, the purity of the extracted DNA is poor. Commercial DNA extraction kits offer a low risk of contamination and they are faster than conventional protocols, but the amount of DNA recovered is highly variable (3). We examined TENT (Tris-EDTANaCl-TritonX100) buffer for DNA extraction from Staphylococcus aureus and DNA purity compared to detergent and kit methods.
In this study, S. aureus ATCC 29247 was cultured on tryptic soy agar (TSA) at 37 °C for 24 h and used in different DNA extraction methods. Pure colonies were suspended in 300 μL of TENT buffer (10 mM Tris-HCl, 0.1 M NaCl, 1 mM EDTA, 5% [v/v] Triton X100, pH 8.0). The cell suspension was boiled at 100 °C and then centrifuged. Supernatant fluid was transferred into a new sterile tube. Subsequently, cold 95% ethanol was added to the supernatant and kept at −20 °C for 20 min. After this stage, the solution was centrifuged. DNA template was dissolved in 50 μl sterile distilled water and stored at −20 °C until PCR amplification. This step was repeated three times and the result were compared.
Manual genomic DNA extraction of S. aureus was performed using detergent method (4). Genomic DNA extract by a kit, the product of the Viogene Company (UK) according to the manufacturer’s instructions. Like before, this step was repeated three times. Purity of extracted DNA was determined based on measurement of OD using Nano Drop instrument (Thermo Scientific). Additionally, the integrity of extracted DNA was evaluated by electrophoresis in 0.8% agarose gel. To determine any inhibitors remained from the extraction procedure, PCR amplification of mecA was conducted. The amplified PCR products were electrophoresed in 1% agarose gel at 120 V for 1 h and lastly stained with KBC (0.5 μg/ml) (Kowsar, Iran) and photographed under UV light.
Concentration and purity of the extracted DNAs was evaluated using Nanodrop (NanoDrop Technologies and the results are shown in Table 1. Electrophoresis of extracted DNAs on 1% agarose gel (Fig. 1) showed that all of the extracted DNAs by three methods have high integrity and perfection. The results of PCR products amplified from extracted DNA using three methods showed that the amplification of mecA gene was successful (Fig. 2).
Table 1:
Performance results of different methods tested based on purity factor and concentration of extracted DNA
| Method | Purity factor1 (A260/280) | Concentration (mg/μL) |
|---|---|---|
| TENT | 18.35 ± 8.55 | 919.0 |
| Detergent | 5.78 ± 2.89 | 286.3 |
| Kit | 1.40 ± 0.68 | 70.1 |
Fig. 1:
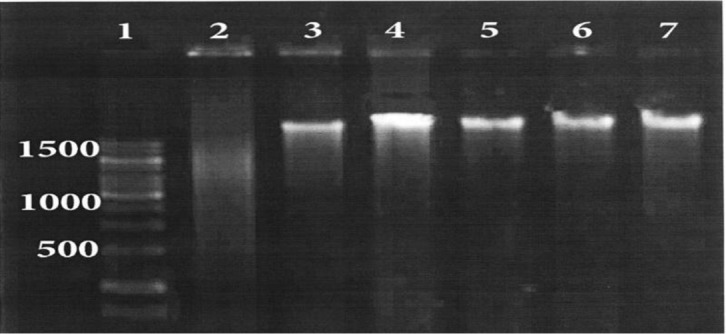
Analysis of extracted genomic DNA from S.aureus on 0.1% agarose gel. Lane 1 is DNA size marker; lane 2, control negative; lane 3, extracted DNA using kit; lane 4 and 5, extracted DNA using TENT; lane 6, 7 extracted DNA using detergent
Fig. 2:
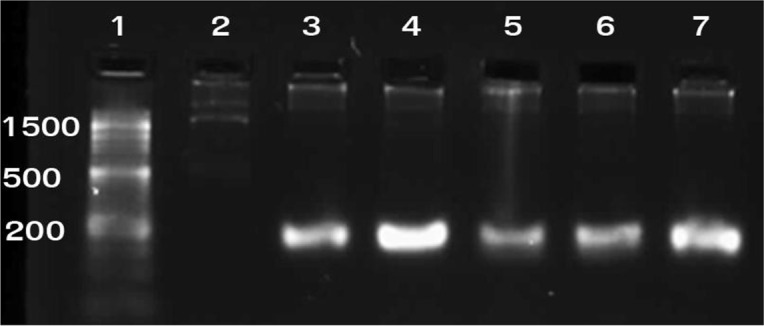
Agarose gel electrophoresis of the PCR-amplified mecA gene. Lanes: 1: 50-bp ladder; 2: Negative control (S. aureus ATCC 8325-4); 3: Positive control (S. aureus strain COL); 4–7: S. aureus isolates showing 162 bp mecA amplicon.
In many studies, various methods including phenol-chloroform, detergent and commercial kits have been conducted for extraction DNA. Each of these methods has advantages and disadvantages. The advantages of phenol-chloroform approach can be pointed to the high purity extracted DNA but it is time-consuming and unsafe to user. Detergent and kit methods are more convenient and faster, but in these methods the purity of the extracted DNA may be low (5).
In current study, the highest concentration of the extracted DNA was associated to TENT method (919μg/μl). This difference in concentration of DNA may be attributed using Triton ×100 along boiling that cause complete lysis of cell wall. Recently, similar buffer solution has been applied for other bacteria, including Clostridium perfringens, Listeria monocytogenes and Nocardia species (6–7).
In our study, TENT buffer showed acceptable and satisfactory results for molecular assay such as PCR. The described method is simple, fast, cost-effective, sensitive, and highly reproducible for DNA extraction from S. aureus, and there is no need for a skillful specialist to perform this method as well as DNA concentration is higher than commercial kits or detergent methods.
Acknowledgements
The authors wish to express their gratitude to research council of Tehran University of Medical Sciences (TUMS), Iran, for financial support [grant number 20105]. The authors declare that there is no conflict of interests.
References
- 1. Hebron HR, Yang Y, Hang J. (2009). Purification of genomic DNA with minimal contamination of proteins. J Biomol Tech, 20 ( 5): 278–281. [PMC free article] [PubMed] [Google Scholar]
- 2. Santos EM1, Paula JF, Motta PM, Heinemann MB, Leite RC, et al. (2010). Comparison of three methods of DNA extraction from peripheral blood mononuclear cells and lung fragments of equines. Genet Mol Res, 9 ( 3): 1591–1598. [DOI] [PubMed] [Google Scholar]
- 3. Hogg GM1, McKenna JP, Ong G. (2008). Rapid detection of methicillin-susceptible and methicillin-resistant Staphylococcus aureus directly from positive BacT/Alert blood culture bottles using real-time polymerase chain reaction: evaluation and comparison of four DNA extraction methods. Diagn Microbiol Infect Dis, 61 ( 4): 446–452. [DOI] [PubMed] [Google Scholar]
- 4. Mousazade Moghadam M, Babavalian H, Mirnejad R, Shakeri F. (2012). Rapid DNA extraction of bacterial genome of Staphylococcus aureus using laundry detergents and assessment of the efficiency of DNA in downstream process using PCR. Golestan Univ Med Lab J, 6 ( 1): 35–42. [Google Scholar]
- 5. Mirnejad R, Babavalian H, Moghaddam MM, Khodi S, Shakeri F. (2012). Rapid DNA extraction of bacterial genome using laundry detergents and assessment of the efficiency of DNA in downstream process using polymerase chain reaction. Afr J Biotechnol, 11 ( 1): 173–178. [Google Scholar]
- 6. Ahsani MR, Mohammadabadi MR, Shamsaddini MB. (2010). Clostridium perfringens isolate typing by multiplex PCR. J Venom Anim Toxins, 16 ( 4): 573–578. [Google Scholar]
- 7. Bafghi MF, Eshraghi SS, Heidarieh P, Habibnia S, Nasab MR. (2014). DNA extraction from nocardia species for special genes analysis using PCR. N Am J Med Sci, 6 ( 5): 231–233. [DOI] [PMC free article] [PubMed] [Google Scholar]


