Abstract
Imaging a material with electrons at near-atomic resolution requires a thin specimen that is stable in the vacuum of the transmission electron microscope. For biological samples, this comprises a thin layer of frozen aqueous solution containing the biomolecular complex of interest. The process of preparing a high-quality specimen is often the limiting step in the determination of structures by single-particle electron cryomicroscopy (cryo-EM). Here we describe a systematic approach for going from a purified biomolecular complex in aqueous solution to high-resolution electron micrographs that are suitable for 3D structure determination. This includes a series of protocols for the preparation of vitrified specimens on various specimen supports, including all-gold and graphene. We also describe techniques for troubleshooting when a preparation fails to yield suitable specimens, and common mistakes to avoid during each part of the process. Finally, we include recommendations for obtaining the highest quality micrographs from prepared specimens with current microscope, detector and support technology.
Keywords: electron cryo-microscopy, single-particle reconstruction, substrates, cryo-EM, electron tomography, protein structure
1. Introduction
In the past three years, spectacular progress has been made in the ability to determine structures by cryo-EM. But the method as a whole is still in its adolescence when compared with established methods like X-ray crystallography. Recent advances, including direct electron detectors with increased quantum efficiency [ref RH ch], easier-to-use microscopes with automated alignment and data collection [ref BC ch], improved software for the classification and reconstruction of density maps [ref SS,NG,SL chs] and more stable and reproducible specimen support technology (Russo and Passmore, 2016a), have coalesced to bring the method out of its infancy. Still, for many projects, the major limiting factor for structure determination is specimen preparation.
The origin of this limitation is two-fold in nature:
-
1)
During the creation of a thin layer of water for vitrification and imaging, specimens are exposed to surfaces and conditions which are very different from the inside of a test tube or cell. The effects of these on the molecules and complexes are not known a-priori, and can be difficult to remedy if destructive to the specimen.
-
2)
Specimen preparation for cryo-EM is a delicate process that still requires skilled handling and careful technique through a number of detailed preparation steps. This often confounds novice and experienced microscopists alike by making it difficult to distinguish problems with the specimen from problems in technique and methods.
Together, these obstacles can slow the progress of a cryo-EM project and prevent structure determination, irrespective of other advances in cryo-EM technology. While an improved understanding of protein-surface interactions as well as new approaches and technology will be necessary to fully remove these obstacles to structure determination, there are a number of techniques currently available to improve specimen preparation in practice. Our intent in this work is to provide a rational and systematic approach to the problem of specimen preparation and help novice microscopists to avoid many of the common pitfalls. We also provide protocols for various parts of the process and make recommendations for microscope and data collection settings to make this as efficient as possible. We do not provide the theoretical and historical background to cryo-EM, as there are several other recent reviews that address this (Cheng, 2015; Henderson, 2015; Nogales and Scheres, 2015; Vinothkumar and Henderson, 2016). The field of cryo-EM is rapidly evolving so methods will continue to advance as the technology improves. Still, the procedures provided here represent a snapshot of the current state-of-the-art, and the systematic approach to specimen preparation delineated below is likely to remain important even in the context of future disruptive advances in the field.
2. A systematic approach to specimen preparation
Cryo-EM specimen preparation, as we consider it here, can be defined in one sentence: It is the process of taking an aqueous sample of a biological material (usually a purified protein complex), applying it to a support structure (grid), reducing its dimension to a layer that is as thin as possible (∼100–800 Å depending on the size of the biological molecule), and then freezing this layer fast enough to prevent the water from crystallising. This process is essentially the same now as when the method was first developed by Dubochet and colleagues in the 1980s (Adrian et al., 1984). Still, as Dubochet recognised even then (Dubochet et al., 1985, 1988), many aspects of this process are problematic because we have a poor understanding, and poor control of the microscopic surfaces, materials and dynamic changes that the purified complexes encounter during their journey from the test tube to the thin layer of vitreous ice. In practice this means that what we see on the grid in the cryomicroscope often bears little resemblance to what we know was in the test tube.
To address this problem in the most efficient way possible, with respect to both the time of the scientist and the use of expensive resources like electron microscopes (see Table 1), each step of the sample preparation process should be assessed. This affords one a much better chance of determining where problems occur and hopefully, of systematically reaching the goal of an excellent specimen that yields a high-resolution structure with a minimum of data processing and effort. The process is outlined in Figure 1, and described in detail below followed by procedures to perform most of the steps listed.
Table 1.
General classes of transmission electron cryomicroscopes
| Microscope class | Typical examples | ∼ marginal cost/day (in 2016 £, including detectors) |
|---|---|---|
| Entry level | FEI T12, JEOL 1400 | 250 |
| Mid-range | FEI F20/Talos, JEOL 2100F | 600 |
| Upper-mid-range | FEI F30/Polara, JEOL 3200FS | 1000 |
| High-end | FEI Titan Krios | 3000 |
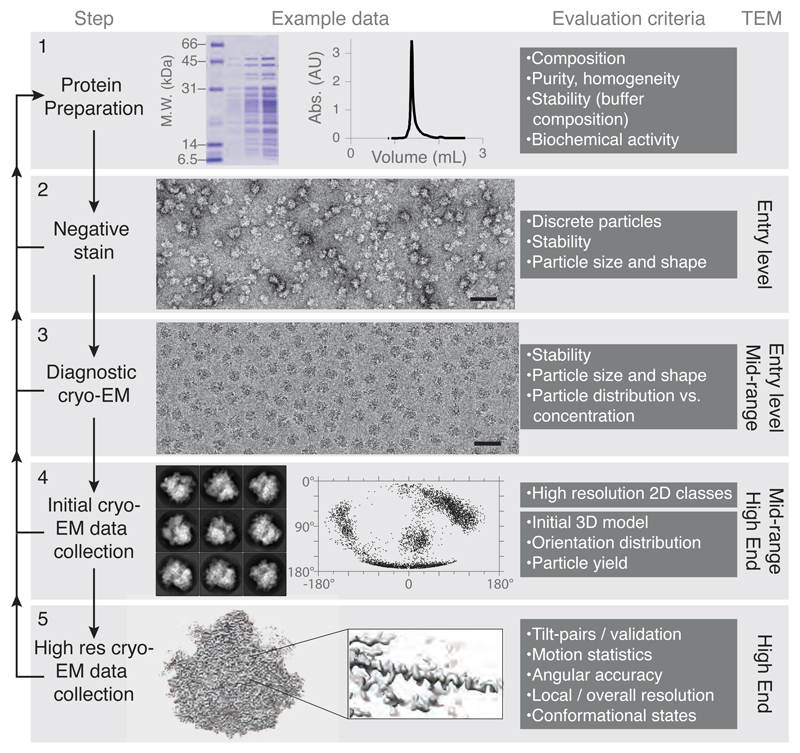
Figure 1. Structure determination by cryo-EM.
A systematic approach to 3D structure determination is shown. In the left column, the major steps are listed. Each step should be performed successively and only after one has been completed successfully should the scientist move onto the next step. In the second column, example data are shown for ribosomes (details in text). Scale bars on the micrographs are 500 Å. Each step should be evaluated with the criteria listed in the third column, returning to earlier steps for troubleshooting. The final column lists the class of electron microscope to be used, as defined in Table 1.
2.1. Protein preparation
The first step in a systematic approach is to evaluate protein composition and homogeneity thoroughly using biochemical methods (Figure 1). Contaminating proteins or degradation products may interfere with complex stability and subsequent computational analysis of the particle images, wasting resources on the more time-consuming and expensive cryo-EM data collection and image processing steps. Specimen homogeneity can be evaluated using SDS-polyacrylamide gel electrophoresis (PAGE) and proteins can be identified using mass spectrometry. A Coomassie blue stained gel of purified ribosomes is shown in Figure 1. It is important to ensure that only a single species is present and this can be judged using various techniques including native-PAGE, size exclusion chromatography (example chromatogram shown in Figure 1), dynamic light scattering (DLS) and size exclusion chromatography coupled to multi-angle light scattering (SEC-MALS). Using these methods to monitor subunit association, sample stability can then be optimised by changing buffer conditions (e.g. salt, pH, detergent). In addition, functional or biochemical assays should be used to test the activity of the protein in a given buffer condition. All of these steps should be done prior to making the first EM specimen, since no EM specimen will be better than the preparation from which it was made.
EM specimens are typically prepared using 3 μl protein solution at a concentration of 0.05 – 5 μM. Thus, it is essential for the protein complex to remain intact at these concentrations. If the dissociation constant (Kd) for the subunits is known, one can calculate whether it is expected to remain intact. Experimentally, one can run the protein complex on a size exclusion column repeatedly, at decreasing concentrations, to ensure it will not dissociate at the concentration required for cryo-EM. If the complex does dissociate, a few options are available. One can work at concentrations above the Kd and adjust the subsequent plasma and blotting conditions to achieve a very thin layer that only allows a monolayer of complex. Alternatively, a chemical crosslinking agent can be used to covalently link the subunits together, thus preventing their dissociation after dilution (Plaschka et al., 2015; Weis et al., 2015). Crosslinking can be performed in solution or within a glycerol or sucrose gradient (GraFix) (Kastner et al., 2008; Stark and Grant, 2010). It is important to minimise (e.g. using optimal protein and crosslinker concentrations) or remove (e.g. using gradients or size exclusion chromatography) aggregates which can occur due to crosslinking of two neighbouring complexes.
2.2. Negative stain
After suitable protein is purified, the next step is to evaluate it using negative stain electron microscopy (Figure 1). In this method, the protein is embedded in a layer of heavy metal salts (e.g. uranium, molybdenum, tungsten) which surrounds the protein like a shell (Brenner and Horne, 1959; Unwin, 1974). Since the density of the stain is about three times higher than the protein, there is excellent contrast even with modest dose on an inexpensive microscope. A negative stain micrograph is shown in Figure 1.
Negative staining is fast - making and imaging a specimen takes around the same time as running a gel - and is best used to rapidly assess various parameters in a preparation like pH, salt and buffer conditions, or the different fractions from a chromatography run. It reveals the solvent-excluded surface and shape of molecules and can therefore be used to evaluate homogeneity and size of proteins as well as the presence of binding partners. A homogenous sample will have discrete particles of uniform size. Averaging of a translationally (but not rotationally) aligned set of negatively-stained particles will provide an estimate of the diameter of the protein. This should be consistent with the size determined by solution methods like DLS or SEC-MALS. If the size and shape of the particles are not consistent, this is a clear sign that something is wrong and further troubleshooting should be undertaken before starting cryo-EM preparations. 3D structures can also be determined from negative stain and these can be used as a starting point for structure determination by cryo-EM, although in practice, this is often not required with newer algorithms for generating initial maps for 3D reconstruction (Elmlund et al., 2013; Tang et al., 2007),[chSS,chSL,chNG]. Methods for preparation of negatively stained specimens have been described previously and we refer the reader to these for the details (Ohi et al., 2004). Even in the era of high-resolution cryo-EM, negative staining remains a valuable method for evaluating protein preparations quickly and cheaply.
2.3. Diagnostic cryo-EM
After one is confident that a given protein preparation is monodisperse, stable in well defined solution conditions, and yields a uniform distribution of discrete particles in negative stain, the specimen can be evaluated by cryo-EM (Figure 1). Initially, cryo-EM should be performed at a diagnostic level. Three properties of the specimen should be evaluated at this point: 1) protein concentration and stability, 2) ice thickness and uniformity across the grid, 3) phase of the ice (should be uniformly amorphous, not crystalline). All three should be consistent enough across the grid to yield several squares appropriate for data collection. An example of a micrograph of a ribosome specimen is shown in Figure 1.
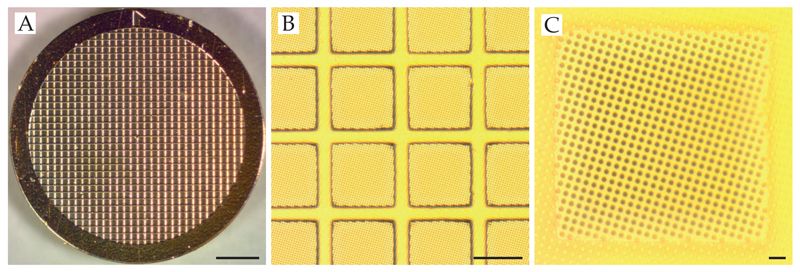
Standard substrates for cryo-EM consist of a 3 mm metal grid which supports a perforated foil (Figure 2) (Russo and Passmore, 2016a). Foils are patterned with a regular array of holes (Ermantraut et al., 1998; Quispe et al., 2007) and can be made from various materials, the most common of which is amorphous carbon (Table 2). We recently showed that making both the grid and the foil from the same material, gold, improves the stability of the support and reduces the movement of the specimen during imaging by more than an order of magnitude, thus improving image quality (Russo and Passmore, 2014c). During vitrification, proteins are preserved in a hydrated, native state in the holes of the foil. The contrast of protein in vitreous ice is low so it is important to ensure that the ice is only slightly thicker than the particle diameter to maximise contrast. A water layer that is too thin will exclude proteins and this is usually apparent from the presence of a circular region of ice near the center of a hole where no particles are present. Given an appropriate concentration of stable complex in the droplet, the particle distribution and ice thickness should be optimised by changing one of a few parameters: 1) the blotting and plasma conditions, 2) adding a small amount of detergents, 3) adding a thin film of amorphous carbon or graphene to the surface of the foil to induce adsorption of particles across the holes. Currently, this remains a trial and error process and so must be done in a way that minimises uncontrolled changes.
Figure 2. Supports for cryo-EM.
Here we show a support comprising a 3 mm metal mesh grid (A) with a perforated gold foil covering the surface of the mesh (B,C). Thin films (∼2–30 Å thick) can be added on top of the perforated foil. Scale bars are (A) 0.5 mm (B) 50 μm and (C) 5 μm.
Table 2.
Specimen support geometries for particular applications
| Mode | Grid | Foil | Film |
|---|---|---|---|
| Negative stain | 400 mesh | none | 50–100 Å am-C |
| Diagnostic cryo | 300 mesh | 1.2/1.3 μm | none / 20 Å am-C |
| Medium-resolution (≥3.5 Å) cryo | 300 mesh | 1.2/1.3 μm | none / 20 Å am-C |
| High-resolution cryo (<3.5 Å) >400 kDa | 300 mesh | 1.2/1.3 μm | none / 20 Å am-C |
| High-resolution cryo (<3.5 Å) <400 kDa | 300 mesh | 1.2/1.3 μm | none / graphene |
| Very high-resolution cryo (<2.8 Å) | 300 mesh | 0.6/1.0 μm | none / graphene |
| Cryo-tomography: cellular (>30 Å) | 200 mesh | 2.0/2.0 μm * | none |
| Cryo-tomography: high-resolution sub-tomogram avg. (<15 Å) | 300 mesh | 1.2/1.3 μm | none / 20 Å am-C |
Larger holes may be used for particularly large specimens.
Proteins often denature when they come into contact with surfaces (Seigel et al., 1997), and several are present during the vitrification process. These include the hydrophobic air-water interface, amorphous carbon support layers, copper, gold and other metals used to create grids, and even the cellulose paper used to blot away the excess liquid. Of these, the air-water interface is likely the most problematic as the requirement for a thin layer of water ice means the particles must be brought in close proximity to one just before freezing. Its hydrophobic nature means it is likely to induce the adsorption, and possible denaturation, of many proteins and complexes (Seigel et al., 1997; Vogler, 2012).
For a given concentration of protein, one can calculate the number of particles that should be found in the thin layer of vitreous ice. For example, for a 1 MDa protein in 800 Å thick vitreous ice, a solution of 2 mg/ml should give approximately 100 particles/μm2. A 250 kDa protein at the same concentration should give approximately 400 particles/μm2. Often, the protein concentration in solution does not match the protein concentration on the support: protein density can be higher than expected if the particles tend to adsorb to the surface, or lower if they tend to repel the surface (or are attracted to the support or other surfaces like the blotting paper instead). Changing the buffer conditions (e.g. changing pH or salt concentration, adding detergents, lipids or other chemical modifying agents like crosslinkers) may change the interaction of proteins with surfaces (Dubochet et al., 1985; Cheung et al., 2013; Glaeser et al., 2016). A support that is too hydrophobic, for example because of insufficient plasma treatment or contamination with hydrocarbons, can strongly adsorb proteins to itself, making the concentration of particles in the holes much less than was in the applied droplet. This is a clear indication that there is a problem with the specimen. The opposite situation can also occur. That is, the particles can adhere to the air-water interface, artificially concentrating them prior to the removal of most of the water and the creation of the thin layer. This leads to a particle concentration in the holes that is much higher than it was in the droplet; also a clear indication that there is a problem with the specimen. Often, the particles adhered to the interface will have low contrast and will be partially or fully denatured. In addition, the particles will appear larger in size than expected as they unfold to varying degrees on the surface (Seigel et al., 1997). This should be resolved before moving on to collect large datasets in cryo.
2.4. Initial cryo-EM data collection
Once protein density and ice thickness/quality have been optimized, an initial data collection should be performed on a mid-range electron microscope (Table 1). The goal is to obtain 2D reference-free class averages which have high-resolution features at the level of secondary structure (alpha-helices or better). Particles should be hand-picked, at least at first, to avoid the problems of template matching and “Einstein from noise” (Henderson, 2013). With modern field emission microscopes and direct detectors, only a few hundred particle images are needed to generate a 2D class of sufficient resolution to see alpha helices. Thus a dataset of a few thousand particles should be sufficient to yield a few high-resolution classes. High-resolution 2D classes are shown for ribosomes in Figure 1. If high-resolution 2D classes are not obtained, the specimen should be re-evaluated and optimized, including modification of protein preparations, buffers, detergents and surface modifications, and the addition of support films like amorphous carbon or graphene. Particles that don’t align well may be partially or fully denatured and could lead to incorrect structures if the subsequent computation and validation is not done with great care.
Many high-resolution structures to date have relied on discarding a large portion of the particles from the initial data collected. The proportion of discarded ‘junk’ particles can vary by orders of magnitude from one specimen to the next. We suggest that many, if not most of these particles are damaged during the sample preparation process. By evaluating specimen preparation using the above criteria, one can optimize the specimen and improve particle yield. This is generally preferable to collecting large datasets and discarding most particles to reach a desired resolution. Improving particle yield will make the entire process more efficient and more likely to succeed.
Once a preliminary dataset is collected that generates suitable 2D classes, a larger dataset with a few hundred micrographs and several tens of thousands of particles is collected on a mid-range or high-end microscope, from which an initial 3D map can be calculated. Several important factors can then be evaluated. First, do the 2D classes show several distinct views of the particle, with self-consistent dimensions, each with secondary structural features? Second, is the orientation distribution sufficient to allow calculation of a 3D structure with isotropic resolution? Figure 1 shows an equal area projection map of the orientation angles of ribosomes relative to an amorphous carbon substrate. Although these ribosomes exhibit preferred orientations, they cover Fourier space sufficiently for a high-resolution structure. If the orientation distribution is not suitable, one can alter buffers, add detergents, change plasma conditions or use an alternative surface, e.g. graphene or amorphous carbon to improve the distribution and promote additional views of the complex. Crosslinking can also alter particle orientations since modification of charged surface amino acids (often lysines) changes the surface properties of molecules and their interaction with the supporting surfaces and the air-water interface (Bernecky et al., 2016). Collecting small datasets will be sufficient to determine if any changes in preparation conditions were effective at improving the orientation distribution.
2.5. High-resolution data collection
If the resolution of the initial 3D reconstruction reaches a reasonable resolution for the number of particles and image acquisition settings, a larger data collection on a high-end electron microscope should be performed with the aim of obtaining a high-resolution structure (Figure 1). What constitutes a “reasonable resolution” will be arbitrary and should ultimately depend on the resolution of the biological question and the state of the technology, but as a guideline, we suggest that currently, sub-nanometer resolution should be routinely possible from 10–50 thousand asymmetric particle images. With this in mind, a large dataset (500–1000 micrographs, ∼24 hours of microscope time) should be collected with the best available microscope: currently this is a 300 keV instrument with one of several commercially available direct electron detectors (Tables 1 and 3). The data collection strategy will depend on the size of the particle, the resolution desired and the particular microscopes available to the microscopist. Our current recommendations for data collection on various specimen types are discussed in §8.
Table 3.
Current electron detectors for cryomicroscopy
| Detector type | Current examples | Pixel pitch (μm) | Num. pixels | Frame rate (Hz) | Recommended flux (optimum [range] e−/px/s) |
|---|---|---|---|---|---|
| Phosphor - CCD | Gatan Orius 830 | 7.4 | 2048×2048 | 1 | - |
| Gatan US1000 | 14 | 2048×2048 | 1.5/15 | - | |
| Phosphor - CMOS | Tietz TemCam | 15.6 | 4096×4096 | 1 | - |
| Gatan OneView | 15 | 4096×4096 | 25 | - | |
| FEI Ceta | 14 | 4096×4096 | 32 | - | |
| Direct integrating | Direct El. DE16* | 6.5 | 4096×4096 | 60 | ∼ 240 |
| Direct El. DE20* | 6.4 | 5120×3840 | 32 | ∼ 100 | |
| FEI Falcon 2* | 14 | 4096×4096 | 18 | 50 [10 – 60] (300 keV) | |
| 40 [8 – 48] (200 keV)† | |||||
| Direct counting | FEI Falcon 3* | 14 | 4096×4096 | 32 | 100 [20 – 120] (300 keV) |
| Gatan K2 | 5 | 3838 x 3710 | 400 | 5 [2 – 8] (300 keV) | |
These detectors can also be operated in electron counting mode but current frame rates make this impractical for normal data collection.
The flux, f0 at one energy can be scaled with reasonable accuracy to other energies relevant to transmission electron microscopy using the equation where β is the ratio of the electron velocity to the speed of light.
All biomolecular complexes exist in multiple conformational states, and for each to be resolved, even more high quality data are required. Many computational algorithms are available to help extract this information from a dataset and we refer the reader to other chapters in this volume for more details [ch SS NG SL]. In some cases, ligands (e.g. small molecules, binding partners, nucleotides) can be added to stabilize particular conformations, or to interrogate a particular biological question related to how a ligand affects the conformational state of the complex. These usually require the collection of additional large datasets to achieve high-resolution after small datasets have been collected to confirm the change in conformation or presence of the ligand of interest.
Finally, but perhaps most importantly, all high-resolution datasets should be collected with validation in mind [ch PR]. In particular, we recommend collecting a small set of tilt-pairs (∼1–2% of the number of micrographs) with every dataset intended for determination of a previously unknown structure. They can be subsequently used to validate the reconstructed density map (Rosenthal and Henderson, 2003; Henderson et al., 2011; Baker et al., 2012; Wasilewski and Rosenthal, 2014), measure the angular accuracy of the projection assignments (Russo and Passmore, 2014b), and determine the absolute hand of the structure (Rosenthal and Henderson, 2003).
By using the systematic approach described above, the microscopist will have the best chance of efficiently going from a protein in solution to a high-resolution structure. In the following section we describe many of the practical methods and details of the processes that are important to successful specimen preparation. This includes a series of protocols that are intended to help guide the reader in the current state-of-the-art, and hone the techniques of new microscopists.
3. Support choice, handling and storage
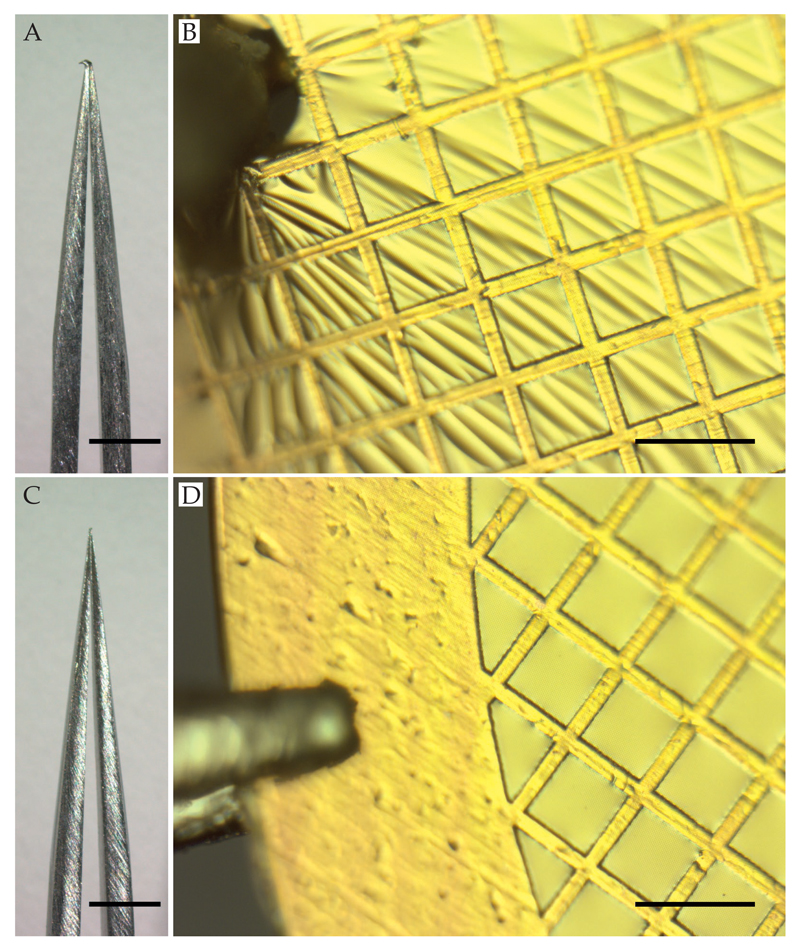
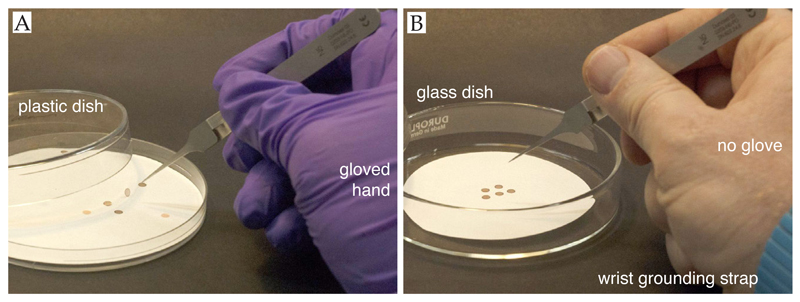
A fundamental but often overlooked factor in preparing high quality specimens for cryo-EM is correct handling and storage of the supports. They are delicate and can easily be damaged or contaminated, compromising later steps in the process and reducing reproducibility. Before starting, it is useful to check supports using an optical microscope and discard any with defects, large areas of broken squares or contamination. The time spent checking supports in a light microscope is trivial compared to subsequent time spent preparing and imaging the specimen. The supports should start, and remain as flat as possible to prevent damage to the foil. Importantly, supports should always be handled with sharp tweezers and picked up by the rim only (Figure 3). We like to use tweezers with a normally closed configuration (e.g. Dumont type N5) as this applies a well-defined and reproducible force to the grid during handling. Static discharge can also damage supports but this can be eliminated by wearing a wrist grounding strap whenever handling supports (Figure 4). We do not recommend wearing a glove on the hand that holds the tweezers as this induces static buildup on the tweezer/support and reduces dexterity. After cleaning or modification, supports should be stored in glass, not plastic, petri dishes and in clean, oil-free dry boxes to keep them clean and dust-free.
Figure 3. Tweezer damage to specimen supports.
Bent tweezers (A) or improper use (B) results in damage to the specimen support. For best results, sharp, straight tweezers (C) should be used and supports should be picked up by the rim only (D). For Panels A and C, the scale bars are 1 mm. For Panels B and D, the scale bars are 100 μm.
Figure 4. Storing grids to avoid contamination and static charge.
Panel A shows bad practice in grid handling: The use of gloves and plastic storage dishes results in the accumulation of static charge. In the image, a charged grid is standing on end. Panel B shows recommended handling procedures including glass containers, no glove on the hand holding the tweezers and a wrist grounding strap to prevent accumulation of charge.
To prevent carry-over of specimens and to remove contamination, grid handling implements, glass slides and glass dishes should be thoroughly cleaned on a regular basis. Glass can be cleaned by sonicating in a high-purity detergent mixture (e.g. 2% Micro-90) and rinsing with ultra-pure, 18 MΩ deionized water. Tweezers can be cleaned after routine use by sonication in alcohol (ethanol or isopropanol) or with more aggressive solvents like chloroform and acetone if they have been contaminated with plastic or oil residues. Note that tweezers used for grid plunging should be decontaminated with ethanol sonication after each distinct specimen, as cross-contamination from one grid to the next via the tweezers is common.
Supports with irregular arrangements (lacey or holey carbon) or regular arrays of holes are available but the latter, e.g. Quantifoil, UltrAuFoil, C-flat, are more reproducible and simplify data collection methods. A detailed discussion of support types can be found in (Russo and Passmore, 2016a). Here we provide recommendations for support geometries for various applications (Table 2). As to the choice of material, we recommend amorphous carbon (am-C) on copper for negative stain, as they are commonly available and inexpensive, and all-gold supports for cryo-EM as they reduce the movement of specimens and improve image quality (Russo and Passmore, 2014c). All-gold supports can be made using the protocols described in (Russo and Passmore, 2016b) or are commercially available from Quantifoil (UltrAuFoil®).
4. Contamination and cleaning
Even under optimal storage conditions, supports may need to be cleaned prior to use. Contamination can arise from particulate matter including flakes of evaporated carbon, residual particles from manufacturing processes or accumulated dust from storage. If these are not removed, they will be resuspended upon application of the aqueous protein solution and may be visible in the vitreous ice, which can interfere with subsequent data processing and analysis. In addition, contamination can arise from organic residue deposited on the surface during handling or storage, and from residual photoresist or other plastics and solvents used during the manufacturing processes. Such surface residues can affect wetting and other material properties of the support, and reduce reproducibility.
Here, we provide a method to clean supports by water and solvent washes prior to use (Protocol 4.1). Ultra-high purity (CMOS grade) solvents are required for cleaning without contamination. We do not recommend cleaning supports by heating as this can compromise the structural integrity of the support.
Warning: The protocols described below involve using high voltage, high temperatures, liquid nitrogen, hazardous and flammable chemicals/gases and equipment under vacuum. They are for use by experienced scientists who know how to handle such equipment and have done all appropriate local safety training and risk assessments, etc. Wear safety glasses, appropriate protective equipment and use at your own risk.
Protocol 4.1. Support cleaning
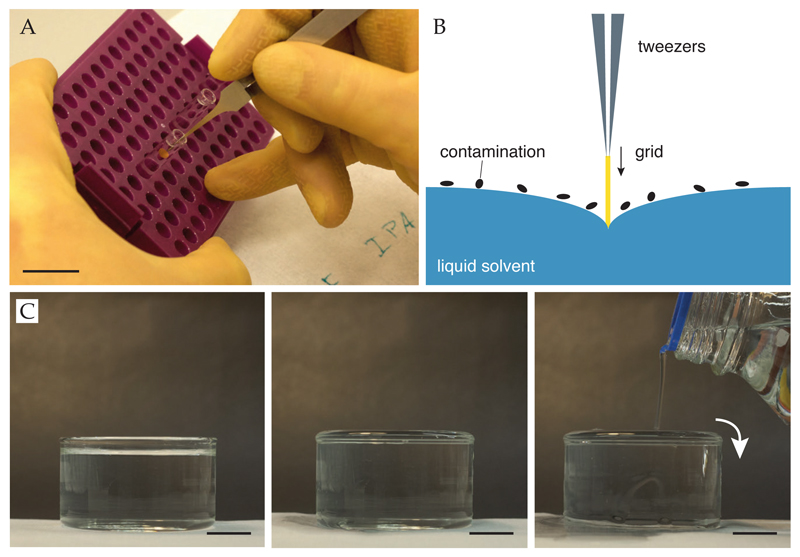
Fill clean, glass crystallization dish (Pyrex, diameter 90 mm, height 50 mm) with deionized water (18 MΩ, filtered, UV treated). Overfill it to break the meniscus at the surface and remove any contaminating surface layers on the water (Figure 5C).
Using small, clean glass test tubes, prepare three solvent washes (Figure 5A): one with 1 ml chloroform (Sigma 650498), one with 1 ml acetone (Sigma 40289) and one with 1 ml isopropanol (Sigma 40301). Use glass pasteur pipettes as the solvents can dissolve and redeposit residue from most plastics.
Pick up a single support using clean tweezers (Dumont N5 or 5) and dunk it into the water at one side of the dish. As the grid enters the water, loose contamination can float off onto the surface of the water. Pulling the grid out in the same place will result in the contamination being redeposited on the grid surface. Move the grid through the water to remove it from the opposite side of the dish. (Ensure it is moved through the water with the thin edge leading to prevent damage to the perforated foil.) Touch edge of grid briefly to filter paper (Whatman No. 1) to remove excess water, but take care not to bend the grid. If not using anti-capillary tweezers, blot between the tines.
Dunk grid sequentially in chloroform, acetone, and isopropanol (prepared above) for 10–20 sec each. It is particularly important that the final rinse step is in the cleanest possible solvent as any residue from it will be deposited on the grid surface. If not using anti-capillary tweezers, blot between the tines after each rinse to remove excess.
Touch edge of grid briefly to filter paper to remove excess isopropanol.
Place grid on filter paper (Whatman No. 1, 70 mm rounds) in clean glass petri dish (Schott, 70 mm), foil side up, to dry. Cover with glass lid to minimize dust accumulation.
Store in the covered glass petri dish after drying but use soon after cleaning.
Figure 5. Removing surface contamination from specimen supports.
Supports can be washed sequentially (A) in chloroform, acetone and isopropanol. Care must be taken to avoid deposition of contamination from the surface of water (or solvents) onto the support. A schematic is shown in panel B. Overfilling of containers can reduce surface contamination, shown in panel C. Scale bars are 20 mm.
5. Continuous films of amorphous carbon and graphene
A continuous film of amorphous carbon or graphene provides an alternative surface for proteins to interact with. In some cases, this improves protein distribution or orientation within the vitreous ice. In addition, lower protein concentrations can sometimes be used because the particles adsorb to the surface before blotting away the excess liquid. Here we provide a protocol for producing thin (20–50 Å) films of amorphous carbon for use as a surface for particle adsorption. The same protocol can be used for thicker films (50–100 Å) of carbon appropriate for transfer to bare grids (no perforated foil) for negative stain.
Protocol 5.1. Amorphous carbon deposition
This protocol is for depositing carbon onto a sheet of mica using an Edwards 306 Turbo coating system equipped with an Inficon crystal thickness monitor with water cooling, but can be adapted to other deposition systems.
Vent the chamber and remove the implosion guard and bell jar.
Put on clean gloves and handle everything inside the chamber with gloves.
Remove the carbon source apparatus, remove the shield and mount a new sharpened carbon rod (high purity graphite <5 ppm impurity), under tension, in the chuck.
Use compressed nitrogen to blow out any bits of carbon or flakes as these can cause a short between the electrodes.
Replace the source in the chamber, with the rod positioned 125 mm from the stage. Finger tighten the nuts (no wrench).
Mica is a multi-layered mineral crystal which is extremely flat (less than 1 nm RMS per mm2) and easy to obtain in large sheets, and so is used as a template surface for carbon deposition. Cleave a piece of mica (e.g. Agar G250-1) in half with a razor blade and position the two sheets, cleaved surface up, on the specimen stage directly beneath the source on top of a fresh piece of filter paper (Whatman no. 1). Mica should be cleaved immediately before coating as the freshly cleaved surface is clean and hydrophilic, but it becomes contaminated (and thus hydrophobic) with time spent in air.
A clean penny or other small piece of metal can be used to hold the mica sheets and filter paper in place during evacuation.
Check the position of the shutter in the open and closed state: closed it should cover the solid angle from the source to the mica and the crystal thickness monitor. In the open state the paths to both the mica and the crystal should be unobstructed and equal in length. Leave in the closed position.
Check the bell jar gasket and base plate for any dust, dirt or flakes of carbon or metal. Clean with lint free paper and methanol if necessary.
Replace the bell jar and implosion guard.
Fill the liquid nitrogen cold trap.
Evacuate the chamber by pressing cycle on the panel.
When the gate valve opens, the pressure should drop rapidly into the 10−5 Torr range. Begin heating the source apparatus by turning on the low tension power supply (LT) and slowly increasing the current to about 1.0 Amp.
Wait 10 min and increase the current to 1.2 Amps.
Wait 5 min and increase to 1.4 Amps.
Leave at 1.4 amps for 20 min. Then shut off. Top up the liquid nitrogen.
Turn on the cooling water to the crystal thickness monitor and check the correct program for carbon is selected. Zero the thickness.
Vacuum should now drop to the 10−7 Torr range in about an hour.
Once vacuum is < 5 × 10−7 Torr, slowly ramp up the current again to 1.6–1.7 Amps over about 5 min, being careful not to let it spark. The pressure should not rise above < 5 × 10−6 Torr as the source is heated.
Once the carbon begins to deposit the current will start to fluctuate. Open the shutter and deposit for 10–20 sec, then close the shutter and turn the current back down slightly.
The crystal will initially go negative as it heats up but after closing the shutter, it will return to a positive value which is the accumulated thickness. This can be repeated until the desired thickness is reached.
When finished evaporating, close the shutter and turn off the power to the source.
Let the chamber cool for at least 30 min before venting.
Vent with dry nitrogen, remove your coated mica and store in a glass Petri dish.
Replace the bell jar and implosion guard, press cycle, wait for the vacuum to reach 200 mTorr and then press seal.
Notes: The crystal thickness monitor should be calibrated according to the manufacturer using an independent thickness measurement method. We have used atomic force microscopy in the past for this purpose (Russo and Passmore, 2014a). Carbon density can vary depending on the quality of your source material but should be approximately 2.2 g/cm3. From start to finish, the process should take less than 4 hours. Unless using a fully dry (no oil) pumping system, overnight pumps are not recommended as oil will back-stream into the chamber when the liquid nitrogen trap is not cold and this will contaminate the chamber and mica with oil.
We previously provided a detailed protocol for transferring amorphous carbon onto supports (Russo and Passmore, 2016b) and we have had good results using this with Quantifoil and all-gold supports. This is reproduced here in Protocol 5.2.
Protocol 5.2. Amorphous carbon transfer onto supports
Wear wrist grounding strap to prevent static damage to supports during handling. Don’t wear a glove in the hand that holds the tweezers, or ground the tweezers directly.
Inspect supports in dissecting microscope and discard any with defects. All grids should be flat, continuous and without dust or lint.
If there is evidence of residual plastic from lithographic processing, they can be cleaned using Protocol 4.1.
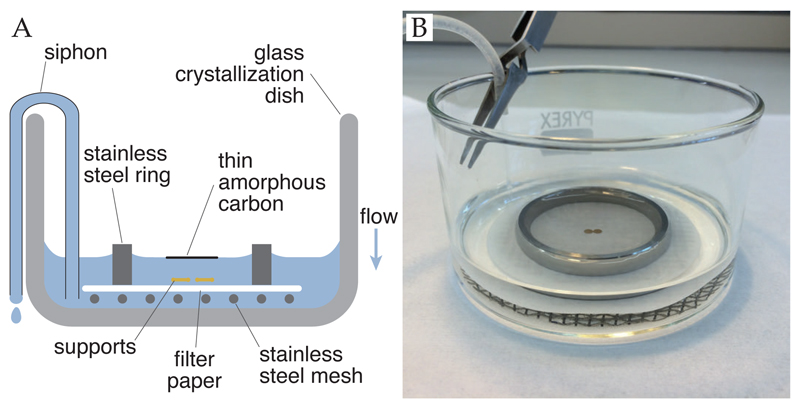
Using gloves to prevent fingerprints and contamination, place filter paper (Whatman No. 1, diameter 70 mm) on a stainless steel mesh circle (diameter 65 mm, made with 0.7 mm wire with 3 mm mesh) inside a stainless steel ring (polished and bevelled edges, 2 mm thick, height 10 mm, diameter 50 mm) in a glass crystallization dish (Pyrex, diameter 90 mm, height 50 mm) (Figure 6) (Russo and Passmore, 2016b).
Fill dish with deionized water (18 MΩ, filtered, UV treated). Overfill it to break the meniscus at the surface and remove any contaminating surface layers on the water (Figure 5C). Pour off excess water so the level is just below the rim of the glass dish.
Using tweezers (Dumont N5 or 5, cleaned by solvent rinse), carefully place grids, foil side up, onto the center of the filter paper. The grids should enter the water slowly and perpendicular to the surface.
Set up a siphon: attach tubing (length 0.6 m, outer diameter 3.2 mm, inner diameter 1.6 mm) to dish with normally closed forceps or a spring clip. Start flow of syphon with syringe and clamp off flow while floating the carbon (next step).
Float carbon off mica by slowly lowering a ≃ 2.5 x 2.5 cm sheet of amorphous carbon coated mica (carbon-side up) into the water at a 20–30° angle. A light can be placed to shine off the surface of the water at a glancing angle, allowing you to see the carbon better.
Use siphon to slowly lower the water level.
Monitor the position of the carbon with respect to the grids and ensure it stays centered by gently nudging with clean tweezers.
Once the carbon has been deposited on the grids and the water level is below the filter paper, carefully lift off the stainless steel ring. Remove the filter paper and mesh together. Place on a dry filter paper and cover with a clean glass petri dish or beaker, tilted slightly to leave room for evaporation. Allow to dry.
When dry, store in a clean glass petri dish until ready to use.
Figure 6. Apparatus for depositing thin films of amorphous carbon on supports.
Panel A shows a cross-sectional diagram of the float chamber. As the water level is lowered, the thin film of amorphous carbon is deposited onto the supports. The stainless steel ring makes a positive meniscus to help lower the carbon film down in the center of the ring. Panel B shows a photo of the apparatus in use. This figure is reproduced from (Russo and Passmore, 2016b).
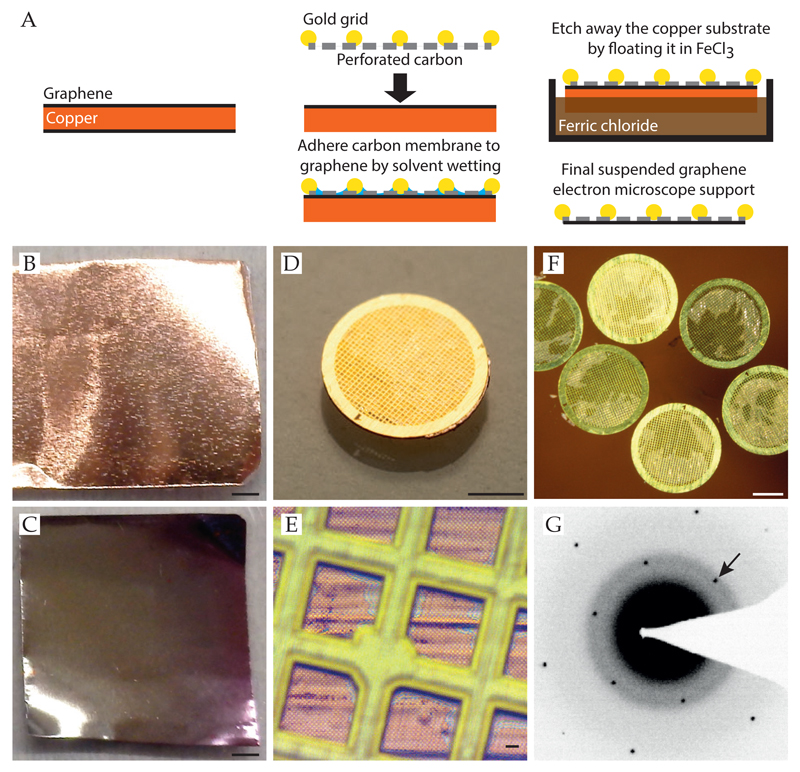
Graphene is superior to amorphous carbon due to its defined structure and conductive properties (Geim, 2009; Pantelic et al., 2010, 2011; Russo and Passmore, 2014a). In addition, it is effectively invisible at the resolutions of interest for cryo-EM, whereas amorphous carbon adds additional background noise that is especially detrimental for smaller proteins. Here, we provide procedures for transferring graphene onto Quantifoil supports (Figure 7, Protocol 5.3) (Regan et al., 2010; Russo and Golovchenko, 2012). Ultra-high purity (CMOS grade) solvents and acids are required for reliable transfers without contamination and a wrist grounding strap should be worn at all times to prevent static damage to supports during handling since graphene films are particularly sensitive to static discharge. All glassware should be cleaned prior to use. A new transfer protocol for all-gold supports is currently under development but we expect that graphene on all-gold supports will become commercially available.
Figure 7. Graphene transfer onto supports with carbon foils.
The process is diagrammed in Panel A. Panels B and C show copper heated to 150°C for 10 min in air, where B is fully covered in graphene so does not oxidise while C has no graphene and turns color due to oxidisation; scale bars are 3 mm. This simple test is used to map the location of the graphene on the foil. Panels D and E show the grid-graphene-copper sandwich, scale bars are 1 mm and 10 μm respectively. Panel F is the sandwich floating in the etchant, where the partially etched grains of copper are visible (scale 1 mm). Panel G is an electron diffraction pattern of suspended graphene with ice, where the arrow points to the 2.1 Å reflection from the graphene lattice.
Protocol 5.3. Graphene transfer onto supports
Inspect freshly cleaned supports (Quantifoil Au 300 1.2/1.3, see Procedure 4.1) in dissecting microscope and discard any with defects.
Graphene is grown on thin sheets of copper using chemical vapor deposition (CVD) and can be purchased from commercial vendors (Graphene Supermarket, Structure Probe Inc., etc.). Punch out 3.2 mm disks from copper/graphene source material where the copper has been checked for the presence of graphene (Figure 7B, C). We use a custom-made mechanical punch but these are also available from Structure Probe, Inc.
Place a support, foil side down, on each disk.
Press together between two clean glass slides and then remove the top glass slide.
Using a clean pipet, add 7 μL of isopropanol (Sigma 40301) to the top of the grid-graphenecopper sandwich and let dry.
Inspect in high-resolution optical microscope to check for adherence of the carbon foil to the copper. The foil changes color when well adhered to the copper surface, (Figure 7E) and only well adhered regions will transfer successfully.
Layout and label seven borosilicate glass crystallization dishes (70 mm diameter) in fume hood.
Fill first crystallization dish 2/3 full with copper etchant containing buffered FeCl3 (Sigma 667528).
Gently place the support disk sandwiches on the surface of FeCl3 by sliding off glass slide. Cover with borosilicate glass Petri dish covers (80 mm) to keep out dust during the etch.
Etch for 20 min in FeCl3 (for 25 μm thick copper).
Fill three dishes 2/3 with 20% HCl (Sigma 40233), 20% HCl and 2% HCl.
Transfer grids to each using a flamed platinum loop (homemade or EMS 70944). Leave floating for 10 min at each step.
Overfill three more dishes with cleanest possible water.
Transfer to each using platinum loop, and rinse for 3–5 min each.
Use loop to transfer and flip, graphene side up, onto filter paper (Whatman No. 1, 70 mm rounds). Let dry in clean borosilicate glass Petri dish (70 mm).
Dispose of acids in appropriate hazardous waste streams.
Store long term in glass in a dry environs free of any type of oils or hydrocarbons. Do not store in any type of plastic container.
6. Surface treatments
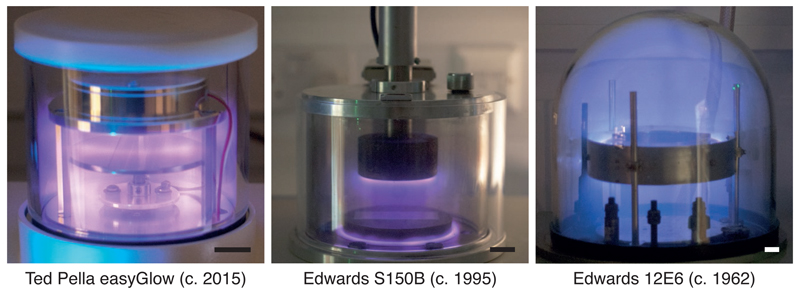
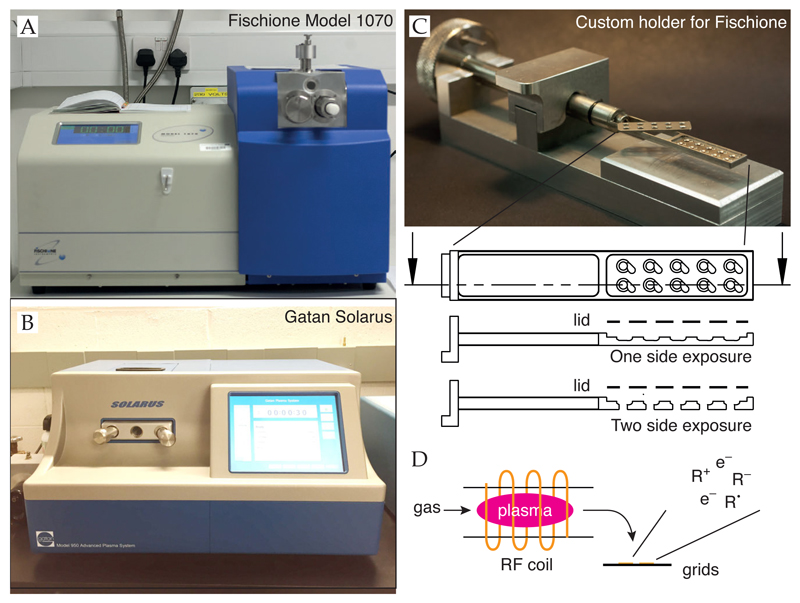
Support surfaces are often hydrophobic and this prevents efficient spreading of aqueous solutions. To control the hydrophilicity, treatment with low-energy plasmas is used (Figure 8). Ions and radicals generated from a low-pressure gas interact with surfaces to remove residual organic contamination and react chemically with the surface to reduce their hydrophobicity. Although plasmas generated from residual air (glow discharging) can be used (Protocol 6.1), we recommend the use of plasmas with controlled composition as this is more reproducible (Protocol 6.4). Plasma treatment is typically performed with argon:oxygen mixtures but hydrogen can also be used (Protocol 6.3). In addition, other molecules (e.g. amylamine) can be introduced to alter the surface and change the orientation distribution of particles adsorbed to the surface (Protocol 6.2) (Miyazawa et al., 1999; da Fonseca and Morris, 2015).
Figure 8. Plasmas generated by glow discharge.
Residual air plasma generation by three different instruments is shown. All are effective in increasing hydrophilicity of support surfaces but have varying degrees of reproducibility and can damage the supports. Note that the plasmas in glow discharge apparatuses are often non-uniform which can vary the exposure dose significantly, even in a single batch. Scale bars are 20 mm.
Protocol 6.1. Glow discharge treatment of supports
Inspect clean supports in dissecting microscope and discard any with defects. All supports should be flat, continuous and without dust or lint.
Place supports in the chamber of a glow discharging apparatus (e.g. Edwards S150B) on a clean glass slide, foil side up, in the center of the stage. Wear gloves when handling anything that will be placed in the chamber.
Pump the chamber to 200 mTorr.
Turn on the HT to 7 kV, current should be 28–30 mA.
Expose the supports for 30 sec.
Turn off the HT and slowly vent the chamber.
Store the treated supports in a clean glass petri dish and use as soon as possible (within 1 hour).
Protocol 6.2. Amylamine plasma treatment of supports
Inspect supports in dissecting microscope and discard any with defects. All supports should be flat, continuous and without dust or lint.
Place supports in the chamber on a clean glass slide, foil side up, in the center of the electrode ring (Figure 8).
A small glass vial containing 0.5 mL of amylamine is placed in the chamber at the edge of the stage.
Cover with the bell jar and pump the chamber to below 400 mTorr.
Turn on the HT and adjust the power to form a uniform plasma (∼ 90 V, 1.5 A for our system).
Expose the supports for 30–60 sec.
Turn off the HT and slowly vent the chamber. Discard the vial in an appropriate waste stream.
Store the treated supports in a clean glass petri dish and use within one hour.
Protocol 6.3. Argon oxygen plasma treatment
Inspect supports in dissecting microscope and discard any with defects. All supports should be flat, continuous and without dust or lint.
We use a Fischione 1070 plasma chamber with a custom suspension holder that holds up to ten supports (Figure 9). In this configuration, the grids are 15 ± 1 cm from the radio frequency coils. It is desirable to always position the grids in the same location relative to the plasma to improve the reproducibility of the exposures from batch to batch. Alternatively, the specimens can be placed directly in the chamber on a clean glass slide, foil side up, on the shelf in the chamber. Plasmas that are not well shielded or not sufficiently low in energy may damage gold supports by sputtering; this should be avoided. Load four supports into the plasma chamber. Everything that goes inside the vacuum chamber should be handled with gloves to prevent contamination and fingerprints.
Evacuate chamber to ≪ 10−4 Torr.
Admit high purity argon and oxygen (BOC 99.9999%) in a ratio of 9:1 to a pressure of 21 mTorr (31.0 SCCM gas flow).
Apply radio frequency plasma with 38 W of forward power and ≤ 2 W of reverse power (∼70% setting on a Fischione 1070) for 10–60 sec (20 sec works well for most supports). The time can be optimized for suitable hydrophilicity (spreading of aqueous solution and ice thickness). For amorphous carbon-containing supports, calibrate the carbon etch rate empirically by performing several plasma treatments of increasing dose and determining when a carbon layer of known thickness is fully etched.
For very thin films of continuous carbon, use a ratio of 19:1 argon:oxygen and 35 W forward power for 5–15 sec.
Vent plasma chamber, remove supports and use within one hour.
Figure 9. Generation of defined plasmas for surface modification.
(A,B) The Fischione Model 1070 and Gatan Solarus generate plasmas with defined compositions. (C) Photo (top) and diagram (bottom) of a custom specimen holder made at MRC LMB. Two different holder designs are shown - one is used for exposure of one side of a support, the other is used for exposure of both sides. The lid is used to prevent the supports from moving during the process. (D) Diagram of plasma generation.
Using a low-energy hydrogen plasma, one can remove contamination and control protein adsorption onto graphene (Russo and Passmore, 2014a). Here we provide the procedure for hydrogen plasma treatment using a Fischione 1070 with a Dominik Hunter model 20H-MD hydrogen generator, attached to one of the input ports of the plasma generator. Hydrogen plasma treatment is stable but should be performed immediately prior to use to minimize accumulation of contamination after modifying the surface.
Protocol 6.4. Hydrogen plasma treatment of graphene supports
Use only ultra-high purity hydrogen (> 99.999% pure), all stainless steel tubing and fittings (no plastic), and ultra-high purity grade regulators.
The distance of the coils to the sample is important since it affects the energy of the hydrogen species. A plasma where atoms and ions strike the surface and recoil, delivering an energy greater than ∼21 eV to the carbon atoms, might cause significant damage to the lattice rather than chemically modifying it. The energy of the plasma vs. distance can be measured with a Langmuir probe to ensure that the energy is below the ∼21 eV sputter threshold at the sample. Alternatively, one can test whether the energy is too high by imaging a graphene-covered support before and after increasing doses of hydrogen plasma to determine whether it remains intact.
Prior to hydrogen plasma treatment, the plasma chamber should be pre-treated. Insert the holder and evacuate the plasma chamber to ≪ 10−4 Torr.
Burn the empty holder for 10 min in 100 % pure hydrogen (70% power which is 35 watts forward, ≪ 2 watts reverse power, 20 SCCM gas flow, pure hydrogen). Vent only just prior to loading.
Mount graphene supports in holder, load, and evacuate the plasma chamber to ≪ 10−4 Torr.
Treat with 5–40 seconds (typically 20 seconds) hydrogen plasma using the same settings.
Carefully remove the supports and use immediately.
7. Vitrification
Vitrification was first developed by Jacques Dubochet and colleagues in the 1980s (Adrian et al., 1984; Dubochet et al., 1988). To successfully make vitreous ice, a thin layer of protein solution must be cooled quickly so that it remains in an amorphous, non-crystalline state. This is usually performed in an apparatus which plunges the specimen into a liquid cryogen (usually ethane or propane) such that it is cooled about 200 K in < 10−4 seconds. For water to vitrify, the temperature has to drop faster than 105 – 106 K/s (Dubochet et al., 1988). Water is a poor thermal conductor so the sample must be less than 3 μm thick. Several designs for manual and semi-automated plungers are available for vitrification of biological specimens, including commercial models from FEI, Leica, EMS and Gatan. These provide controlled temperature and humidity throughout the process, preventing evaporation and making specimen preparation more reproducible. We note that after blotting, even a small amount of evaporation, equivalent to a few hundred monolayers of water, can concentrate the salt and change the pH of a suspended thin layer by a factor of two or more. For example, at 4°C and 90% relative humidity, the evaporation velocity is of order 100 Å/s so in the 2 seconds between the blot and the freeze, a 400 Å film can be concentrated by a factor of two, causing significant osmotic and conformational changes in the specimen. So we also recommend that the relative humidity surrounding the specimen support always be kept at 100% to prevent deleterious changes in the concentration of solutes just prior to freezing. This is easiest to achieve at 4°C, because much less water vapor per unit volume is required to bring the dewpoint to the air temperature, thus preventing evaporation. It is also important to work quickly once sample is applied to the support to minimize the interaction time of proteins with the often destructive, air-water interface.
A detailed discussion of vitrification procedures can be found in (Dobro et al., 2010) and we recommend consulting this review for further information. We include here the standard settings we use as a starting point and other recent work can be consulted for further advice (Thompson et al., 2016), including more advanced techniques like time-resolved cryo-EM (Chen et al., 2015). The following procedure using a Vitrobot (FEI) works well for all-gold and standard Quantifoil supports:
Protocol 7.1. Standard vitrification procedure
Fill cryoplunger reservoir with fresh deionized water (18 MΩ) and equilibrate to 4 °C and 100 % relative humidity.
Ensure Vitrobot tweezers (FEI, Ted Pella 47000-500) are sharp by examining under an optical microscope. Check that they are not bent, and symmetrically aligned in the following way: place on a flat surface and measure the tip to surface distance. Do this for all four sides; it should be the same (within 0.5 mm) for opposing sides.
Clean tweezers and blotting pads with ethanol, and clean the ethane cup, grid box holder and foam dewar with a high purity detergent (2% Micro-90 in Miliq water); rinse with Miliq deionised water and dry completely.
Place new filter paper rounds (Whatman 595) on blotting pads.
After cryo-plunger is equilibrated (for at least 20 min to saturate the water in the filter paper) and just prior to specimen vitrification, cool down the plunging dewar with liquid nitrogen then fill the central cup with ethane (Dobro et al., 2010). Warning Liquid ethane can cause severe burns and blindness if splashed in the eye. Always wear safety glasses when handling.
Check that the temperature of the ethane is just above the melting point, 90 K.
Make supports hydrophilic by plasma treatment, as described above. We emphasize that only clean, flat, intact supports should be used (see above). Bent supports will be further damaged upon cryo-plunging (Figure 10).
Mount a support in clean cryo-plunger tweezers. Use the foot pedal trigger on the Vitrobot to reduce the delay time between steps in the process.
Apply 3 μl protein solution (usually 10–5000 nM) to the foil side. Make sure the pipet tip does not touch the support - only touch the liquid droplet to the surface. Close access port.
-
Blot using the following settings:
Description Manual plunger Vitrobot III Vitrobot IV Temperature 4 °C 4 °C 4 °C Relative humidity 100% 100% 100% Wait time 0 s 0 s 0 s Force setting n/a -2 -20 Blot time 2 + 1 s 4–6 s 2–4 s Drain time n/a 0 s 0 s Settings here are for FEI Vitrobots but are easily adapted to other manual or semi-automated plunge instruments. We do not use a ‘wait time’ or ‘drain time’, in order to speed up the process and minimize contact time with the air-water interface. Note, changing the blot force setting to a more negative value will increase the force.
Plunge into liquid ethane.
Wait a minute for the tweezer to cool after the plunge into the ethane.
Carefully transfer the frozen specimen to a storage box via the cold vapor above the liquid nitrogen.
When releasing the grip of the tweezer, gently let the solidified ethane crack away from the support if present.
Store the supports in covered, labeled grid boxes in a liquid nitrogen storage dewar.
Figure 10. Vitrification and mounting of grids.
(A) Supports that are bent will be damaged upon cryo plunging, resulting in broken foils. An example of a broken gold foil is shown in panel B (scale bar 2 μm). Supports also need to be mounted correctly in microscope cartridges. Panel C shows a support that is incorrectly mounted, and so damaged, in a Krios cartridge. The support in panel D is correctly mounted. Scale bars in panels C and D are 500 μm.
Take care not to bump the support into anything during transfers to storage boxes, mounting in stage cartridges, etc. Any support that is bumped or bent at any point should be discarded.
Generally, the only variables we change to optimize ice thickness are plasma exposure time and blot force. Blot time can also be varied but has a less reproducible effect on ice thickness, particularly for short blot times. If the support is not hydrophilic enough, one usually finds thicker ice in the center of the grid squares because the hydrophobic surfaces repel liquid, causing a drop of solution to accumulate in the center of the square.
8. Data collection
As with most technological devices, there is a tradeoff between ultimate performance of the current microscopes and detectors and the amount of money and effort required to achieve this performance. But since the goal is ultimately to determine structures at resolutions which are sufficient to unambiguously answer biological questions, here we try to make reasonable compromises between the performance of the instrument and the amount and quality of data required to achieve a particular resolution that is appropriate for the stage of structure determination (Figure 1). In general, we think it is better to spend more effort on preparing optimal specimens, as now, and even more so in the future, this is where the biggest differences in the resolution and interpretability of the resulting density maps are likely to come from.
Since biological imaging is ultimately limited by damage to the specimen [ch RG], currently the most important single factor governing the quality of the micrographs is the efficiency of the electron detector [ch RH]. Using the current understanding of detective quantum efficiency (DQE) and published measurements of DQE for the various commercially available detectors (McMullan et al., 2014; Chiu et al., 2015; Kuijper et al., 2015), we have tabulated the parameters and recommended flux of current detectors in Table 3. These, in conjunction with the illumination and specimen movement considerations discussed below, form the basis for our recommended imaging conditions in Table 4. These can be considered a starting point for users to make trade-offs in the amount of data collected vs. resolution or other factors like microscope time, and they will certainly change as the technology continues to develop in the coming years.
Table 4.
Currently recommended data collection settings at the MRC-LMB
| Mode | Source | Microscope type | Energy (keV) | Detector | Pixel size (Å/px) | Flux (e−/Å2/s) | Exp. time (s) | Fluence (e−/Å2) |
|---|---|---|---|---|---|---|---|---|
| Negative stain | W-thermal | Entry level | 80-120 † | CCD | 3.3 | 9 | 2 | 18 |
| Diagnostic cryo | W-thermal | Entry level | 80-120 † | CCD | 3.3 | 9 | 2 | 18 |
| Diagnostic cryo | FEG | Mid-range | 200 | Falcon 2 | 2.1 | 10 | 2 | 20 |
| Medium-resolution (≥3.5 Å) cryo | FEG | Mid-range/High-end | 300 | Falcon 2 | 1.7 | 17 | 3 | 51 |
| High-resolution (≤3.5 Å) ≥400 kDa | FEG | High-end | 300 | Falcon 2 | 1.3 | 28 | 2 | 56 |
| High-resolution (≤3.5 Å) <400 kDa | FEG | High-end | 300 (±5 eV) | K2 | 1.8 | 1.5 | 40 | 60 |
| Very high-resolution (<2.8 Å) | FEG | High-end | 300 (±5 eV) | K2 | 0.90 | 6.2 | 10 | 62 |
| Cryo-tomography cellular (>30 Å) | FEG | High-end | 300 (±5 eV) | K2 | 3.5 ‡ | 0.65 | 1.25 per tilt angle | 100 |
| Cryo-tomography high-resolution sub-tomogram avg. (<15 Å) | FEG | High-end | 300 (±5 eV) | K2 | 2.2 ‡ | 1.5 | 1 per tilt angle | 60 |
DQE is maximum for a phosphor coupled CCD at approximately 80 keV but the effects of specimen charging and mean free path are lower at 120 keV.
Pixel size is limited by flux instead of spatial resolution.
Here we provide a standard protocol for data collection at high-resolution on a Krios with a Falcon 2 detector. The protocol is easily adapted to other specimen and microscope configurations using the data collection settings in Tables 3 & 4.
Protocol 8.1. High-resolution data collection using a Krios / Falcon 2 and all-gold supports
Load specimens in cartridges and mount in autoloader, including a calibration grid (PtIr, graphitized carbon or similar). Discard any that are bent or broken during handling.
Load a specimen in the column and use low mag to check grid is intact and contains several squares with an appropriate thickness of ice. Ice thickness is best judged at low magnification by increasing the defocus to hundreds of micrometers to improve the contrast.
Once a grid is selected for imaging, load the calibration grid and perform standard microscope alignments in low dose exposure mode. On a well aligned and stable Krios these include gun alignment, condenser and objective aperture alignments, beam tilt (coma free alignment), camera gain correction, condenser and objective stigmation. Setup the exposure mode using the parameters in Table 4.
Reload the selected specimen and locate all the squares for data collection using low magnification and high defocus. Using automated software to create a montage of images at low magnification (“atlas”) can be useful for this [ch BC].
All squares for data collection should be free from cracks, crystalline ice and contamination.
Collect a couple of test micrographs to confirm ice phase, thickness, and particle distribution.
Set focus in exposure mode using beam tilt or by looking at the fringes / bright spots at the edge of the hole (Russo and Passmore, 2016b).
Check the illumination geometry of the beam in exposure mode: it should be round, centered on the imaging axis, and centered on the hole while encompassing an annulus of the support around the hole (Figure 11).
Begin collecting data, either manually or using automated data collection software [ch BC] †.
Check the focus and illumination every 20–30 holes. If mounted correctly, focus should vary by <1–2 μm across an 80 μm square.
Collect a few (1–2 % of the total micrographs) tilt-pair micrographs for validation. Tilt angles and exposures of [0,15°] and [1,3 s] respectively, are reasonable for validation.
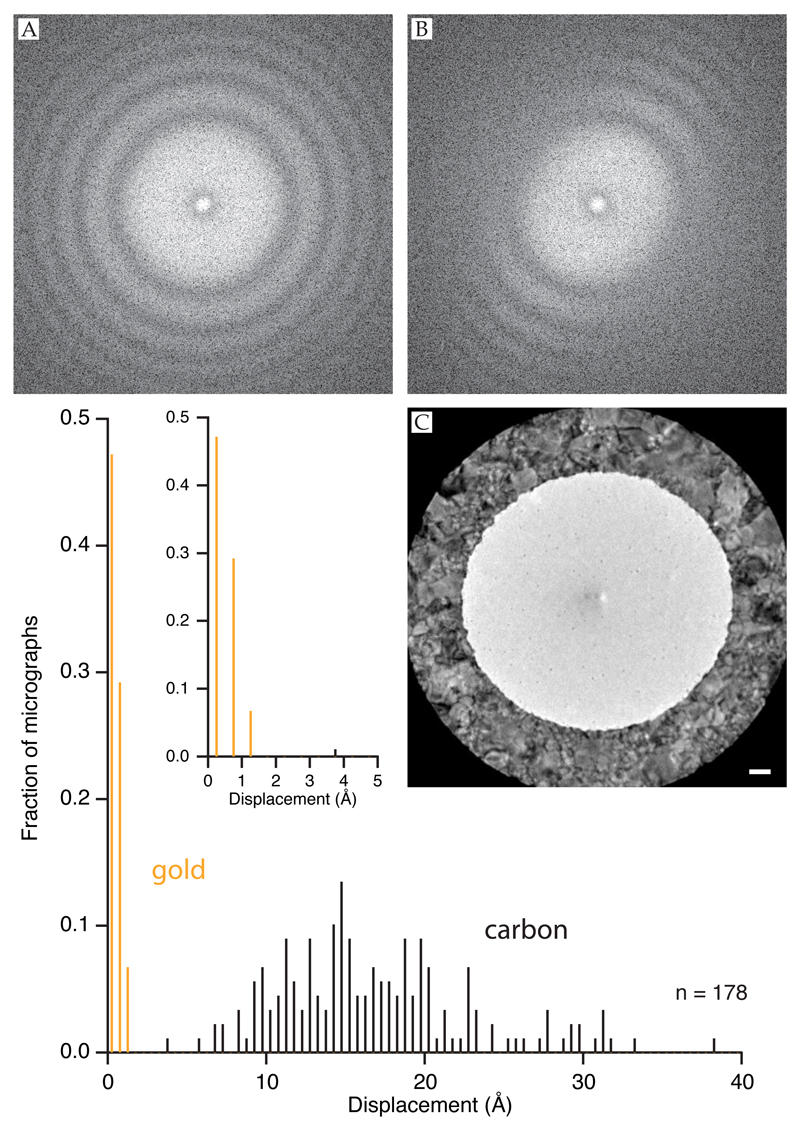
Figure 11. Testing for specimen movement during or after data collection.
Large amounts of specimen movement or stage drift can be detected during data acquisition using real-time fast Fourier transforms (FFTs) of the collected micrographs. FFTs of specimens are shown with no stage drift (A) and 10 Å/sec temperature induced stage drift (B). Micrograph C shows the recommended, symmetric illumination of a frozen specimen suspended across a hole in an all-gold support foil. Histogram is the in-plane movement statistics for 1 second micrographs (16 e−/Å2) on all-gold supports vs. amorphous carbon on gold (Quantifoil) under the same symmetric illumination conditions shown in C. Inset is enlargement of histogram near origin.
† Notes on automated data collection on all-gold supports: For hole recognition using FEI’s EPU program (hole selection), use a slightly smaller template hole then the actual hole size and then readjust to the true size before selecting the illumination geometry (template definition). This helps with hole locating because the gold foils have so much more contrast than the standard carbon foils. The ‘drift’ step is not required on all-gold supports since they do not move during irradiation. Focusing is only required every 20–40 μm.
Every dataset collected using a direct electron detector should be checked to verify that the specimen preparation and illumination conditions are optimized to minimize particle motion. Currently, the simplest way to do this is using a per-micrograph motion correction program like motioncorr (Li et al., 2013) [ch JR]. Each micrograph is collected as a movie where the total dose is subdivided into individual frames. The algorithm is used to determine the overall movement of the specimen in the micrograph with time, and then offset and re-sum the frames thus removing some of the information loss. The program is fast, as it is implemented for a GPU architecture, and therefore can be used to quickly check the overall movement of the specimen support during imaging. Distributions of movement for micrographs in a one second exposure at 16 e−/Å2 (Titan Krios on a Falcon 2 direct electron detector with 1.7 Å pixels) are shown in Figure 11. Both sets were collected with the recommended symmetric conditions. On all-gold supports, the movement is less than 1.5 Å, which is less than one pixel in the image. On standard Quantifoil supports under the same conditions, the per-micrograph movement is an order of magnitude larger. Thus, with ultra-stable supports and modern low-drift microscope stages, we use micrograph motion tracking algorithms to check for incorrect data collection settings, stage drift, or damage to the foil or grid since under normal conditions with ultra-stable supports, the overall movement of the specimen support should be essentially undetectable.
Acknowledgements
The authors thank R. Henderson and G. McMullan for many helpful discussions, S. Scotcher for fabrication of custom instruments, the EM facility at MRC LMB (S. Chen, C. Saava), the Ramakrishan lab for the gift of ribosomes, S. Tan and C. Hill for a critical reading of the manuscript and B. Carragher for the photograph of the Gatan Solarus. CJR and LAP are inventors on a patent filed by the Medical Research Council, UK related to this work, which is licensed to Quantifoil under the trade mark UltrAuFoil®. This work was supported by an Early Career Fellowship from the Leverhulme Trust (CJR), the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007-2013)/ERC grant agreement no. 261151 (LAP) and Medical Research Council grants U105192715 and U105184326.
References
- Adrian M, Dubochet J, Lepault J, McDowall AW. Cryo-electron microscopy of viruses. Nature. 1984;308(5954):32–36. doi: 10.1038/308032a0. [DOI] [PubMed] [Google Scholar]
- Baker LA, Watt IN, Runswick MJ, Walker JE, Rubinstein JL. Arrangement of subunits in intact mammalian mitochondrial ATP synthase determined by cryo-EM. Proceedings of the National Academy of Sciences. 2012;109(29):11675–11680. doi: 10.1073/pnas.1204935109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernecky C, Herzog F, Baumeister W, Plitzko JM, Cramer P. Structure of transcribing mammalian RNA polymerase II. Nature. 2016;529(7587):551–554. doi: 10.1038/nature16482. [DOI] [PubMed] [Google Scholar]
- Brenner S, Horne R. A negative staining method for high resolution electron microscopy of viruses. Biochimica et Biophysica Acta. 1959;34:103–110. doi: 10.1016/0006-3002(59)90237-9. [DOI] [PubMed] [Google Scholar]
- Chen B, Kaledhonkar S, Sun M, Shen B, Lu Z, Barnard D, Lu T-M, Gonzalez RL, Frank J. Structural dynamics of ribosome subunit association studied by mixing-spraying time-resolved cryogenic electron microscopy. Structure. 2015;23(6):1097–1105. doi: 10.1016/j.str.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y. Single-Particle Cryo-EM at Crystallographic Resolution. Cell. 2015;161(3):450–457. doi: 10.1016/j.cell.2015.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung M, Kajimura N, Makino F, Ashihara M, Miyata T, Kato T, Namba K, Blocker AJ. A method to achieve homogeneous dispersion of large transmembrane complexes within the holes of carbon films for electron cryomicroscopy. Journal of Structural Biology. 2013;182(1):51–56. doi: 10.1016/j.jsb.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu P-L, Li X, Li Z, Beckett B, Brilot AF, Grigorieff N, Agard DA, Cheng Y, Walz T. Evaluation of super-resolution performance of the k2 electron-counting camera using 2d crystals of aquaporin-0. Journal of Structural Biology. 2015;192(2):163–173. doi: 10.1016/j.jsb.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Fonseca PC, Morris EP. Cryo-EM reveals the conformation of a substrate analogue in the human 20S proteasome core. Nature Communications. 2015;6:7573. doi: 10.1038/ncomms8573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobro MJ, Melanson LA, Jensen GJ, McDowall AW. Plunge freezing for electron cryomicroscopy. Methods in Enzymology. 2010;481:63–82. doi: 10.1016/S0076-6879(10)81003-1. [DOI] [PubMed] [Google Scholar]
- Dubochet J, Adrian M, Chang JJ, Homo JC, Lepault J, McDowall AW, Schultz P. Cryo-electron microscopy of vitrified specimens. Quarterly Reviews of Biophysics. 1988;21(2):129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- Dubochet J, Adrian M, Lepault J, McDowall A. Emerging techniques: Cryo-electron microscopy of vitrified biological specimens. Trends in Biological Sciences. 1985:143–146. [Google Scholar]
- Elmlund H, Elmlund D, Bengio S. PRIME: probabilistic initial 3D model generation for single-particle cryo-electron microscopy. Structure. 2013;21(8):1299–1306. doi: 10.1016/j.str.2013.07.002. [DOI] [PubMed] [Google Scholar]
- Ermantraut E, Wohlfart K, Tichelaar W. Perforated support foils with pre-defined hole size, shape and arrangement. Ultramicroscopy. 1998;74(1):75–81. [Google Scholar]
- Geim AK. Graphene: status and prospects. Science. 2009;324(5934):1530–1534. doi: 10.1126/science.1158877. [DOI] [PubMed] [Google Scholar]
- Glaeser RM, Han B-G, Csencsits R, Killilea A, Pulk A, Cate JH. Factors that influence the formation and stability of thin, cryo-EM specimens. Biophysical Journal. 2016;110(4):749–755. doi: 10.1016/j.bpj.2015.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R. Avoiding the pitfalls of single particle cryo-electron microscopy: Einstein from noise. Proceedings of the National Academy of Sciences. 2013;110(45):18037–18041. doi: 10.1073/pnas.1314449110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R. Overview and future of single particle electron cryomicroscopy. Archives of Biochemistry and Biophysics. 2015;581:19–24. doi: 10.1016/j.abb.2015.02.036. [DOI] [PubMed] [Google Scholar]
- Henderson R, Chen S, Chen JZ, Grigorieff N, Passmore LA, Ciccarelli L, Rubinstein JL, Crowther RA, Stewart PL, Rosenthal PB. Tilt-pair analysis of images from a range of different specimens in single-particle electron cryomicroscopy. Journal of Molecular Biology. 2011;413:1028–1046. doi: 10.1016/j.jmb.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastner B, Fischer N, Golas MM, Sander B, Dube P, Boehringer D, Hartmuth K, Deckert J, Hauer F, Wolf E, et al. GraFix: Sample preparation for single-particle electron cryomicroscopy. Nature Methods. 2008;5(1):53–55. doi: 10.1038/nmeth1139. [DOI] [PubMed] [Google Scholar]
- Kuijper M, van Hoften G, Janssen B, Geurink R, De Carlo S, Vos M, van Duinen G, van Haeringen B, Storms M. FEIs direct electron detector developments: Embarking on a revolution in cryo-TEM. Journal of Structural Biology. 2015;192(2):179–187. doi: 10.1016/j.jsb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nature Methods. 2013;10(6):584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullan G, Faruqi A, Clare D, Henderson R. Comparison of optimal performance at 300keV of three direct electron detectors for use in low dose electron microscopy. Ultramicroscopy. 2014;147:156–163. doi: 10.1016/j.ultramic.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazawa A, Fujiyoshi Y, Stowell M, Unwin N. Nicotinic acetylcholine receptor at 4.6 Å resolution: transverse tunnels in the channel wall. Journal of Molecular Biology. 1999;288(4):765–86. doi: 10.1006/jmbi.1999.2721. [DOI] [PubMed] [Google Scholar]
- Nogales E, Scheres SH. Cryo-EM: a unique tool for the visualization of macromolecular complexity. Molecular Cell. 2015;58(4):677–689. doi: 10.1016/j.molcel.2015.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi M, Li Y, Cheng Y, Walz T. Negative staining and image classification: powerful tools in modern electron microscopy. Biological Procedures Online. 2004;6(1):23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantelic RS, Meyer JC, Kaiser U, Baumeister W, Plitzko JM. Graphene oxide: a substrate for optimizing preparations of frozen-hydrated samples. Journal of Structural Biology. 2010;170(1):152–156. doi: 10.1016/j.jsb.2009.12.020. [DOI] [PubMed] [Google Scholar]
- Pantelic RS, Suk JW, Magnuson CW, Meyer JC, Wachsmuth P, Kaiser U, Ruoff RS, Stahlberg H. Graphene: Substrate preparation and introduction. Journal of Structural Biology. 2011;174(1):234–238. doi: 10.1016/j.jsb.2010.10.002. [DOI] [PubMed] [Google Scholar]
- Plaschka C, Lariviere L, Wenzeck L, Seizl M, Hemann M, Tegunov D, Petrotchenko E, Borchers C, Baumeister W, Herzog F, et al. Architecture of the RNA polymerase II-Mediator core initiation complex. Nature. 2015;518(7539):376–380. doi: 10.1038/nature14229. [DOI] [PubMed] [Google Scholar]
- Quispe J, Damiano J, Mick SE, Nackashi DP, Fellmann D, Ajero TG, Carragher B, Potter CS. An improved holey carbon film for cryo-electron microscopy. Microscopy and Microanalysis. 2007;13(05):365–371. doi: 10.1017/S1431927607070791. [DOI] [PubMed] [Google Scholar]
- Regan W, Alem N, Alemán B, Geng B, Girit Ç, Maserati L, Wang F, Crommie M, Zettl A. A direct transfer of layer-area graphene. Applied Physics Letters. 2010;96:113102–4. doi: 10.1021/nn100459u. [DOI] [PubMed] [Google Scholar]
- Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. Journal of Molecular Biology. 2003;333(4):721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- Russo CJ, Golovchenko JA. Atom-by-atom nucleation and growth of graphene nanopores. Proceedings of the National Academy of Science. 2012;109(16):5953–5957. doi: 10.1073/pnas.1119827109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo CJ, Passmore LA. Controlling protein adsorption on graphene for cryo-EM using low-energy hydrogen plasmas. Nature Methods. 2014a;11(6):649–652. doi: 10.1038/nmeth.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo CJ, Passmore LA. Robust evaluation of 3D electron cryomicroscopy data using tilt-pairs. J Struct Biol. 2014b;187(2):112–118. doi: 10.1016/j.jsb.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo CJ, Passmore LA. Ultrastable gold substrates for electron cryomicroscopy. Science. 2014c;346(6215):1377–1380. doi: 10.1126/science.1259530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo CJ, Passmore LA. Progress towards an optimal specimen support for electron cryomicroscopy. Curr Opin Struct Biol. 2016a Jan;37:81–89. doi: 10.1016/j.sbi.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo CJ, Passmore LA. Ultrastable gold substrates: Properties of a support for high-resolution electron cryomicroscopy of biological specimens. J Struct Biol. 2016b Jan;193(1):33–44. doi: 10.1016/j.jsb.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seigel RR, Harder P, Dahint R, Grunze M, Josse F, Mrksich M, Whitesides GM. On-line detection of nonspecific protein adsorption at artificial surfaces. Analytical Chemistry. 1997;69(16):3321–3328. doi: 10.1021/ac970047b. [DOI] [PubMed] [Google Scholar]
- Stark H, Grant J. GraFix: Stabilization of fragile macromolecular complexes for single particle cryo-EM. Methods In Enzymology. 2010;481:109. doi: 10.1016/S0076-6879(10)81005-5. [DOI] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. Journal of Structural Biology. 2007;157(1):38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Thompson RF, Walker M, Siebert CA, Muench SP, Ranson NA. An introduction to sample preparation and imaging by cryo-electron microscopy for structural biology. Methods. 2016 doi: 10.1016/j.ymeth.2016.02.017. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unwin P. Electron microscopy of the stacked disk aggregate of tobacco mosaic virus protein: II. the influence of electron irradiation on the stain distribution. Journal of Molecular Biology. 1974;87(4):657–670. doi: 10.1016/0022-2836(74)90076-x. [DOI] [PubMed] [Google Scholar]
- Vinothkumar KR, Henderson R. Single particle electron cryomicroscopy: trends, issues and future perspective. Quarterly Reviews of Biophysics. 2016 doi: 10.1017/S0033583516000068. in press. [DOI] [PubMed] [Google Scholar]
- Vogler EA. Protein adsorption in three dimensions. Biomaterials. 2012;33(5):1201–1237. doi: 10.1016/j.biomaterials.2011.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasilewski S, Rosenthal PB. Web server for tilt-pair validation of single particle maps from electron cryomicroscopy. Journal of Structural Biology. 2014;186(1):122–131. doi: 10.1016/j.jsb.2014.02.012. [DOI] [PubMed] [Google Scholar]
- Weis F, Giudice E, Churcher M, Jin L, Hilcenko C, Wong CC, Traynor D, Kay RR, Warren AJ. Mechanism of eIF6 release from the nascent 60S ribosomal subunit. Nature Structural & Molecular Biology. 2015;22(11):914–919. doi: 10.1038/nsmb.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]