Abstract
The phenylpropanoid metabolic space comprises a network of interconnected metabolic branches that contribute to the biosynthesis of a large array of compounds with functions in plant development and stress adaptation. During biotic challenges, such as insect attack, a major rewiring of gene networks associated with phenylpropanoid metabolism is observed. This rapid reconfiguration of gene expression allows for the prioritized production of metabolites that help the plant solve ecological problems. Phenolamides are a group of phenolic-derivatives that originate from the diversion of hydroxycinnamoyl acids from the main phenylpropanoid pathway after N-acyltransferase-dependent conjugation to polyamines or aryl-monoamines. These structurally diverse metabolites are abundant in reproductive organs of many plants and have recently been shown to play roles as induced defenses in vegetative tissues. In the wild tobacco, Nicotiana attenuata in which the herbivory-induced regulation of these metabolites has been studied, rapid elevations of phenolamide levels that function as induced defenses result from a multi-hormonal signaling network that reshapes connected metabolic pathways. In this review, we summarize recent findings in the regulation of phenolamides obtained by mass spectrometry-based metabolomics and outline a conceptual framework for gene discovery in this pathway. We finally introduce a multifactorial approach useful in deciphering metabolic pathway reorganizations among different tissues in response to stress.
Keywords: Phenolamides, Phenylpropanoid pathway, N-acyltransferase, Metabolomics, Systems biology, Self-organizing maps, Nicotiana attenuata
Introduction
Over the last three decades, it has become abundantly clear that the plethora of small molecules produced by plants which are not directly required for growth play important roles as signal and defense molecules rather than being ‘waste’ products (Berenbaum and Zangerl 2008, Hartmann 2007, Pichersky and Gang 2000, Pichersky and Lewinsohn 2011). In contrast to those derived from ‘primary’ metabolic pathways, the biosynthesis of these metabolites is restricted to selected plant taxa. This suggests that specific biosynthetic pathways have been positively selected throughout the course of evolution in particular plant lineages when a compound or group of compounds addressed specific ecological needs. Consistent with this view, ecological and/or evolutionary insights gained from field studies and lab-based functional genomic analyses has clearly established that a plant’s interaction with insects has sculpted many aspects of plant metabolism. This includes the composition and size of metabolic classes as well as regulatory networks that determine their fluxes (Agrawal 2012, Berenbaum and Zangerl 2008, Prasad et al. 2012). The frequently invoked functional distinction between secondary and primary metabolites is therefore more a reflection of our ignorance of the genes controlling their biosynthesis and their biological function (Pichersky and Gang 2000). Metabolites derived from the shikimate/phenylpropanoid core pathway illustrate how sophisticated rearrangements of central metabolic pathways allow plants to solve ecological challenges imposed by their interactions with insects.
Phenylpropanoid derivatives are found ubiquitously across the plant kingdom and share at least one aromatic hydrocarbon ring with one or more hydroxyl groups attached to it, a common feature derived from the skeleton of phenylalanine (Vogt 2010). Simple hydroxycinnamoyl acids and esters produced by the core part of the shikimate pathway serve, after oxidation, reduction, methylation, decoration with different kinds of small molecules and/or polymerization, as metabolic units for the production of an enormous array of compounds such as, among others, flavonoids, coumarins, lignin and phenolamides. These different metabolic classes play essential roles in development but also as plant defenses against biotic challenges. For example, phenolic-derived floral scents and pigments are essential determinants of a plant’s fertility and outcrossing rates by attracting insect or bird pollinators (Dudareva and Pichersky 2000, Kessler et al. 2013). On the other hand, many structurally different phenolics rapidly accumulate to higher levels as components of an induced defense arsenal against hervivore attack (Karban and Baldwin, 1998), processes involving plant species specific transcriptionally-mediated rearrangements of metabolic pathways (Howe and Jander 2008). Important insights into the structural and regulatory genes of the core phenylpropanoid pathway have been summarized in several review articles (Costa et al. 2003, Tohge et al. 2013, Vogt 2010). In contrast, how stressed cells re-channel metabolic fluxes of the phenylpropanoid core pathway for the production of a specific spectrum of metabolites required to solve ecological problems remains largely unknown. The same holds for the well-studied downstream branches for which some branch-specific transcription factors have been identified.
In this review, we describe recent insights into the regulation of phenolamide production, a pathway that originates from the diversion of hydroxycinnamoyl acids from the main phenylpropanoid pathway after conjugation to polyamine or aryl-monoamine molecules by hydroxycinnamoyl-CoA:amine N-(hydroxycinnamoyl)transferases -- hereafter referred to as N-acyltransferases --(Bassard et al. 2010, Edreva et al. 1998, Facchini et al. 2002). These metabolites,sometimes referred to as phenylamides or more accurately as N-hydroxycinnamoyl-amine conjugates, are a diverse group of phenolic-derived secondary metabolites found in many dicotyledonous as well as monocot plant lineages (Bassard et al. 2010, Edreva et al. 2007, Facchini et al. 2002, Martin-Tanguy et al. 1978, Martin-Tanguy 1985). Organ-specific pools of phenolamides were originally thought to function only in developmental homeostasis, but more recently these products have been shown to accumulate during stress and to function as induced defenses (Demkura et al. 2010, Kaur et al. 2010, Martin-Tanguy 1985, Muroi et al. 2009, Newman et al. 2001). Different plant species have been shown to accumulate phenolamides during insect herbivory, but important advances in the defensive function of these metabolites have been mainly obtained in transgenic Nicotiana attenuata plants for which precise genetic manipulations of phenolamide transcriptional regulation and structural genes were conducted. The dramatic increases in the production of some of these metabolites during insect herbivory in N. attenuata (Gaquerel et al. 2010, Kaur et al. 2010, Onkonkesung et al. 2012) have been shown to (i) reshape many other connected metabolic pathways (Gaquerel et al. 2013), (ii) be indicators of herbivory-induced hormonal signals spreading throughout a plant and (iii) decrease insect performance (Kaur et al. 2010).
In a nutshell, probing this complex metabolic grid may provide an interesting framework for assessing transcriptional and metabolic controls that prioritize the activation of metabolic branches when a plant is attacked by insect herbivores. In the conceptual framework we outline here, understanding metabolite regulation through metabolomics approaches is the first step in the gene discovery process, which we will illustrate with recent work in N. attenuata, a plant with well-known herbivory-induced signaling (Wu and Baldwin, 2010). As the herbivory-specific co-expression patterns among genes shaping phenolamide metabolism deployed throughout the plant are likely organ-specific, we describe a newly developed multifactorial approach for deciphering whole-organism metabolic pathway reorganizations.
Mass spectrometry-based profiling of the phenolamide metabolic grid
Phenolamide structural diversity has been summarized by Bassard et al. (2010). Briefly, phenolamides have been identified in many dicotyledonous plants as well as in monocots, including wheat (Triticum aestivum), barley (Hordeum vulgare), rice (Oryza sativa) and maize (Zea mays) (Bassard et al. 2010, Edreva et al. 2007, Facchini et al. 2002, Martin-Tanguy et al. 1978, Martin-Tanguy 1985). Abundance and organ-specific profiles of phenolamide-producing plant lineages have been carefully documented in several studies (e.g. Martin Tanguy et al. 1978, Martin-Tanguy 1985, 1997). Activated forms of coumaric, caffeic and ferulic acids in combination with either aryl-monoamines (tyramine, tryptamine, anthranilate, etc…) or polyamines (putrescine and spermidine) are the most commonly encountered building blocks. In the case of spermidine-containing metabolites, the phenolamide ‘codebook’ produces mono-, di- and tri-substituted metabolites, which results in a large repertoire of structures that can be additional decorated a posteriori (Fellenberg et al. 2008, 2009, 2012a, Matsuno et al. 2009). The degree of acylation, and in turn, the resulting number of free residual amino groups, determines the phenolamides’ physicochemical properties as well as the biological functions mediated by these metabolites. Representative acylated putrescine and spermidine phenolamide structures are presented in Figure 1.
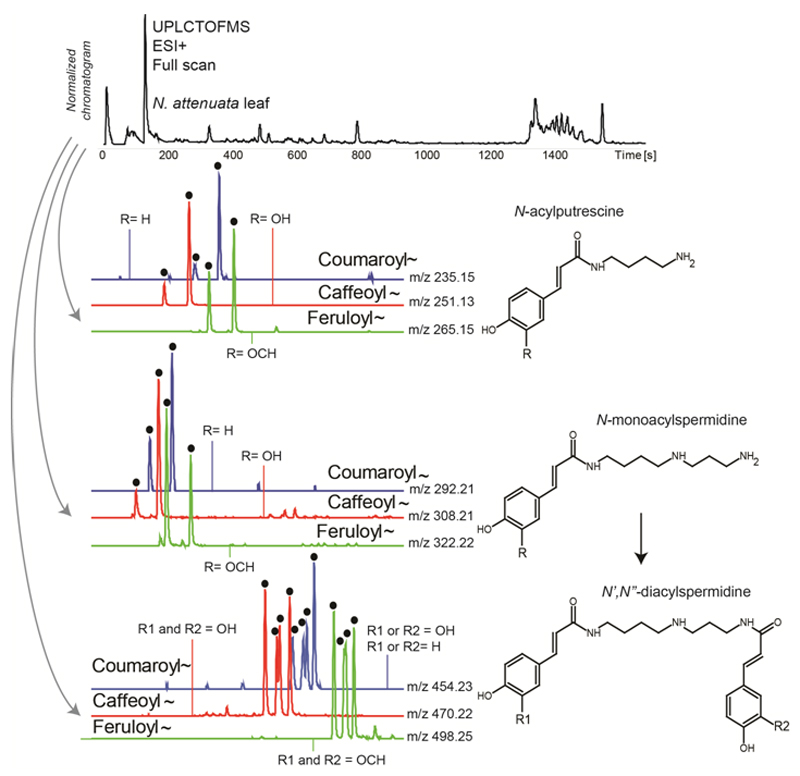
Figure 1. Mass spectrometry-based metabolomics of main phenolamides in N. attenuata leaves.
Phenolamides can readily be analyzed by ultra-high performance liquid chromatography coupled with a mass spectrometer. A representative UHPLC-time-of-flight-mass spectrometry (TOFMS) full scan chromatogram recorded in the positive ionization mode for an extract of an herbivory-induced leaf of Nicotina attenuata is shown. Coumaroyl- (m/z 147.05), caffeoyl- (m/z 163.04), or feruloyl (m/z 177.05) moieties resulting from the cleavage from different core molecules can, for example, be queried rapidly to compute extracted ion currents from the chromatogram. Specific m/z signals corresponding to either coumaroyl-, caffeoyl- and feruloyl-containing mono-acylated putrescine molecules or mono- and diacylated spermidines (N’,N”-coumaroyl,caffeoylspermidine, N’,N”-dicaffeoylspermidine, N’,N”-diferuloylspermidine) can be queried to reveal phenolamide peaks (highlighted with black dots). Representative structures are shown.
Profiling phenolamide levels across tissue types by medium to high-throughput mass spectrometry-based metabolomics has proved decisive in revealing the genomic basis of phenolamide biosynthesis. Ontogenically-tuned increases in phenolamide levels have for instance been reported during herbivory (Kaur et al. 2010, Onkokesung et al. 2012). High levels of polyamine-based phenolamides have long been known as a characteristic feature of developing flower buds of many species and floral organ-specific phenolamide profiles are known to correlate with floral developmental stages (Fellenberg et al. 2009, Fellenberg et al. 2008, Fellenberg et al. 2012a, 2012b, Grienenberger et al. 2009, Matsuno et al. 2009). Phenolamides appear to be absent from mutants that do not flower or produce abnormal flowers suggesting that these metabolites fulfill important roles during normal flower development, however the exact function of these metabolites in these tissues, including the pollen coat, where they can be particularly abundant, remains puzzling.
The modular structure of phenolamides renders them easily tractable for elucidation by mass spectrometry, as spectrometry fragmentation patterns measured with high-resolution and even low-resolution mass spectrometers are frequently highly diagnostic and sufficient for identification of building blocks (Gaquerel et al. 2013, Matsuno et al. 2009, Onkokesung et al. 2012). Many members of the phenolamide library, especially poly-acylated metabolites, undergo fragmentation during in-source ionization so that, among others, coumaroyl- (m/z 147.05), caffeoyl- (m/z 163.04), or feruloyl (m/z 177.05) moieties resulting from cleavage from different core molecules (e.g. polyamine, monoarylamines but also sugars or quinate molecules) can be queried to compute extracted ion currents from mass spectrometry-based chromatograms recorded in profiling mode without bias towards specific metabolic classes (Onkonkesung et al. 2012). More specific m/z signals corresponding to either the mono-acylated spermidine molecules or fragments from polyacylated spermidines (e.g. m/z 308.21 for a mono-caffeoylspermidine molecule or fragment) can be used to visualize acylated spermidine profiles within an extract (Figure 1). To comprehensively mine these data, compound-specific pseudo-spectra collected in the profiling mode can be deconvoluted in an automated fashion by Open Access programs such as the R package CAMERA (Kuhl et al. 2012) as a preliminary step prior to statistical analysis to evaluate the enrichment of phenolamide diagnostic ions (Onkonkesung et al. 2012). The location of the phenolic moieties within the polyamine skeletons cannot be rigorous assigned from mass spectrometry data alone to positions N1, N5 or N10 of spermidine due to rearrangements occurring during fragmentation (Fellenberg et al. 2009, Gaquerel et al. 2010). Characterization of the m/z signals of the phenolamide building blocks make these metabolites amenable for fragmentation rule-based dereplication approaches (the analytical process by which known metabolites are identified from novel matrices based on established analytical rules).
Gene discovery in the phenolamide metabolic grid
The biosynthetic pathways of the amine moieties and of the phenolic blocks from deaminated phenylalanine have been thoroughly reviewed (Bassard et al. 2010, Kusano et al. 2008, Vogt 2010). The conjugation of phenolic units to a polyamine or an aryl monoamine molecule represents the key metabolic entry point in phenolamide biosynthesis, but the N-acyltransferases enzymes catalyzing these important conjugation reactions have remained unknown for many years. Recent functional genomic work on floral organ-specific phenolamide biogenesis has identified a panoply of N-acyltransferases, most belonging to the BAHD gene family (D’Auria 2006), that catalyze phenolamide biosynthesis. Phylogenetic relationships among the characterized hydroxycinnamoyl-CoA:amine transferases and Arabidopsis BAHD genes are presented in Figure 2. Phylogenetic clustering of these genes according to the type of amine molecule used as the acyl acceptor has been reported (Luo et al. 2009), but species-specific divergences are also known among N-acyltransferases acting on the same amine skeleton. Three N-acyltransferases for phenolamide biogenesis that compete for an overlapping set of phenolic acyl donors during herbivory (Figure 3 and Figure 4) have recently been identified in N. attenuata (Onkokesung et al. 2012). AT1 controls the production of N-coumaroylputrescine and N-caffeoylputrescine. N-hydroxycinnamoyl-CoA:putrescine transferase enzymatic activities were first characterized by Negrel et al. (1989, 1991, 1992), from which the formation of diacylated spermidine phenolamides in tobacco was shown to likely be a two-step reaction. DH29 controls the first acylation step on spermidine and silencing its expression disrupts the accumulation of most wound- and herbivory-induced spermidine-based phenolamides. CV86 is involved in the production of certain diacylated spermidine isomers from DH29-dependent mono-acylated spermidines, suggesting that additional N-acyltransferases are required to produce the full spectrum of metabolites found in N. attenuata. These two genes are distantly related to Arabidopsis N-acyltransferases that use spermidine as an acyl acceptor to catalyze in a single gene manner the sequential synthesis of specific polyacylated spermidine-based phenolamides (Luo et al. 2009, Onkokesung et al. 2012).
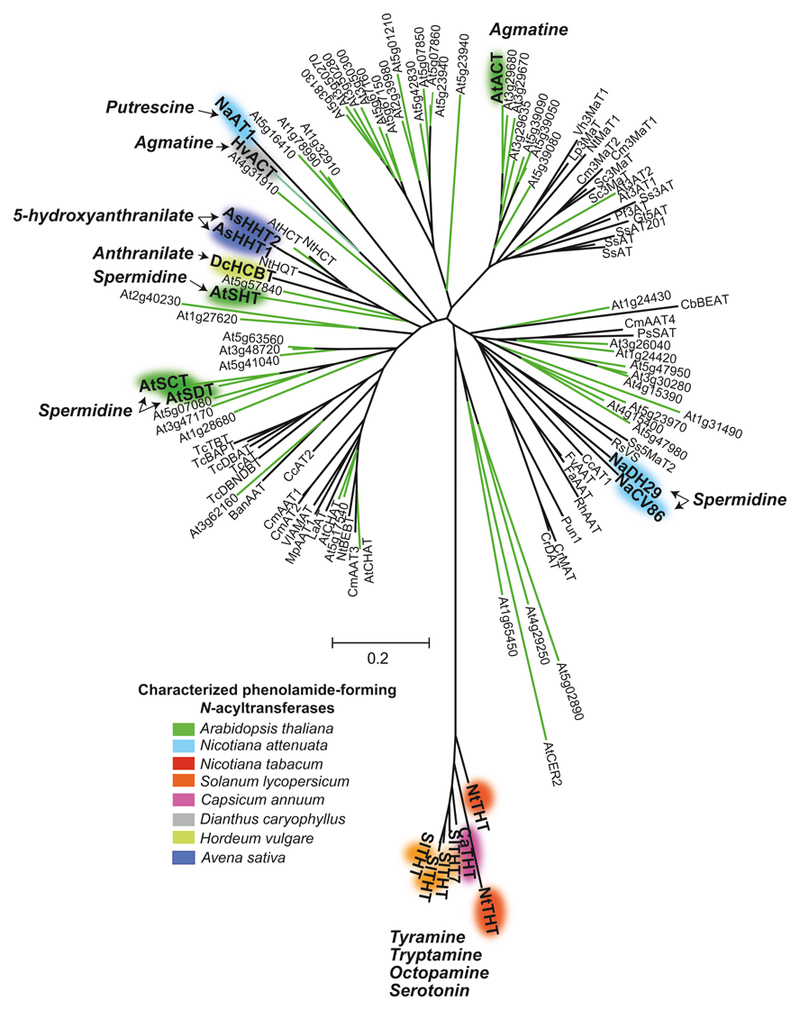
Figure 2. Phylogenetic relationships among phenolamide-forming N-acyltransferases.
A phylogenetic analysis was conducted for Arabidopsis BAHD (green branches of the tree) and functionally characterized N-acyltransferases including phenolamide-forming ones (acyl acceptor indicated) summarized in Bassard et al. (2010) and additional characterized N-acyltransferases reported in Luo et al. (2009). The phylogenetic tree reveals that N. attenuata DH29 and CV86 (highlighted in blue), which control the two-step synthesis of diacylated spermidines, cluster far from the Arabidopsis polyacylated spermidine-forming N-acyltransferases (AtSCT and AtSHT). Phenolamide N-acyltransferases with different acyl acceptor specificities (indicated next N-acyltransferases) are located on different parts of the tree. Sequences were aligned with Muscle and the alignment was trimmed with Gblocks to obtain 133 positions in 16 blocks that were used to calculate the phylogenetic tree using MEGA 4 and the Neighbor-Joining clustering method with 1000 iterations to calculate bootstrap values (Onkokesung et al. 2012). Colored ellipses of the tree connected to gene name in bold -- plant species names are reported in the color key -- denote for characterized phenolamide-forming N-acyltransferases. Plant species names are abbreviated as follows: Arabidopsis thaliana, At; Avena sativa, As; Capsicum annuum, Ca; Catharanthus roseus, Cr; Clarkia breweri, Cb; Curcumis melo, Cm; Dianthus caryophyllus, Dc; Fragaria anassa, Fa; Hordeum vulgare, Hv; Lupinus albus, La; Malus pumila, Mp; Nicotiana attenuata, Na; Nicotiana tabacum, Nt; Papaver somniferum, Ps; Salvia splendens, Ss; Solanum lycopersicum, Sl; Taxum cupsidata, Tc; Vitis labrusca, Vl.
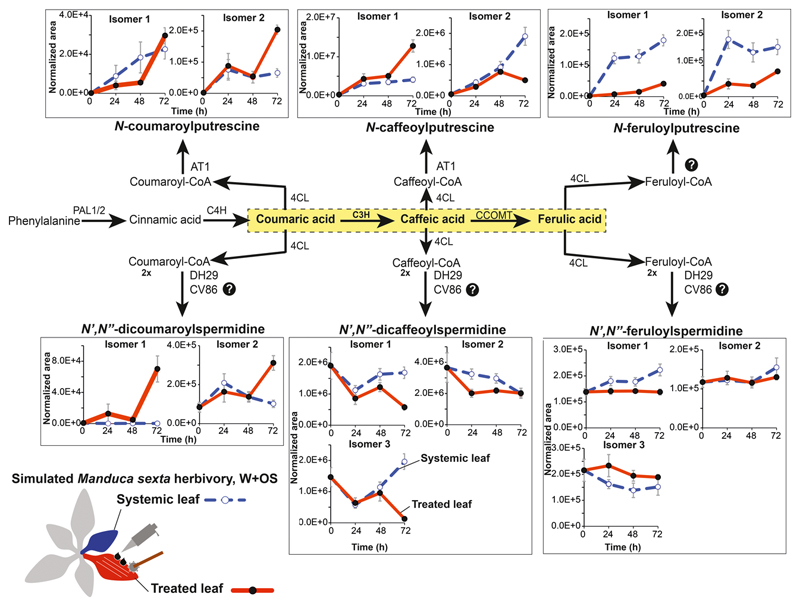
Figure 3. Herbivory-induced changes in N. attenuata leaf phenolamide metabolism.
Gene names and functions are given in the main text. Dark blue lines depict metabolite accumulation patterns in W+OS-treated leaves and red dashed lines depict responses in a systemic leaf from the same plant. Putrescine conjugates show greater induced changes than do spermidine conjugates. Dynamics differ among the predicted isomers.
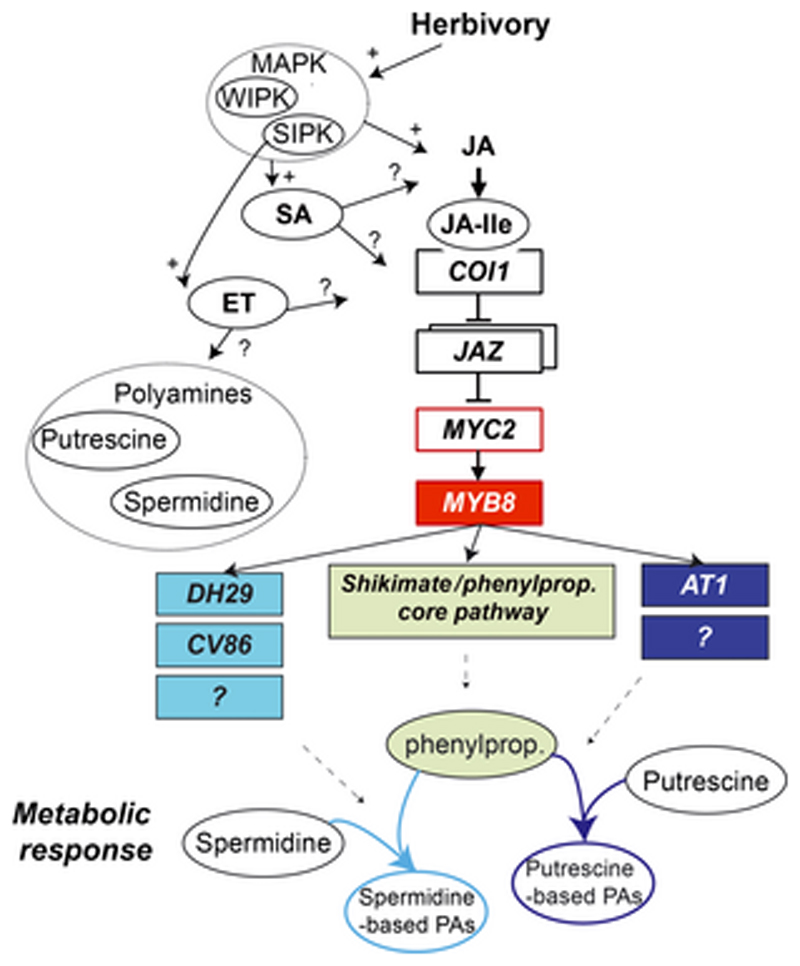
Figure 4. Current view of the regulation of herbivory-induced phenolamide biosynthesis in N. attenuata as mediated by the core jasmonic acid biosynthetic and transcriptional pathway.
MAPK signaling and interactions among other hormonal signaling networks shape the amplitude of the jasmonate bursts and downstream signaling. The role of ethylene in cross-regulating polyamine metabolism has yet to be rigorously investigated. Specific JAZ proteins inhibiting MYB8 transcription are not yet known. MYB8 regulates induced changes in the core phenylpropanoid pathway and DH29, CV86, AT1 and yet unknown phenolamide-forming N-acyltransferases.
Gene and metabolite expression data sets are increasingly being integrated to accelerate gene discovery in this pathway. Previous MS-based metabolomics investigations in Arabidopsis thaliana using a multiple sampling experimental design, publicly available at AtMetExpress webpage (http://prime.psc.riken.jp/lcms/AtMetExpress/) and compatible with the developmental and single-tissue-based experiments available in AtGenExpress that are commonly used by the Arabidopsis community, demonstrated that transcriptional programs largely regulate the tissue-specific production of diverse phytochemicals, with the phenolamides being a case in point (Matsuda et al. 2010).
In this pioneering study, the visualization of gene-to-metabolite co-linearity patterns was enabled by the use of an ‘Electronic Fluorescent Pictogram’ browser (Winter et al. 2007) and of co-expression analysis based on self-organizing maps (Hirai et al. 2004). These tools pave the way for bioinformatics association studies for the discovery of N-acyltransferases with regio-isomer-specific activity for the acylation of spermidine skeletons, as the isomeric profiles of polyacylated spermidines are frequently tissue-specific. Following a similar approach (Ehlting et al. 2008), Matsuno et al. (2009) discovered the role of two tandem duplicated cytochrome P450 genes, CYP98A8 and CYP98A9, arising from successive retroposition and duplication events. These two genes with pollen-specific expression act downstream of spermidine phenolamide-forming N-acyltransferases and control the meta-hydroxylation of the hydroxycinnamoyl moieties of specific phenolamides (Matsuno et al. 2009). Co-expression analysis using these two genes as baits identified an alternative phenylpropanoid pathway specifically supplying hydroxycinnamoyl units for the production of pollen coat phenolamides (Matsuno et al. 2009).
Phenolamide metabolism as part of the anti-herbivore defensive arsenal
Robust elevations in the levels of N-caffeoylputrescine and of certain N’, N”-dicaffeoylspermidine isomers occurring during insect feeding have been used to screen alterations in herbivory-induced signaling networks in leaves of N. attenuata transformants. MS-based metabolomics of the herbivory-regulated metabolome of this plant has shown that almost every aspects of the phenolamide metabolic grid are reconfigured during insect herbivory but these changes occur differently in locally-treated and systemic leaves of the same plant (Figure 3, Gaquerel et al. 2010, Kaur et al. 2010, Onkokesung et al. 2012). Increased phenolamide production following insect herbivory has also been reported in maize (Marti et al. 2013) and pepper (Tebayashi et al. 2007). Wounding a source leaf with a fabric pattern wheel on both sides of the midrib and immediately applying M. sexta oral secretions to the fresh puncture wounds (hereafter referred to as W+OS or OS-elicitation) provides a convenient means of accurately standardizing herbivore elicitation (McCloud and Baldwin 1997) and activating the associated defense/tolerance/escape responses in N. attenuata plants. This procedure recapitulates the major reconfigurations in the phenolamide metabolic nework that are repeatedly induced during insect feeding and allows researchers to conduct replicated time series metabolomics analysis. The resulting metabolic traces can, after appropriate data processing, be used to reconstruct metabolic networks in which phenolamide responses are resolved as the main induced responses.
Most previous analyses on the complete phenolamide profile of N. attenuata leaves have shown that, in line with the amplitude of the responses of their underlying biosynthetic genes, putrescine-based phenolamides exhibit more dramatic responses to the W+OS treatment than do the spermidine conjugates and that the caffeic acid-containing metabolites accumulate to higher levels in N. attenuata leaves than other types of phenolamides (Figure 3). Putrescine-based phenolamides occur at low levels in non-stressed leaves whereas high amounts of developmentally-regulated spermidine conjugates are regularly detected in most vegetative and reproductive tissues (Kaur et al. 2010, Keinanen et al. 2001, Onkokesung et al. 2012). Dynamics of spermidine-based phenolamides are relatively complex, which may not only reflect the multiple metabolic interconnections existing among these different metabolites but also be a signature for their highly specific functions. Only small amounts of mono-acylated spermidine are typically detected during W+OS treatment while these intermediates accumulate during insect feeding, suggesting a rapid conversion into diacylated forms (Onkokesung et al. 2012). Perhaps, as proposed by Bassard et al. (2010), these complex patterns illustrate a plant’s ability to separately control the accumulation of different diacylated spermidine isomers for specific functions in plant defense and/or development. The turnover and interconversions among phenolamides and their respective free precursors remain to be explored and these may also contribute to the changes in metabolite levels seen during herbivory.
Interestingly, levels of some of these spermidine-based phenolamides rapidly decrease following simulated herbivory in N. attenuata such as the unindentifed isomers of N’,N”-dicaffeoylspermidine while others, such as N’,N”-caffeoyl,feruloylspermidine, exhibit inversely correlated accumulation patterns. Several studies have shown that further decoration can be added to phenolic residues when conjugated to polyamines. Notably, Fellenberg et al. (2008) identified an O-methyltransferase from the Arabidopsis CcOAMT gene family which controls the terminal methylation of tri-(5-hydroxyferuloyl)spermidine into N1, N6-di(hydroxyferuloyl)-N10-sinapoylspermidine in the tapetum. Such enzyme-dependent interconversions between pre-existing diacylated spermidine pools (Fellenberg et al. 2008, Matsuno et al. 2009) are most likely shaping the complex dynamics seen in the accumulation of different spermidine-based phenolamides during herbivory and could for instance involve the methylation of N’,N”-dicaffeoylspermidine into N’,N”-caffeoyl,feruloylspermidine.
Regulation via the jasmonate-MYB8 transcriptional module
The accumulation of N-caffeoylputrescine in different solanaceous plants (e.g. Tebayashi et al. 2007) and the more recently characterized profound reconfigurations of most branches of the phenolamide metabolic network of N. attenuata during insect feeding are transcriptionally regulated by the jasmonic acid (JA) signaling pathway (Keinanen et al. 2001, Onkokesung et al. 2012, Paschold et al. 2007, Stitz et al. 2011, Ullmann-Zeunert et al. 2013) (Figure 4). Previous work with transformants or mutants directly impaired in jasmonate accumulation or perception accumulate much lower levels of N-caffeoylputrescine and stress-/herbivory-inducible phenolamides but exhibit less pronounced changes in basal levels of several spermidine-based phenolamides (Onkokesung et al. 2012, Paschold et al. 2007, Ullmann-Zeunert et al. 2013). In general, alterations due to jasmonate signaling deficiency are more pronounced in systemic leaf positions where induced phenolamide accumulation is thought to translate from major transcriptional adjustments initiated by jasmonate-dependent mobile signals transmitted from OS-elicited leaves. Jasmonate-dependent phenolamide accumulation requires the F-box protein COI1. This receptor protein after interaction with jasmonoyl-isoleucine (JA-Ile) targets JAZ transcriptional repressors for degradation by the proteasome, a transcriptional machinery controlling many secondary metabolic pathways (De Geyter et al. 2012). The strict requirement of JA-Ile in this process is clearly discernible in lines ectopically expressing an Arabidopsis jasmonic acid-specific methyltransferase (sJMT) which specifically depletes JA-Ile accumulation (Stitz et al. 2011).
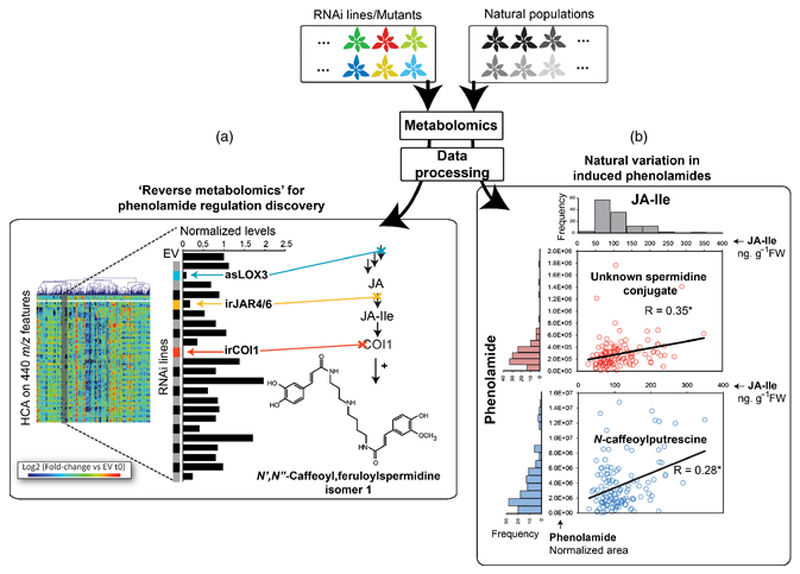
Approaches exploiting natural variations are commonly used in Arabidopsis to infer associations between genes or phytohormone signals, including jasmonates, and quantitative traits of a plant’s phenotype. Recent work on naturally variable traits in N. attenuata populations have highlighted that there exist important variations in the amplitude of the JA and JA-Ile bursts produced after simulated herbivory in these populations (Machado et al. 2013). In panel (b) of Figure 5, we show that herbivory-induced levels of N-caffeoylputrescine, as well as of other phenolamides, vary greatly in native populations when grown under controlled conditions of the laboratory, indicating the existence of genetically-determined variations at the level of the regulation and biosynthesis of these metabolites. These patterns of natural variation are significantly positively correlated with the amplitude of JA-Ile bursts.
Figure 5. Novel approaches based on metabolomics to the discovery of regulatory mechanisms for herbivory-induced changes in phenolamide metabolism.
(a) High-throughput non-targeted metabolite profiling of herbivory-induced changes in a large collection of RNAi transgenic lines reveals regulators of metabolite accumulation. Processed data can be classified using hierarchical clustering and clusters of m/z signals of interest screened across the library of transgenic lines. The jasmonate regulation of N’,N”-caffeoyl,feruloyspermidine is provided as an example. (b) Natural variation in W+OS-induced levels of N-caffeoylputrescine and of an unknown spermidine-based phenolamide in 176 natural accessions of N. attenuata positively correlate with natural variation in the OS-induced JA-Ile bursts.
Jasmonate-dependent activations in phenolamide metabolism do not translate solely from increases in the expression of genes of the phenylpropanoid pathway, most of them being well-known expression markers for wound and jasmonate responses. The expression of N-acyltransferases required for phenolamide production is also controlled by the jasmonate pathway through the transcriptional activity of N. attenuata MYB8 and N. tabacum MYBJS1, members of the R2R3 MYB transcription factor family (Galis et al. 2006, Kaur et al. 2010). The DNA-binding domain of the homologous gene of MYB8 in tobacco, MYBJS1, has been shown to bind to the promoter regions of copies of the PAL gene to regulate the expression of core genes of the phenylpropanoid pathway as well as a few from the polyamine pathway (Galis et al. 2006). Additionally, silencing MYB8 in N. attenuata abolishes herbivory-induced elevations as a result of strong reductions in AT1, DH29 and CV86 expression (Onkokesung et al. 2012). MYB8-silenced plants do not show developmental alterations, indicating that this transcription factor may control herbivory-induced elevations in the phenylpropanoid flux guided towards phenolamide production rather than steady-state parameters of this pathway. In this respect, the current view is that COI1-based perception of JA-Ile alleviates a negative transcriptional control exerted by one or several yet-to-be-characterized JAZ proteins leading to the expression of MYB8 (Figure 4). Recent work suggests that N. attenuata MYC2, a basic helix-loop-helix Leu zipper transcription factor regulating several jasmonate-dependent responses, regulate the expression of MYB8, but only minor alterations of the phenolamide profiles were detected in MYC2-transiently silenced plants (Woldemariam et al. 2013). As already demonstrated for other pathway specific transcription factors (Dal Cin et al. 2011, Mehrtens et al. 2005), the high specificity of MYB8 in regulating phenolamide metabolism opens up interesting perspectives for increasing the rate of gene discovery in this pathway using transcriptional screens.
Herbivory-induced phenolamide profiles reveal interaction between phytohormone signaling pathways and nitrogen metabolism trade-offs
Virtually any signaling nodes influencing jasmonate pools may alter induced phenolamide levels (Heinrich et al. 2013). In this respect, high-throughput metabolomics profiling of transgenic lines with sufficient knowledge regarding disturbed signaling pathways can rapidly contribute to our understanding of phenolamide regulation (Figure 5, panel (a)). This includes the possibility of revisiting how phytohormone crosstalks and downstream transcriptional regulators shape defense metabolite production. The role of ethylene in regulating phenolamide production is particularly noteworthy, because in addition to its signaling function, ethylene biosynthesis connects with the putrescine-to-spermidine conversion (Kumar et al. 1996).
Phenolamide biosynthesis interacts with nitrogen metabolism through their polyamine component (Fellenberg et al. 2012b, Matsuno et al. 2009, Ullmann-Zeunert et al. 2013). Activation of herbivory-induced responses in tobacco plants in which patterns of stress-induced nitrogen accumulation have been tracked in different tissue compartments represents an ideal system for testing the nature of this interaction. The phenolamide profile is also strongly influenced by the soil type used. We have shown that sand-grown plants have their induced spermidine and putrescine-based phenolamide pools replaced by tyramine-based ones (Kim et al. 2011). It is currently unclear whether this metabolic shift is related to the differential nitrogen supplies between these two soils and allocation in the plant (Lou and Baldwin 2004). A recent flux study has demonstrated that elevations in phenolamide levels involves significant trade-offs for nitrogen allocation during insect herbivory (Ullmann-Zeunert et al. 2013). This study is of central importance to understand how nitrogen allocation costs may tune phenolamide metabolism inducibility throughout a plant’s development. Mechanisms behind this metabolic trade-off will be investigated in the near future using transcriptomic approaches such as the one presented in the next section.
New systems-based approaches for the discovery of gene regulation in phenolamide metabolism
As frequently detected in transcriptomic screens, herbivore attack activates specific reorganizations of metabolic pathways which are different between locally-attacked and distal tissues from the same plant (Gulati et al. 2013b, Schittko et al. 2001). Spatially-coordinated modulations in gene expression networks may be a key mechanism to regulate changes in metabolite pools throughout the plant but this remains under-studied. Influential work in the regulation of the glucosinolate biosynthesis and distal networks connected to it, has shown that upstream genes of the pathway with important flux control and which are subjected to intense purifying selection (Olson-Manning et al. 2013) are central in shaping the glucosinolate chemotype according to the ‘genomic context’ or network of genes with which they are co-expressed (Malitsky et al. 2008). Instrumental data analysis tools for mining these gene networks may be found in a series of inspiring recently published ‘evo-devo’ transcriptomic studies. Several recent studies have highlighted the unprecedented perspective into the developmental regulation of genes that appropriate statistical analysis of transcriptomic data-sets provides. For instance, the elegant statistical and data visualization approach developed by Chitwood et al. (2013) has been used to demonstrate that changes in gene networks, rather than sequence divergence patterns, are responsible for the significant anatomical differences between cultivated and wild tomato species. Here we discuss the insights into phenolamide metabolism that resulted from the implementation of such a bioinformatic approach.
‘Interactive effect’ genes are enriched in metabolism-encoded processes
Surprisingly, the fact that most herbivory-inducible secondary metabolites also increase in systemic tissues has hardly been exploited in the context of gene function analysis. The case of phenolamides is particularly germane as the dynamics of these metabolites differ between local and systemic leaf tissues and these differences are known to be essential determinants of systemic defense induction in N. attenuata. Onkokesung et al. (2012) successfully selected N-acyltransferase candidates for the production of phenolamides based on their greater amplification by insects’ OS cues in systemic tissues compared to mechanical wounding. Indeed, most induced defense secondary metabolism genes investigated in N. attenuata in the context of W+OS treatments have their expression amplified by OS-activated mobile signals that are transported into systemic tissues from OS-elicited ones (Gulati et al. 2013b, Kim et al. 2011, Schittko and Baldwin 2003, Schittko et al. 2001). This further underscores that only OS perception, and not mechanical wounding alone, leads to the deployment of robust systemic signals (Gulati et al. 2013a, Gulati et al. 2013b).
Experiments designed to capture the dynamic rewiring of the gene networks that control the spread of herbivory-induced systemic responses often have a complex factorial structure resulting from the different conditions/treatments and tissue types analyzed and necessarily involve time-series analysis. Informed by the targeted interpretation of the metabolic gene regulation presented above, we designed, in a recent study, a dimensionality reduction method based on multifactorial analysis to categorize genes according to their degree of tissue-specificity and responses to W+OS elicitation (Gulati et al. 2013a, Gulati et al. 2013b, Figure 6). The procedure is based on bootstrap-based non-parametric ANOVA models implemented in the R package TANOVA (Zhou and Wong 2011, Zhou et al. 2010). We applied this method to the analysis of a time-course microarray data-sets for tissues collected from control and W+OS treated plants (Kim et al. 2011). When applied to the statistical comparison of gene expression between locally-treated leaves and systemic tissues collected from the same plant, we identified four mutually exclusive groups of genes with different ANOVA structures. We detected ANOVA structures significant for an interactive effect (two leaf positions behaving differently across the time series in response to W+OS elicitation), an additive effect (W+OS-induced responses independent of tissue type), or corresponding to independent effects derived from the main experimental factors (major treatment effects in both treated and untreated tissues or differences in tissue type with no response to treatment) (Gulati et al. 2013b). The interactive gene-set represents 69 % of the non-constantly expressed genes analyzed and is highly enriched in genes involved in metabolic processes. Most processes connected with secondary metabolic pathways map to this group of genes (red sector in Figure 6 panel (c)). Remarkably, genes of the phenylpropanoid and phenolamide pathways are among those exhibiting the largest interactive effects. Additionally, many other metabolic pathways and their transcription factors share similar behavior and have yet to be explored. This necessitates classifying temporal dynamics within this large group of promising genes for metabolic pathway exploration that constitutes the interactive effect group.
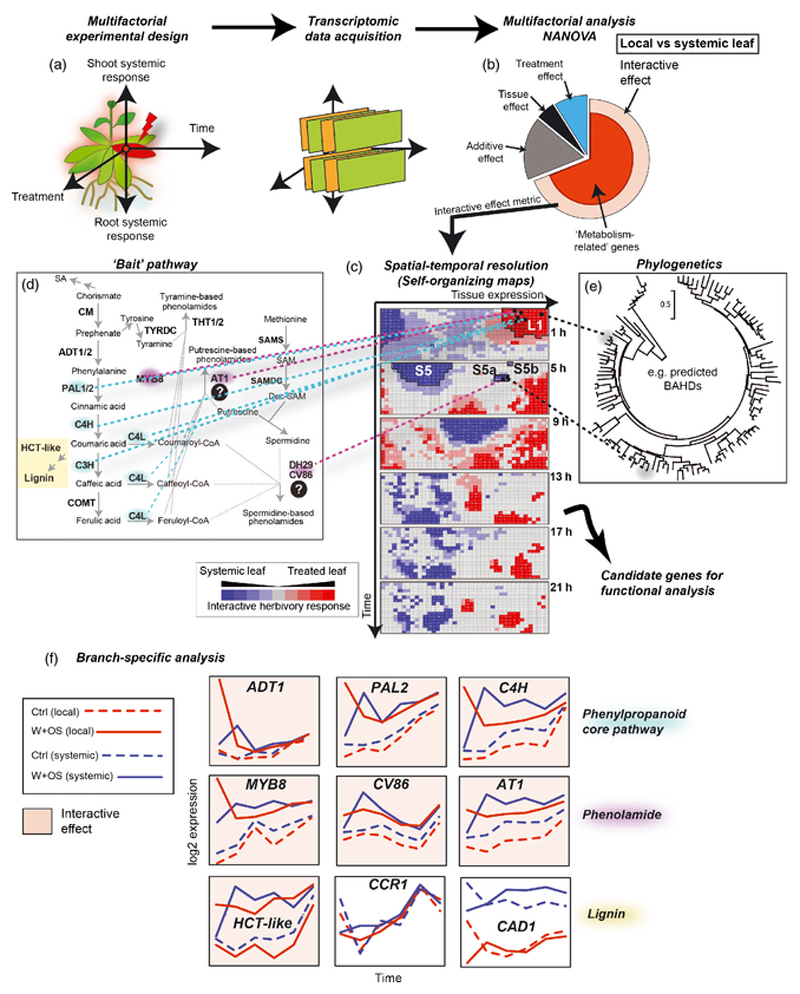
Figure 6. A multifactorial-based coexpression analysis work-flow for delineating systemically-induced secondary metabolic pathways.
A multifactorial analysis work-flow has been developed by Gulati et al. (2013) and its use in delineating genes in the acyclic diterpene glycoside pathway has previously been reported. This strategy is applied to the analysis of multidimensional transcriptomic data-sets acquired from multifactorial experimental designs (different tissue types, treatment, etc…) including time-series experiments (a). Transcriptomic data collected at each time point are combined into a data matrix used for multifactorial analysis. The statistical group corresponding to the interactive effect genes (those genes that respond to the treatment differently according to the tissue type, here locally vs systemically treated leaves) is highly overrepresented with metabolism-related genes (red sector) (b). Self-organizing maps are used to impose structure (c) and to cluster genes within this bin according to their temporal dynamics using a metric derived from the multifactorial analysis. Bait genes (here from the phenylpropanoid and phenolamide pathways) can be localized on the maps to identify clusters of genes of interest (phenylpropanoid genes: L1 for early interactive effects in local leaves; phenolamide genes: L1, S5a and S5b for local and then systemic interactive effects) (d). These clusters of genes can be subsequently mined in accordance with the predictions of phylogenetic relationships (e). (f) Genes from specific branches of the phenylpropanoid space can be classified according to the detection of an interactive effect regulation. Most lignin-related genes, except HCT-Like, do not show interactive effect regulation in response to herbivory, unlike the core phenylpropanoid and phenolamide genes.
Tissue x treatment self-organizing maps visualize the sequential arrangement of metabolic pathways
Metabolic genes belonging to a common biosynthetic pathway tend to be co-regulated as a result of the activation of a robust regulatory system (Saito et al. 2008). Basic statistical approaches used to identify such strong co-expression patterns are however often plagued by the problems of gene prioritization (Bittner et al. 1999, Getz et al. 2000) that arise from conducting clustering analysis of gene expression under all experimental conditions (Swindell 2006). Indeed, patterns revealed by simple co-expression analysis essentially represent the static rewiring of the network, which does not realistically capture the plants’ phenotypic plasticity that results from the ability of cells to activate transient gene associations which represent intermediate biological states. The need for condition-dependent algorithms to resolve functional gene associations which are affected only by a subset of the experimental conditions -- in our study, by the transmission OS-induced signaling to systemic leaf positions -- has been reviewed by Krouk et al. (2013).
We used the time-specific ANOVA coefficients reflecting the degree of significance for the interactive effect between the treatment and leaf positions and scaled them with the difference in amplitudes of responses to OS-elicitation to obtain a metric that characterize the behavior of a given gene in more than one tissue (here two leaf positions). We then applied self-organizing maps (SOMs) to delineate gene network assemblies. SOMs result from an iterative process in which neighboring clusters influence each other. The resulting maps clearly visualize the main expression patterns in the analysis of molecular responses to perturbations (Chitwood et al. 2013, Gulati et al. 2013b, Hirai et al. 2005, Hirai et al. 2004). SOMs colored according to the cluster’s average intensity at each time point are presented in Figure 6. Gene network assemblies along the time-course are visualized by changes in the size of the groups of clusters similarly colored according to the tissue-specificity of gene expression. Of the sequential arrangements of the group of genes termed ‘interactive’ motifs, we isolated one motif (S5) detectable 5h post-elicitation for systemic leaves, which contained an overrepresentation of metabolic pathway-encoded processes (Figure 6 panel (e)). From this motif, we delineated, in a previous study, the acyclic diterpene glycoside pathway, another route leading to the production of anti-herbivore defense metabolites (Heiling et al. 2010). Here we confirm that PAL genes and downstream elements of the phenylpropanoid pathway map into a large interactive motif (L1) rapidly induced in locally treated leaves. Consistent with their induced regulation to supply phenolamide production in these tissues, MYB8, AT1, DH29 and CV86 map on different cells corresponding to interactive effects detected first in local leaves (L1) and then in systemic leaves (S5a and S5b).
In our previous study, a rigorous comparison of Pearson correlation patterns before and after extraction of the interactive effect metric by multifactorial analysis, revealed that the method greatly improves the detection of tight regulation between the phenylpropanoid and its downstream phenolamide branch (Gulati et al. 2013b). We therefore propose that, after delimitation and selection of relevant interactive motifs using bait genes for specific branches of the phenylpropanoid metabolism, SOMs can be quickly mined for phenolamide gene discovery. In Figure 6, the overall workflow is illustrated and phylogenetic relationships between predicted N. attenuata BAHD genes are used as queries for the SOMs. This process based on multidimensional clustering of gene expression is specifically designed to mine enzyme-coding gene families for which substrate specificity and enzymatic functions are not readily predictable from phylogenetic relationships. Finally, the involvement in phenolamide biosynthesis and metabolism of a set of genes analyzed by this method can be tested by transient virus-induced gene silencing (VIGs), a rapid technique with many advantages for screening the role of metabolic genes at the interface between developmental and defense processes (Gaquerel et al. 2013, Galis et al. 2013, Steppuhn et al. 2010). Genes inferred from this analysis in motifs L1, S5a and S5b and exhibiting high sequence similarity with AT1 and CV86 are currently characterized for their respective involvement in the production of N-feruloylspermidine and specific N’, N”-dicaffeoylspermidine isomers.
Plastic gene networks shape developmental vs defensive allocations of phenolic residues to phenylpropanoid sub-branches
Loss-of-function approaches can in some cases highlight complex patterns of ‘metabolic tension’ and feedback regulation existing between interconnected metabolic branches (Vanholme et al. 2012). Our previous study provided support for the existence of an exacerbated competition in the conjugation of phenolic residues to putrescine or spermidine molecules (Onkokesung et al. 2012). Silencing of one acyltransferase enzyme impairs the accumulation of several metabolites while increasing another set of metabolites. Following the same approach, we also uncovered complex interconnections between the lignin and phenolamide pathways by silencing hydroxycinnamoyl-CoA:shikimate/quinate hydroxycinnamoyl transferase-like (HCT-like), an O-acyltransferase catalyzing the production of upstream intermediates in the lignin pathway that branch at the level of the phenylpropanoid pathway (Hoffmann et al. 2004, Hoffmann et al. 2003). Interestingly, HCT-like expression is also controlled by the transcriptional activity of MYB8, but, consistent with its main function in developmentally-controlled lignin deposition, its expression is less pronounced than that of phenolamide biosynthetic genes during herbivory and is also much less affected than these latter genes during herbivory in MYB8-silenced plants (Gaquerel et al. 2013). Widely-targeted metabolomics analysis on plants transiently-silenced for HCT-like revealed large metabolic shifts due to a large diversion of activated coumaric acid units, not channeled into lignin production by the action of HCT-like, into the production of developmentally and herbivory-induced coumaroyl-containing phenolamides (N’, N”-dicoumaroylspermidine, N’, N”-coumaroylputrescine, etc…). The fact that metabolic shifts in the production of unusual coumaroyl-containing phenolamides are largest during herbivory in HCT-like-silenced plants identifies HCT-like as a large effect gene within the gene network underlying phenylpropanoid metabolism plasticity.
Exploring the type of effect revealed by the multifactorial analysis for a given group of genes can be used to track the dynamic behavior of gene expression involved in connected branches of a metabolic pathway. At first blush, this approach could be used to mine the above mentioned interactions between the phenolamide and lignin branches. Interestingly, unlike the phenylpropanoid core pathway and phenolamide genes, preliminary work revealed that most previously characterized lignin biosynthetic genes tested, with the exception of HCT-like, do not exhibit an interactive effect regulation following insect herbivory. Individual expression patterns are presented in Figure 6 panel (f). These patterns suggest that steady-state coordinated expression patterns between the phenylpropanoid and lignin pathways are relaxed after herbivore attack. As a result of the profound reconfigurations of gene expression, tighter co-expression patterns seem to be established between the core phenylpropanoid module and the structural genes of the phenolamide pathway during herbivory compared to control conditions. More research is needed to understand the central function of MYB8 in assembling these coexpression networks between high amplitude regulation genes of the phenylpropanoid and phenolamide pathways in order to prioritize phenolamide production during insect herbivore attack.
Conclusion
The importance of phenolamides as central players in a plant’s defenses is rapidly being recognized. The advances outlined here in understanding the transcriptional regulation and biosynthesis of these metabolites offer new possibilities for manipulating these dynamic phenolamide pools and understanding the many subtle adjustments at the interface between development and stress metabolic responses that determine phenolamine levels. Tissue-specific genetic silencing approaches such as recently established by Schäfer et al. (2013) will likely reveal novel facets to the functions of phenolamide metabolism.
Acknowledgements
We thank Dr. Aura Navarro Quezada for help with the phylogenetic analysis. EG’s research in Heidelberg is funded by a DFG Excellence Initiative grant. EG, JG and ITB are funded by the Max-Planck-Society and EG is additionally funded by Advanced Grant no. 293926 of the European Research Council to ITB.
Contributor Information
Jyotasana Gulati, Email: jgulati@ice.mpg.de.
Ian T. Baldwin, Email: baldwin@ice.mpg.de.
References
- Agrawal AA, Hastings AP, Johnson MT, Maron JL, Salminen J-P. Insect herbivores drive real-time ecological and evolutionary change in plant populations. Science. 338:113–116. doi: 10.1126/science.1225977. [DOI] [PubMed] [Google Scholar]
- Bassard JE, Ullmann P, Bernier F, Werck-Reichhart D. Phenolamides: Bridging polyamines to the phenolic metabolism. Phytochemistry. 2010;71:1808–1824. doi: 10.1016/j.phytochem.2010.08.003. [DOI] [PubMed] [Google Scholar]
- Berenbaum MR, Zangerl AR. Facing the future of plant-insect interaction research: Le Retour a la “Raison d'Etre”. Plant physiology. 2008;146:804–811. doi: 10.1104/pp.107.113472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittner M, Meltzer P, Trent J. Data analysis and integration: of steps and arrows. Nat Genet. 1999;22:213–215. doi: 10.1038/10265. [DOI] [PubMed] [Google Scholar]
- Chitwood DH, Maloof JN, Sinha NR. Dynamic transcriptomic profiles between tomato and a wild relative reflect distinct developmental architectures. Plant physiology. 2013;162:537–552. doi: 10.1104/pp.112.213546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa MA, Collins RE, Anterola AM, Cochrane FC, Davin LB, Lewis NG. An in silico assessment of gene function and organization of the phenylpropanoid pathway metabolic networks in Arabidopsis thaliana and limitations thereof. Phytochemistry. 2003;64:1097–1112. doi: 10.1016/s0031-9422(03)00517-x. [DOI] [PubMed] [Google Scholar]
- Dal Cin V, Tieman DM, Tohge T, McQuinn R, de Vos RCH, Osorio S, Schmelz EA, Taylor MG, Smits-Kroon MT, Schuurink RC, Haring MA, et al. Identification of genes in the phenylalanine metabolic pathway by ectopic expression of a MYB transcription factor in tomato fruit. Plant Cell. 2011;23:2738–2753. doi: 10.1105/tpc.111.086975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Auria JC. Acyltransferases in plants: a good time to be BAHD. Current Opinion in Plant Biology. 2006;9:331–340. doi: 10.1016/j.pbi.2006.03.016. [DOI] [PubMed] [Google Scholar]
- De Geyter N, Gholami A, Goormachtig S, Goossens A. Transcriptional machineries in jasmonate-elicited plant secondary metabolism. Trends in Plant Science. 2012;17:349–359. doi: 10.1016/j.tplants.2012.03.001. [DOI] [PubMed] [Google Scholar]
- Demkura PV, Abdala G, Baldwin IT, Ballare CL. Jasmonate-dependent and -independent pathways mediate specific effects of solar ultraviolet B radiation on leaf phenolics and antiherbivore defense. Plant physiology. 2010;152:1084–1095. doi: 10.1104/pp.109.148999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva N, Pichersky E. Biochemical and molecular genetic aspects of floral scents. Plant physiology. 2000;122:627–633. doi: 10.1104/pp.122.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edreva AM, Velikova VB, Tsonev TD. Phenylamides in plants. Russian Journal of Plant Physiology. 2007;54:287–301. [Google Scholar]
- Ehlting J, Sauveplane V, Olry A, Ginglinger JF, Provart NJ, Werck-Reichhart D. An extensive (co-)expression analysis tool for the cytochrome P450 superfamily in Arabidopsis thaliana. BMC plant biology. 2008;8 doi: 10.1186/1471-2229-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchini PJ, Hagel J, Zulak KG. Hydroxycinnamic acid amide metabolism: physiology and biochemistry. Can J Bot. 2002;80:577–589. [Google Scholar]
- Fellenberg C, Bottcher C, Vogt T. Phenylpropanoid polyamine conjugate biosynthesis in Arabidopsis thaliana flower buds. Phytochemistry. 2009;70:1392–1400. doi: 10.1016/j.phytochem.2009.08.010. [DOI] [PubMed] [Google Scholar]
- Fellenberg C, Milkowski C, Hause B, Lange PR, Bottcher C, Schmidt J, Vogt T. Tapetum-specific location of a cation-dependent O-methyltransferase in Arabidopsis thaliana. Plant Journal. 2008;56:132–145. doi: 10.1111/j.1365-313X.2008.03576.x. [DOI] [PubMed] [Google Scholar]
- Fellenberg C, van Ohlen M, Handrick V, Vogt T. The role of CCoAOMT1 and COMT1 in Arabidopsis anthers. Planta. 2012a;236:51–61. doi: 10.1007/s00425-011-1586-6. [DOI] [PubMed] [Google Scholar]
- Fellenberg C, Ziegler J, Handrick V, Vogt T. Polyamine homeostasis in wild type and phenolamide deficient Arabidopsis thaliana stamens. Frontiers in plant science. 2012b;3:180. doi: 10.3389/fpls.2012.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galis I, Simek P, Narisawa T, Sasaki M, Horiguchi T, Fukuda H, Matsuoka K. A novel R2R3 MYB transcription factor NtMYBJS1 is a methyl jasmonate-dependent regulator of phenylpropanoid-conjugate biosynthesis in tobacco. Plant Journal. 2006;46:573–592. doi: 10.1111/j.1365-313X.2006.02719.x. [DOI] [PubMed] [Google Scholar]
- Galis I, Schuman M, Gase K, Hettenhausen C, Hartl M, Dinh TS, Wu J, Bonaventure G, Baldwin IT. Chapter 9: The use of VIGS technology to study plant-herbivore interactions. In: Becker A, editor. Virus-induced gene silencing: Methods and protocols. 2013. pp. 109–137. [DOI] [PubMed] [Google Scholar]
- Gaquerel E, Heiling S, Schoettner M, Zurek G, Baldwin IT. Development and validation of a liquid chromatography-electrospray ionization-time-of-flight mass spectrometry method for induced changes in Nicotiana attenuata leaves during simulated herbivory. Journal of agricultural and food chemistry. 2010;58:9418–9427. doi: 10.1021/jf1017737. [DOI] [PubMed] [Google Scholar]
- Gaquerel E, Kotkar H, Onkokesung N, Galis I, Baldwin IT. Silencing an N-acyltransferase-like involved in lignin biosynthesis in Nicotiana attenuata dramatically alters herbivory-induced Phenolamide metabolism. PloS one. 2013;8:e62336. doi: 10.1371/journal.pone.0062336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getz G, Levine E, Domany E. Coupled two-way clustering analysis of gene microarray data. Proceedings of the National Academy of Sciences of the U S A. 2000;97:12079–12084. doi: 10.1073/pnas.210134797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienenberger E, Besseau S, Geoffroy P, Debayle D, Heintz D, Lapierre C, Pollet B, Heitz T, Legrand M. A BAHD acyltransferase is expressed in the tapetum of Arabidopsis anthers and is involved in the synthesis of hydroxycinnamoyl spermidines. Plant Journal. 2009;58:246–259. doi: 10.1111/j.1365-313X.2008.03773.x. [DOI] [PubMed] [Google Scholar]
- Gulati J, Baldwin IT, Gaquerel E. An integrative statistical method to explore herbivory-specific responses in plants. Plant signaling & behavior. 2013a;8 doi: 10.4161/psb.25638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulati J, Kim SG, Baldwin IT, Gaquerel E. Deciphering herbivory-induced gene-to-metabolite dynamics in Nicotiana attenuata tissues using a multifactorial approach. Plant physiology. 2013b;162:1042–1059. doi: 10.1104/pp.113.217588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann T. From waste products to ecochemicals: Fifty years research of plant secondary metabolism. Phytochemistry. 2007;68:2831–2846. doi: 10.1016/j.phytochem.2007.09.017. [DOI] [PubMed] [Google Scholar]
- Heiling S, Schuman MC, Schoettner M, Mukerjee P, Berger B, Schneider B, Jassbi AR, Baldwin IT. Jasmonate and ppHsystemin regulate key malonylation steps in the biosynthesis of 17-hydroxygeranyllinalool diterpene glycosides, an abundant and effective direct defense against herbivores in Nicotiana attenuata. Plant Cell. 2010;22:273–292. doi: 10.1105/tpc.109.071449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich M, Hettenhausen C, Lange T, Wunsche H, Fang J, Baldwin IT, Wu J. High levels of jasmonic acid antagonize the biosynthesis of gibberellins and inhibit the growth of Nicotiana attenuata stems. Plant journal. 2013;73:591–606. doi: 10.1111/tpj.12058. [DOI] [PubMed] [Google Scholar]
- Hirai MY, Klein M, Fujikawa Y, Yano M, Goodenowe DB, Yamazaki Y, Kanaya S, Nakamura Y, Kitayama M, Suzuki H, Sakurai N, et al. Elucidation of gene-to-gene and metabolite-to-gene networks in arabidopsis by integration of metabolomics and transcriptomics. The Journal of biological chemistry. 2005;280:25590–25595. doi: 10.1074/jbc.M502332200. [DOI] [PubMed] [Google Scholar]
- Hirai MY, Yano M, Goodenowe DB, Kanaya S, Kimura T, Awazuhara M, Arita M, Fujiwara T, Saito K. Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the U S A. 2004;101:10205–10210. doi: 10.1073/pnas.0403218101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann L, Besseau S, Geoffroy P, Ritzenthaler C, Meyer D, Lapierre C, Pollet B, Legrand M. Silencing of hydroxycinnamoyl-coenzyme A shikimate/quinate hydroxycinnamoyltransferase affects phenylpropanoid biosynthesis. Plant Cell. 2004;16:1446–1465. doi: 10.1105/tpc.020297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann L, Maury S, Martz F, Geoffroy P, Legrand M. Purification, cloning, and properties of an acyltransferase controlling shikimate and quinate ester intermediates in phenylpropanoid metabolism. Journal of biological chemistry. 2003;278:95–103. doi: 10.1074/jbc.M209362200. [DOI] [PubMed] [Google Scholar]
- Howe GA, Jander G. Plant immunity to insect herbivores. Annual Reviews in Plant Biology. 2008;59:41–66. doi: 10.1146/annurev.arplant.59.032607.092825. [DOI] [PubMed] [Google Scholar]
- Kaur H, Heinzel N, Schottner M, Baldwin IT, Galis I. R2R3-NaMYB8 regulates the accumulation of phenylpropanoid-polyamine conjugates, which are essential for local and systemic defense against insect herbivores in Nicotiana attenuata. Plant physiology. 2010;152:1731–1747. doi: 10.1104/pp.109.151738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinanen M, Oldham NJ, Baldwin IT. Rapid HPLC screening of jasmonate-induced increases in tobacco alkaloids, phenolics, and diterpene glycosides in Nicotiana attenuata. Journal of agricultural and food chemistry. 2001;49:3553–3558. doi: 10.1021/jf010200+. [DOI] [PubMed] [Google Scholar]
- Kessler D, Diezel C, Clark DG, Colquhoun TA, Baldwin IT. Petunia flowers solve the defence/apparency dilemma of pollinator attraction by deploying complex floral blends. Ecology Letters. 2013;16:299–306. doi: 10.1111/ele.12038. [DOI] [PubMed] [Google Scholar]
- Kim SG, Yon F, Gaquerel E, Gulati J, Baldwin IT. Tissue specific diurnal rhythms of metabolites and their regulation during herbivore attack in a native tobacco, Nicotiana attenuata. PloS one. 2011;6:e26214. doi: 10.1371/journal.pone.0026214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G, Lingeman J, Colon AM, Coruzzi G, Shasha D. Gene regulatory networks in plants: learning causality from time and perturbation. Genome biology. 2013;14:123. doi: 10.1186/gb-2013-14-6-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhl C, Tautenhahn R, Bottcher C, Larson TR, Neumann S. CAMERA: an integrated strategy for compound spectra extraction and annotation of liquid chromatography/mass spectrometry data sets. Analytical chemistry. 2012;84:283–289. doi: 10.1021/ac202450g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Taylor MA, Arif SAM, Davies HV. Potato plants expressing antisense and sense S-adenosylmethionine decarboxylase (SAMDC) transgenes show altered levels of polyamines and ethylene: Antisense plants display abnormal phenotypes. Plant Journal. 1996;9:147–158. [Google Scholar]
- Kusano T, Berberich T, Tateda C, Takahashi Y. Polyamines: essential factors for growth and survival. Planta. 2008;228:367–381. doi: 10.1007/s00425-008-0772-7. [DOI] [PubMed] [Google Scholar]
- Lou YG, Baldwin IT. Nitrogen supply influences herbivore-induced direct and indirect defenses and transcriptional responses to Nicotiana attenuata. Plant physiology. 2004;135:496–506. doi: 10.1104/pp.104.040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Fuell C, Parr A, Hill L, Bailey P, Elliott K, Fairhurst SA, Martin C, Michael AJ. A novel polyamine acyltransferase responsible for the accumulation of spermidine conjugates in Arabidopsis seed. Plant Cell. 2009;21:318–333. doi: 10.1105/tpc.108.063511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado RA, Ferrieri AP, Robert CA, Glauser G, Kallenbach M, Baldwin IT, Erb M. Leaf-herbivore attack reduces carbon reserves and regrowth from the roots via jasmonate and auxin signaling. New phytologist. 2013 doi: 10.1111/nph.12438. doi: 10.1111. [DOI] [PubMed] [Google Scholar]
- Malitsky S, Blum E, Less H, Venger I, Elbaz M, Morin S, Eshed Y, Aharoni A. The transcript and metabolite networks affected by the two clades of Arabidopsis glucosinolate biosynthesis regulators. Plant physiology. 2008;148:2021–2049. doi: 10.1104/pp.108.124784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Tanguy J. The occurrence and possible function of hydroxycinnamoyl acid amines in plants. Plant Growth Regulator. 1985;3:381–399. [Google Scholar]
- Martin-Tanguy J. Conjugated polyamines and reproductive development: biochemical, molecular and physiological approaches. Physiologia Plantarum. 1997;100:675–688. [Google Scholar]
- Martin-Tanguy J, Cabanne F, Perdrizet E, Martin C. Distribution of hydroxycinnamic acid-amides in flowering plants. Phytochemistry. 1978;17:1927–1928. [Google Scholar]
- Matsuda F, Hirai MY, Sasaki E, Akiyama K, Yonekura-Sakakibara K, Provart NJ, Sakurai T, Shimada Y, Saito K. AtMetExpress Development: A phytochemical atlas of Arabidopsis Development. Plant physiology. 2010;152:566–578. doi: 10.1104/pp.109.148031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuno M, Compagnon V, Schoch GA, Schmitt M, Debayle D, Bassard JE, Pollet B, Hehn A, Heintz D, Ullmann P, Lapierre C, et al. Evolution of a novel phenolic pathway for pollen development. Science. 2009;325:1688–1692. doi: 10.1126/science.1174095. [DOI] [PubMed] [Google Scholar]
- McCloud ES, Baldwin IT. Herbivory and caterpillar regurgitants amplify the wound-induced increases in jasmonic acid but not nicotine in Nicotiana sylvestris. Planta. 1997;203:430–435. [Google Scholar]
- Mehrtens F, Kranz H, Bednarek P, Weisshaar B. The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant physiology. 2005;138:1083–1096. doi: 10.1104/pp.104.058032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muroi A, Ishihara A, Tanaka C, Ishizuka A, Takabayashi J, Miyoshi H, Nishioka T. Accumulation of hydroxycinnamic acid amides induced by pathogen infection and identification of agmatine coumaroyltransferase in Arabidopsis thaliana. Planta. 2009;230:517–527. doi: 10.1007/s00425-009-0960-0. [DOI] [PubMed] [Google Scholar]
- Negrel J. The biosynthesis of cinnamoylputrescines in callus tissue cultures of Nicotiana tabacum. Phytochemistry. 1989;28:477–481. [Google Scholar]
- Negrel J, Javelle F, Paynot M. Separation of putrescine and spermidine hydroxycinnamoyltransferases extracted from tobacco callus. Phytochemistry. 1991;30:1089–1092. [Google Scholar]
- Negrel J, Paynot M, Javelle F. Purification and properties of putrescine hydroxycinnamoyltransferase from tobacco (Nicotiana tabacum) cell suspensions. Plant Physiology. 1992;98:1264–1269. doi: 10.1104/pp.98.4.1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, von Roepenack-Lahaye E, Parr A, Daniels MJ, Dow JM. Induction of hydroxycinnamoyl-tyramine conjugates in pepper by Xanthomonas campestris, a plant defense response activated by hrp gene-dependent and hrp gene-independent mechanisms. Molecular Plant-Microbe Interactions. 2001;14:785–792. doi: 10.1094/MPMI.2001.14.6.785. [DOI] [PubMed] [Google Scholar]
- Olson-Manning CF, Lee CR, Rausher MD, Mitchell-Olds T. Evolution of flux control in the glucosinolate pathway in Arabidopsis thaliana. Molecular biology and evolution. 2013;30:14–23. doi: 10.1093/molbev/mss204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onkokesung N, Gaquerel E, Kotkar H, Kaur H, Baldwin IT, Galis I. MYB8 controls inducible phenolamide levels by activating three novel hydroxycinnamoyl-coenzyme A:polyamine transferases in Nicotiana attenuata. Plant physiology. 2012;158:389–407. doi: 10.1104/pp.111.187229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschold A, Halitschke R, Baldwin IT. Co(i)-ordinating defenses: NaCOI1 mediates herbivore- induced resistance in Nicotiana attenuata and reveals the role of herbivore movement in avoiding defenses. Plant journal. 2007;51:79–91. doi: 10.1111/j.1365-313X.2007.03119.x. [DOI] [PubMed] [Google Scholar]
- Pichersky E, Gang DR. Genetics and biochemistry of secondary metabolites in plants: an evolutionary perspective. Trends in Plant Science. 2000;5:439–445. doi: 10.1016/s1360-1385(00)01741-6. [DOI] [PubMed] [Google Scholar]
- Pichersky E, Lewinsohn E. Convergent Evolution in Plant Specialized Metabolism. Annual Reviews in Plant Biology. 2011;62:549–566. doi: 10.1146/annurev-arplant-042110-103814. [DOI] [PubMed] [Google Scholar]
- Prasad KV, Song BH, Olson-Manning C, Anderson JT, Lee CR, Schranz ME, Windsor AJ, Clauss MJ, Manzaneda AJ, Naqvi I, Reichelt M, et al. A gain-of-function polymorphism controlling complex traits and fitness in nature. Science. 2012;337:1081–1084. doi: 10.1126/science.1221636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Hirai MY, Yonekura-Sakakibara K. Decoding genes with coexpression networks and metabolomics - 'majority report by precogs'. Trends Plant Sci. 2008;13:36–43. doi: 10.1016/j.tplants.2007.10.006. [DOI] [PubMed] [Google Scholar]
- Schafer M, Brutting C, Gase K, Reichelt M, Baldwin I, Meldau S. ‘Real time’ genetic manipulation: a new tool for ecological field studies. The Plant journal. 2013;76(3):506–18. doi: 10.1111/tpj.12301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittko U, Baldwin IT. Constraints to herbivore-induced systemic responses: bidirectional signaling along orthostichies in Nicotiana attenuata. Journal of chemical ecology. 2003;29:763–770. doi: 10.1023/a:1022833022672. [DOI] [PubMed] [Google Scholar]
- Schittko U, Hermsmeier D, Baldwin IT. Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. II. Accumulation of plant mRNAs in response to insect-derived cues. Plant physiology. 2001;125:701–710. doi: 10.1104/pp.125.2.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steppuhn A, Gaquerel E, Baldwin IT. The two alpha-dox genes of Nicotiana attenuata: overlapping but distinct functions in development and stress responses. BMC plant biology. 2010;10:171. doi: 10.1186/1471-2229-10-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitz M, Baldwin IT, Gaquerel E. Diverting the flux of the JA pathway in Nicotiana attenuata compromises the plant's defense metabolism and fitness in nature and glasshouse. PloS one. 2011;6:e25925. doi: 10.1371/journal.pone.0025925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell WR. The association among gene expression responses to nine abiotic stress treatments in Arabidopsis thaliana. Genetics. 2006;174:1811–1824. doi: 10.1534/genetics.106.061374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebayashi SI, Ishihara A, Tsuda M, Iwamura H. Induction of clovamide by jasmonic acid in red clover. Phytochemistry. 2000;54:387–392. doi: 10.1016/s0031-9422(00)00098-4. [DOI] [PubMed] [Google Scholar]
- Tohge T, Watanabe M, Hoefgen R, Fernie AR. The evolution of phenylpropanoid metabolism in the green lineage. Crit Rev Biochem Mol. 2013;48:123–152. doi: 10.3109/10409238.2012.758083. [DOI] [PubMed] [Google Scholar]
- Ullmann-Zeunert L, Stanton MA, Wielsch N, Bartram S, Hummert C, Svatos A, Baldwin IT, Groten K. Quantification of growth-defense trade-offs in a common currency: nitrogen required for phenolamide biosynthesis is not derived from ribulose-1,5-bisphosphate carboxylase/oxygenase turnover. Plant journal. 2013;75:417–429. doi: 10.1111/tpj.12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R, Storme V, Vanholme B, Sundin L, Christensen JH, Goeminne G, Halpin C, Rohde A, Morreel K, Boerjan W. A systems biology view of responses to lignin biosynthesis perturbations in Arabidopsis. Plant Cell. 2012;24:3506–3529. doi: 10.1105/tpc.112.102574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt T. Phenylpropanoid Biosynthesis. Molecular Plant. 2010;3:2–20. doi: 10.1093/mp/ssp106. [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PloS one. 2007;2 doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woldemariam MG, Dinh ST, Oh Y, Gaquerel E, Baldwin IT, Galis I. NaMYC2 transcription factor regulates a subset of plant defense responses in Nicotiana attenuata. BMC plant biology. 2013;13 doi: 10.1186/1471-2229-13-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Baldwin IT. New insights into plant responses to the attack from insect herbivores. Annual Reviews in Genetics. 2010;44:1–24. doi: 10.1146/annurev-genet-102209-163500. [DOI] [PubMed] [Google Scholar]
- Zhou BY, Wong WH. A Bootstrap-Based Non-Parametric Anova Method with Applications to Factorial Microarray Data. Stat Sinica. 2011;21:495–514. [Google Scholar]
- Zhou BY, Xu WH, Herndon D, Tompkins R, Davis R, Xiao WZ, Wong WH, Injury IHR. Analysis of factorial time-course microarrays with application to a clinical study of burn injury. Proceedings of the National Academy of Sciences of the U S A. 2010;107:9923–9928. doi: 10.1073/pnas.1002757107. [DOI] [PMC free article] [PubMed] [Google Scholar]