Abstract
Molecular docking programs are primarily designed to align rigid, drug-like fragments into the binding sites of macromolecules and frequently display poor performance when applied to flexible carbohydrate molecules. A critical source of flexibility within an oligosaccharide is the glycosidic linkages. Recently, Carbohydrate Intrinsic (CHI) energy functions were reported that attempt to quantify the glycosidic torsion angle preferences. In the present work, the CHI-energy functions have been incorporated into the AutoDock Vina (ADV) scoring function, subsequently termed Vina-Carb (VC). Two user-adjustable parameters have been introduced, namely, a CHI- energy weight term (chi_coeff) that affects the magnitude of the CHI-energy penalty and a CHI-cutoff term (chi_cutoff) that negates CHI-energy penalties below a specified value. A data set consisting of 101 protein–carbohydrate complexes and 29 apoprotein structures was used in the development and testing of VC, including antibodies, lectins, and carbohydrate binding modules. Accounting for the intramolecular energies of the glycosidic linkages in the oligosaccharides during docking led VC to produce acceptable structures within the top five ranked poses in 74% of the systems tested, compared to a success rate of 55% for ADV. An enzyme system was employed in order to illustrate the potential application of VC to proteins that may distort glycosidic linkages of carbohydrate ligands upon binding. VC represents a significant step toward accurately predicting the structures of protein–carbohydrate complexes. Furthermore, the described approach is conceptually applicable to any class of ligands that populate well-defined conformational states.
Graphical Abstract
INTRODUCTION
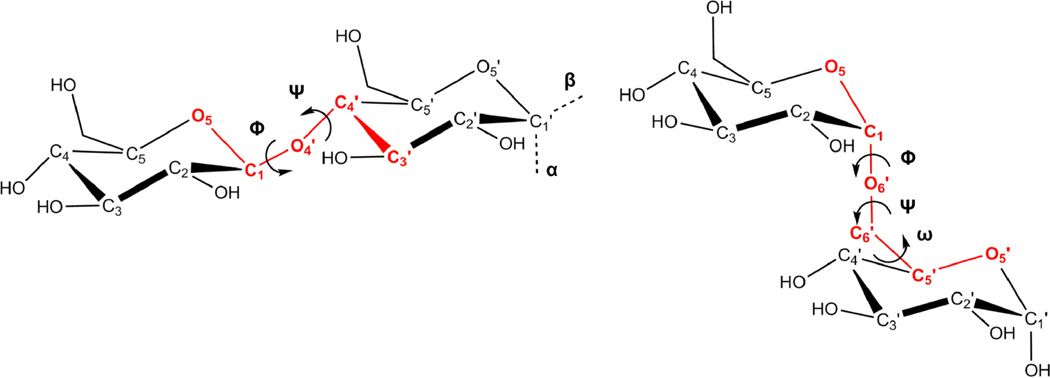
Finite carbohydrate polymers (oligosaccharides, also referred to as glycans) are involved in a range of processes that are critical for proper cellular function.1 Structural characterization of glycans and their binding partners (antibodies, lectins, carbohydrate binding modules (CBMs), enzymes, etc.) has advanced our understanding of these molecular recognition processes; however, obtaining three-dimensional structures of such complexes is particularly challenging due in part to the inherent flexibility of glycans.2 This flexibility stems primarily from the two or three rotatable bonds that connect monosaccharides, known as glycosidic linkages (Figure 1).3 Although glycosidic linkages introduce flexibility, these angles do not adopt random orientations but rather populate a well-defined subset of conformations determined by carbohydrate-specific stereoelectronic and solvent–solute interactions, such as the exo-anomeric and gauche-effects.4
Figure 1.
Disaccharides depicting the φ (O5–C1–Ox′–Cx′), ψ (C1–Ox′–Cx′–Cx−1′) and ω (O6′–C6′–C5′–O5′) glycosidic torsion angles. The C1 atom represents the anomeric carbon, and the orientation of the connecting oxygen determines whether the sugar is α or β.
Because of the challenges faced in determining the structure of carbohydrate–protein complexes using traditional experimental methods, considerable attention has been directed toward the application of theoretical protocols.5 Molecular docking aims to predict various modes of noncovalent interactions between a macromolecule and a ligand, ranking the results based on binding energies or scores.6 In general, the energy functions employed in docking are a summation of contributions from various nonbonded interactions (i.e., electrostatics, van der Waals, hydrogen bonding, and hydrophobic interactions).6,7 These empirical scoring functions are generalized for application to a wide range of ligands, and their ignorance of ligand conformational preferences often leads to unnatural glycosidic angles when applied to glycans.8 These structural distortions can be especially pronounced for large oligosaccharides, which contain a high number of degrees of internal freedom.9
Previous studies of carbohydrate docking have generated customized scoring functions by either recalibration of existing terms10 or the inclusion of additional functions that model specific features of protein–carbohydrate interactions.11 For example, the SLICK scoring function11 within BALLDock12 includes an energy term for CH/π stacking interactions and was calibrated using a set of carbohydrate–lectin complexes. Additionally, it has been shown that inclusion of explicit waters during carbohydrate docking,13 as well as the introduction of scoring functions that favor the placement of carbohydrate hydroxyl groups in water site positions,14 lead to improvements in docking accuracy. Inclusion of the conformational preferences of oligosaccharides offers an additional opportunity to enhance the efficiency and accuracy of carbohydrate docking. Recently, CHI-energy functions have been reported8 that assign relative energies to the torsion angles of the glycosidic linkages. The CHI-energy profiles corresponded with the distribution of glycosidic torsion angles in protein–carbohydrate complexes obtained from the Protein Data Bank (PDB),8 and preliminary results suggested that their inclusion in carbohydrate docking algorithms would be beneficial. The CHI-energy functions would be additive to the scoring function and are therefore transferable between docking packages.
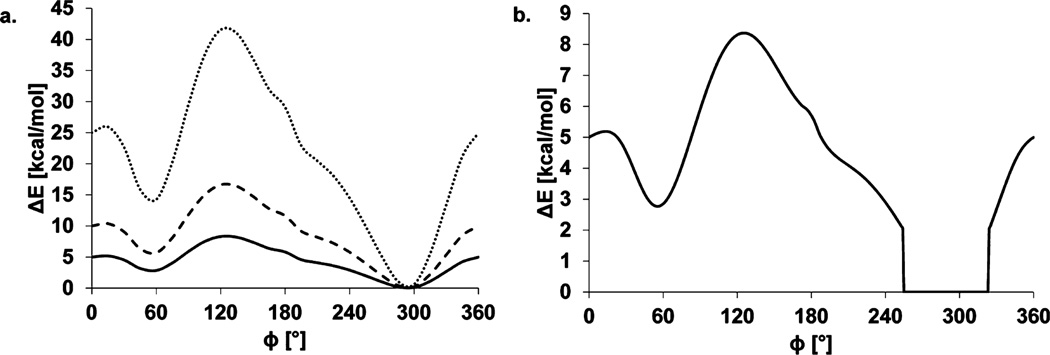
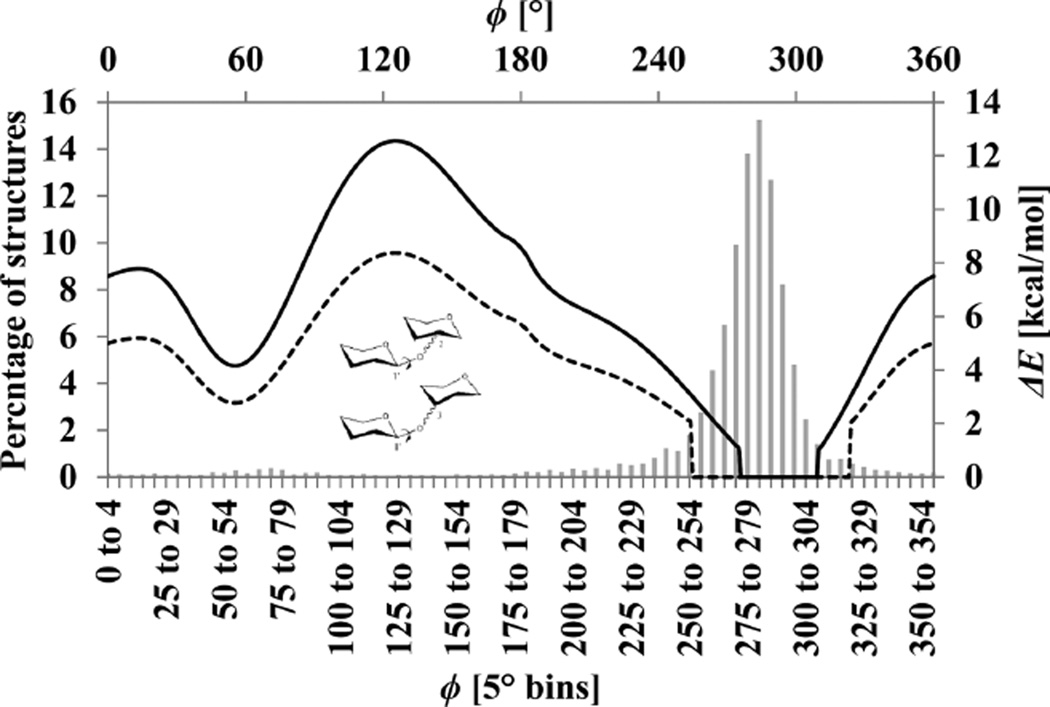
In this work, the CHI-energy functions8 have been incorporated into the AutoDock Vina 1.1.2 (ADV) scoring function9 to create Vina-Carb (VC). Additionally, new CHI-energy functions have been developed to describe the common three-bond (1–6) glycosidic linkage, as well as the 2–3 and 2–6 linkages associated with the biologically important class of monosaccharides known as sialic acids. The CHI-energy is evaluated for each oligosaccharide pose generated by VC and added to the intermolecular interaction energy. Energetically unfavorable oligosaccharide conformations are penalized, and often rejected, within the Metropolis subroutine. The user can control the CHI-energy contribution in VC by adjusting a coefficient term (chi_coeff) and an energy cutoff value (chi_cutoff) (Figure 2). Changing the chi_coeff affects the relative magnitude of the CHI-energy penalty compared to other energy terms within the ADV scoring function. A CHI-energy cutoff eliminates penalties for conformations within chi_cutoff of the minimum CHI-energy. The use of a cutoff term prevents excessive penalization of poses with conformations that deviate from the ideal, for example, due to induced fit.
Figure 2.
(a) Effect of applying CHI-coefficients of 1 (solid line), 2 (dashed line), and 5 (dotted line) to the original VC curve illustrated for the φ-glycosidic angle in a β-linkage. (b) Effect of applying a CHI-cutoff value of 2 to the original CHI-energy curve (VC1|2).
The optimum settings for the CHI-energy functions in VC were determined using a set of carbohydrate co-complexes with antibodies, lectins, and CBMs. Ligand sizes within the Development Set ranged from a disaccharide to an undecasaccharide. A test set consisting of apo-receptors from the Development Set was used to compare the optimized settings of VC against ADV for systems that lack a prearranged binding site. Finally, an application of VC to an enzyme system was demonstrated.
METHODS
File Preparation
Antibody, lectin, and CBM complexes containing carbohydrate ligands were collected from the Protein Data Bank (PDB) and employed as the Development Set for VC (see Table SI-1 for details). When duplicate protein chains were present in the PDB file, the chain with the most well-defined ligand (the ligand with the lowest average B value) was selected (Table SI-3). For the apoprotein structures employed as a Test Set, in the case that multiple protein chains were present, the one with the lowest average B value was selected. For consistency, antibodies were aligned to the Z-axis based on their CDR regions as described previously,8 and the complexes were formatted for docking with AutoDock Tools (ADT).15 During docking, the protein was kept rigid. Unless otherwise noted, the glycosidic linkages and the hydroxyl groups in the oligosaccharide were treated as flexible, while all other bonds were constrained. This protocol was chosen in order to clearly illustrate the impact of the inclusion of CHI-energies on the docking outcome.
Docking Parameters
The dimensions and centers of the grid boxes are described in the SI. The maximum number of poses was limited to 20, and the maximum energy limit set to 10 kcal/mol. The choice of chi_coeff and chi_cutoff in VC is denoted by subscript values. For example, VC performed with a chi_coeff of 2 and chi_cutoff of 4 is indicated as VC2|4.
Analysis
The results of each ADV docking experiment are variable due to the random seed implemented within the genetic algorithm. In order to account for this variation, the results from multiple independent docking experiments were averaged for each system examined. Unless otherwise stated, each root-mean-squared-deviation (RMSD) represents the average result of 10 docking runs. This method of analysis aims to eliminate spurious results, enabling a more accurate comparison between ADV and VC. To increase comparability, the same 10 random seeds generated for each of the 10 ADV docking experiments were employed for the 10 corresponding VC docking runs.
Docking accuracy is determined through two types of RMSD values, namely, those for the ligand pose and the ligand shape; all RMSDs were calculated with respect to the six atoms that define the pyranose ring (typically C1, C2, C3, C4, C5, and O5). The pose RMSD (PRMSD) quantifies the deviation of the docked model from the location of the reference structure relative to the protein surface. In this manner, the PRMSD defines the accuracy of docking the ligand to the receptor. The shape RMSD (SRMSD) compares the docked oligosaccharide conformation to that of the reference structure independent of their locations in space. PRMSDmin(5) and PRMSDmin(20) represent the minimum PRMSD from the top 5 and top 20 ranked models, respectively, averaged across 10 docking runs. The average SRMSD (SRMSDavg) was calculated for each of the 20 models from the 10 docking experiments.
Images of the molecules were prepared using the Visual Molecular Dynamics (VMD) program.16 Unless otherwise noted, the ligands are colored according to the source of the structure; crystal structures are blue, whereas output from ADV or VC are yellow and green, respectively. Additionally, each carbohydrate ring is colored according to whether the CHI-energy penalty is applied to that monosaccharide. CHI-energies are applied to monosaccharides in the 1C4 and 4C1 chair conformations, and these are colored green. Monosaccharides in any other conformations, which would be skipped by VC, are colored red.
CHI-Energy Integration in ADV
Parsing the Ligand
For a glycosidic angle to be identified by the CHI-energy scoring function of VC, the atom names for monosaccharides within the ligand file must follow established convention.1 The carbohydrate ligand file is parsed within parse_pdbqt.cpp, and information about the atoms and residues of the ligand is stored within the data structure ligand_info.
Glycosidic linkages involving (1,2), (1,3), (1,4), (1,6), (2,3), and (2,6) connections are detected by VC. Since the CHI-energy functions were originally developed for chair conformations of oligosaccharide rings, it is also necessary to determine the conformations of the residues comprising the input oligosaccharide ligand before the application of the energy functions.
Determination of Ligand Carbohydrate Ring Conformation
The ring conformations are identified based on a modified version of the Best-Fit-Four-Membered-Plane (BFMP) method.17 Selections made about the appropriate CHI-energy functions to be used for each linkage are stored in the data structures glyco_info and ligand_glyco_info. According to the BFMP method, a carbohydrate ring must fit three criteria in order to be classified as a 1C4 or 4C1 sugar, namely, the internally defined 2d5, 4d1, and 6d3 or 5d2, 1d4, and 3d6 conformations, respectively. Some protein–carbohydrate systems contain sugar rings that are only slightly distorted from the standard 4C1 and 1C4 conformations and still merit application of CHI-energy penalties. To accommodate minor conformational distortions of the carbohydrate ring, a ring is classified as a 1C4 or a 4C1 conformation if any two of the three BFMP criteria are satisfied.
Scoring Individual Ligand Poses
Each docking run consists of a certain number of heuristically determined steps. Each step is characterized by a random perturbation and a local optimization, which is followed by an evaluation of the generated pose. The random perturbation is performed by either transposing or rotating the ligand or by adjusting any of the flexible torsion angles. A new function, eval_chi, has been introduced within model.cpp that calculates the CHI-energy penalty for each ligand pose. This function uses data from ligand_glyco_info to calculate the CHI-energy for every oligosaccharide pose generated. The penalty calculated for each glycosidic torsion angle within eval_chi is modified according to the values of chi_coeff and chi_cutoff. The total CHI-energy of a given oligosaccharide is the summation of the CHI energies for each glycosidic torsion angle comprising the model and is combined with the interaction energy natively calculated by ADV within the function eval_deriv. This composite energy is implemented within the metropolis_accept function in monte_carlo.cpp to calculate the acceptance probability of each ligand pose. A ligand pose with unfavorable glycosidic torsion angles would be penalized by the application of CHI energies, thereby increasing the probability that it would be rejected.
Log File
A VC log file (called VC_log.txt) is written out for each execution of the program and contains information about the glycosidic linkages identified by the program and details about whether CHI-energy penalties were applied to each linkage.
RESULTS AND DISCUSSION
Implementation of the CHI-energy function aims to improve docking accuracy by penalizing unlikely shapes of the oligosaccharide ligand. In order to identify complexes whose docking score might be improved by correcting the ligand shape, each of the crystal structures was initially docked with the glycosidic linkages of the ligand restrained to the angles present in the co-crystal structure. Of the 119 crystal structures selected for evaluation, 12 failed to generate acceptable (PRMSD < 2 Å) poses in this initial evaluation. Failure during this step indicates that even when presented with an ideal ligand shape, other issues prevent ADV from correctly predicting the ligand pose. Therefore, these complexes were eliminated from the analysis.
Optimization of the CHI-Energy Coefficient
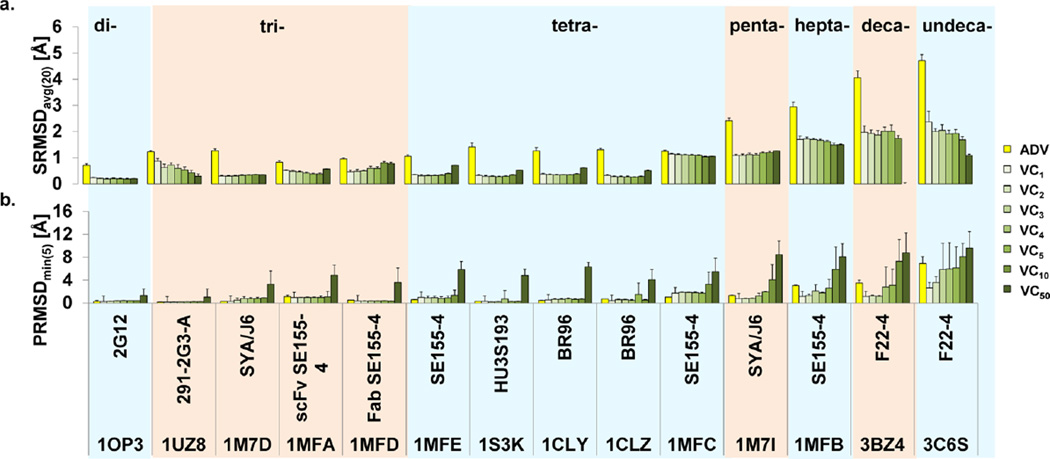
Incorporation of the CHI-energy term into the ADV scoring function immediately improved the carbohydrate conformations over those of the original program (ADV vs VC1 in Figure 3a). However, since the CHI-energy term was developed independently of the ADV scoring function, a range of CHI-energy coefficients was examined (chi_coeff = 1, 2, 3, 4, 5, 10, and 50). The effect of varying the CHI-coefficient for a set of 14 antibody–carbohydrate systems is reported in Figure 2. Each CHI-coefficient produced poses with improved ligand conformations (lower SRMSDavg(20) values) than those from ADV. The CHI-coefficient imposes a higher penalty for torsions outside of the local minima of the CHI-energy curves, thereby attenuating the production of nonideal oligosaccharide conformations during docking. Increasing the magnitude of the CHI-energy contribution generally led to a corresponding decrease in the SRMSDavg(20). This trend was particularly noticeable for systems containing more than five carbohydrate residues due to the increasing number of glycosidic linkages that were affected (Figure 2a). Interestingly, the largest CHI-coefficient (CHI50) increased the SRMSDavg(20) for ligands containing less than four carbohydrate residues. This result reflects a mild induced fit that occurred upon ligand binding, which caused the glycosidic linkages of the crystallized ligand to deviate from the theoretical minima that is over emphasized by CHI50.
Figure 3.
Assessment of docking to 14 antibody systems with ADV and various CHI-energy coefficients of VC: (a) SRMSDavg and (b) PRMSDmin(5).
Notably, the accuracy of the pose (Figure 3b) diminished as the CHI penalty became increasingly large (i.e., VC10 and VC50), despite producing ligand conformations similar to the reference structure (Figure 3a). This suggested a problem associated with pose identification. To demonstrate this, the lowest energy model generated from flexibly docking a large oligosaccharide (from PDBID = 3C6S)18 using VC50 (SRMSD = 1.13 Å; PRMSD = 23.8 Å) was rigidly redocked. Results from 10 docking experiments consistently produced an accurate model with a PRMSDmin(5) of 1.98 Å. Rigidly redocking the ligand allowed the docking scoring function to segregate poses solely based on intermolecular interactions between the protein and ligand. However, during flexible docking, the harsh penalty applied by VC50 eliminated any model that deviated from the minimum of the CHI-energy curve. Since very few of the generated models met this criterion, only those models that were unaffected by the CHI-energy penalty remained, including those positioned incorrectly. The intramolecular forces imparted by a high CHI-energy penalty appear to outweigh contributions from intermolecular interactions between the protein and ligand.
The effect of overweighting the CHI contribution showed that a balance between inter- and intramolecular interactions was required to successfully dock carbohydrate ligands. As a result, lower coefficients of the CHI-energy function produced more accurate models by enabling the generation of favorable glycosidic torsion angles without overwhelming the intermolecular energies associated with ligand binding. The performance of ADV and VC are comparable for systems containing di-, tri-, tetra-, and pentasaccharide ligands; however, VC outperforms ADV with regards to larger oligosaccharide ligands. For example, the PRMSDmin for the three largest oligosaccharides (1MFB,19 3BZ4,18 and 3C6S) generated by VC1 were lower than those produced by ADV by 1.1, 2.0, and 2.3 Å, respectively. Using VC1 and VC2 produced acceptable PRMSDmin(5) poses for 13 out of 14 systems. As a result, only CHI-coefficients of 1 or 2 were considered for subsequent experiments.
Optimization of the CHI-Energy Cutoff
The CHI-energy functions were originally developed by modeling the rotational properties of glycosidic linkages in disaccharide analogs in vacuo. The minima of the CHI-energy curves generally corresponded to crystallographically determined orientations;8 however, oligosaccharides may undergo conformational changes resulting from induced fit that could cause glycosidic linkages to deviate from idealized low energy values. Use of a flat-bottomed CHI-energy potential near the potential energy minimum (as defined by the chi_cutoff term) allows induced fit to occur with no internal energy penalty. Within this region, the pose is scored solely on the basis of the intermolecular interactions dictated by the native ADV scoring function.
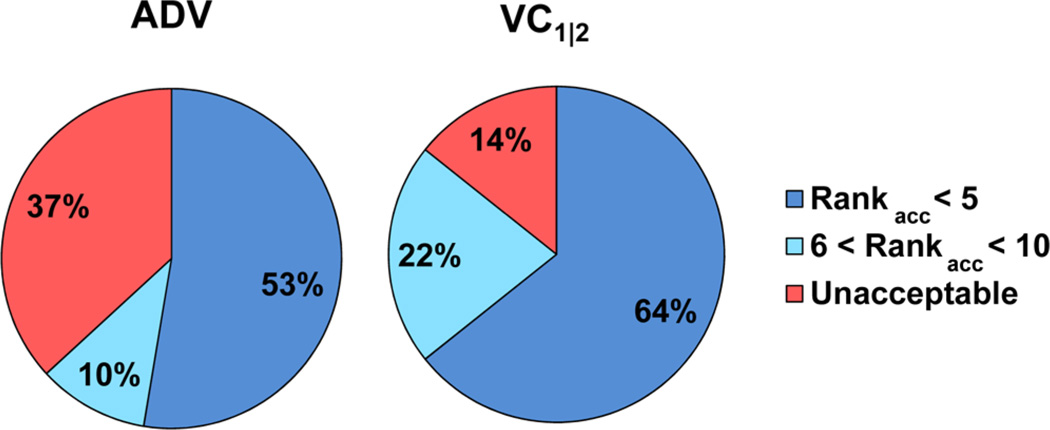
To identify the optimal setting for the chi_cutoff, a range of values were examined from 1 to 5 kcal/mol (Table SI-2). Optimal results were obtained for each CHI-coefficient (VC1 and VC2) using cutoff values of either 1 or 2 kcal/mol (VC1|1, VC1|2, VC2|1, and VC2|2). These four settings of VC generated acceptable binding modes ranked within the top 20 poses for each of the 14 antibody systems and ranked within the top 5 poses for 13 of the 14 antibodies. In order to examine the applicability of VC to protein–carbohydrate complexes other than antibody systems, as well as to further optimize the VC parameters, the study was extended to 93 additional carbohydrate–protein complexes, including CBMs, lectins, and enzymes. The best performance was attained using a CHI-coefficient of 1 and a CHI cutoff of 2 (VC1|2), which generated an acceptable pose among the top five models for 74% of the systems, compared to a 55% success rate for ADV (Table 1).
Table 1.
Comparison between ADV and VC at Four Settings of CHI-Coefficient and CHI-Cutoff
| success ratea [%] | |||||
|---|---|---|---|---|---|
| system types | ADV | VC1|1 | VC1|2 | VC2|1 | VC2|2 |
| antibodies (n = 14) | 79 | 100 | 93 | 100 | 100 |
| lectins (n = 67) | 52 | 61 | 69 | 62 | 62 |
| CBMs (n = 20) | 35 | 50 | 60 | 55 | 45 |
| averages (n = 101) | 55 | 70 | 74 | 72 | 69 |
Success is defined as producing an acceptable binding mode (PRMSDmin(5) < 2 Å).
Similar to the analysis performed in the development of the CHI-energy functions,8 carbohydrate–protein co-crystal structures in the PDB were surveyed using the GlyTorsion tool from www.glycosciences.de20 in order to calculate the percentage of glycosidic linkages exempted from penalization as a consequence of applying VC1|1, VC1|2, VC2|1, and VC2|2. At VC1|2, no CHI-energy penalty would be applied to 87% of glycosidic linkages in the PDB, compared to values of 77%, 62%, and 76% for VC1|1, VC2|1, and VC2|2, respectively (Figure 4). The VC1|2 setting avoids penalizing the widest range of experimentally determined glycosidic torsion angles. Although VC1|2 was selected as default, the alternatives (VC1|1, VC2|1, and VC2|2) were nearly as efficient in binding mode prediction (Table 1); therefore, the CHI-cutoff and CHI-coefficient parameters remain user adjustable.
Figure 4.
Comparison of the VC1|2 (dotted line) and VC2|1 (solid line) CHI-energy curve to the distribution of φ-glycosidic angles in β-glycosidic linkages in carbohydrate cocrystal structures in the PDB. The bottom X-axis and left Y-axis correspond to the histogram that depicts the distribution of PDB structures, while the top X-axis and right Y-axis correspond to the CHI-energy curves.
Although each of the 100 systems passed a positive control evaluation, in which the ligand was successfully docked while held rigidly in its experimental conformation, flexible ligand docking VC1|2 was unable to identify an acceptable pose for 26% of these systems. Challenges that may have prevented VC from identifying correct models are discussed below.
Performance with Ligands Containing (1,6) Linkages
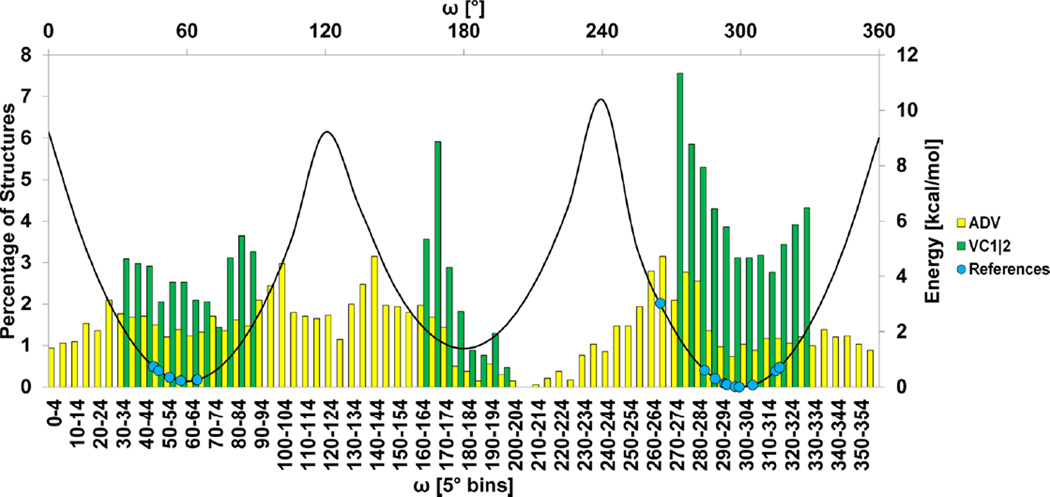
The O6–C6–C5–O5 (ω) angle in 1–6 linkages has unique conformational preferences depending on the configuration of the hydroxyl group at the adjacent C4 position (Figure SI-4.1).4b If O4 is axial, as in galactopyranose, the ω-angle typically populates each of the three staggered rotamers, with a preference for the anti-orientation. In contrast, in gluco- and mannopyranoses, in which O4 is equatorial, the ω-angle preferentially populates the two gauche rotamers. In order to enhance the accuracy of the predicted poses, this gauche effect was explicitly incorporated into VC (Figure 5). The complex interactions with the solvent and neighboring hydroxyl groups that determine the gauche effect in 1–6 linkages are not readily reproduced by gas-phase quantum mechanical calculations,4c and for this reason, the CHI-energy function employed a combination of quadratic energy terms centered at the desired rotamer torsion angles, with relative energies that corresponded approximately to rotamer populations (SI).
Figure 5.
Distribution of ω-angles produced by ADV (yellow) and VC1|2 (green) for 12 test systems containing one or more (1,6) linkages ((1,6) Glc/Man linkages) overlaid against the reference crystal structure ω-angles (blue dots) and the corresponding CHI-energy curve.
The CHI-energy functions for 1–6 linkages were applied to a test set of 12 systems (SI-4). The ability of docking to produce acceptable models among the top five poses for these systems improved from 25% for ADV to 42% with VC1|2 (Table SI-3). The CHI-energies markedly corrected the distribution of ω-angles compared to the structures generated by ADV (Figure 5.).
Performance with Ligands Containing (2,3) and (2,6) Linkages
The glycosidic torsion angles in 2–3 and 2–6 linkages (such as involving sialic acids) are unique in that there is a carboxylate group attached to the anomeric (C2) atom of the linkage. The presence of this ionic moiety complicates quantum-mechanical derivation of the rotational preferences of the φ-angle, and thus, we chose to employ combination of quadratic energy functions (SI) to capture the conformational preferences for this angle. The test set included 15 systems with ligands containing (2,3) glycosidic linkages and eight systems with ligands containing (2,6) glycosidic linkages. ADV succeeded in producing acceptable models among the top five ranked poses in 52% of the test systems, while VC1|2 was able to produce accurate pose predictions among the top five ranked structures in 70% of the test systems. The results for the systems are provided in Table SI-3.
Docking Challenges
Both ADV and VC encountered recurring difficulties that resulted from inherent issues of the docking method. However, in some cases discrepancies between the theoretical and experimental structures appeared to arise from unlikely oligosaccharide conformations in the crystallographic data. In these cases the CHI-energy functions can serve a role to identify putative errors in the experimental structures.
Excessive Carbohydrate–Protein Interactions
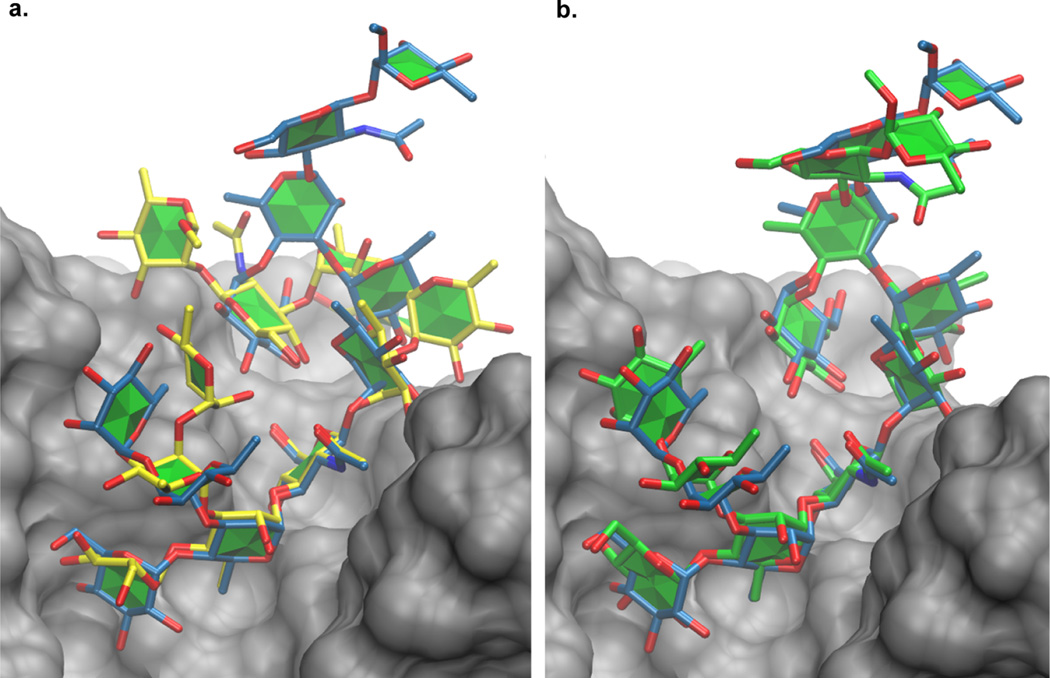
Docking scoring functions favor the formation of intermolecular interactions, with the consequence that it can be difficult for docking to correctly identify ligand poses if a large part of the ligand extends away from the protein surface.21 This can be relevant for some oligosaccharide–protein complexes. For example, both ADV and VC1|2, fail to identify an acceptable pose among the five top-ranked poses when docking the tetrasaccharide ligand to the lectin binding domain of lectinolysin (PDB ID: 4GWI22). Only one residue of the oligosaccharide completely interacts with the protein surface in the crystal structure, and the models produced during docking are unable to reproduce this orientation. Although VC1|2 produced poses similar to the crystal ligand (PRMSD = 2.2 Å), they were ranked lower than other poses that interacted more extensively with the protein surface. One approach to surmount this problem would be to dock only the component of the oligosaccharide that is in direct contact with the protein. The concept of oligosaccharide minimal binding determinant is widespread in glycobiology, where binding motifs are inferred from experimental binding data, such as glycan array screening.23 In the case that the formation of excessive carbohydrate–protein contacts is facilitated by distortion of the ligand conformation, VC improves the likelihood that the non-interacting segment will remain distal from the protein surface. For example, docking results produced by ADV and VC1|2 for the largest oligosaccharide in this test set (PDB ID: 3C6S) are displayed in Figure 6. While those residues that interact with the protein are correctly predicted in both instances, the model produced by VC1|2 better represents the positions of the solvent-exposed residues.
Figure 6.
(a) PRMSDmin(5) pose from ADV compared to the reference ligand (blue). (b) PRMSDmin(5) pose from VC1|2 compared to the reference ligand (blue).
CH-π Stacking
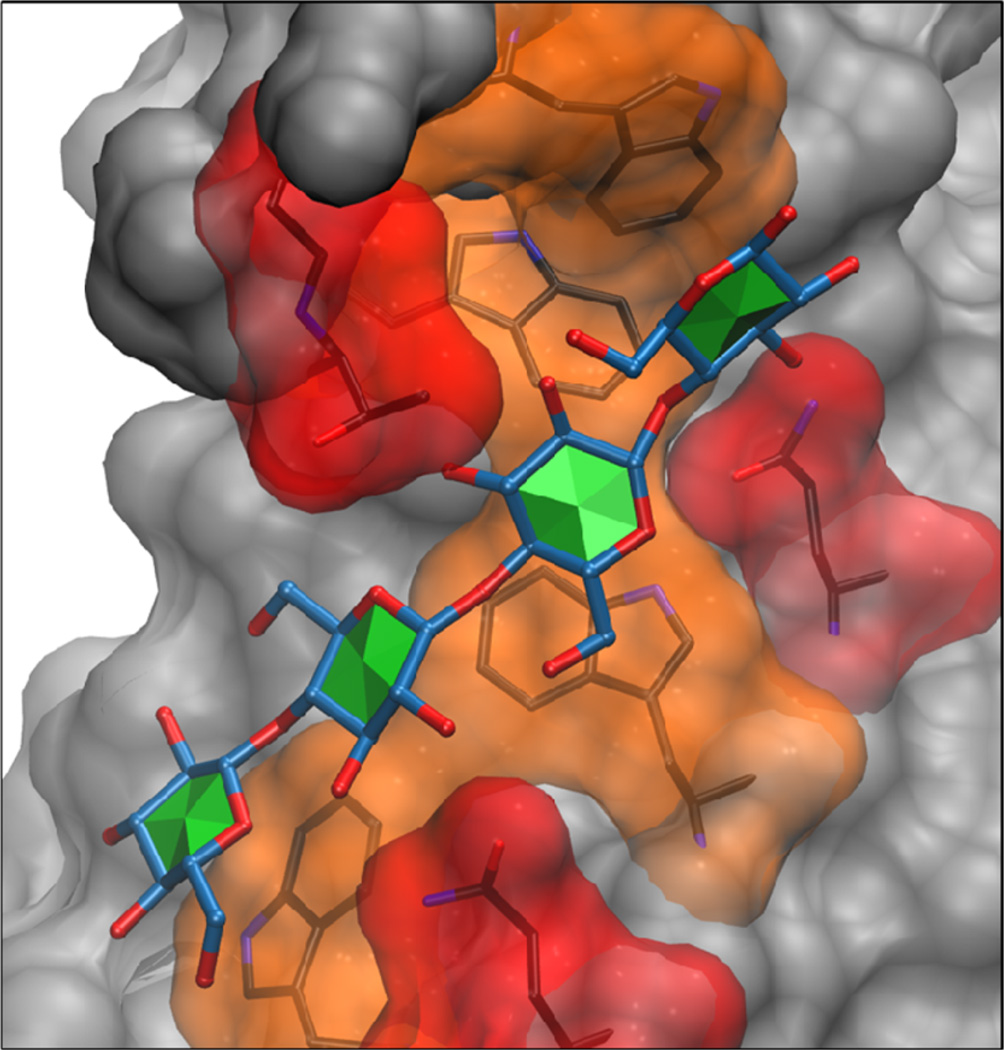
The importance of CH-π stacking between monosaccharides and aromatic rings in carbohydrate binding sites has been demonstrated experimentally;24 however, CH-π stacking interactions are currently omitted from consideration in most docking scoring functions.13,27 As a result, docking algorithms can encounter difficulties when predicting binding modes of oligosaccharides that stack against aromatic amino acids. As an example, the carbohydrate ligand in 4AFD25 stacks against four tryptophan residues (Trp 55, 60, 99, and 108) in the binding groove of the CBM. Neither ADV nor VC accurately predict the binding mode, resulting in high PRMSDmin(5) values of 8.9 and 5.4 Å, respectively (Figure 7).
Figure 7.
Crystal structure of a CBM from endoglucanase Cel5A (PDB ID: 4AFD) is depicted in complex with a tetrasaccharide ligand. All amino acids further than 5 Å away from the ligand are colored gray. Residues within 5 Å are colored orange if they are cyclic and red if acyclic.
Low-Resolution Experimental Data
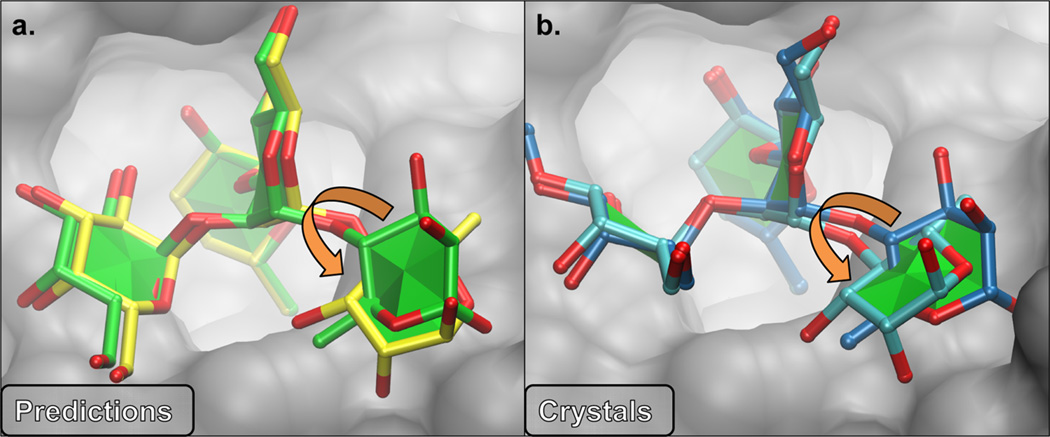
Discrepancies between docked poses and crystallographic data can also arise from errors in experimental structures of the carbohydrate ligands,26 an unfortunate outcome of the lack of adoption of carbohydrate-specific tools for crystallographic data curation.27 For example, the docking of the tetrasaccharide ligand to the Se155-4 antibody (PDB ID: 1MFC19) appeared more challenging for VC than ADV (Figure 3b); however, the results were comparable for three related ligands that have been crystallized with this antibody (PDB ID: 1MFA,28 1MFD,29 and 1MFE30). The ligands in these three systems contain the same core trisaccharide but differ by a rhamnose (Rha) residue. This extra residue is responsible for the difference in PRMSDmin(5) values between VC1|2 and ADV (Figure 8a). While the positions of three of the four residues in the individual structures closely align with one another, the pyranose ring of Rha-524 in the model produced by VC is flipped approximately 180° about the glycosidic ψ-angle, compared to the model produced by ADV. In the reported crystal structure for this complex,19 residue Rha-524 was described as “disordered,” and was placed in both the expected31 and flipped orientation in structures 1MFB and 1MFC, respectively. The ADV orientation more closely aligns with the “flipped” ligand from31 1MFC, giving rise to a low PRMSDmin(5) relative to VC, which predicts the normal conformation to be preferred. Although complexation with the protein may distort the conformation of an oligosaccharide, the preponderance of crystallographic data (Figure 8b) indicates that large distortions, such as the flip of the glycosidic ψ-angle in 1MFC, are rare.8 Thus, there is a role for the CHI-energy functions to aid in crystal structure refinement and/or curation by identifying such distorted glycosidic linkages as high energy. The resolution of each system, as well as average B factors for the ligands, are presented in Table SI-3.
Figure 8.
(a) Models representing the PRMSDmin(5) produced by docking 1MFC with ADV (yellow) and VC at CHI1|2 (green). The primary difference between docked models is a rhamnose (Rha) ring that is flipped around the ψ-angle by approximately 180°, indicated by the orange arrows. (b) Ligands from two crystal structures, 1MFB (dark blue) and 1MFC (light blue), also differ by the orientation of the Rha ring.
Assessment of ADV and VC Using a Test Set of Apoproteins
Cognate docking is useful for determining the ability of the docking algorithm to correctly place the ligand when the binding site is already preordered to receive the ligand; however, if the ultimate goal of docking is to successfully predict protein–ligand interactions in the absence of a preconfigured binding site, it is necessary to assess the performance with apoproteins. Apoprotein crystal structures were available for a subset of the co-complexes studied here and were employed as test cases to compare the performance of ADV and VC1|2. The average difference in amino acid positions between the apoproteins and corresponding cognate proteins for residues within 5 Å of the ligand was 0.77 Å, indicating relatively little induced fit in the protein surface. ADV correctly predicted the binding modes in 35% of the systems, whereas VC1|2 succeeded in 55% of the systems. If the top 20 poses were considered, instead of only the top fve, the success rates for ADV and VC1|2 increased to 55% and 83%, respectively (Table 2). VC1|2 also improved the rankings of these acceptable pose predictions (Figure 9).
Table 2.
Comparison between ADV and VC1|2 for Apoproteins Test Set
| success ratea [%] | ||||
|---|---|---|---|---|
| PRMSDmin(5) | PRMSDmin(20) | |||
| system types | ADV | VC1|2 | ADV | VC1|2 |
| antibodies (n = 7) | 71 | 86 | 71 | 100 |
| lectins (n = 10) | 50 | 50 | 70 | 90 |
| CBMs (n = 12) | 0 | 42 | 33 | 67 |
| averages | 35 | 55 | 55 | 83 |
Success rate is defined as finding an accurate binding mode (PRMSDmin < 2 Å).
Figure 9.
Depiction of the lowest-ranked acceptable pose (PRMSD ≤ 2 Å) produced by ADV and VC1|2 from docking oligosaccharide ligands onto apoprotein structures.
Evaluation of Docking to an Enzyme System Using ADV and Optimized VC
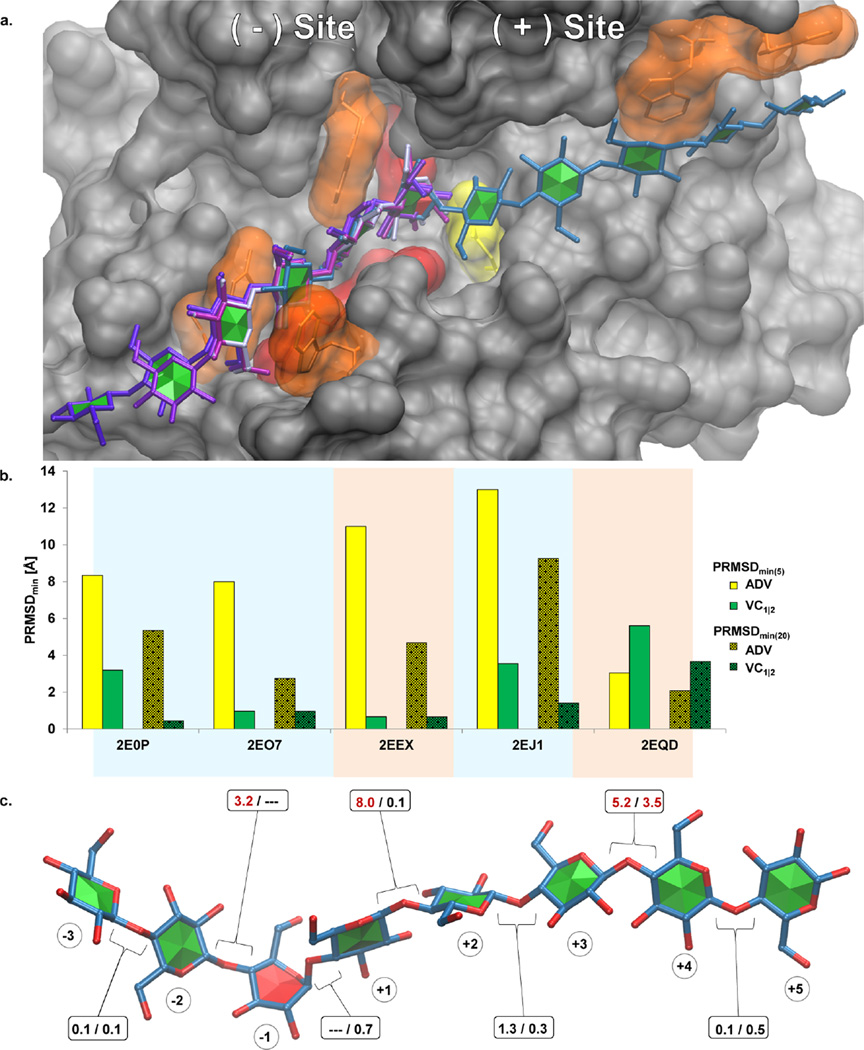
Enzyme active sites often distort monosaccharide ring shapes during catalysis, which makes docking to this class of proteins particularly challenging. As the CHI-energy functions were developed for use with low energy ring conformations, they would not necessarily be applicable to the distorted glycans found in enzyme complexes, and hence, VC is unlikely to offer considerable improvement over ADV when applied to carbohydrate-processing enzymes. An exception to this general statement is for segments of the oligosaccharides extending beyond the active site, in which case the CHI-energy functions in VC should enhance accuracy. A single example of docking to a glycoside hydrolase is presented here in order to demonstrate the potential application of VC to enzymes. Kitago et al.32 produced a series of crystal structures of the cellulase 44A (Cel44A) and a catalytic knockout, in combination with cellulosic fragments. Of the five structures produced, four of the ligands were bound to the (−) site (relative to the catalytic nucleophile), while only one contained a ligand that spanned the entire active site (PDB ID: 2EQD32) (Figure 10a). In that work, a reaction mechanism was proposed in which initial substrate binding enhanced activity through an assortment of interactions with the carbohydrate in the (−) site, while a dearth of interactions in the (+) site was suggested as promoting product release.
Figure 10.
(a) Ligands from five crystal structures (PDB ID: 2E0P, 2EO7, 2EEX, 2EJ1, and 2EQD) of the Cel44A enzyme are superimposed on the protein from PDB ID: 2EQD. Amino acids reported to be involved in substrate binding (N45, R47, W64, W71, W327, W331, E359, and W392) are colored orange or red, depending on whether the residue is cyclic or not. The catalytic residue (Q186) is yellow, and all other amino acids are gray. The active site has been separated into a (−) and (+) sites. The circled values represent the position of each residue relative to the glycosidic linkage that is cleaved during catalysis. The ligands exclusive to the (−) side of the active site are depicted by varying shades of purple. The octasaccharide that extends across both the (−) and (+) sites (2EQD) is colored blue. Each carbohydrate ring is colored according to whether the CHI-energy penalty is applied to the surrounding φ/ψ values, either green or red depending on whether VC is or is not applied, respectively. (b) Representation of the PRMSDmin(5) and PRMSDmin(20) poses from ADV and VC1|2. (c) Glycosidic linkages of the octasaccharide that extends across the active site (2EQD) are labeled according to the penalty received by the CHI-energy curve. Penalties greater than 2 kcal/mol are highlighted in red. VC is not applied to the (−1) residue since it is neither a 4C1 nor 1C4 chair.
VC successfully produced models of the complexes for the four ligands in the (−) site but failed to correctly position the largest ligand that crosses the (+) site (Figure 10b). Although ADV failed to generate a correct model for the ligands bound to the (−) site, it outperformed VC with the ligand that extends across the active site. This result is unsurprising considering the high torsional penalties that would be applied by VC to some of the glycosidic linkages within the crystal structure (Figure 10c). Although VC would not penalize the glycosidic linkage of the (−1) residue due to the nonchair ring conformations, there are other uncommon torsion values in the distal regions of the ligand. For example, the φ-linkage between residues (+1) and (+2) of the reference structure would receive a penalty of 8 kcal/mol from the CHI-energy function, precluding selection of such a model by VC.
CONCLUSIONS
The CHI-energy functions were incorporated into ADV in order to improve the performance of carbohydrate docking. Docking performance was evaluated with 101 antibody, lectin, or CBM systems. Although various CHI-energy coefficients were evaluated, the original energy profiles (chi_coeff = 1) produced accurate models with the highest frequency. An additional term that allows glycosidic torsion angles near the minimum energy to remain unpenalized (chi_cutoff = 2) enhanced the performance. Although these settings have been selected as default values, the variables remain user adjustable. VC1|2 produced accurate docked models for more systems than ADV when docking to either holo- or apoprotein receptors; however, ADV outperformed VC in a few cases where the reference ligands contained high-energy glycosidic linkages. This result suggests that accurately predicting distorted glycosidic linkages, such as those found within the active site of an enzyme, would be difficult for VC. Nevertheless, results from docking to a cellulase demonstrate the potential application of VC toward predicting enzyme–glycan interactions.
There were a few commonalities within the systems that neither ADV nor VC could accurately reproduce. Ligands that partially extend into solution were difficult to reproduce. For these ligands, docking only those parts of the ligand that are expected to interact with the protein may produce better results. A few other systems were identified, which may benefit from a term that accounts for aromatic stacking. Finally, a few low-resolution crystal structures were identified, which contained ambiguous coordinates for the reference ligands, indicating a potential role for the CHI-energy functions as a validation technique for crystallographic models.
Supplementary Material
Acknowledgments
The authors are grateful to the National Institutes of Health (GM094919 (EUREKA), R01 GM100058, and P41 GM103390) for financial support.
Footnotes
ASSOCIATED CONTENT
Supporting Information
- Information as mentioned in the text. (PDF)
Accession Codes
The source code for VC is freely available at www.glycam.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Varki A, Cummings R, Esko J, Freeze H, Hart G, Marth J. Essentials of Glycobiology. New York: Cold Spring Harbor Laboratory Press; 1999. [PubMed] [Google Scholar]
- 2.DeMarco ML, Woods RJ. Structural Glycobiology: A Game of Snakes and Ladders. Glycobiology. 2008;18(6):426–440. doi: 10.1093/glycob/cwn026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Imberty A. Oligosaccharide Structures: Theory Versus Experiment. Curr. Opin. Struct. Biol. 1997;7(5):617–623. doi: 10.1016/s0959-440x(97)80069-3. [DOI] [PubMed] [Google Scholar]
- 4.(a) Lemieux RU, Koto S, Voisin D. The Exo-Anomeric Effect. In: Szarek WA, Horton D, editors. Anomeric Effect. Origin and Consequences. Vol. 87. Washington, DC: American Chemical Society; 1979. pp. 17–29. [Google Scholar]; (b) Wolfe S. Gauche Effect. Some Stereochemical Consequences of Adjacent Electron Pairs and Polar Bonds. Acc. Chem. Res. 1972;5(3):102–111. [Google Scholar]; (c) Kirschner KN, Woods RJ. Solvent Interactions Determine Carbohydrate Conformation. Proc. Natl. Acad. Sci. U. S. A. 2001;98(19):10541–10545. doi: 10.1073/pnas.191362798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grant OC, Woods RJ. Recent advances in employing molecular modelling to determine the specificity of glycan-binding proteins. Curr. Opin. Struct. Biol. 2014;28:47–55. doi: 10.1016/j.sbi.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Halperin I, Ma B, Wolfson H, Nussinov R. Principles of Docking: An Overview of Search Algorithms and a Guide to Scoring Functions. Proteins: Struct., Funct., Genet. 2002;47:409–443. doi: 10.1002/prot.10115. [DOI] [PubMed] [Google Scholar]
- 7.Schulz-Gasch T, Stahl M. Scoring functions for protein–ligand interactions: a critical perspective. Drug Discovery Today: Technol. 2004;1(3):231–239. doi: 10.1016/j.ddtec.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 8.Nivedha AK, Makeneni S, Foley BL, Tessier MB, Woods RJ. Importance of ligand conformational energies in carbohydrate docking: Sorting the wheat from the chaff. J. Comput. Chem. 2014;35(7):526–539. doi: 10.1002/jcc.23517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trott O, Olson AJ. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization and Multithreading. J. Comput. Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laederach A, Reilly PJ. Specific Empirical Free Energy Function for Automated Docking of Carbohydrates to Proteins. J. Comput. Chem. 2003;24(14):1748–1757. doi: 10.1002/jcc.10288. [DOI] [PubMed] [Google Scholar]
- 11.Kerzmann A, Neumann D, Kohlbacher O. SLICK – Scoring and Energy Functions for Protein–Carbohydrate Interactions. J. Chem. Inf. Model. 2006;46(4):1635–1642. doi: 10.1021/ci050422y. [DOI] [PubMed] [Google Scholar]
- 12.Kerzmann A, Fuhrmann J, Kohlbacher O, Neumann D. BALLDock/SLICK: A new method for protein-carbohydrate docking. J. Chem. Inf. Model. 2008;48(8):1616–1625. doi: 10.1021/ci800103u. [DOI] [PubMed] [Google Scholar]
- 13.Samsonov SA, Teyra J, Pisabarro MT. Docking Glycosaminoglycans to Proteins: Analysis of Solvent Inclusion. J. Comput.-Aided Mol. Des. 2011;25(5):477–489. doi: 10.1007/s10822-011-9433-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gauto DF, Petruk AA, Modenutti CP, Blanco JI, Di Lella S, Martí MA. Solvent structure improves docking prediction in lectin–carbohydrate complexes. Glycobiology. 2013;23(2):241–258. doi: 10.1093/glycob/cws147. [DOI] [PubMed] [Google Scholar]
- 15.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. Autodock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexiblity. J. Comput. Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Humphrey W, Dalke A, Schulten K. VMD - Visual Molecular Dynamics. J. Mol. Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 17.Makeneni S, Foley BL, Woods RJ. BFMP: A Method for Discretizing and Visualizing Pyranose Conformations. J. Chem. Inf. Model. 2014;54(10):2744–2750. doi: 10.1021/ci500325b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vulliez-Le Normand B, Saul FA, Phalipon A, Bélot F, Guerreiro C, Mulard LA, Bentley GA. Structures of synthetic Oantigen fragments from serotype 2a Shigella flexneri in complex with a protective monoclonal antibody. Proc. Natl. Acad. Sci. U. S. A. 2008;105(29):9976–9981. doi: 10.1073/pnas.0801711105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cygler M, Wu S, Zdanov A, Bundle DR, Rose DR. Recognition of a carbohydrate antigenic determinant of Salmonella by an antibody. Biochem. Soc. Trans. 1993;21(2):437–441. doi: 10.1042/bst0210437. [DOI] [PubMed] [Google Scholar]
- 20.Lütteke T, Bohne-Lang A, Loss A, Goetz T, Frank M, von der Lieth C-W. GLYCOSCIENCES. de: an Internet portal to support glycomics and glycobiology research. Glycobiology. 2006;16(5):71R–81R. doi: 10.1093/glycob/cwj049. [DOI] [PubMed] [Google Scholar]
- 21.(a) Sternberg MJE. Docking Ligands to Proteins. In: Sternberg MJE, editor. Protein Structure Prediction: A Practical Approach. 1st. Vol. 170. New York: Oxford University Press, Inc.; 1996. pp. 263–290. [Google Scholar]; (b) Schwede T, Manuel CP. Small Molecule Docking. In: Schwede T, Manuel CP, editors. Computational Structural Biology: Methods and Applications. Singapore: World Scientific Publishing Co. Pte., Ltd.; 2008. pp. 469–500. [Google Scholar]
- 22.Lawrence S, Feil S, Holien J, Kuiper M, Doughty L, Dolezal O, Mulhern T, Tweten R, Parker M. Manipulating the Lewis antigen specificity of the cholesterol-dependent cytolysin lectinolysin. Front. Immunol. 2012 doi: 10.3389/fimmu.2012.00330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.(a) Oyelaran O, Gildersleeve JC. Glycan Arrays: Recent Advances and Future Challenges. Curr. Opin. Chem. Biol. 2009;13(4):406–413. doi: 10.1016/j.cbpa.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Taylor ME, Drickamer K. Structural Insights into what Glycan Arrays tell us About how Glycan-binding Proteins Interact with their Ligands. Glycobiology. 2009;19(11):1155–1162. doi: 10.1093/glycob/cwp076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muraki M, Morikawa M, Jigami Y, Tanaka H. The roles of conserved aromatic amino-acid residues in the active site of human lysozyme: a site-specific mutagenesis study. Biochim. Biophys. Acta, Protein Struct. Mol. Enzymol. 1987;916(1):66–75. doi: 10.1016/0167-4838(87)90211-1. [DOI] [PubMed] [Google Scholar]
- 25.Luís AS, Venditto I, Temple MJ, Rogowski A, Baslé A, Xue J, Knox JP, Prates JA, Ferreira LM, Fontes CM, Najmudin S, Gilbert HJ. Understanding how noncatalytic carbohydrate binding modules can display specificity for xyloglucan. J. Biol. Chem. 2013;288(7):4799–4809. doi: 10.1074/jbc.M112.432781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lötteke T, Von der Lieth C-W. pdb-care (PDB CArbohydrate REsidue check): a Program to Support Annotation of Complex Carbohydrate Structures in PDB Files. BMC Bioinformatics. 2004;5(69):1–6. doi: 10.1186/1471-2105-5-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agirre J, Davies G, Wilson K, Cowtan K. Carbohydrate anomalies in the PDB. Nat. Chem. Biol. 2015;11(5):303–303. doi: 10.1038/nchembio.1798. [DOI] [PubMed] [Google Scholar]
- 28.Zdanov A, Li Y, Bundle DR, Deng S-J, MacKenzie CR, Narang SA, Young NM, Cygler M. Structure of a Single-Chain Antibody Variable Domain (Fv) Fragment Complexed with a Carbohydrate Antigen a 1.7-Å Resolution. Proc. Natl. Acad. Sci. U. S. A. 1994 Jul;91:6423–6427. doi: 10.1073/pnas.91.14.6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bundle DR, Baumann H, Brisson J-R, Gagné SM, Zdanov A, Cygler M. Solution Structure of a Trisaccharide-Antibody Complex: Comparison of NMR Measurements with a Crystal Structure. Biochemistry. 1994;33:5183–5192. doi: 10.1021/bi00183a023. [DOI] [PubMed] [Google Scholar]
- 30.Cygler M, Rose DR, Bundle DR. Recognition of a Cell-Surface Oligosaccharide of Pathogenic Salmonella by an Antibody Fab Fragment. Science. 1991;253:442–445. doi: 10.1126/science.1713710. [DOI] [PubMed] [Google Scholar]
- 31.Kirschner KN, Yongye AB, Tschampel SM, González-Outeiriño J, Daniels CR, Foley BL, Woods RJ. GLYCAM06: A Generalizable Biomolecular Force Field. Carbohydrates. J. Comput. Chem. 2008;29(4):622–655. doi: 10.1002/jcc.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kitago Y, Karita S, Watanabe N, Kamiya M, Aizawa T, Sakka K, Tanaka I. Crystal structure of Cel44A, a glycoside hydrolase family 44 endoglucanase from Clostridium thermocellum. J. Biol. Chem. 2007;282(49):35703–35711. doi: 10.1074/jbc.M706835200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.