Abstract
High-throughput sequencing technologies have allowed many gene locus–level molecular biology assays to become genome-wide profiling methods. DNA-cleaving enzymes such as DNase I have been used to probe accessible chromatin. The accessible regions contain functional regulatory sites, including promoters, insulators and enhancers. Deep sequencing of DNase-seq libraries and computational analysis of the cut profiles have been used to infer protein occupancy in the genome at the nucleotide level, a method introduced as ‘digital genomic footprinting’. The approach has been proposed as an attractive alternative to the analysis of transcription factors (TFs) by chromatin immunoprecipitation followed by sequencing (ChIP-seq), and in theory it should overcome antibody issues, poor resolution and batch effects. Recent reports point to limitations of the DNase-based genomic footprinting approach and call into question the scope of detectable protein occupancy, especially for TFs with short-lived chromatin binding. The genomics community is grappling with issues concerning the utility of genomic footprinting and is reassessing the proposed approaches in terms of robust deliverables. Here we summarize the consensus as well as different views emerging from recent reports, and we describe the remaining issues and hurdles for genomic footprinting.
Interactions between DNA-binding proteins and specific sites on the template have been studied for many years via the ‘footprinting’ technique, wherein short regions of the DNA corresponding to a binding motif for the protein are found to be selectively resistant to digestion by nonspecific DNA nucleases. The technique originally displayed these regions as a ‘window of protection’ in a high-resolution sequencing gel. The method has been extensively exploited for in vitro studies with purified proteins and DNA1, as well as for characterization of in vivo binding sites2,3.
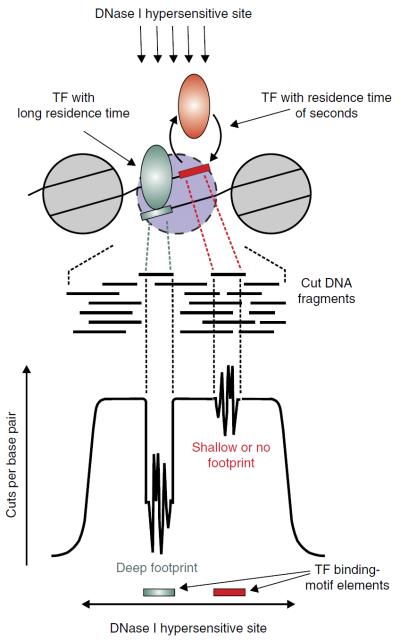
The methodology has now been extended to genome-wide analyses of TF binding events in large cell populations. This approach, termed digital genomic footprinting, involves analysis of deep-sequenced DNase-seq data4 and was initially applied to the yeast Saccharomyces cerevisiae for global identification of TF binding sites in the genome. DNase-seq analysis is most often conducted at a large scale to reveal active regulatory regions, which reside mostly in accessible chromatin and are observed as cell type–specific DNase hypersensitive sites5–8 (DHSs) with sizes varying from 200 bp to 1 kb or larger. In digital genomic footprinting, a higher-resolution computational analysis is performed for each DHS (Fig. 1). In the simplest interpretation of genomic footprinting data, the presence of nuclease protection at a binding motif is assumed to correspond to factor occupancy of that site and protection from attack by steric blockage of the nuclease. Recent findings, however, suggest that the interpretation of footprints is more complex9,10. Here we discuss the current understanding of the biophysical mechanisms involved in footprint detection.
Figure 1.
DHSs versus TF footprints. An accessible regulatory chromatin region is identified as a DHS enriched for sequencing reads in DNase-seq data. In the DHS, one or more narrow regions may be detected as putative TF footprints with evidence of local protection from DNase cleavage. The identity of TFs is inferred from the sequence patterns corresponding to the protected regions. Represented here are two example TFs with different DNA-binding dynamics that influence the degree of protection from DNase cleavage at the binding sites. The DNase cut signatures can be present over motif elements with deep, very shallow or no footprints.
From the yeast proof of principle to mammalian applications
The extension of footprinting to the human genome came a few years after the pioneering yeast study11–14. With the improvement of sequencing technology and the decreasing cost of ultra-deep sequencing, studies were able to demonstrate the feasibility and the potential of genomic footprinting for a mammalian genome. This was not a trivial task, as the human genome (GRCh37 assembly, 3.1 Gb) is 258 times the size of the S. cerevisiae genome (sacCer3 assembly, 12 Mb) in terms of the number of nucleotides. In other words, the sequencing coverage is reduced by 1/258 in principle for the same number of uniquely mapped reads. In addition, a large proportion of mammalian genomes include intergenic regions with sparsely located regulatory sites for which DNase intensities are generally lower than those for promoter-proximal DHSs. The lower numbers of DNase cleavage events in these distal sites make de novo detection of footprints more difficult, as a statistical detection algorithm looks for depletion of cuts relative to the flanking loci. Nonetheless, the first human studies reported examples of TF footprinting on chromatin observed as motif elements protected from DNase cleavage. Despite the sparse distribution of individual cuts over a mammalian genome, clear patterns of protection could be delineated from the DNase cut count profiles averaged over a large number (hundreds or thousands) of cognate binding elements. Concomitantly elevated conservation scores at putative footprints were noted as indirect evidence in support of the functional relevance of detected elements4,12. Although the analyses often focused on descriptive correlations of footprints with occupancy from ChIP-seq data, a rigorous footprint-based prediction of TF binding had been attempted, at least for a few clear-cut cases11,14. The predictive capacity of footprints was objectively assessed for a wider array of TFs in a recent study that included nine TFs representing diverse protein families9.
The early publications on genomic TF footprinting were from the two groups that developed the DNase-based chromatin assay into a genome-wide profiling method; the methodology was not widely adopted in the epigenomics community until 2014. Individual laboratories faced several challenges to the adaptation of DNase-based genomic footprinting, including optimization of the digestion protocol and deep sequencing. The computational handling of deeply sequenced DNase-seq data and proper statistical analysis of footprints necessitate bioinformatics tools. Several computational methods have been introduced in the past few years, some of which are now publicly available9,11,12,15–19. They can be classified broadly into two groups: methods that make predictions only for individual motif elements on the basis of known TF binding specificities11,15,16,18, and methods that can be used to predict footprints on the basis of DNase-seq cut count data and subsequently match the putative footprints with TFs9,12,17,19. The latter approach has the potential to reveal novel TF binding motifs. We refer readers to reports on the comparison of these computational methods9,17 and focus here on the assay-inherent aspects of TF footprinting.
Correspondence between genomic footprints and crystal structures?
One of the features presented by the authors of the first report on digital genomic footprinting is the correspondence between DNase-cleaved nucleotides and those unprotected by bound proteins (and exposed to DNase cleavage) in crystal structures of protein-DNA interactions for a few yeast and human TFs4,12. Such a precise ‘cut signature’ seemed to fit the concept of a protein stably occupying a genomic site, validating the premise of genomic footprinting.
The significance of cut signatures matching the exposed nucleotides in crystal structures was brought into question when DNase-seq data from cleavage of deproteinized, ‘naked’ DNA were generated, providing a proper negative control for protein-induced patterns20. Reanalysis of these data—specifically, of sequence patterns around cleavage sites on naked DNA— revealed the previously unappreciated sequence specificity of DNase-DNA interaction and the consequent bias in DNase-based sampling of chromatin9,10. This newly found property directly violates the original assumption behind DNase-based footprinting, namely, the sequence-nonspecific uniform cutting of DNA in any accessible chromatin. The sequence bias of the DNase enzyme is strong enough to be observed in actual DNase-seq data from chromatin of diverse species9.
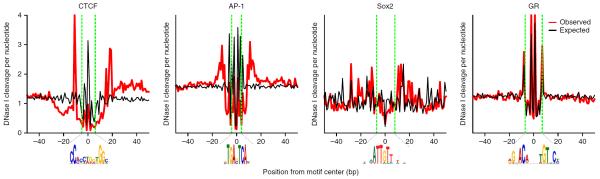
Importantly, the sequence bias has a striking effect on the average cut profile over a specific TF binding motif, which has been the preferred type of data for comparing cut signatures to crystal structures. Several independent groups have reported that the observed cut signatures in motif elements for 15 factors mostly reflect the sequence bias of the DNase enzyme9,10,21 (Fig. 2). Those studies demonstrated that the lack of cutting at the presumably protein-protected nucleotides results instead from the DNA sequence patterns disfavored by DNase I. By the same token, the spikes in the cut signatures arise from the DNA sequences preferably cleaved by DNase I. However, this does not exclude the possibility that the extent to which the sequence bias determines the cut signatures depends on the TF, individual genomic loci or the cell type. Further evidence for enzyme-inherent sequence biases came when other DNA-cleaving enzymes were used to generate genomic data and the resulting cut signatures were compared to those obtained with DNase I9,22. The cut signatures over TF binding motifs were shown to depend on the enzyme used to cleave chromatin DNA, for multiple TFs. Taken together, these findings indicate that the DNase cut signatures over TF binding motifs are highly influenced by the DNA sequence and the choice of cleaving enzyme, and thus contain little information about the identity of the occupying protein or the presence or absence thereof. Therefore, a footprint detection algorithm that relies on the shape of the cut signatures can be expected to be more prone to calling sequence bias–induced false positives.
Figure 2.
Comparison of observed and DNA-intrinsic cut profiles averaged over motif elements bound by TFs. The observed cut profiles were computed using the average raw DNase cut counts over the cognate motif elements bound by the TF (in ChIP-seq peaks). The expected profiles were generated as defined by Stergachis et al.40—that is, by taking the average DNA hexamer frequencies from the cut counts in naked DNA digestion data20 using the hexamers centered at each base-pair position. The CTCF, AP-1 and glucocorticoid receptor (GR) profiles were generated using DNase-seq and ChIP-seq data from mouse mammary cell line 3134 (ref. 8). The Sox2 profile for mouse embryonic stem cells was generated using the ENCODE DNase-seq (University of Washington) data5,46 and ChIP-exo (chromatin immunoprecipitation followed by exonuclease digestion) data36. PWMs of CTCF, AP-1 and GR were derived by motif discovery in ChIP-seq peaks using the MEME software47. The Sox2 PWM was obtained from UniPROBE (http://the_brain.bwh.harvard.edu/uniprobe) using the homologous motif entry (SOX18_PRIMARY) for Sox18. We used FIMO48 with a P value of <10−4 to scan the mouse reference genome (NCBI 37/mm9) and find the motif elements for each PWM.
Depth of footprints and TF binding dynamics
What then might be other relevant features in cut profiles that potentially indicate protein occupancy? A clue was found in the differential ‘depth’ of footprints observed from TFs spanning a wide range of DNA-binding behaviors. The average footprint depth of a protein can be determined from the relative difference between the overall levels of cuts in the DNA motif element versus in the surrounding genomic region (Figs. 2 and 3). We noted that the protein CTCF, frequently used in footprint analyses, has a long DNA residence time23. CTCF indeed confers strong protection from DNase cleavage and deep footprints in cut profiles over bound motif elements, presumably by stably binding to chromatin9,10,14. However, most chromatin factors do not have such long-lasting binding on target DNA elements. A compilation of TFs with in vivo DNA-binding dynamics data available in the literature (Table 1) suggests that their footprint depths correlate with the reported residence time of DNA binding9 (Figs. 2 and 3). The footprints of proteins that bind transiently to chromatin DNA targets are shallow and barely detectable, which is explained by the small fraction of cells in assayed populations that have such proteins bound to their DNA elements. For proteins with in vivo binding residence times of ~10 s, at any given moment the proteins are in the ‘off’ state of the binding cycle in the majority of cells in the assayed population, thereby providing the target DNA with little protection from DNase cleavage. Different microscopy methods have complementary limitations and strengths (Box 1). Fluorescence recovery after photobleaching (FRAP) has been broadly applied to numerous TFs but cannot resolve individual binding sites, whereas single-molecule tracking (SMT) directly measures binding dynamics at individual sites but is technically demanding. The reported in vivo TF binding dynamics are concordant among multiple orthogonal microscopy methods for TFs with available data. Interestingly, a nonmicroscopy method with the capability to resolve binding at specific genomic loci has also shown transient binding of TFs in vivo in yeast cells24.
Figure 3.
DNase sequence-bias-corrected profiles showing the correlation between footprint depth and TF binding residence time in vivo. The TFs are ordered by their reported DNA binding residence times from single-molecule microscopy studies. For each TF, the ratio of observed to expected profiles (Fig. 2) is plotted on the log2 scale. Black dashed lines mark where the observed cut levels are the same as the expected. Dark red dashed lines show the depth of the footprint induced by the TF (the 10% trimmed mean of the log-ratio profile over the motif region extended by 2 bp in both directions). The difference between the two dashed lines in each graph represents the footprint depth, which is smaller for TFs with shorter residence times.
Table 1.
Available data on TF binding dynamics in vivo and footprint depths
| TF | Live cell microscopy data | Epigenomics data |
|---|---|---|
| CTCF | FRAP recovery in ~11 min (ref. 23) | ENCODE ChIP-seq and DNase-seq for multiple cell types46 |
| Rap1 | Slow dynamics estimated from competition ChIP50 | ChIP-seq50 and DNase-seq4 for yeast |
| AP-1 | FRAP recovery of c-Jun AP-1 in ~10 min (ref. 51) | ENCODE ChIP-seq and DNase-seq46 |
| CREB1 | FRAP recovery in ~2 min (ref. 52) | ENCODE ChIP-seq and DNase-seq46 |
| Glucocorticoid receptor (GR) | FRAP, fluorescent correlation microscopy and SMT residence times of ~8 s (refs. 31,33,34) |
ChIP-seq and DNase-seq for mouse mammary epithelial cell line8 |
| Estrogen receptor | FRAP37 and SMT34, similar to GR | ChIP-seq and DNase-seq for MCF-7 (ref. 53) |
| Thyroid receptor | Not available | ChIP-seq and DNase-seq for mouse liver54 |
| Androgen receptor | FRAP recovery in ~40 s (ref. 55) | ChIP-seq56 and DNase-seq10 for LNCaP cell line |
| Mineralocorticoid receptor | FRAP57 and SMT33, similar to GR | ChIP-seq58 and DNase-seq46 for human kidney |
| p53 | SMT residence time of ~4 s (ref. 31) | Cell type–matched ChIP-seq unavailable for published DNase-seq |
| Sox2 | SMT residence time of ~12 s (ref. 36) | DNase-seq46 and ChIP-exo36 in embryonic stem cells |
| NF-κB | FRAP recovery in 20 s, similar to GR38 | Cell type–matched ChIP-seq unavailable for published DNase-seq |
BOX 1. Description of terms.
In SMT (single-molecule tracking) experiments, fluorescently labeled TF molecules in the nuclei of living cells are monitored in successive time frames. On the basis of the dwell times of individual molecules, specifically bound (longer dwell time) and nonspecifically bound (short dwell time) molecules are delineated. Bound fractions and residence times (of specifically bound molecules) are calculated directly from the data.
In FCS (fluorescent correlation spectroscopy), fluctuations of fluorescent intensity in a diffraction-limited spot arise from fluorescently labeled TF molecules entering and leaving the volume during the observation time. The plot of autocorrelation versus time delay is analyzed with a kinetic model of TF binding and diffusion to estimate the in vivo binding parameters.
In FRAP (fluorescence recovery after photobleaching) experiments, fluorophores in a small region inside the nucleus are photo-bleached with a strong laser, and subsequent recovery of fluorescent intensity in the bleached region is monitored. Kinetic models of TF binding and diffusion can be used to estimate in vivo binding parameters such as residence time.
Our finding that some dynamic TFs do not leave footprints9, which is supported by the work of Brown, Liu and colleagues10, is a step toward the reconciliation of divergent views of TF-genome interactions, which have arisen largely from the use of two distinct approaches: the rapidly growing epigenomics research and the technology-intensive live cell microscopy–based analysis of TF dynamics. The explosive accumulation of epigenomic data has been fueled by recent breakthroughs in high-throughput sequencing and has provided insight into the functional regulation of the genome at an unprecedented pace. However, it is often tempting to interpret data without considering the full implications of a major caveat: each chromatin sample comes from a large population of cells, and the inevitable heterogeneity is masked in the aggregated genomic data25,26. Dynamic processes and consequent effects are lost in such bulk assays, and multiple equally plausible scenarios cannot be distinguished27–30. Interpretation of population-based epigenomic data should always take into consideration the fact that individual cells in the population are in different states of any dynamic process at any given moment, and thus the observed profile is an aggregate snapshot of such states.
Footprintless TF binding at cognate motifs is not necessarily weak
Alternative explanations for TF binding sites without footprints have been provided (e.g., these may be low-affinity or weak binding sites)14,21. In some of these discussions, there has been confusion as to what is meant by a “binding site without a footprint”. We define this as a TF binding-motif element in a ChIP-seq peak where no footprints are detected from the DNase-seq data, provided that the ChIP-seq and DNase-seq data are from the same cell type. Such a motif-centric definition excludes indirect binding events, which may indeed produce weaker ChIP signal, as tethered sites do not have cognate binding-motif elements. The relevant question then becomes whether ‘binding strength’ is weaker at motif elements with no footprints than at those accompanying detectable footprints.
A general correlation between footprinting and ChIP-seq signal intensity has been presented visually at the level of motif elements12. However, our quantitative assessment showed that qualitative visual representation and best-fit curves can be misleading and that there is little or no correlation between footprint depth and ChIP-seq signal intensity9. A prime example of this is the nuclear receptor family of proteins with binding residence times of about 10 s at specific DNA targets in vivo31–34 that, according to our report9 and that of Brown, Liu and coworkers10, do not exhibit footprints. Their ChIP-seq signal intensities, however, reach very high values because the rapid sequential binding to DNA is captured during a long cross-linking step, contributing to the ChIP signal24.
Furthermore, functional regulation of target genes by TFs does not require stable long-lived or high-affinity DNA binding35. Numerous studies have shown that transient binding of TFs without footprints leads to robust induction of target genes for a diverse array of TFs31,36–39. These data contradict the notion that TF binding that does not leave footprints is weak or functionally insignificant.
Prerequisites for improved genomic footprinting
Arguably the most ambitious application of the TF occupancy information gleaned from DNase-seq has been the unbiased and comprehensive construction of mammalian TF regulatory networks12,40,41, but several serious problems still need to be solved to ensure the validity and completeness of genomic footprinting data.
Reproducibility of detected footprints: It is still unclear how reproducible detected footprints are from replicate to replicate. Because of the size of a mammalian genome and the far-from-saturation sensitivity permitted by current sequencing depths, the commonly used assessment of reproducibility has not been feasible for DNase-based genomic footprinting. Instead, footprinting has been performed on all available sequencing data pooled, and indirect indicators of reproducibility have then been sought. More confident inferences of TF binding could be made if the reproducibility of the individual footprints were demonstrated clearly.
Complete characterization of genomic occupancy for all proteins: In a recent study of mouse TF footprints, Stergachis et al.40 assessed regulatory networks of factors on the basis of their putative footprints, using essentially the same approach previously applied to the human ENCODE data12. The authors did not address the false negatives arising from the TFs that produced no footprints, claiming that the lack of footprints for nuclear receptors9,10 results from unoccupied sites contributing to the average cut profiles. However, in references9,10 and the extent of footprinting was examined specifically at occupied TF motif sites using cell type–matching ChIP-seq data. In contrast, Stergachis et al.40 attempted to remove false positives from among all the putative TF footprints (obtained without using ChIP-seq data) by filtering out those with cut profiles that were explained by the sequence bias of DNase cleavage in an ad hoc procedure. Their filtered set of footprints included TFs with fast in vivo binding dynamics such as Sox2, raising the possibility that the final set still contained some false positives. Sox2 is a stem cell pluripotency TF with a median DNA binding residence time of 12 s (ref. 36), and we have shown that it leaves no footprint even after correction for DNase cleavage bias9 (Figs. 2 and 3). These findings indicate that conclusions from existing TF network–construction studies using genomic footprints should be interpreted with caution12,40,41. The difficulty, if not impossibility, of obtaining footprints from biologically important TF binding events is a serious impediment to the comprehensive capture of all TF occupancies.
Negative control DNase-seq data from deproteinized DNA: Several groups have documented that the cleavage bias of DNase must be corrected and that the observed pattern of sequence bias depends on the particular DNase-seq protocol used9,10,21. On the basis of these findings, it is recommended that investigators adjust for the cut bias by performing control DNase-seq on naked DNA with their chosen protocol, rather than using published control digestion data. Even with protocol-matching control data, the bias correction is expected to be less reliable for DNA sequences (hexamers) with low genomic occurrences because of insufficient statistics.
Annotation of genome-wide occurrences of TF motifs for all proteins: Once putative footprints are detected as statistically significant local protections, these initially anonymous regions are matched with likely TFs on the basis of the DNA sequences or, if there are no matches, are labeled as novel elements. The matching process depends heavily on the known DNA-binding specificity of TFs. Notably, the binding specificity of some TFs has been reported to be too complex to be accurately captured by a position weight matrix42,43 (PWM), which assumes the independence of nucleotide preferences at different positions within a motif. Nonlinear models have been proposed44, but these alternative approaches have not been explored as tools for inferring TFs for the footprints detected in the DNase cut count data. Moreover, many TFs have poorly characterized binding specificity or no available information. Inaccurate assignment or missing annotation for TF binding elements is an underappreciated caveat that hampers the identification of regulatory relationships between TFs from genomic footprints.
Biologically justifiable procedure for resolving overlapping occurrences of motif elements bound by distinct proteins: We use an example to illustrate the often encountered problem of the same DNA element matching two TF binding motifs (Fig. 4). Many proteins have similar DNA-binding domain structures and consequently have similar PWMs. However, their biological functions may have diverged significantly during evolution, and therefore it is important to distinguish their binding sites in a given cell type. CREB1 and the Jun family of proteins are examples of proteins demonstrating this type of similarity; a commonly used tool for scanning the genome for motif occurrences reads 56% of CREB1 elements and 37% of Jun elements as indistinguishable (Fig. 4).
Figure 4.

The difficulty of assigning TFs on the basis of motif matches. The sets of genome-wide motif matches obtained using two similar but distinct PWMs have extensive overlap. The CREB1 elements were retrieved by scanning the mouse genome mm9 with FIMO and the top enriched PWM in phospho-CREB1 ChIP-seq peaks. The Jun elements were obtained by running FIMO on mm9 using the second enriched PWM from c-Jun ChIP-seq peaks (after the top enriched AP-1 motif)49. The ~159,000 motif elements matching either PWM cannot be matched to CREB1 or Jun dimers because many cell types express both factors and their biological functions are distinct.
What do we know about footprinting with ATAC-seq?
Assay for transposase-accessible chromatin using sequencing (ATAC-seq) has gained momentum recently as a simple assay that can be performed on a small population of cells, making it possible to probe chromatin accessibility in precious samples45. The signal intensities of DNase-seq and ATAC-seq have been shown to be very similar at the level of DHSs. However, the concordance between the two assays has not been examined carefully at the level of TF footprints. Aside from considerations of inter-assay concordance, it is noteworthy that the reproducibility of the more established DNase-based genomic footprinting has not been demonstrated. A direct assessment of reproducibility typically requires an independent data track generated from a separate experiment and comparison of the signal intensity between the two replicates using tools such as correlation coefficients and scatter plots. Instead, indirect methods have been used to argue for the validity and reproducibility of detected footprints12,40. One reason for this is that the currently feasible sequencing depth is still far from the saturating level required for the detection of most TF footprints. The sequencing depth becomes a more serious issue for ATAC-seq data, as a significant fraction of the reads originate from mitochondrial DNA and therefore are discarded before subsequent analyses.
In addition, ATAC-seq data analysis begins with the assumption that the genomic contact point of the transposase is a fixed number of base pairs from the ends of the sequence reads. This has technical implications for mappability normalization and sequence-bias correction, both of which need to be handled with caution. Given the relatively sparse information on the utility of ATAC-seq for genomic footprinting, further validation will be necessary to delineate any assay-specific features and issues.
Concluding remarks
Despite the clear conceptual advantages of DNase-seq over ChIP-based approaches, caveats and limitations should be clarified with respect to its general applicability for genomic footprinting analysis. Unfortunately, in our view, the current analysis method does not deliver the promise of comprehensive de novo construction of TF regulatory networks. There are both technical problems and data-interpretation issues that must be addressed in order for the methodology to become a widely used tool.
In addition to the DNase cut bias, an important limitation of the current genomic footprinting methods should be acknowledged: biologically important TF binding events can be highly dynamic and do not leave TF footprints in chromatin. Results from live cell microscopy studies unequivocally demonstrate that TF-chromatin interactions are dynamic for all the TFs examined so far, including several nuclear receptors, p53, NF-κB, AP-1 and Sox2 (Table 1). Measurement of binding dynamics with advanced live cell microscopy methods for more TFs will be necessary for a better understanding of the relationship between the characteristics of their chromatin binding and the extent of footprints observed by genomic footprinting.
The footprint depth seems largely unaffected by correction of the cut profiles for the sequence bias of DNase cleavage, presumably because the spikes and dips in the cut signature over the motif tend to cancel out and have a negligible effect on the depth of protection (Figs. 2 and 3). This idea is supported by analyses of individual sites showing that adjusting the cut count data for the DNase cleavage bias does not improve the accuracy of footprint-based prediction for TF binding9,21.
Analysis of DNase-seq data for the purpose of TF footprinting must address at least two issues. First, the sequence bias of the enzyme is a prominent background signal at TF motif elements. Appropriate computational algorithms can be used to detect putative footprints after correction for the enzyme bias, in addition to the well-known mappability bias of short reads, which affects all high-throughput sequencing methods. The other issue is an inherent feature of the assay that limits the ability to capture TF occupancy in a comprehensive manner. Recognizing that short-lived chromatin binding may be more a rule than an exception, on the basis of cumulative imaging studies so far, it is fair to speculate that a substantial number of TFs have footprints with depths that are hardly detectable even with the most sophisticated algorithms. Advances could be achieved more quickly if the limitations of the current method were completely recognized and made transparent, so that efforts could be focused on the needed areas. More methodological improvements are necessary to enable an accurate global prediction of TF occupancy in a single analysis framework.
ACKNOWLEDGMENTS
Computationally intensive tasks were performed using the US National Institutes of Health (NIH) Biowulf cluster, a GNU-Linux parallel processing system. We thank the NIH Helix systems staff for the management of this system. This work was supported by the Intramural Research Program of the NIH, National Cancer Institute.
Footnotes
AUTHOR CONTRIBUTIONS
M.-H.S. and G.L.H. conceived the project. S.B. and M.-H.S. performed the analysis. M.-H.S. and G.L.H. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Tullius TD. Physical studies of protein-DNA complexes by footprinting. Annu. Rev. Biophys. Biophys. Chem. 1989;18:213–237. doi: 10.1146/annurev.bb.18.060189.001241. [DOI] [PubMed] [Google Scholar]
- 2.Church GM, Ephrussi A, Gilbert W, Tonegawa S. Cell-type-specific contacts to immunoglobulin enhancers in nuclei. Nature. 1985;313:798–801. doi: 10.1038/313798a0. [DOI] [PubMed] [Google Scholar]
- 3.Jackson PD, Felsenfeld G. A method for mapping intranuclear protein-DNA interactions and its application to a nuclease hypersensitive site. Proc. Natl. Acad. Sci. USA. 1985;82:2296–2300. doi: 10.1073/pnas.82.8.2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hesselberth JR, et al. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat. Methods. 2009;6:283–289. doi: 10.1038/nmeth.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baek S, Sung MH, Hager GL. Quantitative analysis of genome-wide chromatin remodeling. Methods Mol. Biol. 2012;833:433–441. doi: 10.1007/978-1-61779-477-3_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morris SA, et al. Overlapping chromatin remodeling systems collaborate genome-wide at dynamic chromatin transitions. Nat. Struct. Mol. Biol. 2014;21:73–81. doi: 10.1038/nsmb.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.John S, et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sung MH, Guertin MJ, Baek S, Hager GL. DNase footprint signatures are dictated by factor dynamics and DNA sequence. Mol. Cell. 2014;56:275–285. doi: 10.1016/j.molcel.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He HH, et al. Refined DNase-seq protocol and data analysis reveals intrinsic bias in transcription factor footprint identification. Nat. Methods. 2014;11:73–78. doi: 10.1038/nmeth.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pique-Regi R, et al. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res. 2011;21:447–455. doi: 10.1101/gr.112623.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neph S, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mercer TR, et al. The human mitochondrial transcriptome. Cell. 2011;146:645–658. doi: 10.1016/j.cell.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyle AP, et al. High-resolution genome-wide in vivo footprinting of diverse transcription factors in human cells. Genome Res. 2011;21:456–464. doi: 10.1101/gr.112656.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cuellar-Partida G, et al. Epigenetic priors for identifying active transcription factor binding sites. Bioinformatics. 2012;28:56–62. doi: 10.1093/bioinformatics/btr614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo K, Hartemink AJ. Using DNase digestion data to accurately identify transcription factor binding sites. Pac. Symp. Biocomput. 2013;2013:80–91. [PMC free article] [PubMed] [Google Scholar]
- 17.Gusmao EG, Dieterich C, Zenke M, Costa IG. Detection of active transcription factor binding sites with the combination of DNase hypersensitivity and histone modifications. Bioinformatics. 2014;30:3143–3151. doi: 10.1093/bioinformatics/btu519. [DOI] [PubMed] [Google Scholar]
- 18.Sherwood RI, et al. Discovery of directional and nondirectional pioneer transcription factors by modeling DNase profile magnitude and shape. Nat. Biotechnol. 2014;32:171–178. doi: 10.1038/nbt.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piper J, et al. Wellington: a novel method for the accurate identification of digital genomic footprints from DNase-seq data. Nucleic Acids Res. 2013;41:e201. doi: 10.1093/nar/gkt850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lazarovici A, et al. Probing DNA shape and methylation state on a genomic scale with DNase I. Proc. Natl. Acad. Sci. USA. 2013;110:6376–6381. doi: 10.1073/pnas.1216822110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yardimci GG, Frank CL, Crawford GE, Ohler U. Explicit DNase sequence bias modeling enables high-resolution transcription factor footprint detection. Nucleic Acids Res. 2014;42:11865–11878. doi: 10.1093/nar/gku810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grøntved L, et al. Rapid genome-scale mapping of chromatin accessibility in tissue. Epigenetics Chromatin. 2012;5:10. doi: 10.1186/1756-8935-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakahashi H, et al. A genome-wide map of CTCF multivalency redefines the CTCF code. Cell Rep. 2013;3:1678–1689. doi: 10.1016/j.celrep.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poorey K, et al. Measuring chromatin interaction dynamics on the second time scale at single-copy genes. Science. 2013;342:369–372. doi: 10.1126/science.1242369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buenrostro JD, et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523:486–490. doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cusanovich DA, et al. Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science. 2015;348:910–914. doi: 10.1126/science.aab1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Voss TC, Hager GL. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat. Rev. Genet. 2014;15:69–81. doi: 10.1038/nrg3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sung MH, McNally JG. Live cell imaging and systems biology. Wiley Interdiscip. Rev. Syst. Biol. Med. 2011;3:167–182. doi: 10.1002/wsbm.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hager GL, McNally JG, Misteli T. Transcription dynamics. Mol. Cell. 2009;35:741–753. doi: 10.1016/j.molcel.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Voss TC, et al. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell. 2011;146:544–554. doi: 10.1016/j.cell.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morisaki T, Muller WG, Golob N, Mazza D, McNally JG. Single-molecule analysis of transcription factor binding at transcription sites in live cells. Nat. Commun. 2014;5:4456. doi: 10.1038/ncomms5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Royen ME, et al. Androgen receptor complexes probe DNA for recognition sequences by short random interactions. J. Cell Sci. 2014;127:1406–1416. doi: 10.1242/jcs.135228. [DOI] [PubMed] [Google Scholar]
- 33.Groeneweg FL, et al. Quantitation of glucocorticoid receptor DNA-binding dynamics by single-molecule microscopy and FRAP. PLoS ONE. 2014;9:e90532. doi: 10.1371/journal.pone.0090532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gebhardt JC, et al. Single-molecule imaging of transcription factor binding to DNA in live mammalian cells. Nat. Methods. 2013;10:421–426. doi: 10.1038/nmeth.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crocker J, et al. Low affinity binding site clusters confer Hox specificity and regulatory robustness. Cell. 2015;160:191–203. doi: 10.1016/j.cell.2014.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen J, et al. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell. 2014;156:1274–1285. doi: 10.1016/j.cell.2014.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharp ZD, et al. Estrogen-receptor-alpha exchange and chromatin dynamics are ligand- and domain-dependent. J. Cell Sci. 2006;119:4101–4116. doi: 10.1242/jcs.03161. [DOI] [PubMed] [Google Scholar]
- 38.Bosisio D, et al. A hyper-dynamic equilibrium between promoter-bound and nucleoplasmic dimers controls NF-kB-dependent gene activity. EMBO J. 2006;25:798–810. doi: 10.1038/sj.emboj.7600977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McNally JG, Mueller WG, Walker D, Wolford RG, Hager GL. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science. 2000;287:1262–1265. doi: 10.1126/science.287.5456.1262. [DOI] [PubMed] [Google Scholar]
- 40.Stergachis AB, et al. Conservation of trans-acting circuitry during mammalian regulatory evolution. Nature. 2014;515:365–370. doi: 10.1038/nature13972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neph S, et al. Circuitry and dynamics of human transcription factor regulatory networks. Cell. 2012;150:1274–1286. doi: 10.1016/j.cell.2012.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Badis G, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jolma A, et al. DNA-binding specificities of human transcription factors. Cell. 2013;152:327–339. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 44.Weirauch MT, et al. Evaluation of methods for modeling transcription factor sequence specificity. Nat. Biotechnol. 2013;31:126–134. doi: 10.1038/nbt.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gerstein MB, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bailey TL, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Biddie SC, et al. Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol. Cell. 2011;43:145–155. doi: 10.1016/j.molcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lickwar CR, Mueller F, Hanlon SE, McNally JG, Lieb JD. Genome-wide protein-DNA binding dynamics suggest a molecular clutch for transcription factor function. Nature. 2012;484:251–255. doi: 10.1038/nature10985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Malnou CE, et al. Heterodimerization with different Jun proteins controls c-Fos intranuclear dynamics and distribution. J. Biol. Chem. 2010;285:6552–6562. doi: 10.1074/jbc.M109.032680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mayr BM, Guzman E, Montminy M. Glutamine rich and basic region/leucine zipper (bZIP) domains stabilize cAMP-response element-binding protein (CREB) binding to chromatin. J. Biol. Chem. 2005;280:15103–15110. doi: 10.1074/jbc.M414144200. [DOI] [PubMed] [Google Scholar]
- 53.Guertin MJ, Zhang X, Coonrod SA, Hager GL. Transient ER binding and p300 redistribution support a squelching mechanism for E2-repressed genes. Mol. Endocrinol. 2014;28:1522–1533. doi: 10.1210/me.2014-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grøntved L, et al. Transcriptional activation by the thyroid hormone receptor through ligand dependent receptor recruitment and chromatin remodeling. Nat. Commun. 2015;6:7048. doi: 10.1038/ncomms8048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marcelli M, et al. Quantifying effects of ligands on androgen receptor nuclear translocation, intranuclear dynamics, and solubility. J. Cell. Biochem. 2006;98:770–788. doi: 10.1002/jcb.20593. [DOI] [PubMed] [Google Scholar]
- 56.Yu J, et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell. 2010;17:443–454. doi: 10.1016/j.ccr.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tirard M, Almeida OF, Hutzler P, Melchior F, Michaelidis TM. Sumoylation and proteasomal activity determine the transactivation properties of the mineralocorticoid receptor. Mol. Cell. Endocrinol. 2007;268:20–29. doi: 10.1016/j.mce.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 58.Le Billan F, et al. Cistrome of the aldosterone-activated mineralocorticoid receptor in human renal cells. FASEB J. 2015;29:3977–3989. doi: 10.1096/fj.15-274266. [DOI] [PubMed] [Google Scholar]