Abstract
We present the case of a patient with a left frontal glioblastoma with PNET features and hypermutated genotype in the setting of a POLE germline alteration. During standard-of-care chemoradiation, the patient developed a cervical spine metastasis and was subsequently treated with Pembrolizumab. Shortly thereafter, the patient developed an additional metastatic spinal lesion. Using whole exome DNA sequencing and clonal analysis, we report changes in the subclonal architecture throughout treatment. Furthermore, a persistently high neoantigen load was observed within all tumors. Interestingly, following initiation of Pembrolizumab, brisk lymphocyte infiltration was observed in the subsequently resected metastatic spinal lesion and an objective radiographic response was noted in a progressive intracranial lesion suggestive of active CNS immunosurveillance following checkpoint blockade therapy.
Introduction
Glioblastoma is the most aggressive primary brain tumor in adults, with a median survival of 12–14 months following standard-of-care treatment (1). There is growing enthusiasm for treating malignant brain tumors with cancer immunotherapies due to successes in other cancers and the growing realization that the central nervous system (CNS) is not immunoprivileged as has long been held (2). However, it is not clear what biomarkers will predict responses or whether lymphocytes will infiltrate CNS tumors following these therapies.
Work at the nexus of genomics and immunology has established the cancer immunogenomics concept in which expressed nonsilent tumor-specific mutations represent neoantigens recognizable by the immune system (3–5). Because mutational burden is the engine for neoantigen production, several groups have correlated mutational load with response to immunotherapy. Patients with greater neoantigen load exhibited improved clinical responses to checkpoint blockade immunotherapy (6–8).
Unlike carcinogen-induced tumors, glioblastomas typically harbor fewer than 100 exome-wide mutations (9), with only a subset representing candidate neoantigens. However, a subset of hypermutated glioblastomas have been described in which mutational loads can be 10–50 fold higher than average (10, 11). This genotype is observed in approximately 20–30% of recurrent glioblastomas following temozolomide treatment and has been ascribed to acquired somatic mismatch-repair deficiency (12, 13). A hypermutated genotype was also recently described in four adults with newly-diagnosed glioblastomas associated with somatic POLE mutations, each carrying more than 100-fold the average number of mutations (14). However, neither the genomic landscape of an adult glioblastoma arising from a germline POLE mutation nor its recurrent tumors has been reported.
We describe an adult patient with germline POLE deficiency who developed a hypermutated glioblastoma and underwent additional resection of two metachronous spinal metastases. The patient was treated with Pembrolizumab, an inhibitory monoclonal anti-PD-1 antibody, and had evidence of a clinical and immunologic response. Herein, we profile the genomic landscape and remodeling of the subclonal architecture of this hypermutated glioblastoma during the transition from primary tumor to metastases. Moreover, we demonstrate robust immune infiltration of these CNS tumors following Pembrolizumab therapy.
Results
Case Report
A 31 year-old man with a history of colonic polyps presented to the hospital after experiencing a new onset seizure. A brain MRI showed a 5.0 × 5.0 × 6.0 cm enhancing lesion in the left frontotemporal lobe and a T2/FLAIR hyperintense lesion in the frontal horn of the right lateral ventricle (Figure 1A). The patient underwent a left frontotemporal craniotomy for a near-gross total resection (>95%) of the left-sided lesion. Histopathologic analysis revealed a diagnosis of glioblastoma (WHO grade IV) with primitive neuroectodermal tumor (PNET) features (Figure 1A). Immunohistochemical stains for IDH1 R132H and BRAF V600E were negative, and MGMT methylation was equivocal in that 10 of 17 sites were positive at one institution but assessed as indeterminate during central pathology review for clinical trial eligibility at another institution. Given the personal and family history of numerous colonic polyps, the patient underwent analysis for germline DNA mismatch-repair deficiency and tested positive for a POLE mutation encoding the L424V substitution, previously implicated in colorectal cancer susceptibility (15). Genomic profiling of the resected tumor was performed by Foundation Medicine (Cambridge, MA)(16), and of the 315 genes assessed, 165 (52.4%) contained mutations.
Figure 1.
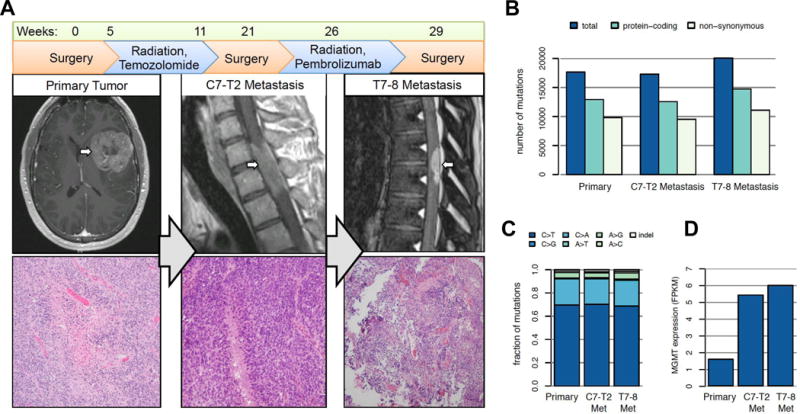
Presentation and progression of glioblastoma with PNET features in an adult patient. A) A left frontoinsular enhancing tumor was identified at presentation and resected. Following concomitant temozolomide and radiation treatment, an intradural/extramedullary C7-T2 metastasis was identified and also resected. Following a course of Pembrolizumab, a second intradural/extramedullary metastasis at T7-8 was resected. All tumors were glioblastoma with PNET features. (B) Mutational burden as detected by whole exome DNA sequencing. Each tumor harbored between 9–11,000 non-synonymous mutations. (C) All tumors carried a nearly identical spectrum of mutations, with the majority C>G alterations. (D) Relative expression of MGMT by RNA-seq across each tumor.
Before starting treatment, a spine MRI to evaluate for metastatic lesions was performed and showed no evidence of disease. The patient received radiation therapy to 60 Gy in 30 fractions over 41 days combined with temozolomide therapy. After chemoradiation treatment, the patient was treated with higher-dose maintenance temozolomide. Four weeks after initiating maintenance temozolomide, the patient developed difficulty walking. Repeat spinal MRI revealed an intradural/extramedullary enhancing lesion from C7-T2 (Figure 1A). He underwent a C7-T2 laminectomy for gross total resection, and histopathologic analysis revealed glioblastoma with PNET features consistent with a “drop” metastasis (Figure 1A). The patient’s leg strength improved, and the C5-T3 region was treated with radiation to 50.4 Gy. Due to a report of clinical responses to anti-PD-1 checkpoint blockade immunotherapy in hypermutated mismatch-repair deficient colon cancers, the patient was started on Pembrolizumab (8). Before treatment, a repeat spine MRI demonstrated 2 intradural enhancing foci dorsal to T7-8 concerning for disease.
Following 2 doses of Pembrolizumab over 3.4 weeks, the patient again developed difficulty walking. Spinal MRI demonstrated increased size of the enhancing T7-8 lesion causing local compression. The patient underwent a T7-8 laminectomy for tumor resection, and histopathologic analysis confirmed a glioblastoma metastasis but with significant inflammatory changes. The patient continued on Pembrolizumab and underwent radiation therapy to the T6-L4 region treating to 45 Gy. The patient’s strength improved to the point that he was walking. Four months after starting Pembrolizumab, a non-enhancing left cerebellopontine angle lesion and several small non-enhancing intraventricular lesions were being monitored.
Glioblastoma from a Germline POLE Mutation
To understand the genomic landscape of glioblastoma arising in the setting of germline POLE mutation, we performed DNA and RNA sequencing on the patient’s pre-treatment frontotemporal glioblastoma and both spinal metastases. All tumors were hypermutated, containing 17,276 to 20,045 mutations, over half of which were non-synonymous (Figure 1B, Supplemental Table 1), and all tumors carried a nearly identical spectrum of mutations with the majority being C>G alterations (Figure 1C, Supplemental Figure 1). Few copy number alterations (CNAs) were observed, and gene fusions were predominantly found in the cervical metastasis (Supplemental Figure 2, Supplemental Tables 2–3). No CNAs or fusions were shared among all three samples suggesting that the few events identified are subclonal. As the patient’s metastases developed after temozolomide treatment, we examined RNA sequencing data to determine whether O6-methylguanine DNA methyltransferase (MGMT), which drives temozolomide resistance, was more highly expressed in post-temozolomide lesions (Supplemental Table 4). Both metastases harbored nearly 4-fold higher MGMT expression compared to the primary tumor, consistent with enrichment of temozolomide-resistant subclones following chemotherapy (Figure 1D). Using previously described molecular subtypes for glioblastoma (20), transcriptome data categorized the primary tumor as most representative of the Classical subtype, whereas the metastases most represented the Mesenchymal subtype (Supplemental Figure 3).
Given the substantial mutational burden, specific driver mutations were difficult to identify. However, we identified 62 DNA repair-related genes containing non-silent somatic mutations within the founder clone (Supplemental Table 5). These included ATM (R457*), MSH5 (P581H), and TP53 (R175C), which are classified as pathogenic in the ClinVar database (21), as well as 34 other gene variants with supporting functional implications from ClinVar, COSMIC, SIFT, or PolyPhen (22–24). There were two additional POLE missense somatic mutations in the founder clone: R793C, predicted to be damaging and previously seen in a stomach carcinoma (25), and V1002A, which is of unknown significance. Mutational signature decomposition links the founder clone to a poorly characterized signature associated with deficient DNA-repair processes, but not specifically with POLE-induced mutations (Supplemental Figures 1 and 3).
Subclonal Evolution of Glioblastoma Metastases Throughout Treatment
Because glioblastomas are heterogeneous (26), we examined the subclonal architecture of the primary brain tumor and the two spinal metastases to determine how it changed over the course of 2 different treatments. Clonal analysis revealed 4 distinct mutation clusters in the treatment naïve brain tumor, of which one was the founder clone (Figure 2A–C, Supplemental Table 6). Comparatively, in the first spinal metastasis, which emerged following concurrent radiation and temozolomide therapy, cluster 4 was absent while cluster 3 became the dominant clone. We also identified new subclonal populations (clusters 5, 6, and 7) that became apparent in this tumor (Figure 2A–C). The second metastasis (after 3 weeks of treatment with Pembrolizumab) lacked clusters 2 and 3, suggesting that it was descended from the primary brain lesion, and not from the first metastasis. This tumor also harbored two unique subclones (clusters 8 and 9). By examining each subclone’s mutation spectrum individually, we found that all subclones have similar mutational signatures that are associated with DNA repair defects (Supplemental Figure 3) but have not been otherwise well characterized (27). Cluster 9, an emergent subclone in the second metastasis, also contains a signature suggesting that around 30% of its mutations may have been temozolomide-induced.
Figure 2.
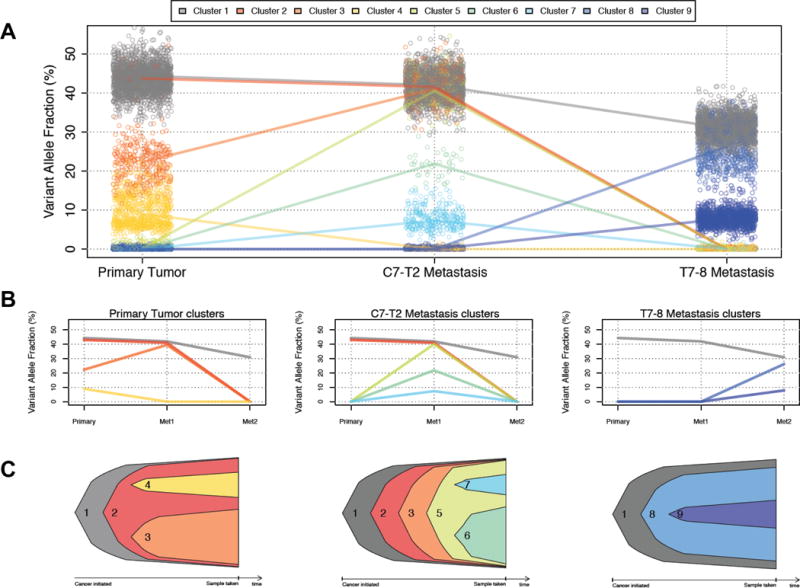
Clonal evolution of a glioblastoma and two metastases. (A) Three-dimensional clustering of variant allele frequencies infers nine distinct clonal populations. Points represent individual single-nucleotide variants, and lines connect the median variant allele frequencies of each cluster. (B) Simplified view showing only subclones present in the primary tumor (left), the C7-T2 metastasis (center) and the T7-8 metastasis (right). (C) Nested view showing the origins of each subclone in the primary tumor (left), the C7-T2 metastasis (center) and the T7-8 metastasis (right).
Neoantigen Landscape in Hypermutated Tumors and Response to Pembrolizumab
As response to checkpoint blockade immunotherapy is likely influenced by the presence of neoantigens (6, 8, 28), we applied a cancer immunogenomics approach to define the expressed neoantigen landscape across all tumor samples (3, 19)(Figure 3A, Supplemental Table 7). By incorporating expression data, the number of candidate neoantigens was reduced substantially. As such, we identified between 2,040 and 3,254 high-quality neoantigenic mutations per sample, with the first metastasis carrying the highest burden (Figure 3A, Supplemental Table 7). Interestingly, the founder clone contained 1,245 expressed candidate neoantigens representing a shared population of potentially immunodominant epitopes (Figure 3A).
Figure 3.
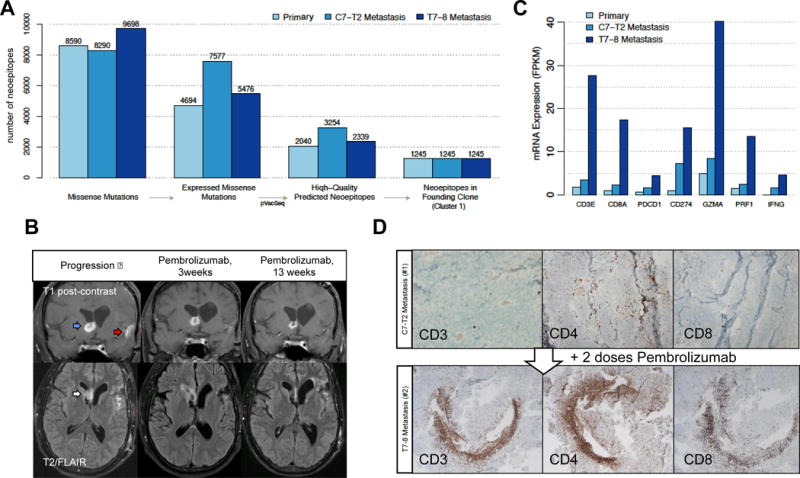
Predicted neoantigen burden and response to checkpoint blockade treatment. (A) Defining immunologically-relevant mutations through successive filtering steps, resulting in only expressed missense mutations that are present in every cell of the tumors and are predicted to create neoantigens. (B) Coronal T1 post-contrast (top) and axial T2/FLAIR (bottom) brain MRI sequences at progression (left panel), 3 weeks after Pembrolizumab initiation (middle panel), and 13 weeks after Pembrolizumab treatment (right panel). On post-contrast images, the right frontal enhancing lesion (blue arrow) decreases in intensity following Pembrolizumab. Enhancement near the site of initial resection (red arrow) disappears. On the T2/FLAIR sequence, the right frontal horn lesion (white arrow) increases in intensity and then decreases. (C) mRNA expression of key immune response-related genes. (D) Tumor-infiltrating lymphocytes following Pembrolizumab treatment. The glioblastoma metastatic specimens before (top panel) and after (bottom panel) Pembrolizumab were stained with antibodies to CD3, CD4, and CD8.
Following progression while on standard adjuvant chemotherapy, the patient was transitioned to Pembrolizumab treatment since responses to checkpoint blockade immunotherapies have been correlated to tumors with hypermutated genomes (8). Of note, prior to Pembrolizumab administration, the right frontal horn lesion included in the prior radiation plan had also progressed (Figure 3B, blue arrow). There was increased T2/FLAIR signal, suggestive of edema and inflammation, noted in this location (Figure 3B, white arrow) and persistent enhancement in the left temporal lobe near the initial resection bed indicating residual disease (Figure 3B, red arrow). Re-irradiation to this site and the entire neuraxis was considered, but the patient elected to first pursue systemic treatment with Pembrolizumab. After 3 weeks of Pembrolizumab, there was decreased enhancement of the right frontal lesion but a perilesional increase in T2/FLAIR signal that included the frontal horn ependyma (Figure 3B, middle panel). After 13 weeks of Pembrolizumab, the right frontal lesion decreased further in size, and no enhancement was observed adjacent to the left-sided resection bed. There was also a corresponding decrease in T2/FLAIR hyperintensity in the right frontal horn compared to previous studies (Figure 3B, right panel).
We next tested the hypothesis that Pembrolizumab treatment would lead to increased cytolytic tumor-infiltrating lymphocytes in CNS tumors using two approaches. First, we assessed the expression of key immune genes within the 3 tumor samples by RNA-seq. Expression of CD3, CD8, Granzyme A (GZMA), and Perforin (PRF1) was significantly elevated in CNS metastatic tissue following Pembrolizumab treatment, and levels of PD-1 (PDCD1), PD-L1 (CD274), and IFN-γ (IFNG) were also increased (Figure 3C, Supplemental Figure 4). Second, immunohistochemistry was performed on the resected metastases taken before and 3 weeks after Pembrolizumab treatment. We identified prominent areas of brisk CD3+, CD4+, and CD8+ T cell infiltrate in the post-Pembrolizumab lesion (Figure 3D). Together, these data show that hypermutated tumors harbor abundant high affinity, expressed neoantigens and that checkpoint blockade in this setting was associated with objective radiographic responses and brisk cytolytic lymphocyte infiltration into CNS tumors.
Discussion
Hypermutated tumors with POLE germline mutations have been described in colorectal cancer patients (15), and somatic alterations in POLE were recently reported in subsets of patients with glioblastomas and endometrial cancers (14, 29). To our knowledge, our report represents a description of only the second reported adult patient with glioblastoma in the setting of germline POLE deficiency (30). POLE encodes the DNA polymerase ε catalytic subunit, which synthesizes and proofreads leading strand DNA. In this report, the germline POLE alteration was likely permissive in creating an unstable genomic context that facilitated additional abundant somatic DNA repair machinery alterations. However, the level of hypermutation makes it difficult to clearly assign causality to mutated POLE alone, based on the observed mutational signatures.
The relationship between neoantigen burden and checkpoint blockade therapy response has been described in other tumor types but has not been reported extensively in the setting of glioblastoma. Colorectal cancer patients harboring high mutational burdens due to mismatch-repair deficiency demonstrated improved clinical responses compared to patients with lower mutational loads (8), a correlation also described in other solid tumors (6, 31). Also, a recent study reported clinical benefit from Nivolumab treatment in two pediatric patients with hypermutated gliomas (32). Herein, the sequential resection of glioblastoma metastases before and after Pembrolizumab treatment provided an opportunity to observe post-treatment lymphocyte infiltration in a CNS tumor, which has not been observed previously following checkpoint blockade. Although it is important to broaden this observation, these data suggest that CNS malignancies may be appropriate targets for checkpoint blockade treatment in defined scenarios and that lymphocytes may infiltrate across the blood-brain/blood-CSF barriers.
These results have several clinical implications. First, diagnostic efforts should be made to identify tumors with highly elevated mutational loads. Extensive genomic characterization of this patient’s tumor was critical in establishing the mutational burden, underscoring the importance of assaying a sufficient number of genes in clinical workflows to identify the hypermutated state. Second, although hypermutation in newly-diagnosed glioblastomas is rare, it is more common in recurrent glioblastomas after temozolomide treatment in which the incidence may approach 20–30% (10, 12, 33). Thus, recurrent tumors could be sampled and tested for a hypermutated genotype, which may confer increased sensitivity to checkpoint blockade treatment. Prospective multi-institutional clinical trials are needed to validate this possibility and determine whether temozolomide-associated hypermutation will be as favorable a biomarker as hypermutation due to heritable or somatic mismatch repair deficiency alone. Third, although this patient’s hypermutated condition was not discovered until after chemoradiation treatment, early identification in other glioblastoma patients prior to concurrent temozolomide and radiation treatment could lead to trials that assess the replacement of alkylating agents with checkpoint blockade for this select subset of patients. This patient’s disease dissemination was likely due both to the aggressive behavior exhibited by a tumor with PNET components as well as the temozolomide resistance that developed once DNA mismatch repair mutations were enriched (12, 13). Finally, clonal analysis of the expressed neoantigen landscape revealed that identification of the founder clone (7), which contains mutations present in all cells, may contribute to personalized cancer vaccine strategies that can broadly target heterogeneous cancers or those with spatially distinct sites of metastatic disease.
Methods
Genomic Characterization
The patient provided written informed consent for tissue banking and genomic characterization to protocol #201111001 approved by the Washington University School of Medicine IRB. DNA exome and RNA sequencing was performed, resulting in over 86× coverage of the normal and over 200× coverage of each tumor. Sequence data was aligned to reference sequence build GRCh37-lite-build37 then variant detection was performed using an ensemble approach followed by statistical and heuristic filtering. Copy number aberrations were detected using VarScan. RNA-seq data was aligned to reference sequence build GRCh37-lite-build37 with Tophat and expression levels were quantified using Cufflinks. Gene fusions were inferred jointly from RNA and DNA using INTEGRATE. The SciClone algorithm (17) was used for clonal inference, followed by reconstruction of tumor phylogeny using clonEvol (18). Mutational signatures were inferred using the deconstructSigs package for R.
Neoantigen Prediction
Neoantigen prediction was performed using the pVacSeq pipeline (19), incorporating 5 MHC-binding prediction algorithms and requiring a minimum IC50 score of 500 nM.
Supplementary Material
Statement of Significance.
It is unclear whether hypermutated glioblastomas are susceptible to checkpoint blockade in adults. Herein, we provide proof-of-principle that glioblastomas with DNA repair defects treated with checkpoint blockade may result in CNS immune activation leading to clinically and immunologically significant responses. These patients may represent a genomically-stratified group for whom immunotherapy could be considered.
Acknowledgments
Grant Support:
This work was supported by NIH grant K08NS092912 (G.P.D.), the Physician-Scientist Training Program at Washington University School of Medicine (T.M.J.), and an endowment to The Elizabeth H. and James S. McDonnell III Genome Institute from the McDonnell family.
Footnotes
Conflict of Interest: The authors have no conflicts of interest to disclose.
Additional methods and detailed version and parameter information are available in the Supplementary Appendix.
References
- 1.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–96. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Dunn GP, Okada H. Principles of immunology and its nuances in the central nervous system. Neuro Oncol. 2015;17(Suppl 7):vii3–vii8. doi: 10.1093/neuonc/nov175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400–4. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348:69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 5.Hacohen N, Fritsch EF, Carter TA, Lander ES, Wu CJ. Getting personal with neoantigen-based therapeutic cancer vaccines. Cancer immunology research. 2013;1:11–5. doi: 10.1158/2326-6066.CIR-13-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–8. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGranahan N, Furness AJ, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351:1463–9. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372:2509–20. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.TCGA Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343:189–93. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim H, Zheng S, Amini SS, Virk SM, Mikkelsen T, Brat DJ, et al. Whole-genome and multisector exome sequencing of primary and post-treatment glioblastoma reveals patterns of tumor evolution. Genome Res. 2015;25:316–27. doi: 10.1101/gr.180612.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cahill DP, Levine KK, Betensky RA, Codd PJ, Romany CA, Reavie LB, et al. Loss of the mismatch repair protein MSH6 in human glioblastomas is associated with tumor progression during temozolomide treatment. Clin Cancer Res. 2007;13:2038–45. doi: 10.1158/1078-0432.CCR-06-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Thuijl HF, Mazor T, Johnson BE, Fouse SD, Aihara K, Hong C, et al. Evolution of DNA repair defects during malignant progression of low-grade gliomas after temozolomide treatment. Acta Neuropathol. 2015;129:597–607. doi: 10.1007/s00401-015-1403-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erson-Omay EZ, Caglayan AO, Schultz N, Weinhold N, Omay SB, Ozduman K, et al. Somatic POLE mutations cause an ultramutated giant cell high-grade glioma subtype with better prognosis. Neuro Oncol. 2015;17:1356–64. doi: 10.1093/neuonc/nov027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palles C, Cazier JB, Howarth KM, Domingo E, Jones AM, Broderick P, et al. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet. 2013;45:136–44. doi: 10.1038/ng.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–31. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller CA, White BS, Dees ND, Griffith M, Welch JS, Griffith OL, et al. SciClone: inferring clonal architecture and tracking the spatial and temporal patterns of tumor evolution. PLoS computational biology. 2014;10:e1003665. doi: 10.1371/journal.pcbi.1003665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–10. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hundal J, Carreno BM, Petti AA, Linette GP, Griffith OL, Mardis ER, et al. pVAC-Seq: A genome-guided in silico approach to identifying tumor neoantigens. Genome Med. 2016;8:11. doi: 10.1186/s13073-016-0264-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862–8. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–11. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–4. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. In: Haines Jonathan L, et al., editors. Current protocols in human genetics. 2013. Chapter 7:Unit7 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas Research N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–9. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aum DJ, Kim DH, Beaumont TL, Leuthardt EC, Dunn GP, Kim AH. Molecular and cellular heterogeneity: the hallmark of glioblastoma. Neurosurgical focus. 2014;37:E11. doi: 10.3171/2014.9.FOCUS14521. [DOI] [PubMed] [Google Scholar]
- 27.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGranahan N, Furness AJ, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351:1463–9. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Gool IC, Eggink FA, Freeman-Mills L, Stelloo E, Marchi E, de Bruyn M, et al. POLE Proofreading Mutations Elicit an Antitumor Immune Response in Endometrial Cancer. Clin Cancer Res. 2015;21:3347–55. doi: 10.1158/1078-0432.CCR-15-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spier I, Holzapfel S, Altmuller J, Zhao B, Horpaopan S, Vogt S, et al. Frequency and phenotypic spectrum of germline mutations in POLE and seven other polymerase genes in 266 patients with colorectal adenomas and carcinomas. Int J Cancer. 2015;137:320–31. doi: 10.1002/ijc.29396. [DOI] [PubMed] [Google Scholar]
- 31.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189–99. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bouffet E, Larouche V, Campbell BB, Merico D, de Borja R, Aronson M, et al. Immune Checkpoint Inhibition for Hypermutant Glioblastoma Multiforme Resulting From Germline Biallelic Mismatch Repair Deficiency. J Clin Oncol. 2016;34:2206–11. doi: 10.1200/JCO.2016.66.6552. [DOI] [PubMed] [Google Scholar]
- 33.Kim J, Lee IH, Cho HJ, Park CK, Jung YS, Kim Y, et al. Spatiotemporal Evolution of the Primary Glioblastoma Genome. Cancer Cell. 2015;28:318–28. doi: 10.1016/j.ccell.2015.07.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


