Abstract
HIV Prevention Trials Network (HPTN) 052 demonstrated that antiretroviral therapy (ART) prevents HIV transmission in serodiscordant couples. HIV from index-partner pairs was analyzed to determine the genetic linkage status of partner infections. Forty-six infections were classified as linked, indicating that the index was the likely source of the partner’s infection. Lack of viral suppression and higher index viral load were associated with linked infection. Eight linked infections were diagnosed after the index started ART: four near the time of ART initiation and four after ART failure. Linked infections were not observed when the index participant was stably suppressed on ART.
Keywords: HIV-1, serodiscordant couples, partner, transmission, linkage, HPTN 052
INTRODUCTION
The HIV Prevention Trials Network (HPTN) 052 trial was a Phase 3, randomized, controlled trial that tested whether early initiation of antiretroviral therapy (ART) prevented sexual transmission of HIV in serodiscordant couples.1,2 Index participants in the early ART arm started ART at study enrollment with baseline CD4 cell counts of 350–550 cells/mm3. Index participants in the delayed ART arm started ART when their CD4 count fell below 250 cells/mm3 or they developed an AIDS-defining illness. The primary endpoints for the trial were genetically-linked partner infections (i.e., infections where the enrolled index participant was the likely source of the partner’s infection). In April 2011, an interim analysis demonstrated that early ART prevented 96% of linked HIV infections and offered health benefits to the individual receiving ART.1,3 After these results were released, ART was offered to all HIV-infected index participants regardless of CD4 cell count. The trial continued until May 2015, to assess the durability of the study intervention. The final intent-to-treat analysis found that early ART prevented 93% of linked HIV infections.2 In this report, we present genetic linkage analysis and characterization of partner infections in HPTN 052, and analysis of the association of demographic, behavioral, and clinical factors with linked HIV infection.
METHODS
Study cohort
HPTN 052 enrolled 1,763 serodiscordant couples at 13 study sites in Africa, Asia, and the Americas (97% heterosexual, April 2005-May 2010).1 ART failure was defined as the first of two consecutive study visits where the index’s viral load was >1,000 copies/mL more than 24 weeks after ART initiation. HIV-uninfected partners were tested for HIV infection throughout the trial, as described;1 partner infections were confirmed at the HPTN Laboratory Center (Baltimore, MD, USA).
Linkage analysis
Genetic linkage of partner infections was determined using phylogenetic and statistical methods described previously.4 Linkage analysis was performed using samples collected from partners with confirmed HIV infection and the corresponding index participants (index-partner pairs); whenever possible, two samples from each individual were analyzed. Samples from random index participants at the same study sites were also analyzed (control samples). Three methods were used for linkage analysis: phylogenetic analysis of HIV pol sequences generated from population-sequencing; Bayesian analysis of pol sequence data; and phylogenetic analysis of env sequences obtained by next generation sequencing (Supplemental Digital Content 1).
HIV drug resistance testing and HIV subtyping
HIV genotyping was performed using the ViroSeq HIV-1 Genotyping System (Celera Diagnostics, Alameda, CA). HIV subtyping was performed using the resulting pol region sequences, as described.4
Analysis of factors associated with linked vs. unlinked partner infections
Associations between linkage status and categorical variables were assessed using Fisher’s exact test. Associations between linkage status and continuous variables were assessed using Wilcoxon’s rank sum test. All p-values are two-sided. Statistical significance was defined as p<0.05. Backwards stepwise multivariate logistic regression was performed using a cutoff of <0.05 to retain the factors considered significant.
Analysis of the timing of partner infections
The timing of partner infections was analyzed in three cases as described5 using HIV RNA testing; serologic assays; and sequence analysis using BEAST (which estimates the time since HIV infection using phylogenetic methods for analysis of sequence data6) and Poisson Fitter (which estimates the time since HIV infection based on the accumulation of neutral mutations in the viral population7).
Informed consent
Ethical review committees at each participating institution approved the HPTN 052 trial (NCT00074581). Written informed consent was obtained from all study participants.
RESULTS
Linkage status of index-partner pairs
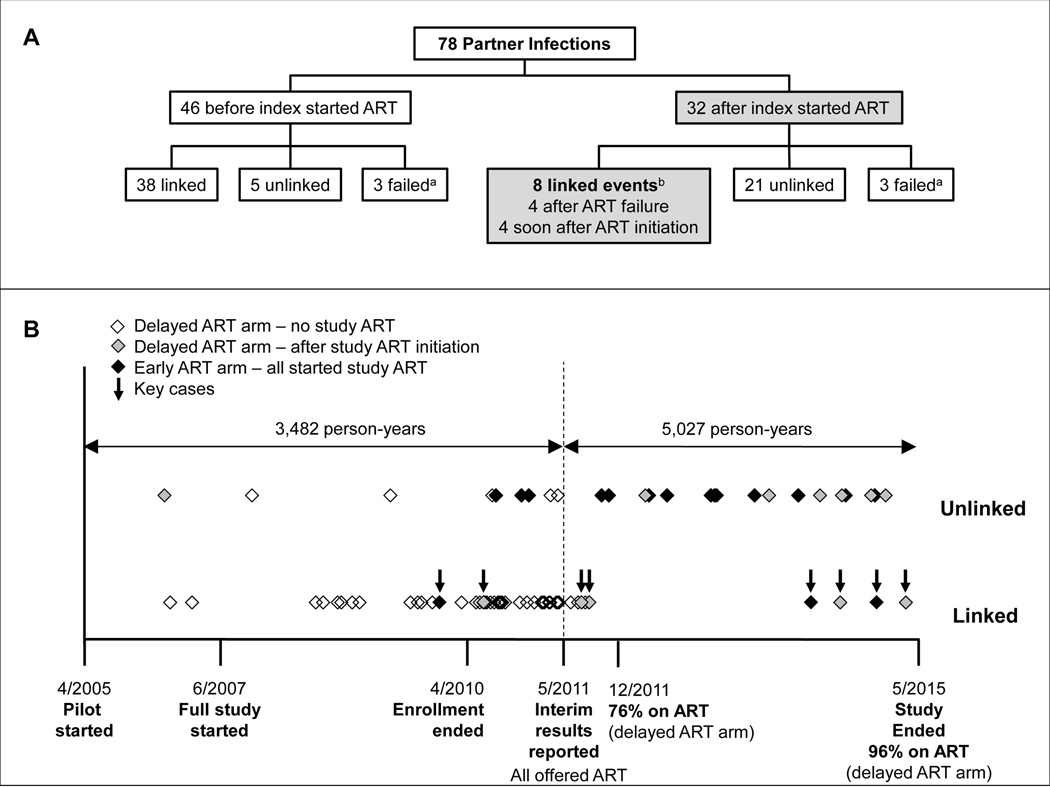
Seventy-eight partner infections were observed over 8,295 person-years of follow-up for uninfected partners, including 6,620 person-years of follow-up for partners after the corresponding index participant started ART. Forty-six infections were diagnosed before the index participant started study ART, and 32 were diagnosed after the index participant started study ART (19 in the early ART arm and 13 in the delayed ART arm, Figure 1A).
Figure 1. Analysis of partner infections in HPTN 052.
(A) The chart shows an overview of the linkage status of partner infections in HPTN 052. Data are presented separately for couples where the partner was diagnosed with HIV infection before the index participant started antiretroviral therapy (ART) in the trial (N=46) or after the index participant started ART in the trial (N=32). Infections were classified as linked if the index participant was the likely source of the partner’s infection, and unlinked if the partner was most likely infected from another source.
(B) Seventy-eight partner infections were identified in the HPTN 052 trial. The dates (month/year) of milestones in the HPTN 052 trial are shown on the X-axis. A vertical dashed line indicates the date that the interim report was released, showing the benefit of early initiation of ART. Horizontal arrows show the number of person-years of follow-up for partners who were HIV-uninfected at study enrollment. Partner infections are represented by diamonds. Data are presented separately for linked and unlinked partner infections. Open diamonds indicate infections in the delayed ART arm of the study that were diagnosed before the index participant started ART. Shaded diamonds indicate infections in the delayed ART arm of the study that were diagnosed after the index participant started ART. Black diamonds indicate infections in the Early ART arm of the study, all of which were diagnosed after the index participant started ART. Eight linked partner infections were diagnosed after the index started ART (Key cases, arrows).
aIn six cases, linkage status could not be determined because HIV RNA in the index and/or partner sample(s) could not be amplified (failed cases). In three cases, HIV RNA could not be amplified because the index was virally suppressed at all study visits (including the enrollment visit). In two of those cases, the index participant was on ART at study enrollment, but did not disclose this to study staff17; in the third case, ARV drugs were not detected in study specimens, indicating that the index participant was most likely an elite controller. In the fourth case, the partner was diagnosed with HIV infection and started ART outside of the study and was virally suppressed at subsequent study visits. It is not clear why amplification was unsuccessful in the other two cases. In those cases, attempts to amplify HIV RNA using a nested PCR method and alternate amplification primers were not successful.
b3 early ART arm; 5 delayed ART arm.
Linkage of partner infections was assessed using phylogenetic and statistical methods (Supplemental Digital Content 1). HIV sequencing and linkage analysis were successful for 72 of the 78 cases; in the other six cases, samples were not available for analysis or amplification failed for either the index or partner samples. Overall, 46 infections were classified as linked and 26 were classified as unlinked (Figure 1A). Figure 1B shows the timing of linked and unlinked infections in each study arm, relative to key study milestones.
In all cases, HIV subtypes were consistent with the subtypes prevalent in the region (Supplemental Digital Content 2A). HIV drug resistance was detected in HIV from the partner in only two of the 46 linked cases; in both cases, the same resistance mutations were detected in the corresponding index participant, suggesting that the resistant virus was transmitted (Supplemental Digital Content 2B). Both of these cases occurred in the delayed ART arm, before the index participant started ART. HIV drug resistance was detected in the partner in only two of the 26 unlinked cases; in these cases, resistance mutations were not detected in HIV from the index participant (Supplemental Digital Content 2B).
Factors associated with linked infections
We analyzed the association of clinical, demographic, and behavioral factors with linked partner infections (Supplemental Digital Content 3). In univariate analyses, linked partner infections were associated with the following characteristics: couple randomized to the delayed ART arm; index participant not on ART at the time of partner diagnosis; higher index viral load at the time of partner diagnosis (index viral load >400 copies/mL or higher log10 viral load); lower index CD4 cell count at the time of partner diagnosis; shorter time between enrollment and partner diagnosis; and fewer sexual partners in the three months prior to diagnosis. Region (Africa vs. Asia/Americas) and index sex were not associated with linked infection.
Backwards stepwise regression models were used for model building in multivariate analysis; separate analyses were used for each measure of index viral load at the time of partner infection (greater or less than 400 copies/mL or log10 viral load). In both models, higher index viral load was strongly associated with linked infection (p=0.0006 and p<0.0001). When index viral load was analyzed as a binary variable (greater or less than 400 copies/mL), randomization in the delayed ART arm and lower index CD4 cell count at the time of partner diagnosis were also associated with linked partner infection (p=0.045 and p=0.033, respectively). When index viral load was analyzed as a continuous variable (median log10 viral load), the only factor significantly associated with linked infection was index viral load at the time of partner diagnosis.
Linked partner infections diagnosed after the index participant started ART
Eight of the 46 linked infections occurred after the index participant started ART (three in the early ART arm; five in the delayed ART arm, Figure 1B, key cases); these eight cases were analyzed in detail. None of the eight partners in these key cases had HIV drug resistance. In four cases, the partner was diagnosed after the index participant failed ART (Supplemental Digital Content 4, Panel A). In three of these four cases, the index was viremic at the time of partner diagnosis. In the fourth case, the index was intermittently viremic prior to the partner diagnosis; in that case, the partner was lost-to-follow up for more than one year; during that time, the partner was diagnosed with HIV infection and had started ART. In the other four cases, the partner was diagnosed with HIV infection shortly after the index participant started ART (Supplemental Digital Content 4, Panel B). In one of those four cases, the index participant did not achieve virologic suppression on ART and was viremic when the partner was diagnosed with HIV infection. In the other three cases (Cases A–C), the index’s viral load was <400 copies/mL when the partner was diagnosed with HIV infection.
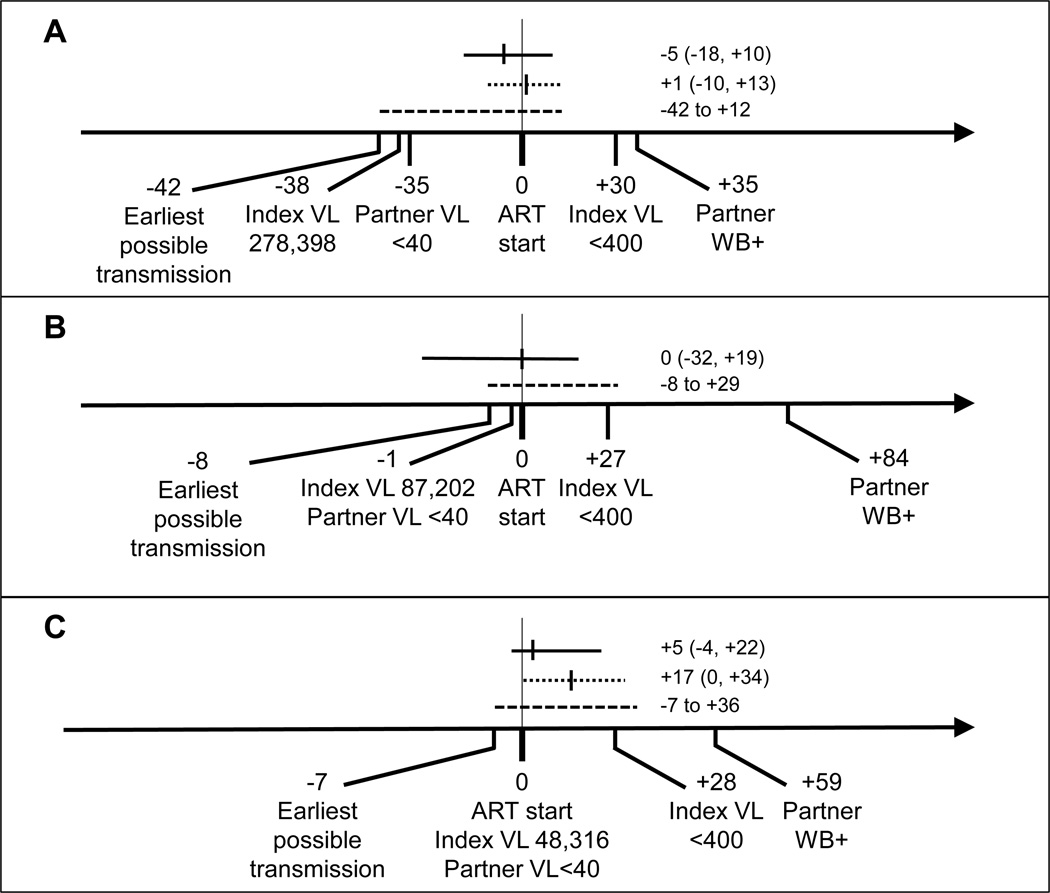
Additional laboratory assessments were performed in Cases A–C to estimate the timing of HIV transmission relative to index ART initiation (Figure 2). Results obtained for two of these cases are described in a previous report5 and are updated here. In all three cases, the analyses indicated that the transmission event occurred either before index ART initiation (when the index was not virally suppressed), or shortly after index ART initiation (most likely before the index was virally suppressed from ART). Of note, in HPTN 052, the cumulative percentage of index participants who achieved viral suppression by 3, 6, 9, and 12 months were 76%, 87%, 90%, and 91%.
Figure 2. Analysis of the timing of linked partner infections that were diagnosed when the index participant was virally suppressed.
The figure shows data for three partner infections where the partner was diagnosed shortly after the index participant started antiretroviral therapy (ART), when the index was virally suppressed. The vertical line at Day 0 indicates the day of that the index participant started ART. Days before the start of ART are indicated with negative numbers; days after the start of ART are shown with positive numbers. Index and partner viral load (VL) values are indicated. The date of the partner’s first positive HIV test is indicated (WB+: Western blot positive). The solid horizontal lines indicate the estimated transmission date with 95% confidence intervals (CI) obtained using BEAST. Dotted lines indicate the estimated transmission date with 95% CI obtained using the Poisson Fitter. Dashed lines indicate the time between the earliest possible transmission date (7 days before the partner’s last test where HIV RNA was undetectable) and the end of the likely transmission period, determined by the Fiebig stage18 of the partner at the first HIV positive visit. (A) In Case A, the partner was diagnosed with HIV infection on Day +35. HIV RNA was undetectable (<40 copies/mL) on Day −35, indicating that the earliest possible transmission date was Day −42 (based on a 7-day eclipse period). The Western blot was positive on Day +35 (Fiebig stage V18), indicating that the transmission event occurred before Day +12. BEAST analysis estimated that the transmission event occurred on Day −5. Poisson fitter analysis estimated that the transmission event occurred on Day +1. (B) In Case B, the partner was diagnosed with HIV infection on Day +84. HIV RNA was undetectable on Day −1, indicating that the earliest possible transmission date was Day −8. The Western blot was positive on Day +84 (Fiebig stage VI18), indicating that the transmission event occurred before Day +29. BEAST analysis estimated that the transmission event occurred on Day 0. Poisson fitter analysis was not performed in this case because of the complexity of the partner’s viral population5. (C) In Case C, the partner was diagnosed with HIV infection on Day +59. HIV RNA was undetectable on Day 0, indicating that the earliest possible transmission date was Day −7. The Western blot was positive on Day +59 (Fiebig stage V18), indicating that the transmission event occurred before Day +36. BEAST analysis estimated that the transmission event occurred on Day +5. Poisson fitter analysis estimated that the transmission event occurred on Day +17.
DISCUSSION
Phylogenetic methods can be used to study HIV transmission networks and hotspots within communities and populations8–10 and to assess the genetic linkage of specific transmission events.4,11 In this study, we used phylogenetic and statistical methods to identify genetically-linked partner infections in the HPTN 052 trial, which were the basis of the primary endpoint analysis for this landmark study.1,2 This report highlights the importance of genetic linkage analysis in evaluating HIV transmission in clinical trials that include serodiscordant couples. Forty-six linked partner infections were observed in the HPTN 052 trial. Only eight of these infections were diagnosed after the index participant started ART. We identified three risk periods for index-to-partner (linked) transmission in couples after the index participant had started ART: (1) near the time of ART initiation, before viral suppression was achieved, (2) after ART initiation if viral suppression did not occur, and (3) after ART failure.
High HIV viral load is a major driver of sexual HIV transmission and is also associated with ART outcomes that are relevant to use of treatment as prevention. In a previous study, we showed that higher baseline viral load in index participants was associated with a longer time to viral suppression and lack of viral suppression 3 or 6 months after ART initiation was associated with a shorter time to ART failure and a higher frequency of ART failure.12 In this report, higher index viral load at study enrollment2 and higher index viral load at the time of partner seroconversion were associated with linked partner infection.
In most cases, several months of ART is required before viral suppression is achieved. More rapid viral suppression is observed with ART regimens that include integrase inhibitors;13,14 however, these regimens may not be available in resource-limited settings. Previous studies have examined the risk of HIV transmission in the months after ART initiation. In a review of six studies of serodiscordant couples, only one linked infection was observed >6 months after ART initiation among 1,672 couples with 2,773 person-years of follow-up.15 In another study, no infections were observed >6 months after ART initiation (over 167 person-years follow-up).16 In HPTN 052, four linked infections were observed >6 months after ART initiation (over 7,032 person-years of follow-up); all four cases occurred long after ART failure. Importantly, we did not observe any linked infections in couples where the index participant was stably suppressed on ART. These data were based on 6,620 person-years of follow-up of partners after index ART initiation.
This report highlights the importance of achieving and maintaining viral suppression when ART is used to reduce the risk of HIV transmission. In this setting, special efforts should be made to minimize HIV transmission risk before the index is virally suppressed, to achieve durable viral suppression on ART, and to identify and address ART failure early.
Supplementary Material
Acknowledgments
Source of Funding
This project was supported by: (1) The HIV Prevention Trials Network (HPTN) sponsored by the National Institute of Allergy and Infectious Diseases (NIAID), the National Institute on Drug Abuse (NIDA), the National Institute of Mental Health (NIMH), and the Office of AIDS Research, of the National Institutes of Health (NIH), Dept. of Health and Human Services (DHHS) [grant numbers UM1AI068613 (HPTN Network Laboratory – Susan Eshleman, PI), UM1AI068617 (HPTN Statistical and Data Management Center – Deborah Donnell, PI), and UM1AI068619 (HPTN Core and Operations Center – Wafaa El-Sadr, PI)], (2) the Division of Intramural Research, NIAID, NIH, and (3) R37-AI44667 (Ronald Swanstrom, PI).
The authors acknowledge the dedication, commitment, and efforts of the entire HPTN 052 team, and acknowledge the invaluable contributions of the participants in the HPTN 052 trial. The authors thank the laboratory staff at Johns Hopkins University, at the Rocky Mountain Laboratories, and at the HPTN 052 study sites for assistance with sample and data management.
AUTHORS AND CONTRIBUTORS
All authors meet the journal’s criteria for authorship. Individual contributions / author roles are listed below.
| Susan H. Eshleman* | Designed the study; analyzed the data; prepared the manuscript |
| Sarah E. Hudelson | Performed HIV genotyping and linkage analysis |
| Andrew D. Redd | Performed next generation sequencing and assisted with linkage analysis |
| Ronald Swanstrom | Responsible for analysis of the timing of infection in selected cases |
| San-San Ou | Statistical Research Associate for HPTN 052 |
| Xinyi Cindy Zhang | Performed statistical analysis |
| Li-Hua Ping | Performed analysis of the timing of infection in selected cases |
| Estelle Piwowar-Manning | HPTN Laboratory Center Quality Assurance / Quality Control Coordinator for HPTN 052 |
| Stephen F. Porcella | Responsible for next generation sequencing |
| Matthew Sievers | Assisted with next generation sequencing |
| Craig A. Martens | Performed next generation sequencing and assisted with linkage analysis |
| Daniel Bruno | Performed next generation sequencing |
| Elena Dukhovlinova | Performed analysis of the timing of infection in selected cases |
| Marybeth McCauley | Senior Study Manager for HPTN 052 |
| Theresa Gamble | Senior Study Manager for HPTN 052 |
| Jessica M. Fogel | Assisted with analysis and presentation of viral load data |
| Devin Sabin | Assisted with analysis of viral load data |
| Thomas C. Quinn | Assisted with analysis of next generation sequencing data |
| Laurence Gunde | HPTN 052 investigator, Blantyre, Malawi |
| Madalitso Maliwichi | HPTN 052 investigator, Lilongwe, Malawi |
| Nehemiah Nhando | HPTN 052 investigator, Harare, Zimbabwe |
| Victor Akelo | HPTN 052 investigator, Kisumu, Kenya |
| Sikhulile Moyo | HPTN 052 investigator, Gabarone, Botswana |
| Ravindre Panchia | HPTN 052 investigator, Soweto, South Africa |
| Nagalingeswaran Kumarasamy |
HPTN 052 site PI, Chennai, India |
| Nuntisa Chotirosniramit | HPTN 052 investigator, Chiang Mai, Thailand |
| Marineide Gonçalves de Melo | HPTN 052 investigator, Porto Alegre RS, Brazil |
| Jose H. Pilotto | HPTN 052 site PI, Rio de Janeiro, Brazil |
| Beatriz Grinsztejn | HPTN 052 site PI, Fiocruz, Rio de Janeiro, Brazil |
| Kenneth H. Mayer | HPTN 052 site PI, Boston, USA |
| Ying Q. Chen | Protocol statistician for HPTN 052 |
| James P. Hughes | Statistician for HPTN 052 linkage analysis |
| Myron S. Cohen | Protocol Chair for HPTN 052 |
Footnotes
A portion of this work was presented at the 8th IAS Conference on HIV Pathogenesis, Treatment and Prevention, July 19–22, 2015, Vancouver, Canada.
This paper was submitted with the permission of the Kenya Medical Research Institute (KEMRI) ethical review committees. The corresponding author had full access to all of the data in the study and had final responsibility for the decision to submit the manuscript for publication.
Conflict of Interests
None of the authors has a financial or personal relationship with other people or organizations that could inappropriately influence (bias) their work. The following relationship is noted: SHE has collaborated with Abbott Diagnostics (distributor of the ViroSeq HIV Genotyping System) on evaluation of HIV-related assays.
Supplemental Digital Content
Supplemental Digital Content 1. Methods used for linkage analysis.
Supplemental Digital Content 2. Characterization of partner infections.
Supplemental Digital Content 3. Analysis of factors associated with linked partner infection.
Supplemental Digital Content 4. Relationship of linked partner infections to index viral load.
Contributor Information
Susan H. Eshleman, Dept. of Pathology, Johns Hopkins Univ. School of Medicine, Baltimore, MD, 21205, USA.
Sarah E. Hudelson, Dept. of Pathology, Johns Hopkins Univ. School of Medicine, Baltimore, MD, 21205, USA.
Andrew D. Redd, Laboratory of Immunoregulation, NIAID, NIH, Baltimore, MD, 21205, USA.
Ronald Swanstrom, Dept. of Biochemistry and Biophysics, Univ. of North Carolina at Chapel Hill, Chapel Hill, NC, 27599, USA.
San-San Ou, Statistical Center for HIV Research and Prevention, Seattle, WA, 98109, USA.
Xinyi Cindy Zhang, Vaccine and Infectious Disease Science Division, Fred Hutchinson Cancer Research Institute, Seattle, WA, 98102, USA.
Li-Hua Ping, Lineberger Comprehensive Cancer Center, Univ. of North Carolina at Chapel Hill, Chapel Hill, NC, 27599, USA.
Estelle Piwowar-Manning, Dept. of Pathology, Johns Hopkins Univ. School of Medicine, Baltimore, MD, 21205, USA.
Stephen F. Porcella, Genomics Unit, Research Technologies Section, Rocky Mountain Laboratories, DIR, NIAID, NIH, Hamilton, MT, 59840, USA.
Matthew F. Sievers, Dept. of Medicine, Johns Hopkins Univ. School of Medicine, Baltimore, MD, 21205, USA.
Craig A. Martens, Genomics Unit, Research Technologies Section, Rocky Mountain Laboratories, DIR, NIAID, NIH, Hamilton, MT, 59840, USA.
Daniel Bruno, Genomics Unit, Research Technologies Section, Rocky Mountain Laboratories, DIR, NIAID, NIH, Hamilton, MT, 59840, USA.
Elena Dukhovlinova, Lineberger Comprehensive Cancer Center, Univ. of North Carolina at Chapel Hill, Chapel Hill, NC, 27599, USA.
Marybeth McCauley, Science Facilitation Department, FHI360, Washington, DC, 20009, USA.
Theresa Gamble, Science Facilitation Department, FHI360, Durham, NC, 27701, USA.
Jessica M. Fogel, Dept. of Pathology, Johns Hopkins Univ. School of Medicine, Baltimore, MD, 21205, USA.
Devin Sabin, Dept. of Pathology, Johns Hopkins Univ. School of Medicine, Baltimore, MD, 21205, USA.
Thomas C. Quinn, Laboratory of Immunoregulation, NIAID, NIH, Baltimore, MD, 21205, USA; Dept. of Medicine, Johns Hopkins Univ. School of Medicine, Baltimore, MD, 21205, USA.
Laurence Gunde, College of Medicine-Johns Hopkins Project, Blantyre, Malawi.
Madalitso Maliwichi, UNC Project, Lilongwe, Malawi.
Nehemiah Nhando, Dept. of Medicine, University of Zimbabwe, Harare, Zimbabwe.
Victor Akelo, Kenya Medical Research Institute (KEMRI)-Centers for Disease Control (CDC), Kisumu, Kenya.
Sikhulile Moyo, Botswana Harvard AIDS Institute, Gaborone, Botswana.
Ravindre Panchia, Soweto HPTN CRS, Chris Hani Baragwanath Hospital, Soweto, 1860, South Africa.
Nagalingeswaran Kumarasamy, YRGCARE Medical Centre, Chennai, 600113, India.
Nuntisa Chotirosniramit, Research Institute for Health Sciences, Chiang Mai University, Chiang Mai, 50200, Thailand.
Marineide Gonçalves de Melo, Hospital Nossa Senhora da Conceição, Porto Alegre RS, 91350-200, Brazil.
Jose Pilotto, Hospital Geral de Nova Iguaçu and Laboratorio de AIDS e Imunologia Molecular (IOC/Fiocruz), Rio de Janeiro, 26030-380, Brazil.
Beatriz Grinsztejn, Instituto Nacional de Infectologia Evandro Chagas-INI-Fiocruz, Rio de Janeiro, 21045-900, Brazil.
Kenneth Mayer, Fenway Health and Infectious Disease Division, The Fenway Institute, Boston, MA, 02115, USA; Dept. of Medicine, Harvard Medical School, Beth Israel Deaconess Medical Center, Boston, MA, 02215, USA.
Ying Q. Chen, Vaccine and Infectious Disease Division and Public Health Science Division, Fred Hutchinson Cancer Research Institute, Seattle, WA, 98109, USA.
James P. Hughes, Dept. of Biostatistics, Univ. of Washington, Seattle, WA, 98195, USA.
Myron S. Cohen, Dept. of Medicine, Univ. of North Carolina at Chapel Hill, Chapel Hill, NC, 27514, USA.
REFERENCES
- 1.Cohen MS, Chen YQ, McCauley M, et al. Prevention of HIV-1 infection with early antiretroviral therapy. N Engl J Med. 2011;365:493–505. doi: 10.1056/NEJMoa1105243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen MS, Chen YQ, McCauley M, et al. Antiretroviral therapy for the prevention of HIV-1 transmission. N Engl J Med. 2016 doi: 10.1056/NEJMoa1600693. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grinsztejn B, Hosseinipour MC, Ribaudo HJ, et al. Effects of early versus delayed initiation of antiretroviral treatment on clinical outcomes of HIV-1 infection: results from the phase 3 HPTN 052 randomised controlled trial. Lancet Infect Dis. 2014;14:281–290. doi: 10.1016/S1473-3099(13)70692-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eshleman SH, Hudelson SE, Redd AD, et al. Analysis of genetic linkage of HIV from couples enrolled in the HIV Prevention Trials Network 052 trial. J Infect Dis. 2011;204:1918–1926. doi: 10.1093/infdis/jir651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ping LH, Jabara CB, Rodrigo AG, et al. HIV-1 transmission during early antiretroviral therapy: evaluation of two HIV-1 transmission events in the HPTN 052 prevention study. PLoS One. 2013;8:e71557. doi: 10.1371/journal.pone.0071557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giorgi EE, Funkhouser B, Athreya G, et al. Estimating time since infection in early homogeneous HIV-1 samples using a poisson model. BMC Bioinformatics. 2010;11:532. doi: 10.1186/1471-2105-11-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poon AF, Joy JB, Woods CK, et al. The impact of clinical, demographic and risk factors on rates of HIV transmission: a population-based phylogenetic analysis in British Columbia, Canada. J Infect Dis. 2015;211:926–935. doi: 10.1093/infdis/jiu560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brenner B, Wainberg MA, Roger M. Phylogenetic inferences on HIV-1 transmission: implications for the design of prevention and treatment interventions. AIDS. 2013;27:1045–1057. doi: 10.1097/QAD.0b013e32835cffd9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poon AF, Gustafson R, Daly P, et al. Near real-time monitoring of HIV transmission hotspots from routine HIV genotyping: an implementation case study. Lancet HIV. 2016;3:e231–e238. doi: 10.1016/S2352-3018(16)00046-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodger AJ, Cambiano V, Bruun T, et al. Sexual activity without condoms and risk of HIV transmission in serodifferent couples when the HIV-positive partner is using suppressive antiretroviral therapy. JAMA. 2016;316:171–181. doi: 10.1001/jama.2016.5148. [DOI] [PubMed] [Google Scholar]
- 12.Fogel JM, Ou S-S, Chen YQ, et al. Identification of factors associated with viral suppression and treatment failure when antiretroviral therapy is used for HIV prevention: Results from the HIV Prevention Trials Network (HPTN) 052 trial [MOPEC417]. Paper presented at: 8th IAS Conference on HIV Pathogenesis, Treatment, and Prevention; July 19–22, 201; Vancouver, Canada. [Google Scholar]
- 13.Markowitz M, Nguyen BY, Gotuzzo E, et al. Rapid and durable antiretroviral effect of the HIV-1 Integrase inhibitor raltegravir as part of combination therapy in treatment-naive patients with HIV-1 infection: results of a 48-week controlled study. J Acquir Immune Defic Syndr. 2007;46:125–133. doi: 10.1097/QAI.0b013e318157131c. [DOI] [PubMed] [Google Scholar]
- 14.Murray JM, Emery S, Kelleher AD, et al. Antiretroviral therapy with the integrase inhibitor raltegravir alters decay kinetics of, HIV, significantly reducing the second phase. AIDS. 2007;21:2315–2321. doi: 10.1097/QAD.0b013e3282f12377. [DOI] [PubMed] [Google Scholar]
- 15.Supervie V, Viard JP, Costagliola D, et al. Heterosexual risk of HIV transmission per sexual act under combined antiretroviral therapy: systematic review and bayesian modeling. Clin Infect Dis. 2014;59:115–122. doi: 10.1093/cid/ciu223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mujugira A, Celum C, Coombs RW, et al. HIV Transmission Risk Persists During the First 6 Months of Antiretroviral Therapy. J Acquir Immune Defic Syndr. 2016 doi: 10.1097/QAI.0000000000001019. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fogel JM, Wang L, Parsons TL, et al. Undisclosed antiretroviral drug use in a multinational clinical trial (HIV Prevention Trials Network 052) J Infect Dis. 2013;208:1624–1628. doi: 10.1093/infdis/jit390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fiebig EW, Wright DJ, Rawal BD, et al. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS. 2003;17:1871–1879. doi: 10.1097/00002030-200309050-00005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.