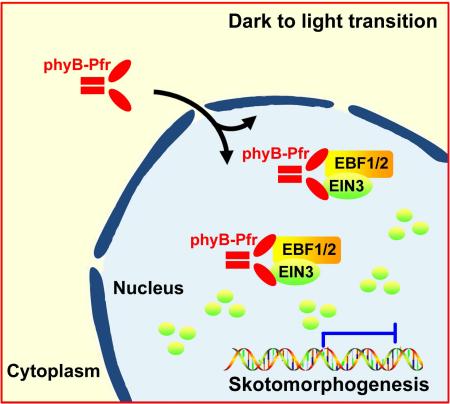
Summary
Plants germinating under subterranean darkness assume skotomorphogenesis, a developmental program strengthened by ethylene in response to mechanical pressure of soil. Upon reaching the surface, light triggers a dramatic developmental transition termed de-etiolation that requires immediate termination of ethylene responses. Here, we report that light-activation of photoreceptor phyB results in rapid degradation of EIN3, the master transcription factor in ethylene signaling pathway. As a result, light rapidly and efficiently represses ethylene actions. Specifically, phyB directly interacts with EIN3 in a light-dependent manner and also physically associates with F-box protein EBFs. The light-activated association of phyB, EIN3, and EBF1/EBF2 proteins stimulates robust EIN3 degradation by SCFEBF1/EBF2 E3 ligases. We reveal that phyB manipulates substrate-E3 ligase interactions in a light-dependent manner, thus directly controlling the stability of EIN3. Our findings illustrate a mechanistic model of how plants transduce the light information to immediately turn off ethylene signaling for de-etiolation initiation.
Keywords: seedling emergence, light signaling, phytochrome B, skotomorphogenesis, photomorphogenesis, ethylene signaling, EBF1 and EBF2, EIN3
Graphical Abstract
Introduction
As sessile organisms, plant growth has evolved to become highly plastic in response to environmental signals. Light is one of the most important environmental factors, serving as the energy source of photosynthesis and a cue for plant developmental programs (Chen et al., 2004; Jiao et al., 2007). The red and far-red wavelength of light is sensed by photoreceptor called phytochromes (phyA to phyE in Arabidopsis), which exist in two reversible forms in plants: the biologically inactive Pr and active Pfr forms (Quail et al., 1995; Schafer and Bowler, 2002). Light-activated phys translocate from the cytoplasm to the nucleus, where they directly interact with a subfamily of bHLH transcription factors, defined as phytochrome-interacting factors (PIFs) (Fankhauser and Chen, 2008; Nagy and Schafer, 2000; Ni et al., 1998; Quail, 2002). Specific binding by active phys induces rapid phosphorylation and degradation of PIFs, resulting in transcriptional changes in target genes (Al-Sady et al., 2006; Leivar and Monte, 2014; Shen et al., 2008). A recent study proposed that photo-activated phyB induces PIF3 phosphorylation to enhance the affinity of PIF3 for the LRB E3 ligases for degradation (Ni et al., 2014), providing a paradigm of how phyB transduces a light signal to alter gene expression.
Ethylene is a gaseous plant hormone regulating a wide range of physiological and cellular activities (Bleecker and Kende, 2000; Ecker, 1995). After being perceived by a family of ER-localized receptors, the ethylene signal is transduced through multi-step cascade to master transcription factors Ethylene-Insensitive 3 (EIN3) and EIN3-Like 1 (EIL1) in the nucleus (Alonso et al., 1999; Chang and Shockey, 1999; Chao et al., 1997; Hua and Meyerowitz, 1998; Kieber et al., 1993). EIN3/EIL1 are targeted for proteasome-mediated degradation by two closely related F-box proteins, EBF1 and EBF2, in the form of SCF E3 ligase complexes (Gagne et al., 2004; Guo and Ecker, 2003; Potuschak et al., 2003). Ethylene inhibits the action of EBF1/EBF2 at multiple levels (e.g. protein stability and translational regulation) to stabilize EIN3/EIL1, causing myriad ethylene responses (An et al., 2010; Ju and Chang, 2012; Li et al., 2015; Merchante et al., 2015; Qiao et al., 2012; Wen et al., 2012).
Terrestrial flowering plants often begin life under the soil. To adapt to the dark subterranean environment, germinating seedlings undergo a program known as skotomorphogenesis (Chory et al., 1989; Von Arnim and Deng, 1996). PIFs were previously identified as key promoters of skotomorphogenesis for seedlings grown in the dark and rapidly degrade upon light exposure (Leivar and Monte, 2014; Leivar et al., 2008). The extend of skotomorphogenic development is responsive to the level of ethylene, which quantitatively exaggerates apical hook formation and represses the opening and expansion of cotyledons to protect seedlings against mechanical injuries in penetrating the soil (Alonso et al., 1999; Ecker, 1995; Harpham et al., 1991; Shi et al., 2016; Zhong et al., 2014). EIN3 has been shown to repress the opening and expansion of cotyledons and promote apical hook formation, maintaining skotomorphogenesis in the dark (Ecker, 1995; Shi et al., 2016). During the emerging process, COP1 and ethylene channel the light and mechanical pressure information of soil overlay to control EIN3 protein levels, respectively (Shi et al., 2016; Zhong et al., 2014). EIN3 thus coordinately regulates seedling hypocotyl growth and cotyledon development in accordance with the soil conditions, ensuring that seedlings successfully emerge from the soil (Shi et al., 2016; Zhong et al., 2014). After growing out of soil to reach the sunlight, dark-grown seedlings undergo a rapid and dramatic developmental transition, termed de-etiolation, which involves apical hook unfolding, opening and expansion of cotyledons, and generation of the photosynthetic machinery to achieve photoautotrophic growth (Lau and Deng, 2012; Von Arnim and Deng, 1996). As EIN3 promotes the adapted phenotypes to the soil environment and antagonizes light-induced responses, its action must be rapidly turned off by light.
In this study, we found that photoreceptor phyB directly targets EIN3 protein for light-induced rapid degradation, relieving its suppressive effects on the morphological changes upon light exposure. Furthermore, phyB, upon light activation, directly interacts with EIN3 and acts as a scaffold to anchor and enhance the interaction of EIN3 and its E3 ligases EBF1/EBF2 in a light-dependent manner. This study defines an instant pathway in which photoreceptor directly promotes degradation of key transcription factors in endogenous hormone signaling to allow a critical developmental transition, de-etiolation.
Results
Photoreceptor phyB interacts directly with EIN3 in a red light-dependent manner
To characterize the role of ethylene in regulating light-induced morphological responses, we examined the effects of ethylene on seedling photomorphogenesis under red light exposure. In the absence of exogenous ethylene treatment, wild-type (WT, Col-0) seedlings displayed a shortened hypocotyl with open and expanded cotyledons (Figure S1A). When grown on medium containing ACC, a precursor of ethylene, the seedling hypocotyl was elongated, while the cotyledons were less expanded and curled (Figure S1A and S1B), suggesting that ethylene inhibits seedling photomorphogenesis in red light. The ethylene-imposed suppression on photomorphogenesis was further mimicked in the constitutive ethylene response mutant ctr1 or when the master transcription factor EIN3 was overexpressed in WT (EIN3ox) (Figure S1A and S1B). In addition, the ein3eil1 mutant showed a shorter hypocotyl and larger cotyledons, which could not be restored by ACC treatment (Figure S1A and S1B). We also investigated the role of ethylene and EIN3 in far-red and blue light. The results showed that neither ACC treatment nor EIN3 alternations could visibly change the hypocotyl lengths (Figure S1C and S1D), suggesting that far-red/blue light repress hypocotyl elongation via other dominant mechanisms. These results indicate that ethylene through EIN3 antagonizes red light actions in seedling photomorphogenesis.
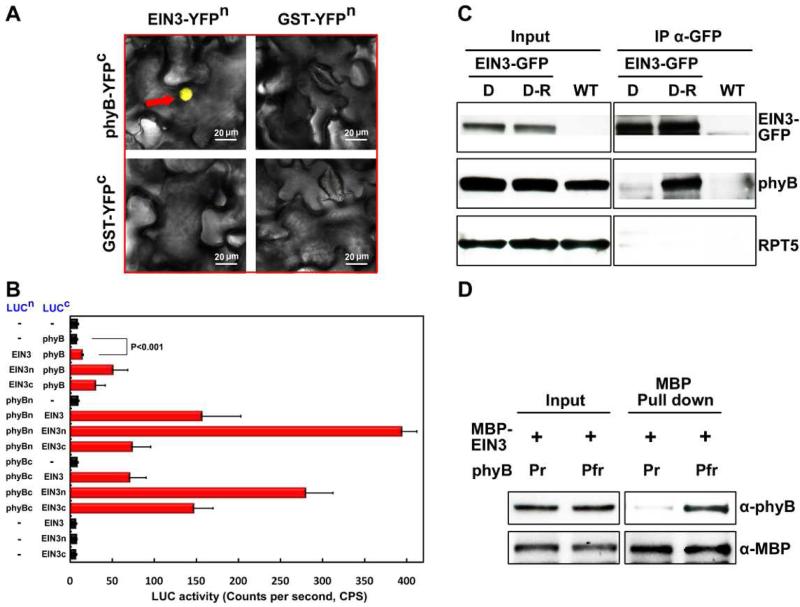
The photoreceptor phyB is known to physically interact with PIF (phytochrome interacting factors) transcription factors to elicit responses to red light (Leivar and Monte, 2014). The phenotypes of ein3eil1 and EIN3ox made us wonder whether EIN3 might be directly targeted by phyB. We performed a bimolecular fluorescence complementation (BiFC) assay in which the full-length sequences of EIN3 and phyB were fused with the split N-terminal and C-terminal portions of YFP (YFPn, YFPc), respectively. When these fusion proteins were co-expressed in tobacco leaves, we observed a strong YFP fluorescence signal (Figure 1A), suggesting that functional YFP was reconstituted via direct interaction of phyB and EIN3 in the nucleus. Co-expression of either EIN3 or phyB with the GST protein, as a control, could not reconstitute a functional YFP (Figure 1A). We next performed the firefly luciferase complementation imaging (LCI) assays in tobacco leaves to investigate which domains are responsible for the phyB-EIN3 interaction. We found that phyB strongly interacted with the N-terminal half of EIN3 (Figure 1B), which is the conserved region among EIN3 and EIN3-like proteins. Within the phyB protein, the N-terminus attaches the chromophore, and the C-terminal half mediates dimerization. Our results showed that EIN3 N-terminus can bind both regions of phyB, but with a preference to the N-terminus of phyB (Figure 1B), which was further supported by the interaction of phyB N-terminus and EIN3 homolog EIL1 (Figure S1E).
Figure 1. Photoreceptor phyB directly binds to the transcription factor EIN3 in a red light-dependent manner.
(A) BiFC assay detecting the interactions of phyB and EIN3. Full-length EIN3 and phyB were fused to the split N-terminal or C-terminal fragment of YFP (YFPn or YFPc), respectively. GST was used as a negative control. Reconstituted functional YFP is indicated with a red arrow. Bar = 20 μm.
(B) LCI assay showing the interactions of EIN3 and phyB in planta. The full-length sequence, the N-terminal or C-terminal fragments of EIN3 (EIN3, EIN3n, EIN3c) and phyB (phyB, phyBn, phyBc) were fused to the split N-terminal and C-terminal fragments of luciferase (LUCn and LUCc). “–“ indicates for the empty vectors as negative controls. C.P.S indicates for counts per second. Mean ± s.d., n=5. Student's t test was used to determine statistical significance.
(C) CoIP assay to detect the phyB-EIN3 association. 35S:EIN3-GFP/ein3eil1 (EIN3-GFP) and Col-0 (WT) seedlings were grown on 1/2 MS medium with 10 μM ACC for 4 days in the dark, and then either maintained in the dark (D) or exposed to red light (D-R) for 30 min. Proteins were then extracted and immunoprecipitated using anti-GFP antibody and immunoblotted using anti-GFP, anti-phyB and anti-RPT5 antibodies.
(D) In vitro pull-down assay for determining the interaction of the inactive Pr or active Pfr form phyB with MBP-EIN3. Recombinant phyB, added with either phycocyanobilin (Pfr) or methanol alone (Pr), was pre-treated with 0.5 hr of red light and then pulled down with MBP-EIN3 proteins. Anti-MBP and anti-phyB were used for immunoblot analysis.
To test whether the phyB-EIN3 interaction is red light dependent, we used transgenic plants over-expressing GFP-tagged EIN3 in an ein3eil1 background (EIN3-GFP) and performed co-immunoprecipitation assays. The seedlings were grown in the dark for 4 days and then either maintained in the dark (D) or exposed to red light (D-R) for 30 min before harvesting. We found that phyB can be strongly co-immunoprecipitated by EIN3-GFP only after red light exposure, while no interaction was detected in the dark (Figure 1C). Conversely, using phyB-GFP as bait in transgenic lines over-expressing phyB-GFP and EIN3-Myc proteins, we found that red light dramatically increased the co-immunoprecipitated levels of the EIN3-Myc protein (Figure S1F). Furthermore, the light-dependent, or the Pfr form-specific, interaction of phyB and EIN3 can be recapitulated in vitro. As shown in Figure 1D, the purified MBP-EIN3 proteins could specifically pull-down the red light-activated Pfr form of phyB, but not the Pr form (Figure 1D). These in vitro and in vivo data demonstrate that phyB directly binds to EIN3 in a red-light dependent manner.
phyB mediates red light-induced rapid degradation of EIN3
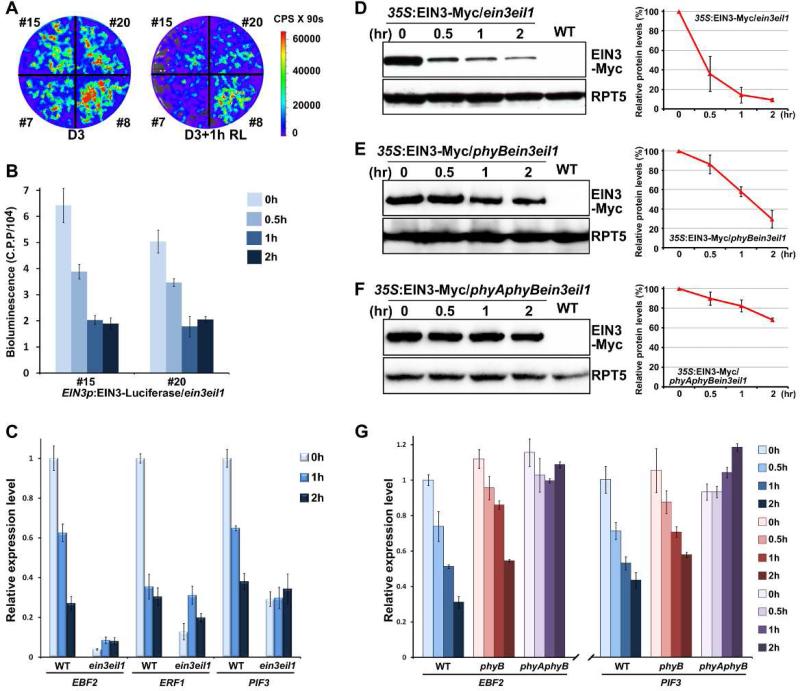
To investigate how light regulates EIN3, we used a transgenic plant expressing an EIN3 native promoter-driven EIN3-luciferase fusion protein in an ein3eil1 background (EIN3p:EIN3-Luciferase/ein3eil1) to monitor the dynamics of endogenous EIN3 proteins in seedlings upon red light exposure. Previous studies have shown that the luciferase activity of 35S:LUC is not affected by light (Shen et al., 2005). The luciferase imaging results showed that EIN3 protein levels were substantially decreased in four independent transgenic lines during dark-to-light transition (Figure 2A). Time-course quantification of luciferase activities in two representative lines showed that EIN3 protein levels in etiolated seedlings declined rapidly upon red light exposure, with a half-time of approximately 30 min, reaching a relatively steady low level within 1 hr (Figure 2B). We further examined the transcription levels of a few genes directly activated by EIN3, such as EBF2, ERF1 and PIF3 (Konishi and Yanagisawa, 2008; Solano et al., 1998; Zhong et al., 2012). The expression of all of these genes in WT was notably down-regulated upon light exposure (Figure 2C), while in ein3eil1, these genes were constitutively repressed in the dark and did not respond to light (Figure 2C). In fact, after 2 hr of light irradiation, the transcription of these genes in WT was decreased to levels similar to those in dark-grown ein3eil1 (Figure 2C). These results suggest that the activity of EIN3 is rapidly suppressed during the dark-to-light transition.
Figure 2. Red light induces rapid degradation of EIN3 through phyA/phyB to repress ethylene-responsive genes.
(A) Bioluminescent images of EIN3p-EIN3-Luciferase/ein3eil1 seedlings. Four independent transgenic lines were grown in the dark for 3 days, and then maintained in darkness (D3) or exposed to red light (D3+1h RL) for 1 hr before photographing. The color-coded bar indicates the intensity of luciferase activity. C.P.S is Counts Per Second.
(B) Quantitative analysis of EIN3-Luciferase bioluminescence with the time course of red light irradiation. Two representative lines of 3-day-old dark-grown EIN3p-EIN3-Luciferase/ein3eil seedlings were subjected to red light irradiation for the indicated times. C.P.P is Counts Per mg of Protein.
(C) qRT-PCR results showing the expression of EIN3 target genes in WT and ein3eil1. The seedlings were grown in the dark for 3 days (0h) and then subjected to red light irradiation for the indicated times. Gene transcription was normalized to PP2A. Mean ±s.d., n=3.
(D-F) Western blot results for determination of the protein levels of EIN3 upon light exposure. Seedlings over-expressing EIN3-Myc in an ein3eil1 (D), phyBein3eil1 (E) or phyAphyBein3eil1 (F) background were grown in the dark for 4 days (0 hr) and then transferred to red light irradiation for the indicated times. WT was used as a negative control. RPT5 was employed as a loading control. Right panels show the quantification analysis of three biological replicates of EIN3-Myc protein levels after normalization to RPT5. The dark EIN3-Myc protein level was set as 100%. Mean ± s.d., n=3.
(G) qRT-PCR results showing the expression of EIN3 target genes in WT, phyB and phyAphyB. Four-day-old etiolated seedlings were transferred to red light irradiation for the indicated times. Gene expressions were normalized to PP2A. Mean ± s.d., n=3.
As qRT-PCR results showed that light did not regulate the EIN3 transcripts of EIN3p:EIN3-Luciferase/ein3eil1 and the expression levels of both EIN3 and EIL1 genes were not significantly altered upon light exposure (Figure S2A and S2B), we next focused on the posttranscriptional regulation of EIN3. Using a transgenic plant constitutively expressing the EIN3-Myc fusion protein in the ein3eil1 mutant (35S:EIN3-Myc/ein3eil1) in which the transcripts of constitutively expressed EIN3-Myc were not affected by light (Figure S2C), we found that the protein levels of EIN3-Myc were rapidly decreased to 40% of its original level within 30 min and reached a steady low level after 1 hr of red light irradiation (Figure 2D). Similarly, the constitutively expressed EIL1-GFP proteins were also rapidly degraded upon red light exposure (Figure S2D).
In order to investigate whether phyB is involved in red light-induced EIN3 protein degradation, we induced the EIN3-Myc transgene into phyBein3eil1 background by genetic crossing. The EIN3-Myc protein in phyBein3eil1 showed a much slower rate of protein degradation, with 80% of the protein remaining after 30 min of red light irradiation (Figure 2E), indicating that phyB is mainly responsible for the rapid decrease of EIN3 proteins. Still, the protein levels of EIN3-Myc in phyBein3eil1 gradually declined to approximately 20% after 2 hr of light exposure (Figure 2E), suggesting that other phytochromes might also function redundantly. Finally, we obtained the EIN3-Myc/phyAphyBein3eil1 homozygous plants. In the absence of both phyA and phyB, the EIN3-Myc proteins were stable, maintaining a 70% of initial protein level after 2 hr of light exposure (Figure 2F). Consistent with the observed dynamics of EIN3 protein levels, the transcription of the EIN3-target genes EBF2 and PIF3 showed a slower rate of decrease in response to red light in phyB mutant, and exhibited almost no decline in the phyAphyB mutant (Figure 2G). Taken together, these results indicate that phyB primarily mediates red light-induced EIN3 protein degradation, along with redundant functioning of phyA.
Previous studies reported that phytochromes are activated by red light and reverted to the inactive form under far-red light irradiation or long-time dark incubation (Rockwell et al., 2006). We found that a short period of red light (Rp) exposure effectively induced the EIN3 protein degradation (Figure S2E), while this Rp induction could be blocked by an immediately subsequent far-red light pulse (Figure S2E). Moreover, when transferred the 4-day old continuous red light grown seedlings to dark incubation for 12 hr, EIN3 proteins were accumulated in the dark-adapted seedlings and could be rapidly degraded upon additional red light exposure (Figure S2F). These data further support the notion that light activation of the photoreceptors were required for the EIN3 protein degradation in red light.
Light triggers degradation of EIN3 via the EBF1/EBF2-mediated 26S proteasome pathway
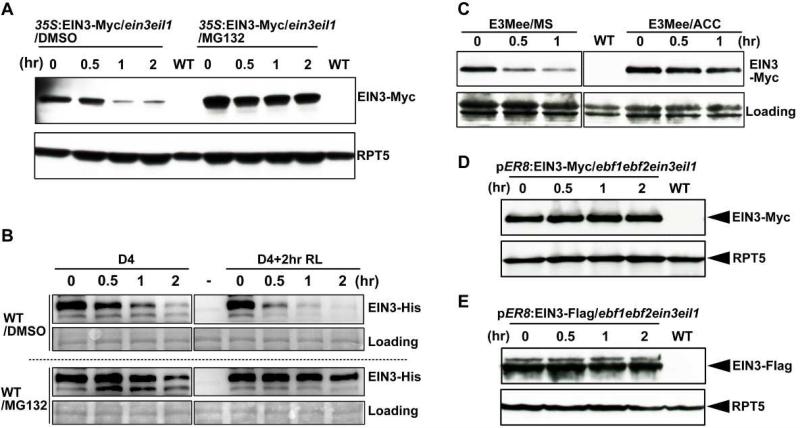
In ethylene signaling pathway, EIN3 is targeted by two F-box proteins, EBF1 and EBF2 (EBF1/EBF2), for proteasomal degradation (Gagne et al., 2004; Guo and Ecker, 2003; Potuschak et al., 2003). We found that light induced EIN3 degradation was also dependent on the 26S proteasome, as the degradation could be prevented by pre-treating the seedling with MG132, a 26S proteasome-specific inhibitor, without altering the EIN3 gene expression (Figure 3A and S3). We further examined the stability of EIN3 protein in a cell-free assay using purified full-length EIN3-His recombinant proteins. The results showed that EIN3 proteins were degraded at a much faster rate in cell extracts from seedlings subjected to 2 hr of red light irradiation compared with etiolated seedlings (Figure 3B), and that the addition of MG132 to the cell extracts largely prevented EIN3 degradation (Figure 3B). These data demonstrate that light-induced EIN3 degradation occurs through the 26S proteasome pathway.
Figure 3. EBF1 and EBF2 mediate light-induced EIN3 protein degradation through the 26S proteasome pathway.
(A) Western blot analysis of EIN3-Myc protein levels in EIN3-Myc/ein3eil1 (E3Mee) seedlings. Four-day-old dark-grown seedlings, pre-treated with MG132 or DMSO alone for 12 h, were exposed to red light for the indicated times. WT was used as a negative control. RPT5 was used as a loading control.
(B) Cell-free degradation assay of recombinant EIN3-His protein. WT seedlings were grown in the dark for 4 days, and then either maintained in the dark (D4) or exposed to red light (D4+2hr RL) for 2 hr before extraction. Equal amounts of recombinant EIN3-His proteins were added to the cell extracts, followed by incubation in darkness for the indicated times. For proteasome inhibitor treatment, 40 μM MG132 was pre-added in the cell extracts. “−” indicates for the no EIN3-His protein control. Ponceau S staining was used as a loading control.
(C) Western blot results showing the effects of ACC treatment on light-induced EIN3-Myc protein degradation. EIN3-Myc/ein3eil1 (E3Mee) seedlings were grown on 1/2 MS medium, with ACC supplemented (ACC) or without supplementation (MS), in the dark for 4 days, and then exposed to red light for the indicated times. WT was used as a negative control. Ponceau S staining was used as a loading control.
(D-E) Western blot results showing the requirements of EBF1 and EBF2 for light-induced EIN3 protein degradation. The seedlings were grown on 1/2 MS medium with 10 μM β-estradiol supplementation in the dark for 4 days, and then transferred to red light irradiation for the indicated times. WT was used as a negative control. RPT5 was used as a loading control.
As EBF1 and EBF2 are substrate receptors of SCF E3 ubiquitin ligase complexes that target EIN3 in response to ethylene status, we sought to determine whether EBF1 and EBF2 are also involved in light induced EIN3 degradation. We grew etiolated seedlings on medium containing ACC, a precursor of ethylene that has been reported to inhibit EBF1 and EBF2. The results showed that ACC treatment suppressed EIN3 degradation in response to light exposure (Figure 3C), suggesting that light-induced EIN3 degradation is likely dependent on the functions of EBF1 and EBF2. To verify this hypothesis, we generated transgenic lines expressing EIN3-Myc or EIN3-Flag fusion proteins under the beta-estradiol-induced promoter (pER8) in an ebf1ebf2ein3eil1 mutant background. We found that lack of EBF1 and EBF2 completely abolished light-induced EIN3 protein degradation in both tagged lines (Figures 3D and 3E). Collectively, we conclude that light induced EIN3 degradation requires the functions of EBF1 and EBF2.
EBF1 and EBF2 physically associate with phyB
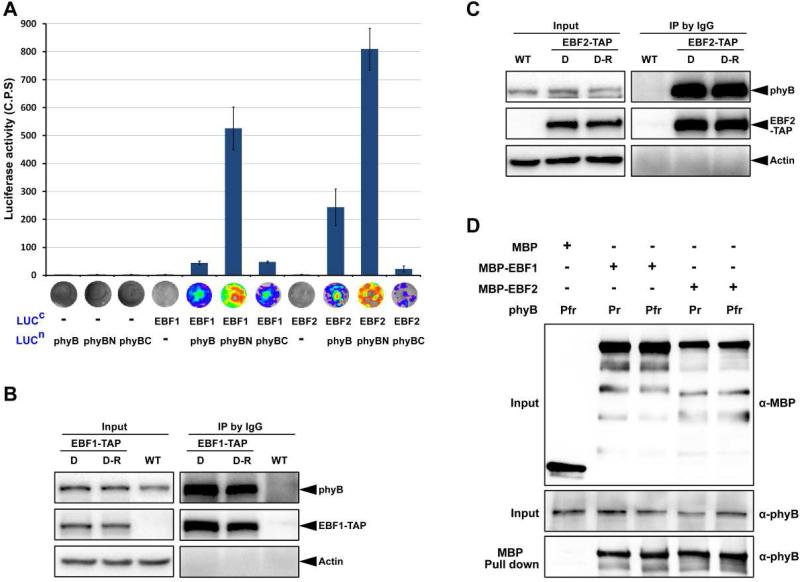
To determine the mechanism underlying light induced EIN3 degradation via EBF1/EBF2, we first examined whether phyB physically interacts with EBF1/EBF2. Firefly luciferase complementation imaging (LCI) assays in tobacco leaves demonstrated that phyB interacts with EBF1 or EBF2 in planta (Figure 4A). Moreover, EBF1 and EBF2 interacted most strongly with phyB N-terminal fragment (Figure 4A). To assess whether the interaction was light dependent, we performed the co-immunoprecipitation assays. Transgenic plants expressing EBF1-TAP or EBF2-TAP proteins were grown in the dark for 4 days and then either maintained in the dark (D) or exposed to red light (D-R) for 30 min before immunoprecipitation. Clearly, the immunoblot results showed that phyB proteins were co-immunoprecipitated by EBF1-TAP or EBF2-TAP at equivalent levels in the dark- and light-treated seedlings (Figures 4B and 4C). In vitro pull-down assays further demonstrated that the recombinant MBP-EBF1 or MBP-EBF2 proteins, but not the MBP protein alone, could pull-down phyB, regardless of the inactive Pr form or red light-activated Pfr form of phyB (Figure 4D). These co-immunoprecipitation and in vitro pull-down data indicate that phyB can physically associate with EBF1/EBF2 proteins, and both activated or inactive forms of phyB are capable of direct interaction with EBF1 and EBF2.
Figure 4. EBF1 and EBF2 directly associate with both the Pr and Pfr forms of phyB.
(A) LCI assay of the interactions of EBF1 and EBF2 with phyB in tobacco leaves. Full-length of EBF1 and EBF2 were fused to the split C-terminal (LUCc) fragments of luciferase. The full-length sequence, N-terminal and C-terminal fragments of phyB (phyB, phyBN, phyBC) were fused to the split luciferase N-terminus (LUCn). Empty vectors were used as negative controls. C.P.S is counts per second. Mean ± s.d., n=5.
(B and C) Co-immunoprecipitation assays for the interactions of EBF1 (B) or EBF2 (C) with phyB. Four-day-old dark-grown WT and transgenic seedlings over-expressing EBF1-TAP or EBF2-TAP seedlings were maintained in the dark (D) or exposed to red light (D-R) for 30 min. Proteins were extracted and immunoprecipitated using IgG Sepharose and immunoblotted using the indicated antibodies. Actin was used as a loading control.
(D) Pull-down assays to examine direct protein interactions in vitro. Recombinant phyB incubated with phycocyanobilin (Pfr) or methanol alone (Pr) was used as prey and pulled down using purified MBP, EBF1-MBP or EBF2-MBP proteins as bait.
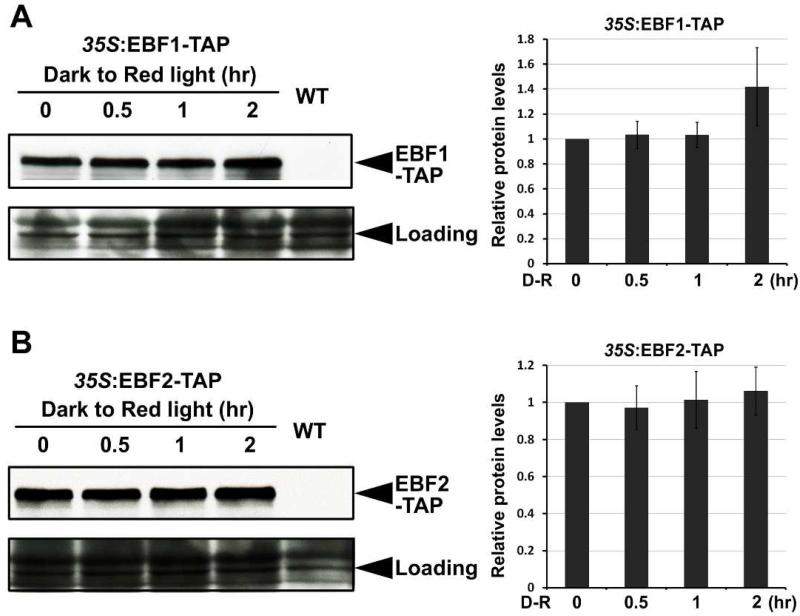
Short periods of strong light exposure do not alter EBF1/EBF2 protein levels
Light-induced degradation of EIN3 by EBF1/EBF2 can be achieved either through elevating EBF1/EBF2 levels or by enhancing EBF1/EBF2 activity towards EIN3. We have recently shown that EBF1/EBF2 levels are regulated by COP1 E3 ligase COP1 in response to dim light during seedling's growth under the soil (Shi et al., 2016). When seedlings constitutively expressing EBF1-TAP or EBF2-TAP proteins were grown for 4 days under a series of light intensities ranging from darkness to strong light, EBF1 and EBF2 levels were found elevated in the light samples compared to the dark ones (Figures S4A and S4B). Indeed, the change of EBF1/EBF2 protein levels were dependent on COP1, since EBF1 and EBF2 proteins exhibited over-accumulation in the cop1 mutant in both dark and light seedlings (Figures S4A and S4B), consistent with the previous report (Shi et al., 2016). Thus, continuous light irradiation leads to increases in EBF1 and EBF2 protein levels through COP1.
We next examined the change in EBF1/EBF2 protein levels during the dark-to-light transition, a condition that mimic natural de-etiolation. Under this condition, we found that both EBF1 and EBF2 were stable and did not exhibit obvious changes within 1 hr of light exposure (Figures 5A and 5B). Quantitative analysis of protein levels showed that after red light exposure for 2 hr, the abundance of EBF1 protein was slightly increased, while EBF2 remained unchanged (Figures 5A and 5B). Therefore, the protein levels of EBF1 and EBF2 were not altered under short periods of light irradiation. Noticeably, more than 80% of EIN3 proteins are rapidly degraded within the first 1 hr during the dark-to-light transition (Figure 2D). This acute light response cannot be explained by the changes of EBF1/EBF2 protein levels, which occurs over a long time scale.
Figure 5. Light exposure does not significantly affect EBF1 and EBF2 protein levels in a short time during the dark-to-light transition.
Western blot results showing EBF1 (A) and EBF2 (B) protein levels during the dark-to-light transition. Four-day-old dark-grown WT and transgenic seedlings over-expressing EBF1-TAP or EBF2-TAP seedlings were exposed to red light for the indicated times. Right panels show the quantification analysis of EBF1 (A, right panel) or EBF2 (B, right panel) protein levels after normalization to RPT5. EBF1-TAP or EBF2-TAP protein levels before light irradiation were set as 1. Mean ± s.d., n=3. RPT5 was used as a loading control.
Light-activated phyB promotes EBF1/EBF2 binding to EIN3
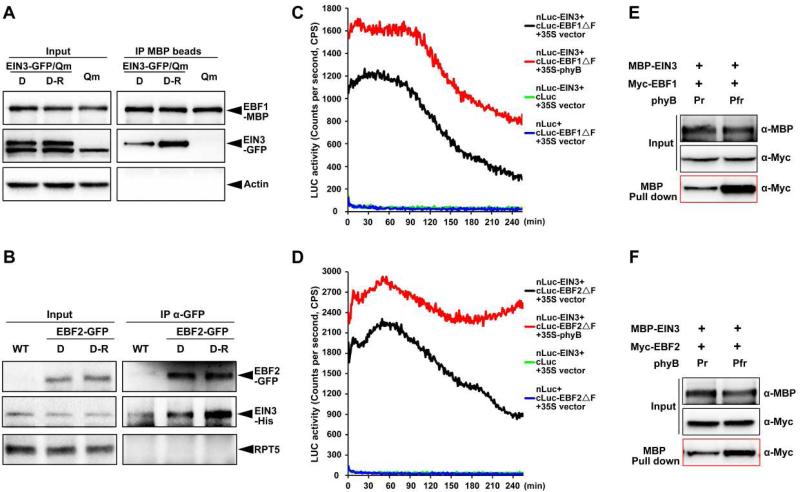
We next investigated whether light affects the binding affinity of EBF1/EBF2 to EIN3. In co-immunoprecipitation assays, 4-day-old dark-grown seedlings expressing EIN3-GFP fusion proteins in an ebf1ebf2ein3eil1 background were maintained in darkness or subjected to red light irradiation for 30 min, and cell extracts of which were mixed with equal amount of recombinant EBF1-MBP proteins. Precipitation of EBF1 using MBP beads pulled down substantially higher amount of EIN3-GFP in light-treated samples than the dark samples (Figure 6A). Conversely, we grew transgenic plants expressing EBF2-GFP in the dark for 4 days with an additional 30 min of dark or red light treatment, and added equal amount of purified EIN3-His proteins to the cell extracts. After immunoprecipitation using a GFP antibody, much more EIN3-His proteins were co-precipitated by EBF2-GFP in the red light-treated seedlings compared to the dark samples (Figure 6B). These findings indicate that light promotes the interactions between EBF1/EBF2 and EIN3 in seedlings.
Figure 6. Red light-activated phyB directly promotes the interaction of EIN3 with EBF1 and EBF2.
(A) Semi-in vivo co-immunoprecipitation assays of EIN3 and EBF1 proteins. Transgenic plants overexpressing EIN3-GFP in an ein3eil1ebf1ebf2 (EIN3-GFP/Qm) background and ein3eil1ebf1ebf2 (Qm, negative control) seedlings were grown in the dark for 4 days, and then maintained in the dark (D) or exposed to red light (D-R) for 30 min before extraction. Equal amounts of recombinant EBF1-MBP proteins were added to the cell extracts and immunoprecipitated with MBP beads.
(B) Semi-in vivo co-immunoprecipitation assay of EIN3 and EBF2 proteins. Four-day-old dark-grown WT and transgenic seedlings overexpressing EBF2-GFP were maintained in the dark (D) or exposed to red light (D-R) for 30 min before harvesting. Equal amounts of recombinant EIN3-His proteins were added to the cell extracts and immunoprecipitated with GFP antibodies.
(C and D) LCI assays to detect the effects of phyB on the interactions of EIN3 with EBF1 or EBF2 in Arabidopsis protoplasts. Full-length EIN3 was fused to the split N-terminal (nLuc) fragment of luciferase. EBF1 or EBF2 with a deletion in the F-box domain was fused to the split luciferase C-terminus (cLuc). Full-length phyB was driven by the 35S promoter. Empty vectors were used as negative controls. The plasmids in each combination were transiently co-transformed into Arabidopsis ein3eil1ebf1ebf2 protoplasts. C.P.S is counts per second.
(E and F) In vitro pull-down assays to examine the effects of phyB on the interaction of EIN3 with EBF1 or EBF2. Recombinant EBF1-Myc or EBF2-Myc was incubated with purified EIN3-MBP. An equal amount of phyB (as Pfr or Pr form) was added in each reaction, and MBP beads were used for precipitation.
Given the direct interactions of phyB with both EBF1/EBF2 and EIN3, we performed Co-IP assay to examine the association of phyB, EBF1/EBF2, and EIN3 proteins in vivo. We used the EIN3-GFP/EBF1-TAP or EIN3-GFP/EBF2-TAP transgenic plants and blocked EIN3 degradation by treating with proteasomal inhibitor MG132. The results showed clearly that both EIN3 and phyB proteins were co-precipitated with EBF2 at the same time (Figure S5), indicating the existence of phyB-EBF2-EIN3 tripartite complex in planta. However, the EIN3 proteins were rarely detected in the EBF1 Co-IP assay (Figure S5). The possibility is that EBF1-TAP protein levels are much lower than EBF2-TAP in the transgenic plants, which can be observed in the input anti-Myc lane (Figure S5). We next sought to determine whether phyB regulates EBF1/EBF2 binding to EIN3. To prevent EIN3 degradation, we used F-box domain-deleted fragments of EBFs, EBF1ΔF or EBF2ΔF, and performed LCI assays in Arabidopsis ein3eil1ebf1ebf2 mutant protoplasts. Our results demonstrated that the luciferase activity was reconstituted by coexpressing nLuc-EIN3 with either cLuc-EBF1ΔF or cluc-EBF2ΔF in protoplasts (Figures 6C and 6D), suggesting that EIN3 directly interacts with EBF1ΔF and EBF2ΔF. Moreover, when phyB was added to the coexpression system, luciferase activity was notably elevated, indicating that phyB enhanced the interactions of EBF1/EBF2 and EIN3 in vivo (Figures 6C and 6D). Furthermore, we performed in vitro pull-down assays using recombinant Myc- EBF1/EBF2 and MBP-EIN3 with an equal amount of the Pr or Pfr form of phyB. Compared with inactive Pr phyB, much stronger interactions between EBF1/EBF2 and EIN3 were found when red light-activated Pfr phyB was added to the reaction systems (Figures 6E and 6F). Collectively, these data reveal that high fluence light rapidly shuts down EIN3 by photo-activated phyB directly gluing EIN3 to E3 ligases EBF1/EBF2.
Negative regulation of EIN3 by phyB is critical for the de-etiolation process upon light exposure
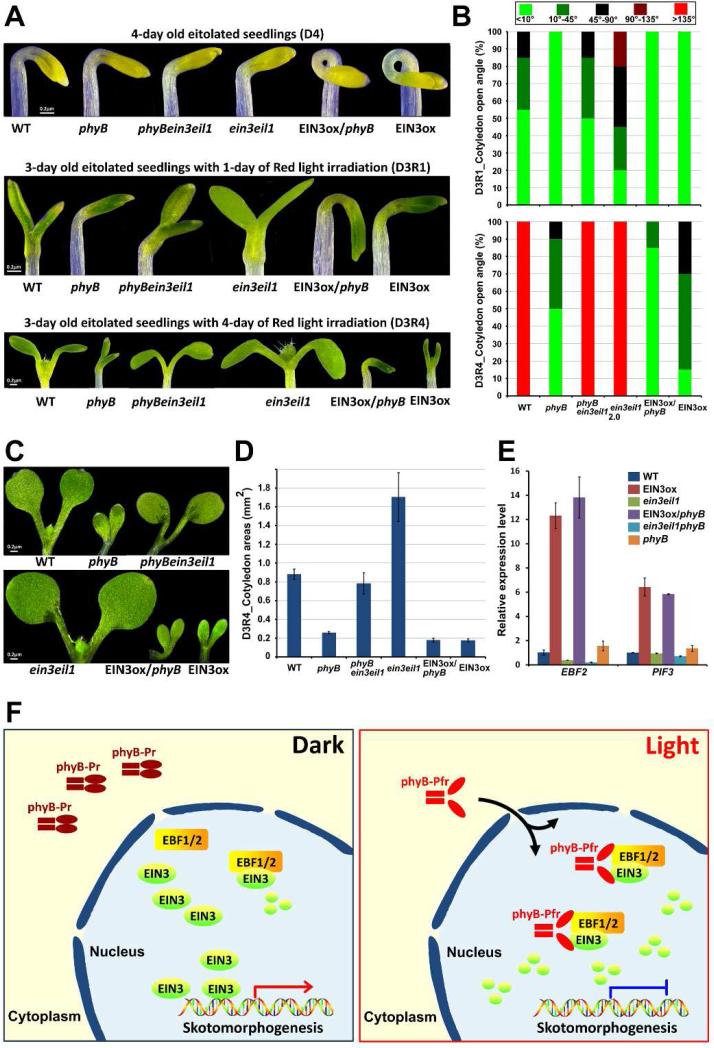
phyB plays an important role in de-etiolation of subterranean dark-grown seedlings upon light exposure, a critical transition to photoautotrophic growth (Quail et al., 1995; Schafer and Bowler, 2002). Upon transfer of etiolated seedlings to red light, phyB mutants exhibited defects in cotyledon separation and expansion compared to WT (Figures 7A-D). Notably, the cotyledons of ein3eil1 opened faster, while those of EIN3ox opened more slowly than WT (Figures 7A and 7B). By measuring the cotyledon areas of the seedlings at day 3 of growth under red light after transferred from darkness, we found that the cotyledons of ein3eil1 were dramatically expanded, while EIN3ox displayed very little growth in cotyledons (Figure 7C and 7D). These results indicate that EIN3 negatively regulates red light-induced cotyledon separation and expansion. Lack of EIN3 (ein3eil1) causes a red-light hypersensitive response characterized by exaggerated expansion and faster opening, while gain-of-function EIN3ox displays a hypo-photomorphogenic phenotype in cotyledon.
Figure 7. Light induces EIN3 degradation via phyB to remove EIN3-maintained skotomorphogenesis for de-etiolation initiation.
(A and B) Images of seedling cotyledon phenotypes (A), and the angle of cotyledon opening (B). The seedlings were grown in the dark for 3 days and then maintained in the dark for 1 day (D4) or exposed to red light for 1 (D3R1) or 4 days (D3R4).
(C and D) Images of cotyledons (C) and the areas (D) of a single cotyledon. The seedlings were grown on 1/2 MS medium in the dark for 3 days and then exposed to red light for an additional 4 days (D3R4). Mean ± s.d., n>20.
(E) qRT-PCR results showing the expression of light-regulated EIN3 target genes in WT, EIN3ox, ein3eil1, EIN3ox/phyB, ein3eil1phyB and phyB. The seedlings were grown in the dark for 3 days and then subjected to additional red light exposure for 2 hr. Gene transcription was normalized to PP2A. Mean ±s.d., n=3.
(F) A working model depicting how phyB directly transduces the light signal to turn off EIN3-maintained skotomorphogenesis. Red light activates phyB, leading to its translocation from the cytoplasm to the nucleus, where phyB directly binds to both EIN3 and its E3 ligases EBF1/EBF2, enhancing the interactions of EBF1/EBF2 and EIN3 to rapidly degrade EIN3. As a result, light, acting through the photoreceptor, directly abolishes the action of EIN3, eliminating EIN3's repression on de-etiolation.
We conducted a genetic interaction experiment for phyB and EIN3 by crossing ein3eil1 and EIN3ox into phyB, respectively. In the dark, where phyB is naturally inactive, phyBein3eil1 and EIN3ox/phyB seedlings looks similar to ein3eil1 and EIN3ox, respectively (Figures 7A and 7B). Upon transfer to red light, phyBein3eil1 largely restored the cotyledon phenotypes of phyB to WT-like (Figures 7A-D). On the other hand, the EIN3ox/phyB seedlings displayed a closed and much less expanded cotyledons (Figures 7A-D). Furthermore, we examined the expression levels of two light-regulated EIN3 target genes EBF2 and PIF3 in the 3-day old etiolated seedlings when transferred to red light for 2 hr. Consistently, these gene expression in EIN3ox/phyB were similar to that of EIN3ox (Figure 7E), and EIN3/EIL1 mutation (ein3eil1) in phyB background rescued the high expression levels of EBF2 and PIF3 inphyB mutant (Figure 7E). Taken together, these results support the idea that EIN3 suppresses light responses, particularly the cotyledon opening and expansion, while phyB acts to eliminate EIN3 activity upon light activation.
Discussion
Light and mechanical pressure are two major environmental signals in regulating seedling emergence from the soil. Buried seedlings adopt the skotomorphogenesis strategy to grow under the soil (Von Arnim and Deng, 1996; Wei et al., 1994), where gaseous hormone ethylene is proportionally produced in response to the physical barrier to promote skotomorphogenesis and facilitate soil emergence (Goeschl et al., 1966; Shi et al., 2016; Zhong et al., 2014). Upon reaching the surface, light triggers de-etiolation, as characterized by cotyledon opening, greening, and expansion, which marks a critical transition to photosynthetic growth (Huq et al., 2004; Von Arnim and Deng, 1996; Wei et al., 1994). For buried seedlings emerging from the soil, this transition is rapid and dramatic, requiring rapid termination of skotomorphogenesis.
EIN3 is a master transcription factor in repressing seedling photomorphogenesis upon the initial dark-to-light transition
COP1 is a well-documented central repressor of photomorphogenesis, evidenced by the fully open and expanded cotyledons of cop1 mutant in the dark (Deng et al., 1992). Our recent study showed that over-expressing EIN3 alone can suppress the constitutive photomorphogenic phenotypes of cop1, resulting in closed and unexpanded cotyledons as those of WT (Shi et al., 2016; Zhong et al., 2009). Moreover, EIN3 has been shown to directly activate HSL1 transcription to enable apical hook formation (An et al., 2012), and act downstream of COP1 to suppress protochlorophyllide biosynthesis (Zhong et al., 2009). In this study, we reveal that the etiolated seedlings of ein3eil1 mutant are hypersensitive, while the EIN3ox are extremely insensitive to light in cotyledon opening and expansion. Furthermore, mutation of EIN3/EIL1 can largely restore the insensitive phenotypes of phyB mutant in response to red light exposure. Collectively, EIN3/EIL1 act as master transcription factors to antagonize light-induced morphological responses, suppressing seedling photomorphogenesis.
When growing under the soil, EIN3 protein levels are quantitatively elevated in response to the mechanical pressure of soil covering, so that the extent of skotomorphogenic development could be accordingly modulated in response to the soil conditions (Shi et al., 2016; Zhong et al., 2014). In addition to EIN3, a group of bHLH transcription factors named PIFs has previously been identified to promote skotomorphogenesis (Leivar et al., 2008; Shin et al., 2009). The fact that ein3eil1 cannot fully restore the insensitive phenotypes of phyB in response to light suggests that EIN3/EIL1 likely cooperate with other factors, such as PIFs, in suppressing the de-etiolation process. PIFs and EIN3 represent the two types of transcription factors mediating skotomorphogenesis, and their functions interconnect with each other in multiple ways. PIF1 was found to function cooperatively with EIN3 in regulating the greening process of etiolated seedlings (Zhong et al., 2009). EIN3 directly activates PIF3 transcription to promote seedling hypocotyl elongation in light (Zhong et al., 2012). Consistently, we found that the expression levels of PIF3 in WT are reduced to that in ein3eil1 mutant upon light when EIN3 proteins are degraded (Figure 2C). Moreover, over-expressing PIF5 in the dark largely increases ethylene production and stabilizes EIN3 to cause ethylene responses (Khanna et al., 2007). Given the essential roles of PIFs and EIN3 in promoting skotomorphogenesis, it will be of great interest to investigate how PIFs are modulated in the soil and what the relationship of PIFs and EIN3 is in regulating seedling emergence.
Plants emerging from subterranean darkness utilize two different mechanisms to inactivate EIN3
In a recent study, we identified an E3 ubiquitin ligase cascade in which COP1 maintains skotomorphogenesis by degrading EBF1/EBF2 and stabilizing EIN3 in subterranean darkness (Shi et al., 2016). This mechanism is responsive to dim light in the soil, which gradually suppress COP1 to decrease EIN3 accumulation as the seedlings slowly approach the surface. Here, we report that plants utilize a different mechanism for light-induced rapid de-etiolation, which operates when seedlings break the soil and are exposed to high fluence light. In this mechanism, photoreceptor phyB directly stimulates the degradation of EIN3 by recruiting and affixing EIN3 to EBF1/EBF2, substrate receptors of SCF ubiquitin E3 ligase (Figure 7F). This instant mode of direct phyB-EIN3 pathway is supported by several lines of evidence: (1) EIN3 accumulates in etiolated seedlings and is rapidly degraded upon light exposure within 1 hr; (2) phyB and EBF1/EBF2 are required for EIN3 degradation in light; (3) Light-activated phyB specifically interacts with EIN3; (4) The levels of EBF1/EBF2 do not change under a short period of light exposure; (5) phyB directly associates with EBF1/EBF2 and markedly enhances the interactions of EBF1/EBF2 and EIN3 proteins.
Together with previous studies, our findings show that EIN3 proteins undergo light-induced degradation through at least two mechanisms. The COP1-EBF-EIN3 pathway fine-tunes EIN3 levels through COP1-controlled EBF1/EBF2 stability, by which seedlings adjust subterranean growth according to gradual changes of dim light in the soil. The direct phyB-EIN3 pathway, on the other hand, represents a robust instant mode that operates to quickly shut down the ethylene responsive genes when seedlings emerge into light environment. These two pathways synergistically halt ethylene responses of the emerging seedlings to achieve de-etiolation.
Light-triggered termination of ethylene signaling is an integral part for initiating the de-etiolation switch
In the soil, seedlings undergo skotomorphogenesis to push its shoot apical tip up through the soil, and produce the gaseous hormone ethylene in response to the physical barrier. Ethylene quantitatively enhances apical hook formation and suppresses cotyledon growth to increase the soil penetration ability (Alonso et al., 1999; Harpham et al., 1991; Shi et al., 2016; Zhong et al., 2014). Given the strong effects of ethylene, upon breaking the soil, plants have to immediately terminate the ethylene signaling to initiate de-etiolation: i.e. unhook the apical shoot to allow the growth of true leaves, remove the repression on cotyledon development, and lift the inhibition of photosynthetic gene expression. Thus, de-etiolation must involve cross-talks of light and hormonal signaling. Our studies unequivocally demonstrate that terminating ethylene signaling is an integral part of de-etiolation. Naturally, the switch of de-etiolation is triggered when the emergent seedlings sense a burst of light irradiation and a simultaneous acute decrease of ethylene due to its evaporation into open air. At molecular level, this study reveals an instant mode in which photoreceptor phyB directly targets EIN3, a master transcription factor in hormonal pathways, to rapidly end ethylene repression of photomorphogenesis upon dark-to-light transition.
phyB acts as a light-reversible “molecular glue” that promotes target protein degradation by enhancing E3 ligase binding
The red and far-red light signals that trigger the de-etiolation switch are perceived by the phytochrome family of photoreceptors (phys) (Quail et al., 1995; Schafer and Bowler, 2002). phyB is critical for photomorophogenesis as the constitutively active phyBY276H transgenic seedlings displaying photomorophogenic phenotypes in the dark (Su and Lagarias, 2007). How phys transduce the information provided by light to modulate growth and developmental responses is a central question in plant photobiology, and has been extensively studied in past decades. Thus far, photo-activated phys are found to directly interact with and induce PIFs phosphorylation via unknown kinase(s), allowing PIFs to be targeted by their E3 ligases (Leivar and Monte, 2014). Recently, LRB was identified as an ubiquitin E3 ligase that concurrently degrades phyB and PIF3 (Christians et al., 2012; Ni et al., 2014). This mechanism has been postulated to function predominantly to desensitize the phyB signal (Ni et al., 2014).
Our findings provide mechanistic insights into how phys transduce light signals to cause rapid degradation of downstream transcription factors. Different from the phyB-LRB-PIF3 pathway, phyB directly enhances the binding of transcription factor EIN3 to its E3 ligases EBF1/EBF2 by acting as a “molecular glue”, resulting in EIN3 degradation mediated by SCFEBF1/EBF2. Interestingly, EIN3 specifically interacts with the light-activated Pfr form of phyB, while EBF1/2 physically associate with both the inactive Pr and activated Pfr phyB. This scheme would ensure the signal dependence for light, while the physical association of phyB with EBF1/EBF2 could increase the efficiency of EIN3 degradation.
Ubiquitin E3 ligase-dependent proteolysis controls diverse signal-dependent processes in plants. Auxin stimulates the interactions between the F-box protein TIR1 and its substrate AUX/IAA, leading to ubiquitination of AUX/IAA by SCFTIR1 (Gray et al., 2001; Tan et al., 2007). Similar scheme also applies to jasmonic acid induced ubiquitination of JAZ proteins by SCFCOI1 (Sheard et al., 2010). In the case of GA signaling, the binding of GA to its receptor GID1 changes the conformation of the receptor that enhances the binding of DELLA and its F-box protein, resulting in ubiquitination of DELLA by SCFSly1/2 (Murase et al., 2008). Analogous to the GA signaling, we show here that photo-activated chromophore, similar to hormone molecules, causes a conformational change of photoreceptor phyB that enhances the binding of EIN3 to its F-box proteins EBF1/EBF2, leading to EIN3 ubiquitination by SCFEBF1/EBF2. With this study, light signaling can now join various plant hormonal pathways in utilizing a familiar scheme of ligand activated receptor-substrate-SCF in signal sensing and relay, a successful signal transduction strategy that has prevailed in plants.
Experimental Procedures
Plant material and growth conditions
The wild-type (WT) Arabidopsis plants used in this study were of the Columbia-0 (Col-0) ecotype unless otherwise specified. EIN3ox, ein3eil1, phyA, phyB, 35S:EIN3-GFP/ein3eil1, EIN3p-EIN3-Luciferase/ein3eil1, 35S:EIN3-Myc/ein3eil1, 35S:EBF1-TAP/Col-0, 35S:EBF2-TAP/Col-0, 35S:EBF2-GFP/Col-0, and 35S:EIL1-GFP/Col-0 were previously reported (An et al., 2010; Rubio et al., 2005; Shi et al., 2016; Stepanova et al., 2005; Zhong et al., 2014; Zhong et al., 2009). Multiple mutants were generated through crossing, and homozygous lines were confirmed through genotyping.
Seedlings were grown on 1/2 Murashige and Skoog (MS) medium. For chemical treatments, 10 μM ACC or 10 μM β-estradiol was supplied in the medium, unless otherwise specified. Seedlings were treated with a 50 μM MG132 or DMSO solution added to the 1/2 MS medium 12 hours prior to harvesting. Approximately 60 μmolm−2s−1 of red light was used for light irradiation treatments, and approximately 10 μmolm−2s−1 of far-red light or 15 μmolm−2s−1 of blue light were used for seedling growth under continuous light. Arabidopsis plants were grown under long-day-photoperiod white light at 22°C, and tobacco plants were grown under long-day-photoperiod white light at 26°C. For phenotype analysis, 3-day-old dark-grown seedlings were exposed to red light for 1 or 4 days. The angle between two cotyledons and the cotyledon area were recorded using Image J software. At least 20 cotyledons were measured in each set of experiments.
Immunoblot assays
Seedlings were ground to powder in liquid nitrogen, and total proteins were extracted in buffer (50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.25% Triton X-100, 1 mM PMSF). For immunoblot detection, anti-Myc (Abcam, 1:1000), anti-Actin (Sigma, 1:3000), anti-His (Cell signaling, 1:1000), anti-MBP (NEB, 1:1500), anti-GFP (Abmart, 1:800), anti-phyB (1:1000), and anti-RPT5 (1:1000) antibodies were used.
Co-immunoprecipitation and pull-down assays
Co-immunoprecipitation assays were performed as previously described (Shi et al., 2013). Purified recombinant proteins (1 μg) were added to 500 μg of total extracted proteins, and the solution was incubated at 4°C for 1 hr with gentle rotation before precipitation. After immunoprecipitation, the beads were washed with lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.1% Tween 20, and 13 Roche protease inhibitor cocktail) 4 times, and collected for immunoblotting detection. In the western blot detection, the Input and IP samples were run on different gels due to the different film exposure time.
Pull-down assays were carried out as previously described (Shi et al., 2016). The indicated in vitro-expressed recombinant proteins were added to the binding buffer (20 mM Tris-HCl, pH=7.5, 150 mM NaCl and 1 mM EDTA), followed by incubation at 4°C for 1 hr with gentle rotation. Amylose resins (20 μl) were then added to the solution, followed by incubation for an additional 1 hr to precipitate the MBP-containing proteins. The resins were finally washed and collected for immunoblotting.
RNA extraction and qRT-PCR
Seedlings were harvested and ground to powder in liquid nitrogen. The spectrum Plant Total RNA Kit (Sigma) was used to extract total RNA, and 2 μg of the RNA was employed to synthesize cDNA using the SuperScript III First-Strand Synthesis System (Invitrogen). Real-time PCR was performed on the ABI Fast 7500 Real-Time system using SYBR Green Mix (Takara). All qRT-PCR experiments were biologically repeated three times, and representative results are shown.
Cell-free degradation and BiFC assays
Cell-free degradation and BiFC assays were carried out as previously described (Shi et al., 2016; Shi et al., 2015; Shi et al., 2013).
Luciferase bioluminescence imaging and luciferase activity measurements
Luciferase bioluminescence imaging was carried out as previously described with minor modifications (Shi et al., 2015; Shi et al., 2013). A 1 mM D-luciferin potassium salt solution (BIOTIUM) was sprayed onto seedlings, followed by incubation in the dark for 10 min. Images of luciferase bioluminescence were obtained using a Xenogen IVIS Spectrum imaging system (Caliper).
Twenty seedlings were harvested and ground to powder in liquid nitrogen. The Firefly Luciferase Assay Kit (BIOTIUM) was used to measure luciferase activity according to the manufactory's instructions.
LCI assay in tobacco leaves and Arabidopsis protoplasts
The Luciferase Complementation Imaging (LCI) assay was carried out in tobacco leaves and Arabidopsis protoplasts as previously described (Chen et al., 2008; Li et al., 2015), with minor modifications. Agrobacterium bacteria containing individual construct were suspended in the infiltration medium to a final concentration of OD600 = 0.4, and the two constructs in each pair were mixed equally. To avoid the bias of transfection efficiency, we used five individual tobacco plants. Biological triplicates were performed for every assay and the representative results were presented. Protoplasts were lysed from flat leaves of 4-week-old ein3eil1ebf1ebf2 mutant plants according to the protocol provided by Jen Sheen's laboratory (http://genetics.mgh.harvard.edu/sheenweb/). Then, 10 μg (<10 μl) of each plasmid was transferred to the protoplasts. The resultant luciferase activity was detected using a BERTHOLD TECHNOLOGIES Multimode Reader LB942.
Supplementary Material
Highlights.
Photoreceptor phyB directly interacts with EIN3 in a red light-dependent manner
phyB mediates red light-induced rapid degradation of EIN3 protein via EBF1/EBF2
phyB enhances E3 ligase binding in a light-dependent manner
phyB-EIN3 module transduces light signal to immediately turn off ethylene responses
Acknowledgements
We greatly appreciate Prof. Hongwei Guo from Peking University for his valuable suggestions and for generously providing us the EBF1/EBF2-TAP and EBF2-GFP plant seeds. This work was supported by grants from the National Key Research and Development Program of China (2016YFA0502900), Natural Science Foundation of China (31330048, 31621001) and US NIH (GM047850); H.S. was supported by a China Postdoctoral Science Foundation Grant (2015T80014).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
S.Z. and X.W.D. conducted the research; S.Z. and H.S. designed the experiments; H.S., X.S., R.L., and C.X. performed the experiments; S.Z., X.W.D., H.S. and N.W. analyzed data; and S.Z., H.S., N.W. and X.W.D. wrote the paper.
References
- Al-Sady B, Ni W, Kircher S, Schafer E, Quail PH. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell. 2006;23:439–446. doi: 10.1016/j.molcel.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science. 1999;284:2148–2152. doi: 10.1126/science.284.5423.2148. [DOI] [PubMed] [Google Scholar]
- An F, Zhang X, Zhu Z, Ji Y, He W, Jiang Z, Li M, Guo H. Coordinated regulation of apical hook development by gibberellins and ethylene in etiolated Arabidopsis seedlings. Cell Res. 2012;22:915–927. doi: 10.1038/cr.2012.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An F, Zhao Q, Ji Y, Li W, Jiang Z, Yu X, Zhang C, Han Y, He W, Liu Y, et al. Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell. 2010;22:2384–2401. doi: 10.1105/tpc.110.076588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleecker AB, Kende H. Ethylene: a gaseous signal molecule in plants. Annu Rev Cell Dev Biol. 2000;16:1–18. doi: 10.1146/annurev.cellbio.16.1.1. [DOI] [PubMed] [Google Scholar]
- Chang C, Shockey JA. The ethylene-response pathway: signal perception to gene regulation. Curr Opin Plant Biol. 1999;2:352–358. doi: 10.1016/s1369-5266(99)00004-7. [DOI] [PubMed] [Google Scholar]
- Chao QM, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR. Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell. 1997;89:1133–1144. doi: 10.1016/s0092-8674(00)80300-1. [DOI] [PubMed] [Google Scholar]
- Chen HM, Zou Y, Shang YL, Lin HQ, Wang YJ, Cai R, Tang XY, Zhou JM. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiology. 2008;146:368–376. doi: 10.1104/pp.107.111740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Chory J, Fankhauser C. Light signal transduction in higher plants. Annual Review of Genetics. 2004;38:87–117. doi: 10.1146/annurev.genet.38.072902.092259. [DOI] [PubMed] [Google Scholar]
- Chory J, Peto C, Feinbaum R, Pratt L, Ausubel F. Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light. Cell. 1989;58:991–999. doi: 10.1016/0092-8674(89)90950-1. [DOI] [PubMed] [Google Scholar]
- Christians MJ, Gingerich DJ, Hua Z, Lauer TD, Vierstra RD. The Light-Response BTB1 and BTB2 Proteins Assemble Nuclear Ubiquitin Ligases That Modify Phytochrome B and D Signaling in Arabidopsis. Plant Physiol. 2012;160:118–134. doi: 10.1104/pp.112.199109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng XW, Matsui M, Wei N, Wagner D, Chu AM, Feldmann KA, Quail PH. COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell. 1992;71:791–801. doi: 10.1016/0092-8674(92)90555-q. [DOI] [PubMed] [Google Scholar]
- Ecker JR. The ethylene signal transduction pathway in plants. Science. 1995;268:667–675. doi: 10.1126/science.7732375. [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Chen M. Transposing phytochrome into the nucleus. Trends in Plant Science. 2008;13:596–601. doi: 10.1016/j.tplants.2008.08.007. [DOI] [PubMed] [Google Scholar]
- Gagne JM, Smalle J, Gingerich DJ, Walker JM, Yoo SD, Yanagisawa S, Vierstra RD. Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proc Natl Acad Sci U S A. 2004;101:6803–6808. doi: 10.1073/pnas.0401698101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeschl JD, Rappaport L, Pratt HK. Ethylene as a factor regulating the growth of pea epicotyls subjected to physical stress. Plant Physiol. 1966;41:877–884. doi: 10.1104/pp.41.5.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature. 2001;414:271–276. doi: 10.1038/35104500. [DOI] [PubMed] [Google Scholar]
- Guo H, Ecker JR. Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell. 2003;115:667–677. doi: 10.1016/s0092-8674(03)00969-3. [DOI] [PubMed] [Google Scholar]
- Harpham NVJ, Berry AW, Knee EM, Rovedahoyos G, Raskin I, Sanders IO, Smith AR, Wood CK, Hall MA. The Effect of Ethylene on the Growth and Development of Wild-Type and Mutant Arabidopsis-Thaliana (L) Heynh. Annals of Botany. 1991;68:55–61. [Google Scholar]
- Hua J, Meyerowitz EM. Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell. 1998;94:261–271. doi: 10.1016/s0092-8674(00)81425-7. [DOI] [PubMed] [Google Scholar]
- Huq E, Al-Sady B, Hudson M, Kim C, Apel K, Quail PH. Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science. 2004;305:1937–1941. doi: 10.1126/science.1099728. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Lau OS, Deng XW. Light-regulated transcriptional networks in higher plants. Nat Rev Genet. 2007;8:217–230. doi: 10.1038/nrg2049. [DOI] [PubMed] [Google Scholar]
- Ju C, Chang C. Advances in ethylene signalling: protein complexes at the endoplasmic reticulum membrane. AoB Plants. 2012;2012:pls031. doi: 10.1093/aobpla/pls031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R, Shen Y, Marion CM, Tsuchisaka A, Theologis A, Schafer E, Quail PH. The basic helix-loop-helix transcription factor PIF5 acts on ethylene biosynthesis and phytochrome signaling by distinct mechanisms. Plant Cell. 2007;19:3915–3929. doi: 10.1105/tpc.107.051508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- Konishi M, Yanagisawa S. Ethylene signaling in Arabidopsis involves feedback regulation via the elaborate control of EBF2 expression by EIN3. Plant J. 2008;55:821–831. doi: 10.1111/j.1365-313X.2008.03551.x. [DOI] [PubMed] [Google Scholar]
- Lau OS, Deng XW. The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 2012;17:584–593. doi: 10.1016/j.tplants.2012.05.004. [DOI] [PubMed] [Google Scholar]
- Leivar P, Monte E. PIFs: Systems Integrators in Plant Development. Plant Cell. 2014;26:56–78. doi: 10.1105/tpc.113.120857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH. Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol. 2008;18:1815–1823. doi: 10.1016/j.cub.2008.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Ma M, Feng Y, Li H, Wang Y, Ma Y, Li M, An F, Guo H. EIN2-Directed Translational Regulation of Ethylene Signaling in Arabidopsis. Cell. 2015;163:670–683. doi: 10.1016/j.cell.2015.09.037. [DOI] [PubMed] [Google Scholar]
- Merchante C, Brumos J, Yun J, Hu Q, Spencer KR, Enriquez P, Binder BM, Heber S, Stepanova AN, Alonso JM. Gene-Specific Translation Regulation Mediated by the Hormone-Signaling Molecule EIN2. Cell. 2015;163:684–697. doi: 10.1016/j.cell.2015.09.036. [DOI] [PubMed] [Google Scholar]
- Murase K, Hirano Y, Sun TP, Hakoshima T. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature. 2008;456:459–U415. doi: 10.1038/nature07519. [DOI] [PubMed] [Google Scholar]
- Nagy F, Schafer E. Nuclear and cytosolic events of light-induced, phytochrome-regulated signaling in higher plants. Embo Journal. 2000;19:157–163. doi: 10.1093/emboj/19.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH. PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell. 1998;95:657–667. doi: 10.1016/s0092-8674(00)81636-0. [DOI] [PubMed] [Google Scholar]
- Ni W, Xu SL, Tepperman JM, Stanley DJ, Maltby DA, Gross JD, Burlingame AL, Wang ZY, Quail PH. A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science. 2014;344:1160–1164. doi: 10.1126/science.1250778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potuschak T, Lechner E, Parmentier Y, Yanagisawa S, Grava S, Koncz C, Genschik P. EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell. 2003;115:679–689. doi: 10.1016/s0092-8674(03)00968-1. [DOI] [PubMed] [Google Scholar]
- Qiao H, Shen Z, Huang SS, Schmitz RJ, Urich MA, Briggs SP, Ecker JR. Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science. 2012;338:390–393. doi: 10.1126/science.1225974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail PH. Phytochrome photosensory signalling networks. Nat Rev Mol Cell Biol. 2002;3:85–93. doi: 10.1038/nrm728. [DOI] [PubMed] [Google Scholar]
- Quail PH, Boylan MT, Parks BM, Short TW, Xu Y, Wagner D. Phytochromes: photosensory perception and signal transduction. Science. 1995;268:675–680. doi: 10.1126/science.7732376. [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Su YS, Lagarias JC. Phytochrome structure and signaling mechanisms. Annual Review of Plant Biology. 2006;57:837–858. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio V, Shen Y, Saijo Y, Liu Y, Gusmaroli G, Dinesh-Kumar SP, Deng XW. An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. Plant J. 2005;41:767–778. doi: 10.1111/j.1365-313X.2004.02328.x. [DOI] [PubMed] [Google Scholar]
- Schafer E, Bowler C. Phytochrome-mediated photoperception and signal transduction in higher plants. Embo Reports. 2002;3:1042–1048. doi: 10.1093/embo-reports/kvf222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheard LB, Tan X, Mao H, Withers J, Ben-Nissan G, Hinds TR, Kobayashi Y, Hsu FF, Sharon M, Browse J, et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature. 2010;468:400–405. doi: 10.1038/nature09430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Moon J, Huq E. PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. Plant J. 2005;44:1023–1035. doi: 10.1111/j.1365-313X.2005.02606.x. [DOI] [PubMed] [Google Scholar]
- Shen H, Zhu L, Castillon A, Majee M, Downie B, Huq E. Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell. 2008;20:1586–1602. doi: 10.1105/tpc.108.060020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Liu R, Xue C, Shen X, Wei N, Deng XW, Zhong S. Seedlings Transduce the Depth and Mechanical Pressure of Covering Soil Using COP1 and Ethylene to Regulate EBF1/EBF2 for Soil Emergence. Curr Biol. 2016;26:139–149. doi: 10.1016/j.cub.2015.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Wang X, Mo X, Tang C, Zhong S, Deng XW. Arabidopsis DET1 degrades HFR1 but stabilizes PIF1 to precisely regulate seed germination. Proc Natl Acad Sci U S A. 2015;112:3817–3822. doi: 10.1073/pnas.1502405112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Zhong S, Mo X, Liu N, Nezames CD, Deng XW. HFR1 Sequesters PIF1 to Govern the Transcriptional Network Underlying Light-Initiated Seed Germination in Arabidopsis. Plant Cell. 2013;25:3770–3784. doi: 10.1105/tpc.113.117424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Kim K, Kang H, Zulfugarov IS, Bae G, Lee CH, Lee D, Choi G. Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc Natl Acad Sci U S A. 2009;106:7660–7665. doi: 10.1073/pnas.0812219106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao QM, Ecker JR. Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes & Development. 1998;12:3703–3714. doi: 10.1101/gad.12.23.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Hoyt JM, Hamilton AA, Alonso JM. A Link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell. 2005;17:2230–2242. doi: 10.1105/tpc.105.033365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YS, Lagarias JC. Light-independent phytochrome signaling mediated by dominant GAF domain tyrosine mutants of Arabidopsis phytochromes in transgenic plants. Plant Cell. 2007;19:2124–2139. doi: 10.1105/tpc.107.051516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- Von Arnim A, Deng XW. Light Control of Seedling Development. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:215–243. doi: 10.1146/annurev.arplant.47.1.215. [DOI] [PubMed] [Google Scholar]
- Wei N, Kwok SF, von Arnim AG, Lee A, McNellis TW, Piekos B, Deng XW. Arabidopsis COP8, COP10, and COP11 genes are involved in repression of photomorphogenic development in darkness. Plant Cell. 1994;6:629–643. doi: 10.1105/tpc.6.5.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X, Zhang C, Ji Y, Zhao Q, He W, An F, Jiang L, Guo H. Activation of ethylene signaling is mediated by nuclear translocation of the cleaved EIN2 carboxyl terminus. Cell Res. 2012;22:1613–1616. doi: 10.1038/cr.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Shi H, Xue C, Wang L, Xi Y, Li J, Quail PH, Deng XW, Guo H. A molecular framework of light-controlled phytohormone action in Arabidopsis. Curr Biol. 2012;22:1530–1535. doi: 10.1016/j.cub.2012.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Shi H, Xue C, Wei N, Guo H, Deng XW. Ethylene-orchestrated circuitry coordinates a seedling's response to soil cover and etiolated growth. Proc Natl Acad Sci U S A. 2014;111:3913–3920. doi: 10.1073/pnas.1402491111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Zhao M, Shi T, Shi H, An F, Zhao Q, Guo H. EIN3/EIL1 cooperate with PIF1 to prevent photo-oxidation and to promote greening of Arabidopsis seedlings. Proc Natl Acad Sci U S A. 2009;106:21431–21436. doi: 10.1073/pnas.0907670106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.