Abstract
Background
Understanding the dynamics of HIV across anatomic compartments is important to design effective eradication strategies. Here, we evaluated viral trafficking between blood and semen during primary HIV infection (PHI) in six antiretroviral-naïve men who have sex with men (MSM).
Methods
Deep sequencing data of HIV env were generated from longitudinal blood plasma, peripheral blood mononuclear cells, and seminal plasma samples. The presence or absence of viral compartmentalization was assessed using tree-based Slatkin-Maddison and distance-based Fst methods. Phylogeographic analyses were performed using a discrete Bayesian asymmetric approach of diffusion with Markov jump count estimation to evaluate the gene flow between blood and semen during PHI. Levels of DNA from human herpesviruses and selected inflammatory cytokines were also measured on genital secretions collected at baseline to evaluate potential correlates of increased viral migration between anatomical compartments.
Results
We detected varying degrees of compartmentalization in all six individuals evaluated. None of them maintained viral compartmentalization between blood and seminal plasma throughout all of the analyzed time points. Phylogeographic analyses revealed that the HIV population circulating in blood plasma populated the seminal compartment during the earliest stages of infection. In our limited dataset, we found no association between local inflammation or herpesvirus shedding at baseline and viral trafficking between semen and blood.
Conclusions
The early spread of virus from blood plasma to genital tract and the complex viral interplay between these compartments suggest that viral eradication efforts will require monitoring viral subpopulations in anatomical sites and viral trafficking during the course of infection.
Keywords: Bayesian inference, phylogeography, compartmentalization, Markov, seminal plasma, PBMC
INTRODUCTION
Within the first weeks of infection, HIV rapidly disseminates through the body and establishes cellular HIV reservoirs [1]. Understanding how HIV migrates, colonizes, and adapts to various anatomic compartments (e.g. blood plasma, peripheral blood mononuclear cells [PBMCs], and the male genital tract) during the course of infection will likely be important for the development of effective eradication strategies [1, 2].
Viral compartmentalization within anatomic regions can occur as a consequence of tissue-specific genetic differentiation and restricted viral migration between anatomic sites or tissues [3]. Such compartmentalization impacts HIV-associated pathogenesis and is associated with neurocognitive disease [4, 5], development of drug resistance, and sexual transmission [6–12]. For example, the male genital tract represents a unique compartment, which can be distinct from blood, including differences in drug resistance mutations, viral replication [13–16], and differential virus evolution [17–19] in response to local environment factors [20–24] and antiretroviral pharmacokinetics [25–27]. While these unique genotypic and phenotypic characteristics might have crucial implications for efficient HIV sexual transmission [6–12], semen-specific variants residing in these “sanctuaries” may also play an important role in reseeding the blood.
Here we analyzed deep sequencing data obtained from longitudinally collected samples from individuals with recent HIV infection to characterize the dynamics of viral spread between the blood and genital tract during early HIV infection.
MATERIAL AND METHODS
Ethics Statement
This study was approved by the UCSD Human Research Protections Program. All study participants provided voluntary, written informed consent before any study procedures were undertaken.
Participants, Samples, and Clinical Laboratory Tests
Six individuals with recent HIV infection participating in the San Diego Primary Infection Resource Consortium [28] were enrolled in this study. Prior to the initiation of antiretroviral therapy (ART), longitudinal samples of blood plasma (n=25), seminal plasma (n=15) and peripheral blood mononuclear cells (PBMC) (n=9) were collected with a median of 5 time-points per individual (range: 3–7). HIV DNA levels were quantified in PBMCs and HIV RNA levels were quantified in both blood and seminal plasma [29] (Table 1 and Figure S1). The estimated dates of infection (EDI) were determined using a series of well-defined stepwise rules to characterize stages of infection based on serologic and virologic criteria, as described by Le et al. [30] and summarized in Supplementary Table S1.
Table 1.
Study Participants.
| Subjects | Age (years) | Time between BL and EDI (days) | Number of time points | Sampling Period (days) | BL Blood HIV RNA Level (Log10 cp/mL)§ | BL Semen HIV RNA Level (Log10 cp/mL)§ | BL CD4 T cell (count/mL)§ |
|---|---|---|---|---|---|---|---|
| S1 | 32 | 20 | 3 | 84 | 5.1 | 3.62 | 337 |
| S2 | 23 | 87 | 5 | 2,009 | 5.42 | 3.75 | 563 |
| S3 | 23 | 84 | 4 | 182 | 5.03 | 3.25 | 638 |
| S4 | 38 | 40 | 5 | 78 | 6.06 | 5.63 | 620 |
| S5 | 41 | 77 | 3 | 177 | 3.33 | 2.8 | 638 |
| S6 | 25 | 169 | 7 | 345 | 4.62 | 3.45 | 851 |
| Median | 28 | 81 | 5 | 180 | 5.07 | 3.54 | 629 |
Levels at baseline.
BL: Baseline; EDI: Estimated date of Infection
Next Generation Sequencing and Sequence Analyses
HIV RNA was extracted from blood and seminal plasma and HIV DNA was extracted from 5 million frozen PBMCs using standard methods [29]. Deep sequencing of PCR-amplified env C2-V3 (HXB2 coordinates 6928–7344) was performed using the Roche 454 FLX Titanium platform (Basel, Switzerland) and sequence haplotypes were generated as described in supplement (Table S2). For each sample, we computed the mean of all pairwise Tamura-Nei 93 distances between reads with at least 100 overlapping base pairs to measure the mean pairwise diversity [31].
To evaluate if sampled viral populations within each individual were under selective pressure, we used the Fast Unconstrained Bayesian AppRoximation (FUBAR) program [32]. This method exploits several computational shortcuts to speed up the detection of positive or purifying selection, leading to improved robustness against model misspecification and permitting the analysis of large data sets for which selection analysis was previously intractable [32]. A posterior probability threshold p of 0.9 was defined to detect codon sites under positive and purifying selection.
Viral compartmentalization and population subdivision for each individual dataset was assessed by a stringent multi-level approach including FST [33] and Slatkin Maddison (SM) assessments of compartmentalization [34] implemented in the HyPhy software package [35]. Briefly for the FST, we compute the fixation index [33], defined as , where πI is the estimate of mean pairwise intra-compartment Tamura Nei (TN93) genetic distance [36], and πD is its inter-compartment counterpart. Both quantities were computed by comparing all reads from two different compartments, subject to the requirement that they share at least 100 aligned nucleotide positions. Statistical significance was derived via a 1,000-fold population-structure randomization/permutation test. For a given sample to be considered compartmentalized, we required that: 1) FST test was statistically significant with a p-value of less than 0.05, (2) the FST p-value remained significant when the haplotype frequency was ignored, (i.e. all haplotypes were assigned a relative weight of 1), and (3) the Slatkin-Maddison test for compartmentalization, based on the inferred maximum likelihood phylogeny, was also statistically significant.
Phylogeographic Analyses
The resulting collection of haplotypes obtained from each sample was used to reconstruct the spatial dynamics of HIV-1 across compartments. Briefly, we employed a Bayesian discrete phylogeographic approach [37] and an MCMC framework as implemented in BEAST v1.8.1 with BEAGLE [38, 39]. We applied a discretized gamma distribution (GTR + 4Γ) to account for among-site rate variation. Time scales of the trees were calibrated with the sampling dates available. We specified a uncorrelated lognormal (UCLN) molecular clock that allows rates to vary among the branches of the inferred phylogenies to infer the timescale of HIV evolution for each individual [40], with a gamma distribution prior. A Bayesian skyline tree prior was used as a coalescent demographic model. Markov chain Monte Carlo simulations were run for 250 million steps, sub-sampling parameters every 50,000 steps. Convergence of the chains was inspected using Tracer.v.1.6. Maximum clade credibility trees were obtained with TreeAnnotator v1.8.1 and visualized by using FigTree 1.4.2 [38]. To obtain the expectations for the location state transitions, we estimated Markov jump counts [41] along the branches of the posterior tree distribution [42].
History of viral movements and estimated percentage of HIV-1 migration events from blood to semen and conversely from semen to blood were obtained using a “robust counting” procedure as implemented in BEAST package v1.8.1 [38]. Briefly, this method allows estimation of the expected number of location changes along the branches of a posterior tree distribution, and can be used to investigate intra-host HIV spatiotemporal dynamics [2]. Here, we estimated the location state transition rates between compartments using a randomized sub-sample of an equal number of blood plasma and seminal sequences to avoid potential bias in spatial inference estimates that may arise from over-sampling one location. Blood and seminal plasma sequences from each available time point were retained in order to maximize temporal evolutionary signal. To improve robustness, the analysis was repeated three times with a new random sub-sample each time. Posterior mean percentage and 95% Bayesian Credible Intervals (BCIs) for the proportion of migration events from and to blood plasma were obtained. A full description of these analyses is provided in the supplementary materials and in our previous manuscript [2].
Cytokines/chemokines quantification
Selected markers of genital inflammation and cellular trafficking (monocyte chemotactic protein [MCP]-1, interleukin [IL]-6, tumor necrosis factor [TNF]-α, Interferon-γ, regulated on activation normal T cell expressed and secreted [RANTES], and Interferon-γ induced protein [IP]-10) were measured at baseline in seminal plasma using a bead-based multiplex array [23, 43].
Herpesvirus DNA and HIV RNA extraction and quantification from seminal plasma
We used quantitative RT-PCR to measure levels of HIV RNA at each time-point. Additionally, DNA levels from various Human Herpes Viruses (HHV) in seminal plasma were measured at baseline (i.e. CMV, EBV, herpes simplex viruses (HSV) types 1 and 2, and HHV types 6, 7, and 8) [43, 44].
Statistics
Statistical analyses were performed using SAS (version 9.4) and Graph-Pad Prism 6.0c software (GraphPad Software, Inc., San Diego, CA). We performed comparisons of inferred numbers of migration events across compartments between groups using the Fisher-exact test. Evaluation of correlations between continuous variables was performed using the Spearman rank correlation.
RESULTS
Study Cohort
HIV-infected participants (n=6) were all men who have sex with men (MSM) recently infected with HIV-1 subtype B. Participants had a mean age of 30 years, and no reported needle-sharing exposures. Baseline blood and semen samples were collected at a median of 81 days after the EDI (interquartile range [IQR], 35–108 days). Additional longitudinal samples were collected serially for a median of 180 days (IQR, 82-761 days) post baseline, with a median of 4.5 time points per participant (IQR, 3–5.5). The median blood and seminal plasma viral load at baseline were 5.07 (IQR, 4.30–5.58) and 3.54 (IQR, 3.14–4.22), respectively and the median CD4+ count was 629 cells/ μL (IQR, 506–691). Characteristics and demographics of the subjects are summarized in Table 1 and Figure S1.
Viral characteristics in blood and the male genital tract
To compare the diversity of HIV-1 env between the male genital tract, peripheral blood plasma, and PBMCs, we calculated the mean pairwise diversity in samples from all three compartments. There was no significant difference in overall nucleotide diversity between sequences sampled from blood plasma, seminal plasma and PBMC across the six individuals at any time point. Using a mixed effects model to examine viral diversity in relation to time from the estimated date of infection within each individual, we found a significant increase in viral diversity over the course of infection in the HIV RNA population in seminal plasma (p=0.03) but not in blood plasma (Figure Suppl.2).
At baseline, we detected evidence of viral compartmentalization between the blood and genital compartments in two out of six participants (subjects S3 and S6). Interestingly, both individuals lost compartmentalization at later time points, 182 days (subject S3) and 457 days (subject S6) post infection, respectively. The remaining four participants were not compartmentalized at baseline but exhibited significant signal for compartmentalization between blood and semen at one or more post-baseline time points (Table 2). There was no association between the time from EDI and the presence of viral compartmentalization between blood and seminal plasma. None of the six individuals maintained viral compartmentalization between blood and seminal plasma throughout all of the analyzed time points.
Table 2.
Compartmentalization Analysis for partial env HIV RNA in blood and seminal plasma and HIV DNA in PBMC
| Subject | Days from EDI | Location | Fst test* | P value | Compartmentalization | |
|---|---|---|---|---|---|---|
| S1 | 20 | BP | PBMC | 0.01 | 0.09 | NO |
| PBMC | SP | 0.05 | 0.06# | NO | ||
| BP | SP | −0.09 | 1.00 | NO | ||
| 104 | PBMC | SP | 0.00 | 0.16 | NO | |
| BP | SP | 0.92 | 0.01 | YES | ||
| BP | PBMC | 0.92 | 0.01#§ | NO | ||
| S2 | 87 | BP | SP | −0.03 | 0.01 | NO |
| 289 | BP | SP | 0.26 | 0.01 | YES | |
| S3 | 98 | PBMC | SP | 0.49 | 0.01 | YES |
| BP | SP | 0.52 | 0.01 | YES | ||
| BP | PBMC | 0.00 | 0.01 | NO | ||
| 182 | BP | SP | −0.06 | 0.85 | NO | |
| S4 | 40 | BP | SP | −0.15 | 1.00 | NO |
| BP | PBMC | −0.37 | 1.00 | NO | ||
| PBMC | SP | 0.13 | 0.01#§ | NO | ||
| 47 | BP | SP | 0.96 | 0.01 | YES | |
| 75 | BP | SP | 0.34 | 0.01 | YES | |
| 118 | BP | PBMC | 0.21 | 0.01#§ | NO | |
| S5 | 77 | BP | SP | −0.05 | 0.03 | NO |
| BP | PBMC | −0.37 | 1.00 | NO | ||
| PBMC | SP | −0.24 | 1.00 | NO | ||
| 162 | BP | SP | 0.97 | 0.01 | YES | |
| 254 | BP | SP | 0.17 | 0.01 | YES | |
| S6 | 177 | BP | SP | 0.10 | 0.01 | YES |
| BP | PBMC | −0.01 | 0.01 | NO | ||
| PBMC | SP | 0.06 | 0.01 | YES | ||
| 457 | BP | SP | −0.17 | 0.96 | NO | |
Compartmentalization is called when all tests indicate compartmentalization [see text]
Indicates tests which become significant (or not significant) if copy numbers are ignored during FST calculations.
Indicates tests, which become insignificant under the phylogeny-based Slatkin-Maddison test [see text]. Statistical significance was derived via a 1,000-fold population-structure randomization/permutation test.
BP: Blood Plasma, SP: Seminal Plasma; PBMC: Peripheral Blood Mononuclear Cells; EDI: estimated date of infection.
While, FST could assume negative values, none of the samples with negative FST could be called compartmentalized.
As viral evolution in distinct anatomic compartments may be differentially affected by selection, we evaluated codon sites under positive and purifying selection within blood and seminal plasma for each participant. Overall, the number of sites under positive and diversifying selection were higher in blood and seminal plasma compared to PBMC (Table Suppl.3) and the the sites identified to be under positive and purifying selection varied between individuals and across compartments (data not shown).
HIV Phylodynamics between Blood and Seminal plasma
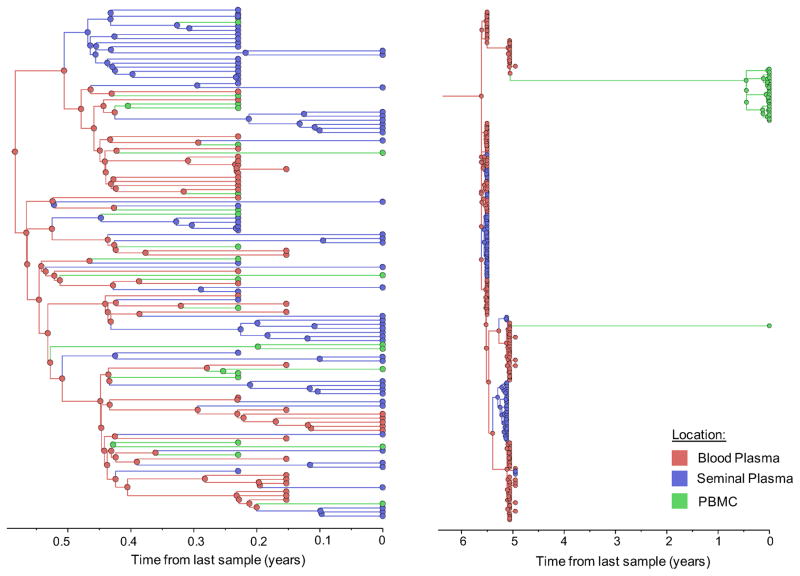
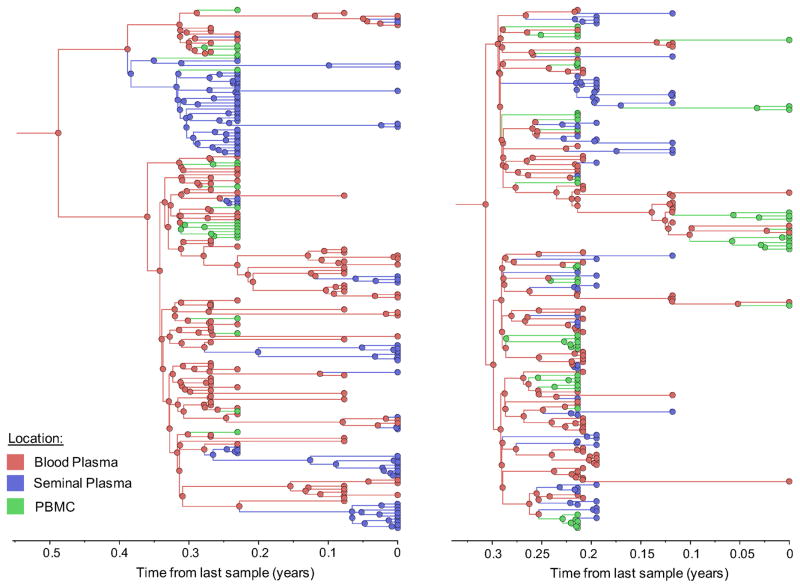
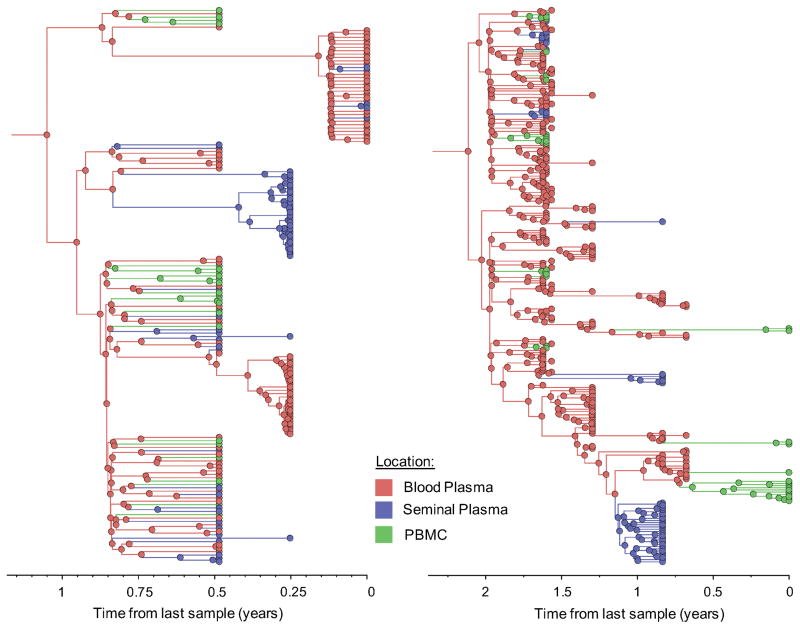
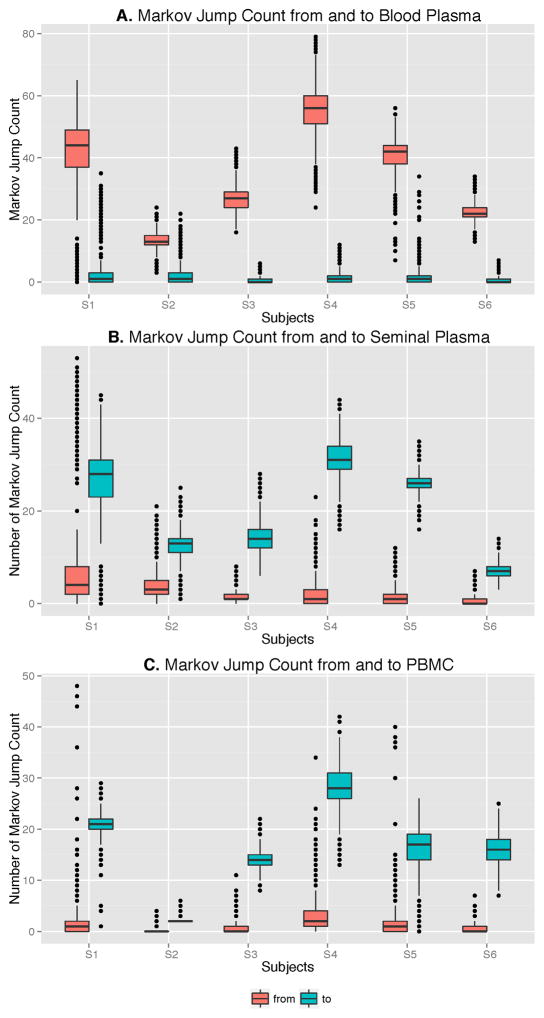
To further characterize the viral gene flow between blood and semen, we applied a Bayesian discrete phylogeographic approach to our sequence datasets [37]. The inferred phylogenies and the distribution of node states across the Markov Chain Monte Carlo (MCMC) analyses all suggested that ancestral sequences within the tree originated from blood plasma and were followed by spread to the PBMCs and genital tract (Figure 1, colored nodes). We then used Markov jump counts to provide a quantitative estimate of gene flow between each compartment. Using random but evenly distributed sub-samples to reduce sampling bias, we found a significantly higher number of migration events originating from blood plasma toward both seminal plasma and PBMC for all individuals (p<0.01) (Figure 2, Table Suppl.4). Gene flow (i.e. the expected number of location state transitions along branches of the tree [41]) from blood plasma to the genital tract and from blood plasma to PBMCs represented a mean of 47.0% (range: 29.6–53.6%) and 38.0% (range:3.4–65.3%) of the total viral movement across the 3 compartments, respectively.
Figure 1. Time-scaled phylogeographic reconstruction of partial env sequences for the 6 individuals.
Branches and nodes are colored by the compartment of origin. HIV RNA sequences from blood plasma and seminal plasma, and HIV DNA sequences from PBMCs are depicted in red, blue and green respectively. These trees represent the inferred phylogenetic relationships of haplotype sequences. The x axis represents time in year before the latest sampling date (in year). Time scales of the trees were calibrated with the sampling dates available. A bayesian skyline tree prior was used as a coalescent demographic model under an uncorrelated lognormal (UCLN) molecular clock that allows rates to vary among the branches of the inferred phylogenies to infer the timescale of HIV evolution for each individual [40], with a gamma distribution prior. Analyses were made with BEAST v1.8.1 with BEAGLE [38, 39] (see Methods section).
Figure 2. Boxplot depicting the number of Markov Jumps from and to Blood plasma (A), Seminal Plasma (B) and PBMC (C) for the 6 participants.
Location state transitions between compartments were estimated by Markov jump counts [41]. Viral migration from (red) and to (green) blood plasma (A), seminal plasma (B) and Peripheral Blood Mononuclear Cells (C) are presented.
Correlates of viral diversity and gene flow
To explore possible inflammatory mechanisms connecting viral evolution and gene flow between blood and genital compartment, we measured levels of selected cytokines and chemokines (i.e., MCP-1, IL-6, TNF-α, Interferon-γ, RANTES, and IP-10) in seminal plasma at baseline (Table Suppl.5). After adjustment for multiple comparisons, in our limited dataset, we found no association between viral diversity or viral migration between semen and blood and the presence of HHV or increased levels of cytokines in the genital tract at baseline. (Table Suppl.6).
DISCUSSION
A better characterization of the viral dynamics of HIV in the genital tract is important not only to inform eradication strategies, but also to direct strategies to reduce infectiousness and reduce the risk of transmission. Previous works have reported compartmentalization of HIV between blood and semen and described the potential implications for HIV transmission and drug resistance [6–11]. In our study, we found evidence of viral compartmentalization between the blood (PBMC or plasma) and seminal plasma in six HIV-infected men during the earliest phase of HIV infection. Interestingly none of the six individuals maintained viral compartmentalization between blood and seminal plasma throughout the entire study period. This is consistent with previous studies showing that compartmentalization of HIV variants can be a transient phenomenon [45–47], revealing the complex viral dynamics between blood and the genital tract as reviewed in [48]. Similar to these human studies, variation was also seen in non-human primate models: studies in recently infected macaques showed both evidence of mixing of the SIV population between blood and the genital tract during acute SIV infection [49] and genital tract sequestration following resolution of primary viremia and during chronic infection [50–51]. The absence of compartmentalization between viral populations in the blood and the genital tract can be the consequence of ongoing viral trafficking or limited divergence rates over the course of infection. Here, we found increasing compartmentalization between viral populations of the blood and seminal plasma over time for four out of six individuals (66.7%). The delayed appearance of viral segregation together with the significant increase in viral diversity in semen during the course of HIV infection might be a consequence of ongoing viral replication under selective pressures unique to the genital tract. Consequently, we evaluated the potential selective pressure by comparing the site under positive and purifying selection in blood and seminal plasma. While the small number of participants evaluated and the short segment of HIV-1 env used represent an important limitation, we found that the number and the nature of sites identified to be under positive and purifying selection varied between individuals and across compartments. This is consistent with previous findings showing distinct local selective pressures that may be contributing to viral evolution and segregation of HIV variants in the genital tract [9, 13–19].
To further understand mechanisms underlying viral compartmentalization, we applied a discrete diffusion approach to phylogenies of all compartment-specific HIV sequence data to characterize the gene flow between blood and semen. This method permits to quantify the migration between compartments by estimating the Markov jump count of location state transitions along tree branches. Similar to our previous report [2], we found evidence of reciprocal migrations between the blood and genital tract but in this study, gene flow predominantly originated from blood plasma (Figure 2), illustrating the complex intermixing of HIV subpopulations within the host. These findings are also consistent with a cross-sectional study of five HIV-infected men, which showed evidence of viral compartmentalization between the blood (PBMC or plasma) and the genital tract, and a predominant viral gene flow from blood plasma to genital tract using a cladistic measure of gene flow (Slatkin and Maddison Test) [52]. Together these results illustrate that viral migration between anatomic compartments is a complex and dynamic bidirectional process [2].
Previous reports have demonstrated that semen and blood are two distinct immunological compartments with distinct cytokine profiles [23, 24, 53]. Local immune activation and T cell activation may provide an environment that can support virus replication with unique selective pressures [53], giving rise to a compartmentalized viral population. Consistent with this idea of local immune activation, a previous study of 16 men with chronic HIV infection demonstrated high levels of MCP-1 in the semen, but found no correlation with the viral compartmentalization [46]. Here we measured seminal levels of MCP-1, IL-6, TNF-α, Interferon-γ, RANTES, and IP-10 and HHV DNA at baseline. While we found no significant association between viral trafficking and local inflammation or the presence of HHV at baseline after adjustment for multiple comparisons, the limited sample size and the lack of longitudinal measures of these markers of inflammation and infection prevented any robust conclusion.
There are several important limitations of this study. Primarily, the small number of subjects evaluated is an important limitation. However, it is generally difficult to obtain paired genital and blood samples from recently infected individuals in a longitudinal fashion, and so while our sample population is small, the study represents one of the largest such datasets in HIV infected individuals. Our data were also generated with first generation deep sequencing technology targeting a small C2-V3 env fragment, limiting our ability to delve into the selection pressures driving compartmentalization. Nevertheless, the phylogenetic signal present in this segment was enough to identify viral compartmentalization, and assess viral gene flow across compartments. Similar to most studies of viral dynamics in humans, sampling bias may have affected our observations. Viral populations are dynamic while sampling is static, and samples were not taken at the same point during infection for all individuals. Further limitations to our study include that there are inherent differences in physiology and evolutionary rates of archived viral DNA and viral RNA, and that there may be sampling biases between HIV DNA and HIV RNA populations, given that HIV RNA populations are usually more uniform than HIV DNA populations, at any one sampling timepoint; however, such sampling biases are limited in this study by the fact that the samples analyzed are from early infection, when overall body viral diversity is limited [54]. Finally, our analysis might be affected by measurement bias, since haplotype reconstruction may affect the proportionality of viral variants, although the relative abundance of haplotypes is usually preserved [55].
In summary, we used newly state-of-the-art Bayesian phylogeographic methods to evaluate the dynamics and determinants of HIV compartmentalization between blood and the male genital tract. Despite these listed limitations, our results are compatible with early spread of virus predominantly from blood plasma to genital tract, with consequent establishment of a local viral population, followed by intra-compartment evolution driven by specific local selective pressures and immune environment. It will be important to monitor these dynamics during HIV cure studies using latency reversing agents to determine if this viral compartment can act as a source for viral rebound.
Supplementary Material
Acknowledgments
Source of Funding.
This work was supported by the Department of Veterans Affairs and grants from the National Institutes of Health AI093163, AI036214, AI027763, MH101012, TR000098-04AI100665, MH097520, DA034978, AI027763, AI068636, MH081482, AmFAR grant 108537, the California HIV/AIDS Research Program (CHRP) Grant F13SD321, the James B. Pendleton Charitable Trust and the Bettencourt-Schueller Foundation.
Footnotes
Conflicts of Interest
These findings have not been previously presented or published.
The authors have no conflicts of interest to declare.
References
- 1.Stekler J, Sycks BJ, Holte S, Maenza J, Stevens CE, Dragavon J, et al. HIV Dynamics in Seminal Plasma during Primary HIV Infection. AIDS Research and Human Retroviruses. 2008;24:1269–1274. doi: 10.1089/aid.2008.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chaillon A, Gianella S, Wertheim JO, Richman DD, Mehta SR, Smith DM. HIV migration between blood and cerebrospinal fluid or semen over time. The Journal of Infectious Diseases. 2014;209:1642–1652. doi: 10.1093/infdis/jit678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nickle DC, Shriner D, Mittler JE, Frenkel LM, Mullins JI. Importance and detection of virus reservoirs and compartments of HIV infection. Current opinion in microbiology. 2003;6:410–416. doi: 10.1016/s1369-5274(03)00096-1. [DOI] [PubMed] [Google Scholar]
- 4.Cowley D, Gray LR, Wesselingh SL, Gorry PR, Churchill MJ. Genetic and functional heterogeneity of CNS-derived tat alleles from patients with HIV-associated dementia. Journal of neurovirology. 2011;17:70–81. doi: 10.1007/s13365-010-0002-5. [DOI] [PubMed] [Google Scholar]
- 5.Smith DM, Zárate S, Shao H, Pillai SK, Letendre SL, Wong JK, et al. Pleocytosis is associated with disruption of HIV compartmentalization between blood and cerebral spinal fluid viral populations. Virology. 2009;385:204–208. doi: 10.1016/j.virol.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eron JJ, Vernazza PL, Johnston DM, Seillier-Moiseiwitsch F, Alcorn TM, Fiscus SA, et al. Resistance of HIV-1 to antiretroviral agents in blood and seminal plasma: implications for transmission. Aids. 1998;12:F181–189. doi: 10.1097/00002030-199815000-00003. [DOI] [PubMed] [Google Scholar]
- 7.Smith DM, Wong JK, Shao H, Hightower GK, Mai SH, Moreno JM, et al. Long-term persistence of transmitted HIV drug resistance in male genital tract secretions: implications for secondary transmission. J Infect Dis. 2007;196:356–360. doi: 10.1086/519164. [DOI] [PubMed] [Google Scholar]
- 8.Zhu T, Wang N, Carr A, Nam DS, Moor-Jankowski R, Cooper DA, et al. Genetic characterization of human immunodeficiency virus type 1 in blood and genital secretions: evidence for viral compartmentalization and selection during sexual transmission. Journal of Virology. 1996;70:3098–3107. doi: 10.1128/jvi.70.5.3098-3107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pillai SK, Good B, Pond SK, Wong JK, Strain MC, Richman DD, et al. Semen-specific genetic characteristics of human immunodeficiency virus type 1 env. Journal of virology. 2005;79:1734–1742. doi: 10.1128/JVI.79.3.1734-1742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delwart EL, Mullins JI, Gupta P, Learn GH, Jr, Holodniy M, Katzenstein D, et al. Human immunodeficiency virus type 1 populations in blood and semen. Journal of virology. 1998;72:617–623. doi: 10.1128/jvi.72.1.617-623.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paranjpe S, Craigo J, Patterson B, Ding M, Barroso P, Harrison L, et al. Subcompartmentalization of HIV-1 quasispecies between seminal cells and seminal plasma indicates their origin in distinct genital tissues. AIDS research and human retroviruses. 2002;18:1271–1280. doi: 10.1089/088922202320886316. [DOI] [PubMed] [Google Scholar]
- 12.Zhu T, Mo H, Wang N, Nam DS, Cao Y, Koup RA, et al. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]
- 13.Sheth PM, Kovacs C, Kemal KS, Jones RB, Raboud JM, Pilon R, et al. Persistent HIV RNA shedding in semen despite effective antiretroviral therapy. Aids. 2009;23:2050–2054. doi: 10.1097/QAD.0b013e3283303e04. [DOI] [PubMed] [Google Scholar]
- 14.Lorello G, la Porte C, Pilon R, Zhang G, Karnauchow T, MacPherson P. Discordance in HIV-1 viral loads and antiretroviral drug concentrations comparing semen and blood plasma. HIV Med. 2009;10:548–554. doi: 10.1111/j.1468-1293.2009.00725.x. [DOI] [PubMed] [Google Scholar]
- 15.Politch JA, Mayer KH, Welles SL, O’Brien WX, Xu C, Bowman FP, et al. Highly active antiretroviral therapy does not completely suppress HIV in semen of sexually active HIV-infected men who have sex with men. Aids. 2012;26:1535–1543. doi: 10.1097/QAD.0b013e328353b11b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gianella S, Smith DM, Vargas MV, Little SJ, Richman DD, Daar ES, et al. Shedding of HIV and human herpesviruses in the semen of effectively treated HIV-1-infected men who have sex with men. Clinical Infectious Diseases: An Official Publication of the Infectious Diseases Society of America. 2013;57:441–447. doi: 10.1093/cid/cit252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tirado G, Jove G, Kumar R, Noel RJ, Reyes E, Sepulveda G, et al. Differential virus evolution in blood and genital tract of HIV-infected females: evidence for the involvement of drug and non-drug resistance-associated mutations. Virology. 2004;324:577–586. doi: 10.1016/j.virol.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 18.Delwart EL, Mullins JI, Gupta P, Learn GH, Jr, Holodniy M, Katzenstein D, et al. Human immunodeficiency virus type 1 populations in blood and semen. J Virol. 1998;72:617–623. doi: 10.1128/jvi.72.1.617-623.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Byrn RA, Kiessling AA. Analysis of human immunodeficiency virus in semen: indications of a genetically distinct virus reservoir. J Reprod Immunol. 1998;41:161–176. doi: 10.1016/s0165-0378(98)00056-4. [DOI] [PubMed] [Google Scholar]
- 20.Zhu T, Wang N, Carr A, Nam DS, Moor-Jankowski R, Cooper DA, et al. Genetic characterization of human immunodeficiency virus type 1 in blood and genital secretions: evidence for viral compartmentalization and selection during sexual transmission. J Virol. 1996;70:3098–3107. doi: 10.1128/jvi.70.5.3098-3107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vernazza PL, Eron JJ, Cohen MS, van der Horst CM, Troiani L, Fiscus SA. Detection and biologic characterization of infectious HIV-1 in semen of seropositive men. Aids. 1994;8:1325–1329. doi: 10.1097/00002030-199409000-00017. [DOI] [PubMed] [Google Scholar]
- 22.Ping LH, Cohen MS, Hoffman I, Vernazza P, Seillier-Moiseiwitsch F, Chakraborty H, et al. Effects of genital tract inflammation on human immunodeficiency virus type 1 V3 populations in blood and semen. Journal of virology. 2000;74:8946–8952. doi: 10.1128/jvi.74.19.8946-8952.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lisco A, Munawwar A, Introini A, Vanpouille C, Saba E, Feng X, et al. Semen of HIV-1-infected individuals: local shedding of herpesviruses and reprogrammed cytokine network. J Infect Dis. 2012;205:97–105. doi: 10.1093/infdis/jir700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vanpouille C, Introini A, Morris SR, Margolis L, Daar ES, Dube MP, et al. Distinct cytokine/chemokine network in semen and blood characterize different stages of HIV infection. Aids. 2016;30:193–201. doi: 10.1097/QAD.0000000000000964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trezza CR, Kashuba AD. Pharmacokinetics of antiretrovirals in genital secretions and anatomic sites of HIV transmission: implications for HIV prevention. Clin Pharmacokinet. 2014;53:611–624. doi: 10.1007/s40262-014-0148-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fletcher P, Herrera C, Armanasco N, Nuttall J, Shattock RJ. Short Communication: Limited Anti-HIV-1 Activity of Maraviroc in Mucosal Tissues. AIDS Res Hum Retroviruses. 2016;32:334–338. doi: 10.1089/aid.2015.0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Else LJ, Taylor S, Back DJ, Khoo SH. Pharmacokinetics of antiretroviral drugs in anatomical sanctuary sites: the male and female genital tract. Antivir Ther. 2011;16:1149–1167. doi: 10.3851/IMP1919. [DOI] [PubMed] [Google Scholar]
- 28.Butler DM, Delport W, Kosakovsky Pond SL, Lakdawala MK, Cheng PM, Little SJ, et al. The origins of sexually transmitted HIV among men who have sex with men. Science translational medicine. 2010:2. doi: 10.1126/scitranslmed.3000447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gianella S, Mehta SR, Strain MC, Young JA, Vargas MV, Little SJ, et al. Impact of seminal cytomegalovirus replication on HIV-1 dynamics between blood and semen. Journal of medical virology. 2012;84:1703–1709. doi: 10.1002/jmv.23398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le T, Wright EJ, Smith DM, He W, Catano G, Okulicz JF, et al. Enhanced CD4+ T-cell recovery with earlier HIV-1 antiretroviral therapy. The New England journal of medicine. 2013;368:218–230. doi: 10.1056/NEJMoa1110187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamura K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Molecular Biology and Evolution. 1992;9:678–687. doi: 10.1093/oxfordjournals.molbev.a040752. [DOI] [PubMed] [Google Scholar]
- 32.Murrell B, Moola S, Mabona A, Weighill T, Sheward D, Kosakovsky Pond SL, et al. FUBAR: A Fast, Unconstrained Bayesian AppRoximation for Inferring Selection. Molecular Biology and Evolution. 2013;30:1196–1205. doi: 10.1093/molbev/mst030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hudson RR, Slatkin M, Maddison WP. Estimation of levels of gene flow from DNA sequence data. Genetics. 1992;132:583–589. doi: 10.1093/genetics/132.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Slatkin M, Maddison WP. A cladistic measure of gene flow inferred from the phylogenies of alleles. Genetics. 1989;123:603–613. doi: 10.1093/genetics/123.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pond SLK, Frost SDW, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics (Oxford, England) 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 36.Chen W, Wang H. Variance estimation for nucleotide substitution models. Mol Phylogenet Evol. 2015;90:97–103. doi: 10.1016/j.ympev.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 37.Lemey P, Rambaut A, Drummond AJ, Suchard MA. Bayesian phylogeography finds its roots. PLoS Computational Biology. 2009:5. doi: 10.1371/journal.pcbi.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suchard MA, Rambaut A. Many-core algorithms for statistical phylogenetics. Bioinformatics (Oxford, England) 2009;25:1370–1376. doi: 10.1093/bioinformatics/btp244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biology. 2006:4. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Minin VN, Suchard MA. Counting labeled transitions in continuous-time Markov models of evolution. Journal of mathematical biology. 2008;56:391–412. doi: 10.1007/s00285-007-0120-8. [DOI] [PubMed] [Google Scholar]
- 42.O’Brien JD, Minin VN, Suchard MA. Learning to count: robust estimates for labeled distances between molecular sequences. Molecular biology and evolution. 2009;26:801–814. doi: 10.1093/molbev/msp003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gianella S, Smith DM, Daar ES, Dube MP, Lisco A, Vanpouille C, et al. Genital Cytomegalovirus Replication Predicts Syphilis Acquisition among HIV-1 Infected Men Who Have Sex with Men. PLoS One. 2015;10:e0130410. doi: 10.1371/journal.pone.0130410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gianella S, Massanella M, Richman DD, Little SJ, Spina CA, Vargas MV, et al. Cytomegalovirus Replication in Semen is Associated with Higher Levels of Proviral HIV DNA and CD4+ T cell Activation during Antiretroviral Treatment. J Virol. 2014;88:7818–7827. doi: 10.1128/JVI.00831-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bull ME, Heath LM, McKernan-Mullin JL, Kraft KM, Acevedo L, Hitti JE, et al. Human immunodeficiency viruses appear compartmentalized to the female genital tract in cross-sectional analyses but genital lineages do not persist over time. The Journal of Infectious Diseases. 2013;207:1206–1215. doi: 10.1093/infdis/jit016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anderson JA, Ping L-H, Dibben O, Jabara CB, Arney L, Kincer L, et al. HIV-1 Populations in Semen Arise through Multiple Mechanisms. PLoS pathogens. 2010:6. doi: 10.1371/journal.ppat.1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gupta P, Leroux C, Patterson BK, Kingsley L, Rinaldo C, Ding M, et al. Human immunodeficiency virus type 1 shedding pattern in semen correlates with the compartmentalization of viral Quasi species between blood and semen. The Journal of infectious diseases. 2000;182:79–87. doi: 10.1086/315644. [DOI] [PubMed] [Google Scholar]
- 48.Houzet L, Matusali G, Dejucq-Rainsford N. Origins of HIV-infected leukocytes and virions in semen. J Infect Dis. 2014;210(Suppl 3):S622–630. doi: 10.1093/infdis/jiu328. [DOI] [PubMed] [Google Scholar]
- 49.Fieni F, Stone M, Ma ZM, Dutra J, Fritts L, Miller CJ. Viral RNA levels and env variants in semen and tissues of mature male rhesus macaques infected with SIV by penile inoculation. PLoS One. 2013;8:e76367. doi: 10.1371/journal.pone.0076367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whitney JB, Hraber PT, Luedemann C, Giorgi EE, Daniels MG, Bhattacharya T, et al. Genital Tract Sequestration of SIV following Acute Infection. PLoS Pathogens. 2011:7. doi: 10.1371/journal.ppat.1001293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Le Tortorec A, Le Grand R, Denis H, Satie A-P, Mannioui K, Roques P, et al. Infection of semen-producing organs by SIV during the acute and chronic stages of the disease. PloS One. 2008:3. doi: 10.1371/journal.pone.0001792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Diem K, Nickle DC, Motoshige A, Fox A, Ross S, Mullins JI, et al. Male genital tract compartmentalization of human immunodeficiency virus type 1 (HIV) AIDS research and human retroviruses. 2008;24:561–571. doi: 10.1089/aid.2007.0115. [DOI] [PubMed] [Google Scholar]
- 53.Olivier AJ, Masson L, Ronacher K, Walzl G, Coetzee D, Lewis DA, et al. Distinct cytokine patterns in semen influence local HIV shedding and HIV target cell activation. J Infect Dis. 2014;209:1174–1184. doi: 10.1093/infdis/jit649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shankarappa R, Margolick JB, Gange SJ, Rodrigo AG, Upchurch D, Farzadegan H, et al. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. Journal of Virology. 1999;73:10489–10502. doi: 10.1128/jvi.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jayasundara D, Saeed I, Maheswararajah S, Chang BC, Tang SL, Halgamuge SK. ViQuaS: an improved reconstruction pipeline for viral quasispecies spectra generated by next-generation sequencing. Bioinformatics (Oxford, England) 2014 doi: 10.1093/bioinformatics/btu754. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.