Abstract
Changes in telomere length are associated with degenerative diseases and cancer. Oxidative stress and DNA damage have been linked to both positive and negative alterations in telomere length and integrity. Here we examined how the common oxidative lesion 8-oxo-7,8-dihydro-2′-deoxyguanine (8-oxoG) regulates telomere elongation by telomerase. When present in the deoxynucleoside triphosphate pool as 8-oxodGTP, telomerase utilization of the oxidized nucleotide during telomere extension is mutagenic and terminates further elongation. Depletion of the enzyme that removes oxidized dNTPs, MTH1, increases telomere dysfunction and cell death in telomerase positive cancer cells harboring shortened telomeres. In contrast, a pre-existing 8-oxoG within the telomeric DNA sequence promotes telomerase activity by destabilizing G-quadruplex structure in the DNA. We show that the mechanism by which 8-oxoG arises in the telomere, either by insertion of oxidized nucleotides or by direct reaction with free radicals, dictates whether telomerase is inhibited or stimulated and thereby, mediates the biological outcome.
Telomeres cap chromosome ends and are essential for genome stability, cell proliferation and human health. Dysfunctional critically short telomeres trigger cell senescence or apoptosis, which drives aging-related degenerative pathologies and loss of regenerative capacity1,2. Telomeres shorten progressively with cell division due to the “end replication problem”. To compensate, telomerase lengthens the telomeres by adding GGTTAG repeats3. In humans telomeres consist of approximately 1,600 TTAGGG duplex repeats and terminate in a single strand overhang4. Telomerase is expressed in human germ and stem cells, and is upregulated in 90% of cancers to enable continued cell proliferation4,5. Thus, telomerase regulation is a critical determinant of degenerative diseases and cancer.
Oxidative stress contributes to the pathogenesis of numerous human diseases including cancer, and results from an imbalance between the production of reactive oxygen species (ROS) and cellular antioxidant defenses. The G-rich content of TTAGGG repeats renders telomeres highly susceptible to oxidative damage, and oxidative stress accelerates telomere shortening6,7. ROS results from normal oxygen metabolism and environmental exposures, and is elevated at sites of chronic inflammation8,9. Free radical reaction with DNA generates chemical alterations, including the common lesion 8-oxo-7,8-dihydro-2′-deoxyguanine (8-oxoG). Oxidative lesions in telomeric DNA are associated with changes in telomere length and integrity6,7,10.
ROS also react with free nucleotide pools, and recent studies underscore the importance of oxidized deoxynucleoside triphosphates (dNTP) in regulating genome stability and cell survival. Free dNTPs are more susceptible to oxidation than duplex DNA11, and insertion of oxidized nucleotides into the genome during replication leads to mutations and cell death12–14. Nudix hydrolase 1 (NUTD1 or MTH1) converts 8-oxodGTP to 8-oxodGMP to prevent utilization during DNA synthesis15. MTH1 upregulation occurs frequently in various cancers16,17. Cancer cell lines are addicted to MTH1 due partly to dysfunctional redox regulation18,19, and are more sensitive to MTH1 inhibitors than normal cells14,17. However, the impact of oxidized dNTPs on telomere maintenance and integrity is unknown.
During base excision repair, 8-oxoguanine DNA glycosylase (OGG1) removes 8-oxoG from the genome20 when the damaged base pairs with cytosine, but not when present in single stranded DNA or in G-quadruplex structures21. Remarkably, an unbiased screen in yeast for genes that alter telomere length revealed that ogg1 deletion strains have longer telomeres than wild type22, and this lengthening depends partly on telomerase23. Ogg1−/− mice also show longer telomeres in vivo, but culturing cells from these mice in pro-oxidant conditions induces accelerated telomere shortening10. Thus, changes in cellular oxidative state influence telomere length. This paradox of 8-oxoG in telomeres promoting telomere lengthening, whereas oxidative stress causes telomere shortening, suggests a level of complexity inherent in ROS-induced DNA damage that remains currently unresolved.
Here we examine how the oxidized base 8-oxoG regulates telomerase activity when present in either the telomeric overhang or within the dNTP pool. We show that 8-oxodGTP incorporation by telomerase is a chain terminator, thereby preventing telomere restoration and promoting cell death. In stark contrast, the presence of a pre-existing 8-oxoG in the telomeric overhang enhances telomerase activity by destabilizing the G-quadruplex structure in the folded overhang. Therefore, we demonstrate that 8-oxoG has a dual role in inhibiting or stimulating telomerase depending on whether the free dNTPs are oxidized and inserted during extension, or the telomeric DNA is oxidized directly by free radicals.
Results
8-oxodGTP insertion is a telomerase chain terminator
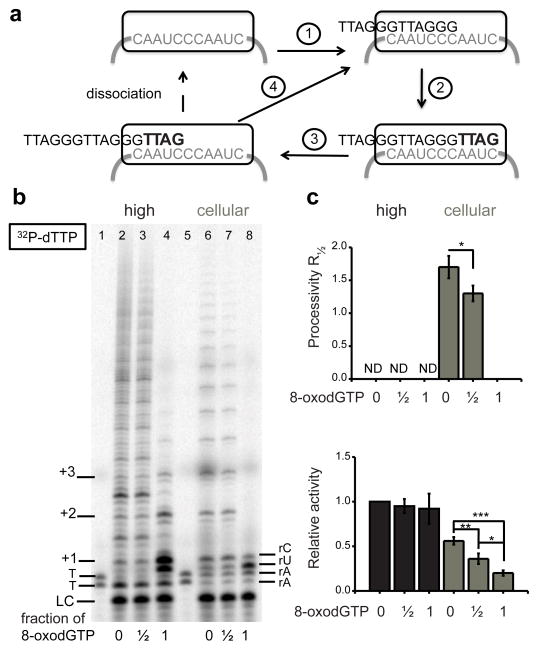
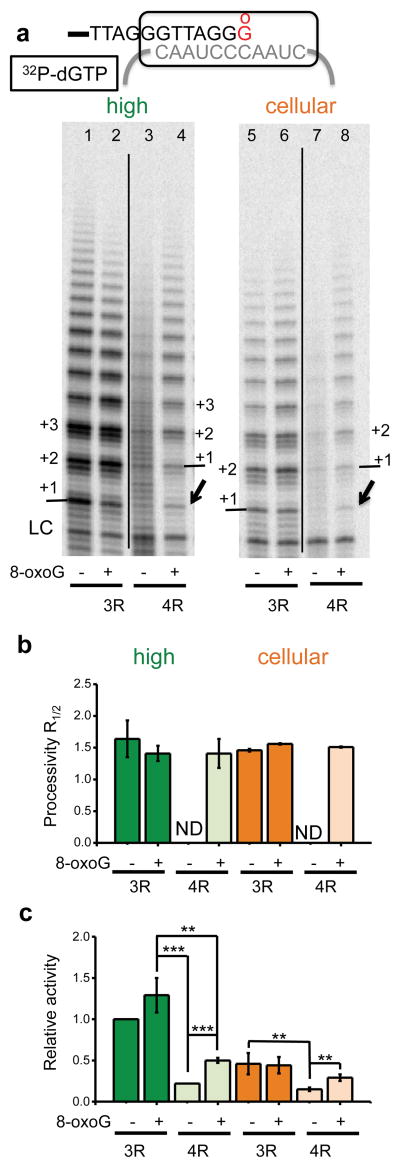
We tested whether telomerase can introduce an 8-oxoG into telomeric DNA via utilization of 8-oxodGTP during telomere elongation. The protein component (TERT) catalyzes reverse transcription of an integral 11 nucleotide RNA template housed within the RNA component (TR) to add repeats to the overhang3. Fig. 1a summarizes the catalytic steps. 1) The overhang anneals to the RNA template to form a short DNA-RNA hybrid that primes TERT-mediated DNA synthesis, 2) telomerase elongates the overhang governed by its template, 3) telomerase translocates and realigns with the template to prime synthesis, and 4) cycling back to elongation generates the characteristic 6-nt product pattern. The number of repeats added prior to telomerase dissociation defines processivity24.
Figure 1.
8-oxodGTP is a telomerase chain terminator. (a) Telomerase catalytic cycle. Blue indicates newly added nucleotides and grey indicates the telomerase template. Numbers represents steps in the cycle. (b) Telomerase reactions were conducted using (TTAGGG)3 primer (3R) with high dNTPs (500 μM dGTP, 500 μM dATP, and 2.9 μM dTTP) (lanes 2 – 4) or cellular dNTPs (37 μM dTTP, 24 μM dATP, 29 μM dCTP, 5.2 μM dGTP) (lanes 6 – 8) and 0.3 μM 32P-α-dTTP. dGTP and 8-oxodGTP were mixed at 1:0, 1:1, and 0:1 ratios for a total of 500 μM or 5.2 μM unlabeled guanine nucleotide. Products were separated on denaturing gels. The LC was a 32P end-labeled 18-mer oligonucleotide. Numbers on the left show number of added repeats, and letters on right indicate template base. (c) Total products were normalized to the LC and used to calculate processivity and relative activity. Bars represent the mean ± sd from three independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001, two-tailed Student’s t test. N.D. = not determined. Uncropped gel image is shown in Supplemental Data Set 1.
To probe for 8-oxodGTP incorporation we used the standard telomerase substrate of three TTAGGG repeats (3R), and examined activity of immunopurified FLAG-tagged telomerase overexpressed in human HEK 293T cells. This well characterized super-telomerase extract exhibits similar kinetic properties as endogenous telomerase25. Reactions were conducted with widely used high dNTP concentrations and with cellular dNTP concentrations26. Utilizing cellular concentrations is biologically relevant and important because dNTP pool balance and concentrations affect the accuracy of DNA polymerases27. Telomerase was similarly processive with both dNTP mixes, but product yield was lower with the cellular dNTP mix as observed in reactions with 32P-α-dGTP (Supplementary Fig. 1). We switched to radiolabelled dTTP for reaction with 8-oxodGTP. Reactions with the high dNTP mix showed aberrant product termination after dTTP incorporation due to the imbalance with low dTTP. Never-the-less, increasing substitution of dGTP with 8-oxodGTP dramatically inhibited telomerase extension (Fig. 1b and Supplementary Fig. 1). The pattern of hexameric repeat addition was restored in reactions with the cellular dNTP mix, and inhibitory effects of 8-oxodGTP were even more obvious (Fig. 1b–c). Complete substitution of dGTP with 8-oxodGTP generated a prominent band representing termination after incorporation opposite template U, and a weak band representing incorporation opposite rC. These data suggest that telomerase can incorporate 8-oxodGTP opposite rC, albeit poorly, and that further extension from an 8-oxodGTP:C base pair is greatly inhibited.
For the 3R substrate the first repeat addition occurs opposite rArArUrC prior to translocation, so if 8-oxodGTP is incorporated opposite rC, then further extension requires immediate translocation (Fig. 1a, step 3). Therefore, the problem may arise with translocation rather than extension. The GG(TTAGGG)2TTAG substrate allows for 8-oxodGTP incorporation opposite cytosine (rCrCrArArUrC) and extension prior to translocation. Increasing substitution of 8-oxodGTP for dGTP led to dramatic telomerase inhibition, with no product detected in reactions lacking dGTP (Supplementary Fig. 2). This confirms that telomerase extension following 8-oxodGTPincorporation is highly unfavorable, and the lack of extension is not due to translocation inhibition.
Telomerase incorporation of 8-oxodGTP is mutagenic
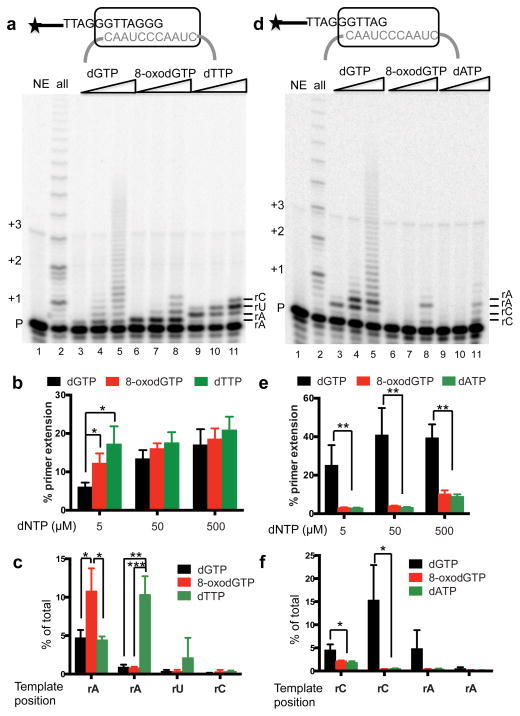
DNA polymerases can insert 8-oxodGTP opposite cytosine or adenine, although steric clashes of 8-oxodGTP:C base pair favor insertion opposite A12,28. The accuracy of telomerase is not well established raising the possibility that incorporation opposite rC might have resulted from erroneous dATP, dTTP or dCTP insertion, rather than 8-oxodGTP. To definitively test whether telomerase can incorporate 8-oxodGTP, and whether this incorporation is error prone, we radiolabeled the primer and added increasing concentrations of only dGTP or 8-oxodGTP. Reaction with a single dNTP type is a well-established method for defining polymerase fidelity (i.e. selectivity for correct over incorrect or damaged dNTPs) and dNTP affinity29–31. Surprisingly, mutagenic incorporation opposite rA occurs even at low 5 μM dGTP, and run-on addition occurs at high dGTP (Fig. 2a). 8-oxodGTP shows significantly more (50%) misinsertion opposite rA compared to dGTP at 5 μM, but minimal extension to the next template rA even at high amounts (Figs. 2b–c). At low dNTP (5 μM) the total primers extended with 8-oxodGTP was statistically similar to extension with correct dTTP, but extension to the next rA was significantly higher for dTTP compared to mutagenic dGTP or 8-oxodGTP (Figs. 2b–c). All products (one nucleotide added and longer) require incorporation opposite the first rA. Next, we tested incorporation opposite two adjacent template rCs. Again, we observed run-on addition of dGTP incorporation beyond rC, but poor incorporation and extension with 8-oxodGTP and mutagenic dATP (Figs. 2d–f). These data indicate that telomerase can utilize 8-oxodGTP during DNA synthesis, but that it preferentially misincorporates 8-oxodGTP opposite rA versus rC, and that further extension is greatly inhibited.
Figure 2.
Telomerase preferentially incorporates 8-oxodGTP opposite rA. (a) and (d) Telomerase activity assays were conducted using 5 nM 32P-end labeled (TTAGGG)3 (a) or (GGTTAG)3 (d) primers. Reactions contained cellular dNTPs (lane 2) or increasing amounts (5, 50, 500 μM) of dGTP (lanes 3 – 5), 8-oxodGTP (lanes 6 – 8), dTTP (lanes 9 – 11, panel a) or dATP (lanes 9 – 11, panel d). Products were separated on denaturing gels. Numbers on the left show number of added repeats, and letters on right indicate template base. (b) and (e) The percent of total primers extended for each reaction. (c) and (f) The percent of primers terminated at each template position as a function of total radioactivity in the lane. Bars represent the mean ± sd from three independent reactions. * p < 0.05, ** p < 0.01, *** p < 0.001, two-tailed Student’s t test. N.D. = not determined. Uncropped gel images are shown in Supplemental Data Set 1.
Telomere dysfunction due to oxidized dNTPs
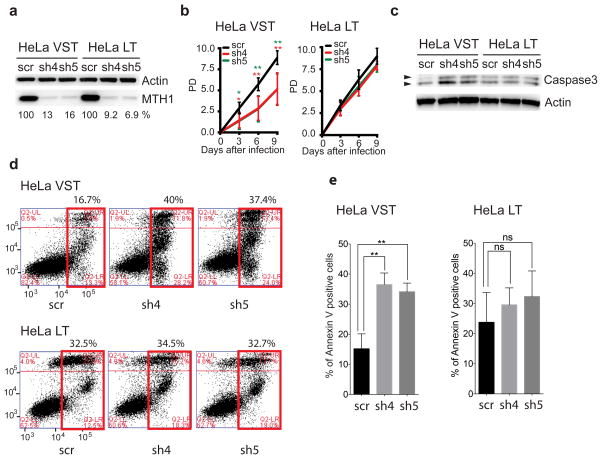
Elevating oxidized dNTPs via MTH1 depletion or inhibition causes 8-oxoG incorporation into the genome and cell death or senescence14,32. Cancer cells are thought to exhibit greater sensitivity to MTH1 inhibition compared to normal cells due to a higher pro-oxidant state, but another notable difference is that most cancer cell lines depend on telomerase for proliferation5. Since 8-oxodGTP is a telomerase chain terminator, we reasoned that cancer cells with critically short telomeres would be the most vulnerable to telomerase inhibition. To test this, we depleted MTH1 in HeLa LT cells harboring long telomeres (~27 kb) and HeLa VST cells possessing very short telomeres (~3.7 kb)33,34 (Supplementary Fig. 3a). Transduction of the cells with lentivirus expressing two distinct shRNAs targeting MTH1 (sh4 and sh5), followed by selection with puromycin, suppressed MTH1 expression by ~80 to 90% (Fig. 3a). MTH1 depletion increased nuclear 8-oxoG staining in both cell lines (Supplementary Fig. 3c) consistent with previous reports14,32. This shows an increase in total cellular 8-oxoG levels in nucleotide pools and incorporation into DNA.
Figure 3. Cells with shortened telomeres are hypersensitive to oxidized nucleotides.
HeLa cell lines with very short telomeres (VST) or long telomeres (LT) were analyzed 3 days after transduction with lentivirus expressing a non-targeting shRNA (scr) or different shRNAs against MTH1 (sh4 and sh5). (a) Immunoblot with antibodies against MTH1 or actin. The percent MTH1 expression relative to the control was calculated. (b) Population doubling values are the mean ± s.d. from four independent experiments. * p<0.05, ** p<0.01, One-way ANOVA with Tukey’s honest significance difference. (c) Analysis for apoptosis by immunoblotting for caspase 3 activation. (d) Flow cytometry results of annexin V/PI staining. The numbers above the red box indicate percent of total annexin V-FITC positive cells (both early and late apoptosis). (e) Quantitation of percent annexin V-FITC positive cells. * p<0.05, ** p<0.01, two-tailed Student’s t test. Uncropped blot images are shown in Supplemental Data Set 1.
While oxidized dNTPs significantly inhibited proliferation of the HeLa VST cells, the HeLa LT were largely unaffected (Fig. 3b). The difference in sensitivity was not due to differences in expression of the ROS-scavenging enzymes catalase or SOD (Supplementary Fig. 3b). Decreased proliferation was due to apoptosis as indicated by elevated annexin V staining and caspase 3 cleavage in the MTH1 depleted HeLa VST cells, relative to controls, which continued to day 6 post infection (Fig. 3c–e and Supplementary Fig. 4a–b). In contrast, MTH1 depletion in HeLa LT cells did not induce apoptosis compared to controls (Fig. 3c–e and Supplementary Fig. 4a–b).
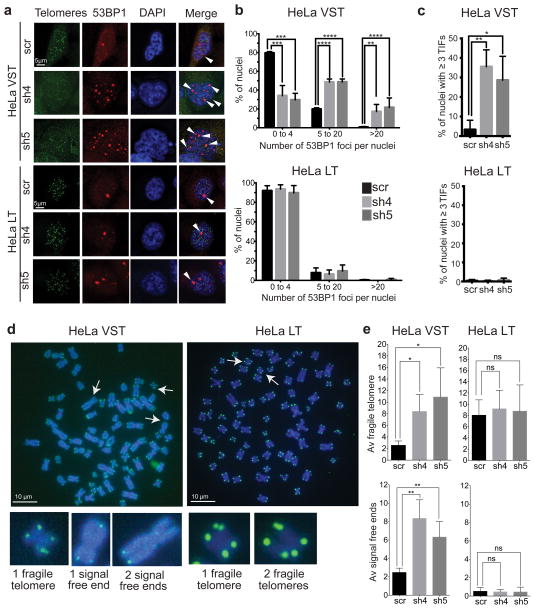
Next we examined whether elevating oxidized dNTPs impacts telomere integrity. MTH1 depletion significantly increased 53BP1 foci formation in HeLa VST cells, as reported for other cell lines14,32, but not in HeLa LT cells (Figs. 4a–c). 53BP1 is a DNA damage response protein that localizes to DNA double strand breaks or dysfunctional telomeres, therefore, 53BP1 foci at telomeres are often referred to as TIFs (telomere dysfunction induced foci)35. MTH1 depletion in HeLa VST cells induced a 6-fold increase in cells displaying three or more TIFs and in the average TIFs per nuclei, whereas HeLa LT cells were unaffected (Fig. 4c and Supplementary Fig. 3e). The number of TIFs in HeLa VST is likely underestimated given that their very short telomeres are difficult to visualize. MTH1 depletion did not induce obvious TRF2 displacement from the telomeres compared to controls (Supplementary Fig. 3d), consistent with a lack of induced telomere fusions (Fig. 4d). Rather, MTH1 depletion significantly increased chromatids lacking telomere staining (signal free ends) in HeLa VST cells compared to controls, but not in HeLa LT cells (Fig. 4e). MTH1 depletion also increased fragile telomeres, manifested as chromatid ends containing multiple telomeric signals, in the HeLa VST cells (Fig. 4d–e). Interestingly, the control HeLa VST cells showed more signal free ends (or critically short telomeres) compared to HeLa LT, but the control HeLa LT cells contained more fragile telomeres. The latter may be related to reduced occupancy of TRF1 in cells with very long telomeres36, since TRF1 prevents telomere fragility by promoting replication37. We passaged survivors from MTH1 depletion to test for effects on telomere shortening rates, but found MTH1 expression recovered to about 30% in both cell lines, and almost completely in the sh5 HeLa VST (Supplementary Fig. 3f). This suggests strong selective pressure for MTH1 activity upon continued passaging at 20% oxygen. Our results indicate that in telomerase expressing cancer cells, the presence of shortened telomeres influences sensitivity to the adverse effects of oxidized dNTPs on cell survival and telomere integrity.
Figure 4.
Oxidized nucleotides induce telomere defects in cells with shortened telomeres. HeLa cell lines with very short telomeres (VST) or long telomeres (LT) were analyzed 3 days after tansduction with lentivirus expressing a non-targeting shRNA (scr) or different shRNAs against MTH1 (sh4 and sh5). (a) 53BP1 foci (red) were visualized by immunofluorescence and telomeric foci were detected by fluorescence in situ hybridization (green). Additional immunostaining of telomeric RAP1 protein was required in HeLa VST cells to amplify the telomere signal. 53BP1 foci at telomeres appear yellow (white arrow). (b) The number of 53BP1 foci per nuclei after binning. Bars represent the mean ± sd from 3 independent experiments (100 – 150 cells per condition). ** p<0.01, *** p<0.001, **** p<0.0001, One-way ANOVA with Tukey’s honest significance difference. (c) The percent of nuclei showing ≥ 3 telomeric 53BP1 foci per nuclei (TIFs). Bars are the mean ± s.d. from 3 independent experiments. * p<0.05, One-way ANOVA. (d) Representative metaphase chromosomes of telomere FISH from HeLa VST and HeLa LT cells expressing sh5 against MTH1. Images of fragile telomeres and signal free ends are shown. (e) Quantitation of telomere aberrations, average number per metaphase. Bars represent the mean ± sd from 3 independent experiments (at least 15–20 metaphases per condition).* p<0.05, ** p<0.01, two-tailed Student’s t test.
To further examine a role for telomerase activity in mediating sensitivity to oxidized dNTPs, we compared in side-by-side experiments MTH1 depletion in primary BJ skin fibroblasts to BJ-hTERT fibroblasts that express exogenous telomerase. Consistent with previous work, we find that MTH1 depletion primarily induces cell senescence in fibroblasts, rather than apoptosis32 (Supplementary Figs. 4c and Supplementary Fig. 5c). However, the BJ-hTERT cells were more sensitive compared to the telomerase negative BJ cells, as evidenced by a greater induction of senescence associated beta-galatosidase activity and p53 expression, and reduction in cell proliferation compared to controls (Supplementary Fig. 5). These data suggest that telomerase activity modulates cellular sensitivity to oxidized dNTPs.
Telomerase extension from a pre-existing 8-oxoG terminus
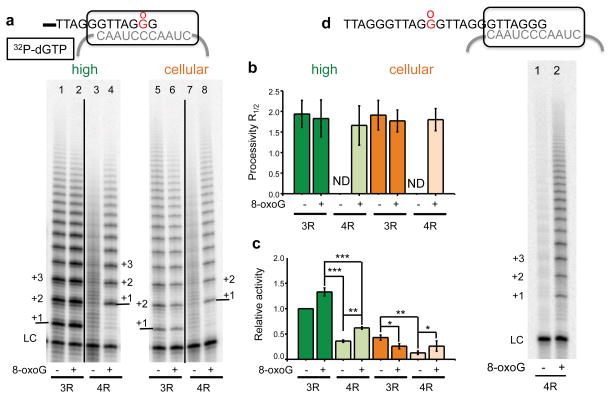
8-oxoG can arise in the telomeric overhang by telomerase incorporation of 8-oxodGTP or by direct free radical reaction with guanines in the overhang. We tested whether telomerase can extend an overhang with a pre-existing terminal 8-oxoG by using substrates of three (3R) and four (4R) TTAGGG repeats with a 3′ G or 8-oxoG (Fig. 5a). Telomeric overhangs in cells consist of 8–30 repeats, and thus, can fold into G-quadruplex (GQ) structures which impede telomerase loading38–41. The 3R substrate lacks sufficient sequence to form GQ and allows testing for 8-oxoG effects in the absence of structure, whereas the 4R substrate enables testing for 8-oxoG effects in the context of biologically relevant structure. The terminal 8-oxoG substitution in 3R minimally impacted telomerase processivity and total activity (Fig. 5), in stark contrast to experiments with 8-oxodGTP (Fig. 1). Telomerase also extends 3R primers when the terminal 8-oxoG aligns with rA in the template, with no significant decrease in efficiency (Supplementary Fig. 6a–c). These data suggest that a pre-existing terminal 8-oxoG does not impair telomerase loading. The 4R substrate is poorly extended by telomerase due to stable GQ folding (Fig. 5a). Remarkably, the presence of a single 8-oxoG caused a dramatic restoration of processivity and activity compared to the unmodified 4R substrate (Figs. 5a–c). These data show that a pre-existing terminal 8-oxoG minimally affects overhang annealing to the telomerase template in the absence of structure, but greatly enhances telomerase loading on the GQ forming substrate.
Figure 5.
Telomerase elongation of primers with a terminal 8-oxoG. (a) Telomerase reactions were conducted using primers containing three (3R) or four (4R) TTAGGG repeats, with a 3′ terminal G or 8-oxoG (Supplementary Table 1, oligos #3, 4, 6 and 7). Reactions contained high (lanes 1 – 4) or cellular (lanes 5 – 8) dNTP concentrations and 0.3 μM 32P-α-dGTP. Telomerase products were separated on denaturing gels. The LC was 32P end-labeled 36-mer oligonucleotide. Arrow points to a product from degraded primer. Numbers with plus sign indicate number of added repeats. (b) and (c) Products were normalized to the LC and used to calculate processivity and relative activity. Bars represent the mean ± sd from three independent reactions. ** p < 0.01, *** p < 0.001, two-tailed Student’s t test. N.D. = not determined. Uncropped gel image is shown in Supplemental Data Set 1.
Use of the 4R substrate revealed a prominent band migrating coincidently with an unextended 4R molecule that was not visible for the 3R substrate due to overlap with the loading control (Fig. 5a, arrow). This product resulted from telomerase extension of a degraded oligonucleotide shortened to (TTAGGG)3TTA. This suggests that with a terminal 8-oxoG:rC base pair the enzyme is in a non-productive extension complex that promotes the reverse polymerase reaction, as described for HIV-RT42. The telomerase preparations lack detectable contaminating exonuclease (Supplementary Fig. 7a). However, we observed enhanced degradation products on primers terminating in 8-oxoG compared to unmodified primers in telomerase reactions containing only dGTP (Supplementary Fig. 7b–c). This agrees with our results that extension following 8-oxodGTP incorporation opposite rC is unfavorable (Fig. 1). Primer degradation also occurred, but was not enhanced, when the primer 8-oxoG aligned with template rA (Supplementary Fig. 6d–e). This agrees with increased efficiency of telomerase 8-oxodGTP incorporation opposite rA versus rC (Fig. 2), and suggests the 8-oxoG:rC base pair is more distorting in the telomerase active site compared to 8-oxoG:rA.
8-oxoG restores telomerase activity by disrupting G-quadruplex structure
Primer degradation and removal of the pre-existing terminal 8-oxoG (Fig. 5), complicates interpretation of an 8-oxoG role in telomerase loading and extension. To circumvent this we prepared 3R and 4R substrates with 8-oxoG substituted for the middle G of the last repeat. Primer degradation products were not observed, indicating the 8-oxoG remained intact and the adjacent terminal G:rC base pair was efficiently extended (Fig. 6a, lanes 5 and 8). For the 3R substrate, the middle 8-oxoG did not affect processivity, but decreased relative activity under cellular dNTP conditions (Figs. 6b–c). Telomerase was less sensitive to 8-oxoG with the artificially high dNTPs. These studies indicate that a middle 8-oxoG may alter optimal telomerase annealing with the 3R overhang, but does not interfere with extension from the G:rC base pair.
Figure 6. 8-oxoG restores telomerase activity on quadruplex folded overhangs.
(a) Telomerase reactions were conducted using primers containing three (3R) or four (4R) TTAGGG repeats, with a middle G or 8-oxoG in the terminal repeat (Supplementary Table 1, oligos #3, 5, 6 and 8). Reactions contained high (lanes 1–4) or cellular (lanes 5–8) dNTP concentrations and 0.3 μM 32P-α-dGTP. Products were separated on denaturing gels. The LC was a 32P end-labeled 36-mer oligonucleotide. Numbers with plus sign indicate number of added repeats. (b) and (c) Products were normalized to the LC, and used to calculate processivity (b) and relative activity (c). Bars represent the mean ± sd from three independent reactions. * p < 0.05, ** p < 0.01, *** p < 0.001, two-tailed Student’s t test. N.D. = not determined. (d) Telomerase reactions were conducted with 4R primers containg a middle G or 8-oxoG in the second repeat (Supplemental Table 1, oligos #11 and 6). Uncropped gel images are shown in Supplemental Data Set 1.
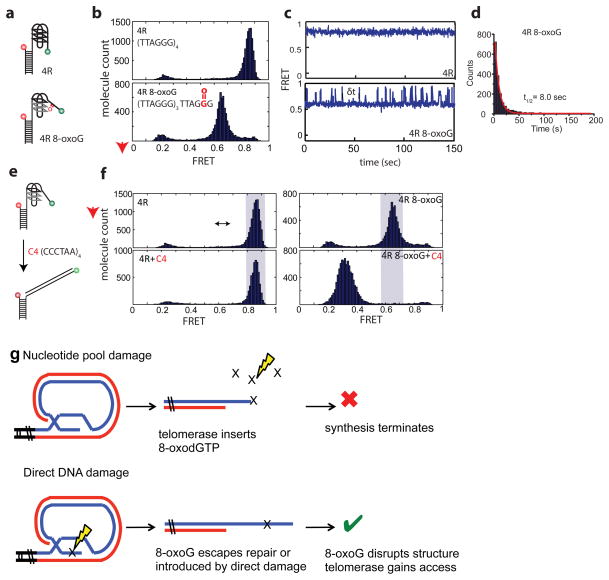
Similar to the terminal 8-oxoG, a middle 8-oxoG in the GQ forming substrate (4R) dramatically restored processive elongation, and significantly increased telomerase activity compared to the unmodified substrate (Fig. 6a–c). We obtained the same result when an 8-oxoG was inserted in the second telomeric repeat, distal from alignment with the RNA template (Fig. 6d). To determine the mechanism for enhanced activity, we used a single molecule approach that enables probing of the GQ structure with high spatial, temporal and molecular resolution. Bulk phase biochemical studies indicate that telomeric DNA with 8-oxoG can fold into GQ, but the lesion reduces the GQ melting temperature21,43. We prepared a single molecule fluorescence energy transfer (smFRET) reporter of telomeric GQ folding39. The two dyes located at both ends of the GQ forming DNA segment with four TTAGGG repeats yielded a high FRET peak of 0.8 in 100 mM KCl, indicative of compact GQ folds (Fig. 7a)39,44. However, the FRET peaks for the molecules with a middle 8-oxoG in the terminal repeat shifted to a lower value, indicating a less tightly folded state (Fig. 7b). Representative single molecule traces of the 4R substrate display stable high FRET, whereas approximately 25% of the 8-oxoG molecules exhibit FRET fluctuations indicating structural dynamics (Fig. 7c). Dwell time based on δt collected from over three hundred smFRET traces revealed a t1/2 of 8 seconds (Fig. 7d). To test accessibility of the substrates we applied a (CCCTAA)4 complementary oligonucleotide, and found this shifted the FRET histogram to a low FRET peak for the 4R 8-oxoG substrate, but not for the unmodified substrate (Figs. 7e–f). These studies strongly suggest that 8-oxoG restores telomerase activity on GQ forming substrates by destabilizing the structure, thereby increasing telomerase accessibility and loading.
Figure 7.
8-oxoG increases the dynamics of telomeric G-quadruplex structures. (a) Schematic of FRET-DNA constructs for the 4R substrate with a middle G or 8-oxoG in the terminal repeat (oligos #6 and 8, Supplementary Table 1) with FRET pair dyes Cy3 and Cy5 attached. (b) Single-molecule frequency histograms revealing FRET peaks. (c) Representative single-molecule traces. (d) The dwell time (t1/2) was calculated for the 4R substrate containing the 8-oxoG lesion. (e) Schematic of complementary oligonucleotide (C4) binding to the unfolded repeat. (f) Frequency count histograms before and after binding of the C4 olignucleotide to the unmodified 4R and the 8-oxoG containing 4R substrates. (g) Model for dual roles of 8-oxoG in regulating telomerase. Oxidation of nucleotide pools generates 8-oxodGTP, which is a telomerase chain terminator. Conversely, oxidation of the telomeric overhang promotes telomerase loading by destabilizing G-quadruplex structure.
Discussion
The excess of free radicals under oxidative stress conditions elevates oxidation of DNA and of the free nucleotide pools. Here we demonstrate that 8-oxoG can arise in telomeres by telomerase incorporation of 8-oxodGTP, in addition to the established mechanism of telomeric DNA reaction with free radicals45. Our studies reveal a dual role for 8-oxoG in regulating telomerase elongation of telomeres (Fig. 7g). We propose that this duality leads to the paradoxical observations that under normoxic conditions unrepaired 8-oxoG lesions promote telomere lengthening, whereas oxidative stress and pro-oxidant conditions promote telomere dysfunction or shortening6,10.
We demonstrate that 8-oxodGTP promotes chain termination following insertion by telomerase. This result infers that perturbations at the primer terminus after damaged nucleotide incorporation prevent alignment of the catalytic groups to support subsequent insertion and extension. Mechanistic insight into the structural changes within the active site can be gained from DNA polymerase studies that utilized 8-oxoG in the template or an 8-oxodGTP inserted at the primer termini. These crystallographic studies indicate that 8-oxoG promotes adverse changes to the nucleoside sugar pucker and phosphate backbone arising from the clash at the adducted oxygen (O8). Insertion of 8-oxodGTP leads to an overall instability at the primer terminus and a loss of base pairing interactions12. The changes likely promote misalignment of the primer terminus following 8-oxodGTP insertion, which then inhibits extension. This phenomena is the basis of chain terminating analogs, which also inhibit telomerase46.
The result that telomerase could extend a primer with a pre-existing terminal 8-oxoG (Fig. 5) was surprising, given that 8-oxodGTP incorporation is chain terminating (Fig. 1). There are several explanations. First, previous studies reported a telomerase associated nuclease can remove 3′ blocking nucleotides, possibly by the reverse polymerase reaction known as pyrophosphoroylsis as described for HIV-RT42,47,48. The appearance of an extended degraded primer for substrates containing a terminal 8-oxoG (Fig. 5), suggests removal of the blocking nucleotide prior to telomerase elongation in reactions with normal dNTPs. However, if this nuclease removes 8-oxodGMP after incorporation, the presence of 8-oxodGTP in the reaction would allow re-incorporation, setting up a futile cycle. Detection of nuclease activity depends on telomerase ability to elongate the substrate47,48 (Supplementary Figs. 6–7), suggesting these activities are tightly linked. Second, telomerase primer utilization may be more flexible than dNTP utilization. Previous work showed telomerase elongates a primer with 3′ non-telomeric sequence or a 3′dideoxynucleotide, but terminates following ddNTP incorporation47,48. This is highly reminiscent of our result that telomerase elongates primers with a 3′ 8-oxoG, but fails to extend following 8-oxodGTP incorporation. The earlier studies provide evidence for a conserved telomerase endonuclease activity that can cleave primers47,48.
The observation that 8-oxoG incorporation terminates telomerase elongation makes several predications about the impact of elevating oxidized dNTPs on telomere integrity. First, this should inhibit telomere maintenance. Following MTH1 depletion, the HeLa VST cells showed an average of 3.5 telomeric foci colocalized with 53BP1 (Supplementary Fig. 3e), which is impressive considering telomeres in these cells are less than 0.005% of the genome. The TIFs are likely underestimated given that very short telomeres are difficult to detect. In agreement, MTH1 depletion also increased telomere loss in HeLa VST cells (Figs. 4d–e). While reliable methods for precisely measuring 8-oxodGTP levels are lacking, even trace amounts of 8-oxodGTP (<1% of dGTP) cause DNA polymerase γ to generate mutations49. Replicative polymerases have much lower error rates (~10−6) than telomerase (~10−3)50,51, and the observation that high fidelity polymerases utilize 8-oxodGTP in cells16,28 predicts that telomerase will as well. Second, MTH1 depletion should accelerate telomere shortening. Unfortunately, the recovery of MTH1 expression during continued cell culturing precluded the ability to determine effects on telomere shortening, and attests to selective pressure for MTH1 expression during passaging at 20% oxygen. Third, the deleterious effects of MTH1 depletion should depend partly on telomerase activity. BJ-hTERT fibroblasts are more sensitive than telomerase deficient BJ cells (Supplementary Fig. 5). Previous studies also reported that telomerase deficient primary cells were less sensitive to MTH1 inhibitors than cancer cell lines, which were almost all telomerase positive14. Tumors frequently have shortened telomeres despite the presence of telomerase52. Whether the telomerase negative U2OS cancer cell line, which maintains telomeres by ALT (alternative lengthening of telomeres), is sensitive to MTH1 inhibitors is controversial14,53. Collectively, these studies indicate that dependence on robust telomerase activity, may partly explain why cancer cells are more sensitive to oxidized dNTPs than normal cells.
The finding that cancer cells with short telomeres are more sensitive to MTH1 depletion than cells with long telomeres, suggests a difference in the dependence on telomerase activity for short-term viability. In agreement, previous studies found that cancer cells with short telomeres were more sensitive to telomerase inhibition, compared to cancer cells with long telomeres, and that apoptosis induction was rapid within 2–4 days41,54, similar to MTH1 depletion in HeLa VST cells. This timing is insufficient to allow enough cell divisions for significant telomere shortening, supporting the model that cells with short telomeres are replicating with critically short telomeres and require telomerase activity at each cycle for viability54. In agreement, control HeLa VST cells have more telomeric signal free ends compared to HeLa LT (Fig. 4e). Similar to our results with MTH1 depletion, treating telomerase positive cancer cells with the analog 6-thio-2′-deoxyguanosine (6-thio-dG) also induces rapid cell death and acute telomere dysfunction55. Normal telomerase deficient cells are less sensitive to 6-thio-dG, similar to oxidized dNTPs14,55. Collectively, these results indicate that cancer cells with limited telomere reserves are vulnerable to acute telomerase inhibition by the incorporation of chain-terminating nucleotides.
Our result that 8-oxoG destabilizes G-quadruplex in the telomeric overhang and enhances telomerase accessibility (Figs. 5–7), offers an explanation for the findings that unrepaired 8-oxoG lesions in OGG1 deficient mice and yeast promote telomere lengthening in vivo or cell culturing at 3% oxygen10,23. Although OGG1 cannot remove 8-oxoG in single stranded DNA, it can when the overhang pairs with duplex DNA in the t-loop/D-loop structure56. Interestingly, culturing the OGG1 deficient cells at 20% oxygen promotes telomere shortening and aberrations10, suggesting in pro-oxidant conditions MTH1 levels in normal cells is insufficient to sanitize dNTP pools32. Our studies indicate that the elevated oxidized dNTPs caused by oxidative stress likely contributes to telomere shortening in the OGG1 deficient cells cultured under pro-oxidant conditions, and overrides any benefits of 8-oxoG destabilization of telomeric G-quadruplex.
Our results indicate that in addition to MTH1 addiction, cancer cells with short telomeres may also be highly dependent on telomerase for viability and telomere stability in the short term. In contrast, cancer cells with long telomeres are largely insensitive to elevated oxidized dNTPs and MTH1 depletion. While incorporation of oxidized nucleotides might also impact shelterin binding or t-loop assembly, such effects are inconsistent with the lack of sensitivity to MTH1 depletion observed in HeLa cells with long telomeres. Thus, the analysis of telomere length in telomerase positive tumors may predict which tumors would be responsive to MTH1 inhibition. A recent study reported that human U2OS and SW480 cell lines were insensitive to MTH1 depletion53, further suggesting that numerous cellular factors influence sensitivity to oxidized dNTPs. For example, MTH1 depletion in cells expressing oncogenic RAS suppresses transformation and tumorigenesis17,57. Finally, antioxidant therapy promotes metastasis of human melanoma in mouse models, suggesting that oxidative stress may inhibit metastatic progression in vivo58. Our studies provide evidence that oxidative stress-induced damage of dNTP pools inhibits the ability of telomerase to maintain telomeres for sustained proliferation of malignant cells harboring critically short telomeres.
ONLINE METHODS
Cell culture and lentiviral infection
HEK-293T cells, BJ skin fibroblasts (CL-5222) and BJ-5ta skin fibroblasts expressing hTERT (CRL-4001) were from ATCC, and HeLa VST and HeLa LT cells were a generous gift from Dr. Roderick O’Sullivan (University of Pittsburgh). Cells were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 50 units/ml penicillin, and 50 units/ml streptomycin (Gibco) at 37°C in humidified chambers with 5% CO2 and 20% O2. The Gibco FBS was replaced with characterized FBS from Hyclone for culturing BJ and BJ-hTERT cells. Mycoplasma testing is done routinely by DAPI staining, and followed up PCR assay when required (VenorTM GeM Mycoplasma Detection Kit, Sigma-Aldrich). MTH1 knock down cell lines were established by overnight transduction with lentivirus expressing shRNAs (sh4 or sh5) targeting MTH1 transcript or a non-targeting scramble shRNA (scr). Selection of transducted cells was performed with 1.5 μg/ml puromycin (HeLa VST and HeLa LT) or 0.75 μg/ml puromycin (BJ and BJ-hTERT) after 24 h recovery. Lentiviral particles were generated by co-transfection of 4 plasmids [Control plasmid (pLKO.1-SCRshRNA-Puro) or one of the five different MTH1-specific shRNA expressing plasmids, pLKO.1-shRNA-MTH1.1 - MTH1.5) together with pMD2.g (VSVG), pVSV-REV and pMDLg/pRRE] into 293-FT cells using TransIT-X2® Dynamic Delivery System (Mirus Bio LLC) with support from the University of South Alabama Mitchell Cancer Institute (USA/MCI) Gene Expression, Editing and Discovery (GEED) Lab. The lentivirus were further concentrated with a Lenti-X Concentrator (Clontech) as per the manufacturers instructions. Five different shRNAs targeting the MTH1 gene were initially screened and Clone IDs (sh#4) NM_002452.3-96s1c1 and (sh#5) NM_002452.3-96s21c1 (Sigma) were the most effective (data not shown). The collection and isolation of lentiviral particles and transduction of cells described in the text was performed as described previously59.
Telomerase preparation
Telomerase was immunopurified as previously described39. Cells expressing hTR and 3xFLAG-tagged hTERT were harvested 48 hours after transfection and lysed in CHAPS lysis buffer (10 mM Tris-HCl, 1 mM MgCl2, 1 mM EDTA, 0.5% CHAPS, 10% glycerol, 5 mM β-mercaptoethanol, 120 U RNasin plus (Promega), 1 μg/ml each of pepstatin, aprotinin, leupeptin and chymostatin, and 1 mM AEBSF) for 30 min at 4°C. Sigma Anti-FLAG M2 affinity gel agarose beads (75 μl of a 1:1 slurry) were pre-washed with 1x human telomerase buffer (50 mM Tris-HCl, pH 8, 50 mM KCl, 1 mM MgCl2, 1 mM spermidine, 5 mM β-mercaptoethanol) with 30% glycerol and mixed with 500 μl of cell lysate for 3 hours at 4°C with rotation. Beads were harvested by centrifugation at 3,500 rpm for 1 min, washed three times with 1x telomerase buffer with 30% glycerol, flash frozen as a 1:1 slurry in 1x telomerase buffer with 30% glycerol, and stored at −80°C.
Telomerase activity assay
Telomerase overexpression in HEK-293T cells was achieved by transient transfection with plasmids expressing hTR and 3xFLAG-tagged hTERT (gift from Thomas Cech, University of Colorado). Cells were grown to 90% confluency and transfected using Lipofectamine 2000 Reagent (Invitrogen), and telomerase was immunopurified as previously described39. Unmodified oligonucleotides were from IDT and oligonucleotides with 8-oxoG were from Midland Certified Reagents Company (Supplementary Table 1). The telomerase assay was as previously described39,60. Briefly, reactions (20 μl) contained 1x human telomerase buffer, 1 μM oligonucleotide substrate, 0.3 μM of 3,000 Ci/mmol 32P-α-dGTP or 32P-α-dTTP (Perkin Elmer) and dNTP (Invitrogen) mix as indicated in the figure legend. Reaction with high dNTPs contained either (500 μM dTTP, 500 μM dATP, 2.9 μM dGTP and 0.3 μM 32P-α-dGTP) or (500 μM dGTP, 500 μM dATP, 2.9 μM dTTP and 0.3 μM 32P-α-dTTP). Reaction with cellular dNTPs contained 37 μM dTTP, 24μM dATP, 29 μM dCTP, 5.2 μM dGTP and 0.3 μM 32P-α-dGTP or 32P-α-dTTP as indicated. Reactions containing 8-oxodGTP (TriLink Biotechnologies) substituted for dGTP are indicated in the figure legends. For reaction with end-labeled primers, the primers were labeled with 32P-γ-ATP (Perkin Elmer) and 10 U Optikinase (Affymetrix) according to the manufacturers’ protocols. Reactions contained 1x telomerase buffer, 5 nM of 32P end-labeled primer, and cellular dNTP mix or either dGTP, 8-oxodGTP, dTTP or dATP (5.2 μM, or 50 μM or 500 μM) as indicated in the figure legend. Reactions were started by adding 6 μl of immunopurified telomerase slurry, incubated for 1 hr at 30°C, and then terminated with 2 μl of 0.5 μM EDTA and heat inactivated at 65°C for 20 min. For reactions with unlabeled primers, 8.0 fmol of 32P end-labeled loading control (LC) was added to the inactivated reaction prior to purification with an Illustra microspin G-25 column (GE Healthcare). After adding equal volume of loading dye the samples were heat denatured and then loaded onto a 10% denaturing acrylamide gel (8M urea, 1x TBE) for electrophoresis. Samples were imaged using a Typhoon phosphorimager (GE Healthcare). Telomerase processivity and relative telomerase activity was quantitated using ImageQuant and normalized to the LC as described60. For reactions with end-labeled primers the products were quantitated with ImageQuant by measuring the intensity of each product band and dividing by the total radioactivity in the lane.
Annexin V/propidium iodide apoptosis assay
The assay was conducted using the Annexin V-FITC Apoptosis Detection Kit (Abcam) according to the manufacturer’s directions. Cells were seeded at 4×105 cells in 60 mm dishes at least 24 hours prior to infection with lentivirus. On day three post lentiviral infection cells were trypsinized and counted. 3×105 cells were harvested by centrifugation for 5 min at 1,200 rpm, and then washed once with PBS. Cells were resuspended in 500 μl binding buffer (provided in the kit) containing 5 μl Annexin V-FITC and 5 μl of propidium iodide (50 μg/ml). Cells were incubated in the dark for 5 min at room temperature. FL1 and FL2 emission was measured using a Accuri C6 flow cytometer after applying color compensation.
Western blots
Cells were resuspended in 400mM NaCl buffer and whole cell extracts were prepared by freeze and thaw cycles followed by 15 min centrifugation at 14000 rpm. Protein amounts were measured using Bradford reagent (Sigma). MTH1 (1:5000, Abcam), β-actin (1:10,000, Sigma), caspase 3 (1:500, Cell Signaling), SOD (1:2000, Abcam, ab13498), catalase (1:2000, Abcam, ab16731) and p53 (1:1000, Santa Cruz, DO-1) antibodies were used.
Immunofluorescence and fluorescence in situ hybridization
Cells were grown on Mattek glass bottom dishes and fixed in 4% formaldehyde for 10 min at room temperature, except for TRF2 immuno-staining cells were fixed on ice in 2% formaldehyde. After three PBS washes, cells were washed three times for 5 min each in PBS containing 0.2% Triton X-100, and blocked for 1 hour in blocking solution (10% goat serum, 1% BSA in PBS). Mouse monoclonal 53BP1 antibody (1:500 dilution, Millipore) or TRF2 monoclonal antibody (1:250 dilution, 4A794.15, Novus) was added and incubated at 4°C overnight. For the HeLa VST cells the 53BP1 antibody mix also contained the rabbit RAP1 polyclonal antibody (1:500 dilution, Bethyl laboratories), to amplify the telomere signal. Cells were washed in PBS three times for 10 min each at room temperature with mild shaking and incubated with secondary antibody (1:1000 dilution, anti-mouse Alexa 594 for HeLa LT, a mix of anti-mouse Alexa 594 and anti-rabbit Alexa488 for HeLa VST, or 1:500 dilution, anti-mouse Alexa 594 for TRF2 immuno-staining) for 1 hour. After three PBS washes, cells were fixed in methanol/acetone (v/v) at −20°C for 10 min or in 2% formaldehyde for 10 min at room temperature for TRF2 immuno-staining, and dehydrated in 70%, 90% and 100% ethanol for 5 min. 100 mg/ml telomeric PNA probe (H2N-Lys-(AATCCC)3-FITC, PNA Innovations61) was diluted 1:500 in hybridization buffer (70% formamide, 10mM Tris HCl pH 7.5, 1x Maleic Acid buffer, 1x MgCl2 buffer) and boiled for 5 min at 95°C. Samples were incubated for 10 min on a hot plate at 75°C and then at room temperature for 2 h in the dark. After two washes in hybridization wash buffer (70% formamide, 10mM Tris HCl pH 7.5), slides were mounted with antifade gold containing DAPI. Imaging acquisition was performed with Nikon Ti inverted fluorescence microscope. Z stacks of 0.2 μm thickness were captured and images were deconvolved using NIS Elements Advance Research software algorithm. Widefield imaging with deconvolution, in this case using blind, interative methods, offered the advantage of minimizing signal loss and was critical for imaging the dim shortened telomeres in the HeLa VST cells.
Chromosome metaphase preparation and telomere FISH
Telomere FISH of metaphase chromosome preparations was carried out as described previously. Briefly, three days following lentiviral transduction, cells were treated with 0.05 μg/ml colcemid for 2 hours. Cells (1 × 106 at a density of 1 × 105 cell/ml) were harvested and incubated with 75 mM KCl hypotonic buffer for 7 min at 37°C, following by fixation in methanol acetic acid (3:1). Samples were stored at −20°C. Lysed cells were dropped onto water-coated glass microscope slides and dried at room temperature overnight. The slides were then fixed in 4% formaldehyde, washed with PBS and then treated with 0.25 mg/ml RNaseA for 15 min at 37°C. Next the slides were incubated with 1 mg/ml Pepsin in 0.01 N HCl for 15 minutes at 37 °C. Fixation and washing were repeated. Slides were then dehydrated in successive ethanol solutions of 70%, 90%, and 100% for 5 minutes each, and allowed to dry over night or for at least 3 hours on at room temperature. Telomere FISH with a FITC conjugated telomeric PNA probe was conducted, and images were captured and analyzed as described above.
Measurement of colocalized 53BP1 and telomeric foci
The number of 53BP1 foci and intersection with telomeres were counted per nuclei using NIS Element Advanced Research software (Nikon) after deconvolution. Briefly, the measurement feature of the software was used to create binary layers based on intensity and defining binary objects corresponding to 53BP1 foci and telomere foci. An intensity threshold was set up for an image from the control experiment (scr) for each channel (threshold FITC for telomeres and threshold Cy3 for 53BP1) and held constant for analyzing images from the MTH1 depleted cells (sh4 and sh5). The binary objects corresponding to areas smaller than 0.05 μm were discarded. The intersection tool was then used to create a third binary layer corresponding to the 53BP1 binary objects overlapping with telomere binary objects. Each nuclei were isolated using the region of interest (ROI) identification tools, based on DAPI staining, and the number of 53BP1 foci and intersections per ROI were exported in excel for data batch analysis.
Immunodetection of 8oxoG by immunofluorescence
Nuclear 8oxoG immuno-detection was performed as described previously62 using monoclonal mouse antibody (MAB3560, Millipore) diluted at 1:50 in goat anti mouse IgM.
Southern blot
Measurement of telomere length in HeLa VST and HeLa LT was performed as described in63,64 with slight modification. Briefly, 3 μg of genomic DNA were digested with a cocktail of 4 restriction enzymes (RsaI, AluI, HindIII, MnlI) for 16 h at 37°C and resolve on 0.8% agarose gel for 16 h 30 min (HeLa VST) or 24 h (HeLa LT). The agarose gel was dried for 2 hours at 50°C and radioactive telomeric probe hybridized over night at 42°C.
Senescence associated beta-galactosidase Assay
The Cellular Senescence Assay Kit (Cell Biolabs) was used to detect of senescence associated beta-gal activity in cells three days post lentiviral infection. The staining was carried out in 35 mm dishes overnight at 37°C as per the manufacturers instructions. For quantification at least 100 cells, spanning 4 to 5 microscopy fields were scored for staining using a 20x objective and a light microscope.
Preparation of oligonucleotides for SM FRET
DNA oligonucleotides used in single molecule fluorescence experiments were 5′ TGG CGA CGG CAG CGA GGC (TTA GGG)4 -Cy3 (oligonucleotide #6, Supplementary Table 1), TGG CGA CGG CAG CGA GGC (TTAGGG)3TTAG(8-oxo-dG)G-Cy3 (oligonucleotide #7, Supplementary Table 1) and Cy5- GCC TCG CTG CCG TCG CCA-Biotin (18 mer, oligonucleotide #12). Cy3 labeled 4R and Cy5 labeled 18 mer were purchased from IDT (Coralville, IA). 4R 8-oxoG was purchased from The Midland Certified Reagent Company (Midland, TX) with an amino modifier C7 at the 3′ end and labeled by reacting with Cy3 maleimide (GE Healthcare). Briefly, 3.3mM Cy3 maleimide was incubated with 40 μM DNA in 100 mM sodium bicarbonate overnight at room temperature. The excess Cy3 was removed by ethanol precipitation twice. The partial duplex DNA molecules, 4R and 4R 8-oxoG, were prepared by mixing a 3′ Cy3 sequence (#6 or #7) with 18mer #12 at a molar ratio of 1:1.5 in 20 mM Tris-HCl pH 7.5, 100 mM KCl. The mixtures were incubated at 95 °C for 2 min then slowly cooled to room temperature in a rate of 2 °C per minute.
Single Molecule FRET Experiments
Oligonucleotide preparation are described in Supplemental Material. Single molecule FRET measurement were carried out on quartz slides (Finkenbeiner) with glass coverslips, which coated with polyethylene glycol (PEG). The slides and coverslips were cleaned and treated with methanol, acetone, potassium hydroxide, burned, treated with aminosilane, and coated with a mixture of 97% mPEG (m-PEG-5000, Laysan Bio, Inc.) and 3% biotin PEG (biotin-PEG-5000, Laysan Bio, Inc). Annealed partial duplex DNA molecules were immobilized on the PEG passivated surface via biotin-neutravidin interaction. Excess donor molecules were washed away with 10 mM Tris pH 7.5, 100 mM KCl with an oxygen scavenging system (0.5% glucose, 10 mM 6-hydroxy-2,5,7,8-tetramethylchromane-2-carboxylic (Trolox), and 1 mg/ml glucose oxidase and 4 μg/ml catalase). All measurements were carried out at room temperature. Widefield prism-type total internal reflection fluorescence (TIRF) microscopy was used with a solid-state 532 nm laser to generate an evanescent field of illumination. Fluorescence signals were separated by a 630-nm dichroic mirror and sent to a charge-coupled device (CCD) camera. Data were recorded with a time resolution of 100 ms as a stream of imaging frames and analyzed by scripts written in Matlab. FRET histograms were generated using over 6,000 molecules and were fitted to Gaussian distributions with unrestrained peak center position using Origin 2016. Dwell times were measured by the time a molecule spends at low FRET state. The dwell time histograms were generated from more than 300 dynamic smFRET traces in at least three separated experiments. The first six transitions were collected from each trace.
Statistics
Statistical analyses were performed with R statistical computing language and Prism 6 (GraphPad Software Inc.). For Figures 3b, 4b and 4c, a one-factor ANOVA was used to determine whether the differences between cells expressing a scramble (scr) shRNA and an shRNA targeting MTH1 mRNA (sh4 and sh5) were significant at a 99% confidence level (Tukey’s honest significance test). For Figures 1, 3e, 4e, 5 and 6, statistical significance was calculated using two-tailed unpaired Student’s t test at a 95% confidence level.
Supplementary Material
Acknowledgments
We thank Drs. Simon Watkins and Claudette St. Croix at the University of Pittsburgh Center for Biological Imaging for assistance with fluorescence imaging, Dr. Shikhar Uttam for help with statistical analyses, Dr. Tatiana Moiseeva for assistance with annexin V/PI FACS assay, and Drs. Roderick O’Sullivan and Bennett Van Houten for critical reading of the manuscript. We also thank Dr. Thomas Cech and Arthur Zaug (University of Colorado) for reagents and assistance with the telomerase assays. Dr. Roderick O’Sullivan (University of Pittsburgh) generously provided the HeLa VST and HeLa LT cell lines. This work was supported by NIH grants R01ES022944, R21AG045545 and R21ES025606 (to P.L.O.), and American Cancer Society RSG-12-066-01-DMC and NIH 1DP2GM105453 (to T. L, G. K. and S. M.), and NIH grant CA148629 and Abraham A. Mitchell Distinguished Investigator fund (R.W.S). This project used the UPCI CTIF and CF that are supported in part by award P30CA047904.
Footnotes
AUTHOR CONTRIBUTIONS
E.F., J.L., and A.B. performed biochemical and cellular experiments, analyzed the data and prepared figures. T.L. and G.M. performed all the SM FRET studies. J. Li and R.W.S. provided lentivirus for MTH1 depletion experiments and R.W.S. provided helpful discussion. E.F., B.F, S.M., and P.L.O. designed experiments, analyzed the data and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
RWS is a scientific consultant for Trevigen, Inc. The other authors declare no competing financial interests.
DATA AVAILABILITY STATEMENT
Any Supplementary Information and Source Data files are available in the online versions of this manuscript. All data supporting the findings of this study are available from the corresponding author upon reasonable request.
References
- 1.Jaskelioff M, et al. Telomerase reactivation reverses tissue degeneration in aged telomerase-deficient mice. Nature. 2011;469:102–106. doi: 10.1038/nature09603. nature09603 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armanios M, Blackburn EH. The telomere syndromes. Nat Rev Genet. 2012 doi: 10.1038/nrg3246. nrg3246 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 4.Hockemeyer D, Collins K. Control of telomerase action at human telomeres. Nat Struct Mol Biol. 2015;22:848–852. doi: 10.1038/nsmb.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim NW, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 6.von Zglinicki T. Oxidative stress shortens telomeres. Trends Biochem Sci. 2002;27:339–344. doi: 10.1016/s0968-0004(02)02110-2. S0968000402021102 [pii] [DOI] [PubMed] [Google Scholar]
- 7.Jurk D, et al. Chronic inflammation induces telomere dysfunction and accelerates ageing in mice. Nat Commun. 2014;2:4172. doi: 10.1038/ncomms5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lonkar P, Dedon PC. Reactive species and DNA damage in chronic inflammation: reconciling chemical mechanisms and biological fates. Int J Cancer. 2011;128:1999–2009. doi: 10.1002/ijc.25815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poljsak B, Fink R. The Protective Role of Antioxidants in the Defence against ROS/RNS-Mediated Environmental Pollution. Oxidative medicine and cellular longevity. 2014;2014:671539. doi: 10.1155/2014/671539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z, et al. Characterization of oxidative guanine damage and repair in mammalian telomeres. PLoS Genet. 2010;6:e1000951. doi: 10.1371/journal.pgen.1000951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamiya H, Kasai H. Formation of 2-hydroxydeoxyadenosine triphosphate, an oxidatively damaged nucleotide, and its incorporation by DNA polymerases. Steady-state kinetics of the incorporation. J Biol Chem. 1995;270:19446–19450. doi: 10.1074/jbc.270.33.19446. [DOI] [PubMed] [Google Scholar]
- 12.Freudenthal BD, et al. Uncovering the polymerase-induced cytotoxicity of an oxidized nucleotide. Nature. 2015;517:635–639. doi: 10.1038/nature13886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsuzuki T, et al. Spontaneous tumorigenesis in mice defective in the MTH1 gene encoding 8-oxo-dGTPase. Proc Natl Acad Sci U S A. 2001;98:11456–11461. doi: 10.1073/pnas.191086798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gad H, et al. MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool. Nature. 2014;508:215–221. doi: 10.1038/nature13181. [DOI] [PubMed] [Google Scholar]
- 15.Sakumi K, et al. Cloning and expression of cDNA for a human enzyme that hydrolyzes 8-oxo-dGTP, a mutagenic substrate for DNA synthesis. J Biol Chem. 1993;268:23524–23530. [PubMed] [Google Scholar]
- 16.Speina E, et al. Contribution of hMTH1 to the maintenance of 8-oxoguanine levels in lung DNA of non-small-cell lung cancer patients. J Natl Cancer Inst. 2005;97:384–395. doi: 10.1093/jnci/dji058. [DOI] [PubMed] [Google Scholar]
- 17.Patel A, et al. MutT Homolog 1 (MTH1) maintains multiple KRAS-driven pro-malignant pathways. Oncogene. 2015;34:2586–2596. doi: 10.1038/onc.2014.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liou GY, Storz P. Reactive oxygen species in cancer. Free Radic Res. 2010;44:479–496. doi: 10.3109/10715761003667554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szatrowski TP, Nathan CF. Production of large amounts of hydrogen peroxide by human tumor cells. Cancer Res. 1991;51:794–798. [PubMed] [Google Scholar]
- 20.Wallace SS. Base excision repair: a critical player in many games. DNA Repair (Amst) 2014;19:14–26. doi: 10.1016/j.dnarep.2014.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou J, Liu M, Fleming AM, Burrows CJ, Wallace SS. Neil3 and NEIL1 DNA glycosylases remove oxidative damages from quadruplex DNA and exhibit preferences for lesions in the telomeric sequence context. J Biol Chem. 2013;288:27263–27272. doi: 10.1074/jbc.M113.479055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Askree SH, et al. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc Natl Acad Sci U S A. 2004;101:8658–8663. doi: 10.1073/pnas.0401263101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu J, Liu Y. Deletion of Ogg1 DNA glycosylase results in telomere base damage and length alteration in yeast. EMBO J. 2010;29:398–409. doi: 10.1038/emboj.2009.355. emboj2009355 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt JC, Cech TR. Human telomerase: biogenesis, trafficking, recruitment, and activation. Genes Dev. 2015;29:1095–1105. doi: 10.1101/gad.263863.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xi L, Cech TR. Inventory of telomerase components in human cells reveals multiple subpopulations of hTR and hTERT. Nucleic Acids Res. 2014;42:8565–8577. doi: 10.1093/nar/gku560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Traut TW. Physiological concentrations of purines and pyrimidines. Mol Cell Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 27.Sohl CD, Ray S, Sweasy JB. Pools and Pols: Mechanism of a mutator phenotype. Proc Natl Acad Sci U S A. 2015;112:5864–5865. doi: 10.1073/pnas.1505169112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katafuchi A, Nohmi T. DNA polymerases involved in the incorporation of oxidized nucleotides into DNA: their efficiency and template base preference. Mutat Res. 2010;703:24–31. doi: 10.1016/j.mrgentox.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 29.Hsu GW, Ober M, Carell T, Beese LS. Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase. Nature. 2004;431:217–221. doi: 10.1038/nature02908. [DOI] [PubMed] [Google Scholar]
- 30.Bertram JG, Oertell K, Petruska J, Goodman MF. DNA polymerase fidelity: comparing direct competition of right and wrong dNTP substrates with steady state and pre-steady state kinetics. Biochemistry. 2010;49:20–28. doi: 10.1021/bi901653g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomlinson CG, et al. Two-step mechanism involving active-site conformational changes regulates human telomerase DNA binding. Biochem J. 2015;465:347–357. doi: 10.1042/BJ20140922. [DOI] [PubMed] [Google Scholar]
- 32.Rai P, et al. Continuous elimination of oxidized nucleotides is necessary to prevent rapid onset of cellular senescence. Proc Natl Acad Sci U S A. 2009;106:169–174. doi: 10.1073/pnas.0809834106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crabbe L, Cesare AJ, Kasuboski JM, Fitzpatrick JA, Karlseder J. Human telomeres are tethered to the nuclear envelope during postmitotic nuclear assembly. Cell Rep. 2012;2:1521–1529. doi: 10.1016/j.celrep.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Sullivan RJ, et al. Rapid induction of alternative lengthening of telomeres by depletion of the histone chaperone ASF1. Nat Struct Mol Biol. 2014;21:167–174. doi: 10.1038/nsmb.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. S0960982203005426 [pii] [DOI] [PubMed] [Google Scholar]
- 36.Takai KK, Hooper S, Blackwood S, Gandhi R, de Lange T. In vivo stoichiometry of shelterin components. J Biol Chem. 2010;285:1457–1467. doi: 10.1074/jbc.M109.038026. M109.038026 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sfeir A, et al. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. S0092-8674(09)00721-1 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Biffi G, Tannahill D, McCafferty J, Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hwang H, et al. Telomeric overhang length determines structural dynamics and accessibility to telomerase and ALT-associated proteins. Structure. 2014;22:842–853. doi: 10.1016/j.str.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 41.Cookson JC, et al. Pharmacodynamics of the G-quadruplex-stabilizing telomerase inhibitor 3,11-difluoro-6,8,13-trimethyl-8H-quino[4,3,2-kl]acridinium methosulfate (RHPS4) in vitro: activity in human tumor cells correlates with telomere length and can be enhanced, or antagonized, with cytotoxic agents. Mol Pharmacol. 2005;68:1551–1558. doi: 10.1124/mol.105.013300. [DOI] [PubMed] [Google Scholar]
- 42.Meyer PR, Matsuura SE, So AG, Scott WA. Unblocking of chain-terminated primer by HIV-1 reverse transcriptase through a nucleotide-dependent mechanism. Proc Natl Acad Sci U S A. 1998;95:13471–13476. doi: 10.1073/pnas.95.23.13471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vorlickova M, Tomasko M, Sagi AJ, Bednarova K, Sagi J. 8-oxoguanine in a quadruplex of the human telomere DNA sequence. FEBS J. 2012;279:29–39. doi: 10.1111/j.1742-4658.2011.08396.x. [DOI] [PubMed] [Google Scholar]
- 44.Tippana R, Xiao W, Myong S. G-quadruplex conformation and dynamics are determined by loop length and sequence. Nucleic Acids Res. 2014;42:8106–8114. doi: 10.1093/nar/gku464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oikawa S, Tada-Oikawa S, Kawanishi S. Site-specific DNA damage at the GGG sequence by UVA involves acceleration of telomere shortening. Biochemistry. 2001;40:4763–4768. doi: 10.1021/bi002721g. bi002721g [pii] [DOI] [PubMed] [Google Scholar]
- 46.Hukezalie KR, Thumati NR, Cote HC, Wong JM. In vitro and ex vivo inhibition of human telomerase by anti-HIV nucleoside reverse transcriptase inhibitors (NRTIs) but not by non-NRTIs. PLoS One. 2012;7:e47505. doi: 10.1371/journal.pone.0047505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oulton R, Harrington L. A human telomerase-associated nuclease. Mol Biol Cell. 2004;15:3244–3256. doi: 10.1091/mbc.E04-03-0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huard S, Autexier C. Human telomerase catalyzes nucleolytic primer cleavage. Nucleic Acids Res. 2004;32:2171–2180. doi: 10.1093/nar/gkh546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pursell ZF, McDonald JT, Mathews CK, Kunkel TA. Trace amounts of 8-oxo-dGTP in mitochondrial dNTP pools reduce DNA polymerase gamma replication fidelity. Nucleic Acids Res. 2008;36:2174–2181. doi: 10.1093/nar/gkn062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kunkel TA. DNA replication fidelity. J Biol Chem. 2004;279:16895–16898. doi: 10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- 51.Kreiter M, Irion V, Ward J, Morin G. The fidelity of human telomerase. Nucleic Acids Symp Ser. 1995:137–139. [PubMed] [Google Scholar]
- 52.Bisoffi M, Heaphy CM, Griffith JK. Telomeres: prognostic markers for solid tumors. Int J Cancer. 2006;119:2255–2260. doi: 10.1002/ijc.22120. [DOI] [PubMed] [Google Scholar]
- 53.Kettle JG, et al. Potent and Selective Inhibitors of MTH1 Probe Its Role in Cancer Cell Survival. J Med Chem. 2016;59:2346–2361. doi: 10.1021/acs.jmedchem.5b01760. [DOI] [PubMed] [Google Scholar]
- 54.Zhang X, Mar V, Zhou W, Harrington L, Robinson MO. Telomere shortening and apoptosis in telomerase-inhibited human tumor cells. Genes Dev. 1999;13:2388–2399. doi: 10.1101/gad.13.18.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mender I, Gryaznov S, Dikmen ZG, Wright WE, Shay JW. Induction of telomere dysfunction mediated by the telomerase substrate precursor 6-thio-2′-deoxyguanosine. Cancer Discov. 2015;5:82–95. doi: 10.1158/2159-8290.CD-14-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rhee DB, Ghosh A, Lu J, Bohr VA, Liu Y. Factors that influence telomeric oxidative base damage and repair by DNA glycosylase OGG1. DNA Repair (Amst) 2011;10:34–44. doi: 10.1016/j.dnarep.2010.09.008. S1568-7864(10)00313-7 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Giribaldi MG, Munoz A, Halvorsen K, Patel A, Rai P. MTH1 expression is required for effective transformation by oncogenic HRAS. Oncotarget. 2015;6:11519–11529. doi: 10.18632/oncotarget.3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Piskounova E, et al. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature. 2015;527:186–191. doi: 10.1038/nature15726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fouquerel E, et al. ARTD1/PARP1 negatively regulates glycolysis by inhibiting hexokinase 1 independent of NAD+ depletion. Cell Rep. 2014;8:1819–1831. doi: 10.1016/j.celrep.2014.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Latrick CM, Cech TR. POT1-TPP1 enhances telomerase processivity by slowing primer dissociation and aiding translocation. EMBO J. 2010;29:924–933. doi: 10.1038/emboj.2009.409. emboj2009409 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pham HH, et al. Cooperative hybridization of gammaPNA miniprobes to a repeating sequence motif and application to telomere analysis. Org Biomol Chem. 2014;12:7345–7354. doi: 10.1039/c4ob00953c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ohno M, Oka S, Nakabeppu Y. Quantitative analysis of oxidized guanine, 8-oxoguanine, in mitochondrial DNA by immunofluorescence method. Methods Mol Biol. 2009;554:199–212. doi: 10.1007/978-1-59745-521-3_13. [DOI] [PubMed] [Google Scholar]
- 63.Parikh D, Fouquerel E, Murphy CT, Wang H, Opresko PL. Telomeres are partly shielded from ultraviolet-induced damage and proficient for nucleotide excision repair of photoproducts. Nat Commun. 2015;6:8214. doi: 10.1038/ncomms9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Herbert BS, Shay JW, Wright WE. Analysis of telomeres and telomerase. Curr Protoc Cell Biol. 2003;Chapter 18(Unit 18):16. doi: 10.1002/0471143030.cb1806s20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.